Abstract
Human glioblastoma (GBM) cells are notorious for their resistance to apoptosis-inducing therapeutics. We have identified lanatoside C as a sensitizer of GBM cells to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–induced cell death partly by upregulation of the death receptor 5. We show that lanatoside C sensitizes GBM cells to TRAIL-induced apoptosis in a GBM xenograft model in vivo. Lanatoside C on its own serves as a therapeutic agent against GBM by activating a caspase-independent cell death pathway. Cells treated with lanatoside C showed necrotic cell morphology with absence of caspase activation, low mitochondrial membrane potential, and early intracellular ATP depletion. In conclusion, lanatoside C sensitizes GBM cells to TRAIL-induced cell death and mitigates apoptosis resistance of glioblastoma cells by inducing an alternative cell death pathway. To our knowledge, this is one of the first examples of use of caspase-independent cell death inducers to trigger tumor regression in vivo. Activation of such mechanism may be a useful strategy to counter resistance of cancer cells to apoptosis.
Keywords: cardiac glycoside, glioblastoma, lanatoside C, non-apoptotic cell death, TRAIL
Glioblastoma (GBM) is the highest-grade (grade IV) glioma and is the number one cause of primary brain tumors.1 Because of the complexity of this disease, its high invasiveness, and the notorious resistance of GBM cells to apoptosis and conventional therapies, new treatment paradigms are highly desirable. One potent cancer-specific agent is the tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), also known as APO2L or TNFSF10.2 TRAIL binds and activates the death receptors TRAIL-R1 (DR4) and TRAIL-R2 (DR5)3 present on cancer cells, resulting in recruitment of the adapter Fas-associated death domain (FADD) and pro-caspases 8 and 10 to their death-inducing signaling complex (DISC), subsequently resulting in the activation of the caspase-induced apoptosis pathway.4 Although TRAIL has been shown to be a promising therapeutic for various types of tumors, considerable numbers of cancer cells, including GBM cells, are resistant to TRAIL-induced apoptosis.5–8
Cell death, defined as the irreversible loss of plasma membrane integrity, can be classified into 3 types on the basis of morphological criteria:9 apoptosis, autophagy, and necrosis. Over the past decade, cumulative evidence suggested that necrosis could also occur in a programmed fashion, notably on activation of the death-domain receptors (DRs), while caspase activity is blocked.10,11 This additional type of cellular demise has been termed “necroptosis.”12 A clear distinction between apoptosis and necroptosis can be made because the latter induces necrotic cell morphology, does not involve caspase activation, and is independent of the Bcl2 family of apoptotic regulators.12
In this study, we show that lanatoside C sensitizes glioblastoma cells to TRAIL both in cultured cells and in subcutaneous xenografts in vivo. Moreover, we show that lanatoside C on its own kills GBM cells by activating a caspase-independent cell death pathway. Activation of this alternative cell death pathway may be a useful strategy to counter apoptosis resistance in cancer cells and may be further exploited for the development of novel cancer therapeutics.
Materials and Methods
Reagents
The following reagents were purchased from Sigma-Aldrich: 3-methyladenine, bafilomycin A1, lanatoside C, and ouabain. Lanatoside C was reconstituted at 10 mg/mL (equivalent to 10 mM) in dimethyl sulfoxide (DMSO). Z-VAD-FMK, staurosporine, and rapamycin were purchased from Calbiochem. Recombinant human TRAIL (GF092) was purchased from Chemicon International. Recombinant Human TRAIL R2/TNFRSF10B Fc Chimera and Q-VD-OPh were obtained from R&D systems. Necrostatin-1 (LDN-57446) was a kind gift from Dr Greg Cuny (Laboratory for Drug Discovery in Neurodegeneration, Harvard NeuroDiscovery Center).
Cell Culture
293T, HF19 primary human fibroblast cells from a normal donor (a kind gift from Dr Breakefield at the Massachusetts General Hospital), U87, Gli36, U251, and primary GBM cells dissociated from tumor sections of different GBM patients were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Sigma), 100 U of penicillin, and 0.1 mg/mL streptomycin (Sigma) at 37°C in a 5% CO2 humidified incubator. To stably express Gluc and the cyan fluorescent protein (CFP), cells were transduced with CSCW-Gluc-IRES-CFP lentivirus vector, as described elsewhere.13
Gaussia Luciferase Assay in Culture
Cells were plated in 96-well plates at a density of 3000–10 000 cells/well. Aliquots of the cell-free conditioned medium (20 µL) were transferred to a clean, white 96-well plate and subjected to Gluc assay by injecting different concentrations of coelenterazine (Prolume Ltd./Nanolight; dissolved in acidified methanol [12 mM stock solution] and further diluted in phosphate buffer saline (PBS) [work solution]) or GAR-1 reagent (Targeting Systems) and acquiring photon counts using a luminometer (Dynex). The signal was measured for 4 s and integrated over 2 s.
In vivo Tumor Model and Bioluminescence Imaging
All animal studies were approved by the Massachusetts General Hospital Review Board. Initially, the maximum tolerance dose (MTD) of lanatoside C was assessed. Groups of 5 nude mice were injected with escalating doses of this drug (range, 1–60 mg/kg body weight) over 5 days. Control groups received the maximum amount of the vehicle DMSO. We found that the MTD of lanatoside C was 10 mg/kg body weight. One million U87-Gluc-CFP cells were implanted subcutaneously in the flanks of nude mice. One week later, mice were divided into groups of 5 mice that received intraperitoneal (i.p.) injection of either phosphate buffered saline, TRAIL (250 µg/kg body weight), lanatoside C (6 mg/kg body weight) or similar doses of both lanatoside C and TRAIL. These injections were repeated daily over 10 days. Tumor volume was monitored by both Gluc-blood assay and Gluc in vivo bioluminescence imaging, as described elsewhere.14,15 This experiment was repeated 2 times to obtain statistical significance.
DNA and siRNA Transfection
Cells were plated in 6-well plates (0.5 × 106), and 24 h later, transfection was performed using 2 µg of plasmid DNA and Lipofectamine 2000 (Invitrogen), in accordance with the manufacturer's instructions. U87-Gluc-CFP cells were transfected with pBABe empty vector, pBABe-Bcl2, pBABe-Bcl-xL, or EGFP-LC316 (Addgene plasmid 11 546). To generate stable cell lines, cells were selected using hygromycin B (Invitrogen), 150 ng/mL. For siRNA experiments, U87 cells were trasfected with 100 nM of All Stars siRNA control (Qiagen) or BECN1 siRNA (Sigma) using Lipofectamine 2000.
Western Blot Analysis
Immunoblot analysis was performed as described elsewhere.13 Frozen tumor samples were disrupted by homogenization and sonication. The following antibodies were used: β-actin (Millipore), cleaved caspase 3, BID (Cell Signaling), caspase 8, PARP, bax (BD Transduction Laboratories), Bcl2, and BECN-1 (Sigma). Western-blot bands were quantified using Image J software.
ATP-Based Cell Viability Assay
Quantification of cell death and ATP detection in metabolically active cells were performed using the bioluminescence-based CellTiter-Glo assay from Promega, as described in the manufacturer's manual.
Bioluminescent Caspase Assay
Cells plated in 96-well plates were treated accordingly. At different time points, caspase 3, 7, and 8 activity was measured using the caspase 3/7-Glo and caspase 8-Glo assay from Promega, in accordance with the manufacturer's recommendations. For in vivo caspase measurement, 0.25 µg of tumor lysates were loaded in 96-well white opaque plates (in triplicates) before adding the caspase 3/7 Glo substrate.
Human Apoptosis Polymerase Chain Reaction (PCR) Array
Cells were plated in 12-well plates at 1 × 106 cells/well and subjected to various treatments 24 h later. Total RNA isolation, reversetranscription, and real-time PCR were performed using the Human Apoptosis RT2 Profiler™ PCR Array kit (SABiosciences) as recommended by the manufacturer. Real-time PCR was performed in an ABI PRISM 7000 Sequence Detection System Thermal Cycler (Applied Biosystems).
Flow Cytometric Analysis
After 24 h of drug treatment, cells were stained with phycoerythrin-labeled DR4 and DR5 antibodies (R&D Systems) and analyzed by FACS using a BD FACS caliber flow cytometer (BD Biosciences). To measure the mitochondrial membrane potential (MMP), we used JC-1 (Invitrogen). Cells were treated with 10 µg/mL JC-1 for 10 min at 37°C followed by FACS analysis.
Transmission Electron Microscopy
Cells were treated for 6 h in a 6-well plate then fixed for 1 h at RT in a mixture of 2.5% glutaraldehyde and 1.25% paraformaldehyde. Cells were then embedded in epoxy resin. Sections were visualized using a Tecnai G2 Spirit BioTWIN transmission electron microscope.
Statistical Analysis and Half-Maximal Inhibitory Concentration (IC50) Calculation
IC50 values were calculated using SigmaPlot by best fitting the dose-response curve using the Four-Parameter Logistic Function with Variable Slope model. The significance levels (by the Student's t test) were calculated using standard spread sheets.
Results
Lanatoside C Sensitizes GBM Cells to TRAIL-Induced Cell Death
Glioma cells are notorious for their resistance to TRAIL.17–19 We first confirmed this by testing the effect of TRAIL on 2 primary GBM cells, as well as U87 and Gli36 glioma cell lines. Both primary and U87 cells showed high resistance to TRAIL, whereas Gli36 cells were found to be semi-resistant to it (Supplementary Material, Fig. S1). Through drug screening, we found that the family of cardiac glycosides sensitizes glioma cells to TRAIL-induced cell death.20 Upon validation, we found that lanatoside C showed synergistic effect with TRAIL on various GBM cells, including U87, Gli36, and primary GBM cells, without significant toxicity on normal human fibroblast cells (Fig. 1A).
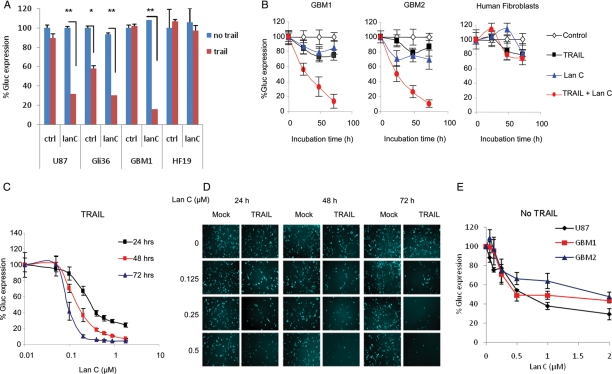
Fig. 1.
Effect of lanatoside C and tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) on glioblastoma multiforme (GBM) cells. (A) Validation of lanatoside C on U87 and Gli36 cell lines, GBM1 primary cells, and primary human fibroblasts (HF19), all expressing Gluc-CFP. Cells were treated with lanatoside C (0.25 µM) and/or TRAIL (50 ng/mL), and Gluc activity was measured after 48 h. (B) Two different primary GBM cells dissociated from different patients and primary human fibroblasts were treated with 0.25 µM of lanatoside C and 50 ng/mL TRAIL. Aliquots of the conditioned medium were assayed for Gluc activity over time. (C) Half-maximal inhibitory concentration (IC50) of lanatoside C in combination with TRAIL. Primary GBM cells were treated with different concentrations of lanatoside C and TRAIL (50 ng/mL). Gluc activity was assayed in conditioned medium at 24, 48, and 72 h. (D) CFP fluorescence images of GBM cells treated with different doses of lanatoside C and/or TRAIL at 24, 48, and 72 h. (E) Effect of lanatoside C alone on GBM cells. Various GBM cells were treated in a 96-well plate with various amounts of lanatoside C. Gluc activity was assayed 48 h later. Data are presented as the percentage of Gluc expression in which the control untreated sample is set at 100%. The average of experimental triplicates ± standard deviation is shown. *P≤ .05, by the Student's t test. **P≤ .001, by the Student's t test.
Lanatoside C was further analyzed in the presence or absence of TRAIL using different doses and at different time points on primary GBM cells dissociated from tissue sections of 2 other patients. Initially, GBM cells expressing Gluc-CFP were treated with 0.25 µM of lanatoside C in the presence or absence of 50 ng/mL TRAIL, and the conditioned media was assayed for Gluc activity over time. At 48 h, we observed 70%–80% decrease in Gluc activity and, therefore, cell viability in both primary GBM cell lines without much effect on normal fibroblasts (Fig. 1B). Dose curves of lanatoside C showed >90% sensitization of primary GBM cells to TRAIL, as evaluated by the Gluc assay (Fig. 1C). These results were confirmed by CFP fluorescent microscopy (Fig. 1D). Interestingly, at the 0.5 µM dose, lanatoside C itself was toxic to GBM cells (Fig. 1E). These results were consistent throughout all validation experiments. The IC50, defined as the concentration of lanatoside C that gives a 50% decrease in Gluc expression in the presence of TRAIL versus the control, was found to be 0.22 µM at 24 h, and it decreased to 0.13 µM at 48 h and to 0.09 µM at 72 h (Fig. 1C). On the other hand, the IC50 of lanatoside C alone on GBM cells was ∼0.5 µM at 48 h after treatment (Fig. 1E).
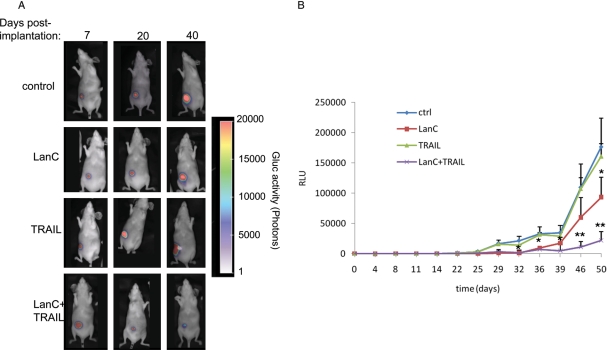
To evaluate the effect of TRAIL and/or lanatoside C in vivo, we initially performed dose-escalation studies of this drug in nude mice to obtain the MTD, which was found to be 10 mg/kg body weight. U87-Gluc-CFP glioma cells were implanted subcutaneously in the flanks of nude mice. One week later, mice were divided into 4 groups that received i.p. injection of either vehicle (DMSO in PBS; control), TRAIL (250 µg/kg body weight), or lanatoside C (6 mg/kg body weight) or similar doses of both lanatoside C and TRAIL (n = 10). These injections were repeated once per day over 10 days. Tumor volume was monitored by assaying 5 µL aliquots of blood for Gluc activity twice per week and was correlated with in vivo bioluminescence imaging once per week. TRAIL alone did not show any effect on tumor growth; however, lanatoside C and TRAIL treatment resulted in tumor regression for >40 days (Fig. 2A and B). At day 40, tumors in mice treated with both lanatoside C and TRAIL were >85% smaller than control tumors. Interestingly, lanatoside C treatment resulted in a slower rate of tumor growth compared with control treatment, confirming our culture findings and previous work showing that cardiac glycosides have an anti-tumor effect (Fig. 1).21,22 We therefore decided to study the mechanism by which lanatoside C is either (1) sensitizing GBM cell to TRAIL-induced apoptosis or (2) killing GBM cells on its own.
Fig. 2.
Effect of lanatoside C and/or tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) on glioblastoma multiforme (GBM) tumors in vivo. U87 glioma cells expressing Gluc were implanted subcutaneously in nude mice. One week later, mice were intraperitoneally injected once per day over 10 days with either DMSO vehicle (control), TRAIL (250 µg/kg body weight), lanatoside C (6 mg/kg body weight), or similar doses of both lanatoside C and TRAIL. (A) In vivo Gluc bioluminescence imaging was performed once per week after intravenous injection of coelenterazine, and photon counts were acquired using a CCD camera. (B) Before and at different time points (twice per week) after treatment, 5 µL of blood was drawn and assayed for Gluc activity. Shown is a representative mouse from each group (A) and the mean ± standard deviation for Gluc blood assay from each group (B; n = 10).
Lanatoside C Upregulates Death Receptor DR5
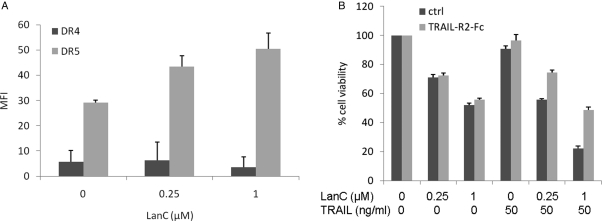
Cardiac glycosides have been reported to sensitize non-small cell lung cancer cells by upregulating DR4 and DR5.23 To verify whether this is also valid for GBM cells, we analyzed the levels of DR4 and DR5 on the cell surface of U87 cells on treatment with lanatoside C using FACS analysis. Although DR4 levels were not altered after treatment, an increase in cell-surface expression of DR5 could be detected. At the lower dose (0.25 µM) of lanatoside C, a ∼1.5-fold increase in DR5 cell surface expression was observed, compared with a 1.75-fold increase at 1 µM (Fig. 3A).
Fig. 3.
(A) Lanatoside C upregulates DR5 on GBM cells. U87 cells were treated with 0.25 or 1 µM of lanatoside C. Sixteen hours later, cells were labeled with anti DR4- or DR5-PE–conjugated antibodies followed by FACS analysis. Data are shown as mean fluorescence intensity (MFI) ± standard deviation. (B) U87-Gluc-CFP cells were pretreated with tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–R2-Fc chimeric protein (300 ng/mL) followed by lanatoside C in the presence or absence of TRAIL. Cell viability was assayed using CellTiter-Glo 24 h later. Data are presented as the percentage of cell viability in treated versus untreated samples and are shown as the mean of biological triplicates ± standard deviation.
To investigate whether this enhanced cell death is mediated through DR5, we used a blocking soluble Fc chimeric protein (TRAIL-R2-Fc) comprising the TRAIL-R2 (DR5) extracellular domain fused to the human IgG1 Fc.24 TRAIL-R2-Fc did not protect from lanatoside C–induced cell death but totally abrogated the TRAIL and lanatoside C combined effect (Fig. 3B). These results show that DR5 is needed and represents an apical point in lanatoside C and TRAIL sensitization.
Lanatoside C Induces a Necrosis-Like, Caspase-Independent Cell Death
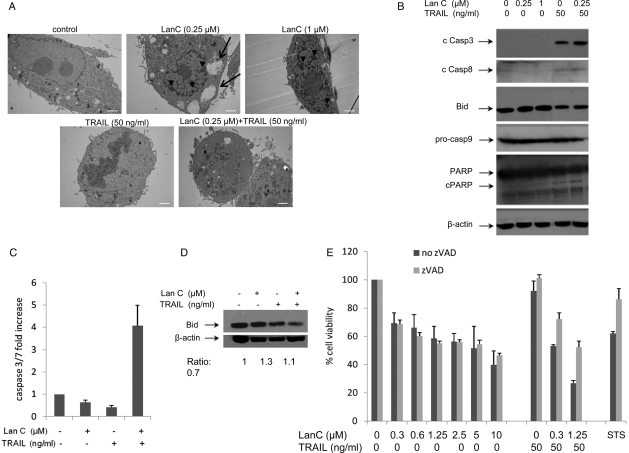
To investigate the mechanism by which lanatoside C was killing GBM cells, we first analyzed the morphology of GBM cells after treatment with this cardiac glycoside and/or TRAIL. Surprisingly, cells treated with lanatoside C alone showed necrotic features characterized by the condensation and segmentation of the chromatin; these features were more severe with the higher dose of the drug (1 µM; Fig. 4A). Vacuolization was also observed in the cytosol of lanatoside C–treated cells, as well as visible damage to cell organelles. On the other hand, in the TRAIL-treated cells, condensation and shrinkage of the nucleus was detected—a typical feature of apoptotic cell death. Cells treated with lanatoside C combined with TRAIL showed a total loss of nuclear membrane (Fig. 4A).
Fig. 4.
Lanatoside C induces caspase-independent cell death. (A) U87 cells were treated for 6 h with lanatoside C (0.25 and 1 µM), tumor necrosis factor–related apoptosis-inducing ligand (TRAIL; 50 ng/mL), or a combination of both and visualized using transmission electron microscopy (scale bar, 2 µm; original magnification, ×4000). Arrowheads indicate condensed chromatin; arrows indicate vacuoles present in the cytosol. (B) U87 cells treated for 16 h with lanatoside C (0.25 or 1 µM) in the presence or absence of TRAIL (50 ng/mL) were harvested and analyzed by western blotting using cleaved caspases 3 and 8, BID, PARP, and pro-caspase 9 as well as β-actin specific antibodies. (C and D) U87 subcutaneous tumor xenografts received injections of DMSO, lanatoside C, TRAIL, or a combination of lanatoside C and TRAIL. Tumors were dissociated, and lysates were analyzed using caspase 3/7-Glo assay (C) or for BID expression using western blotting (D). The ratio of BID to β-actin is shown below the blot, with values normalized to the control sample. (E) U87 cells were treated with either lanatoside C and TRAIL or staurosporine (STS; positive control) in the presence or absence of the caspase inhibitor zVAD-FMK (100 µM, with 1 h of pretreatment). Cell viability was assayed 24 h later using CellTiter-Glo.
To understand the cell death mechanism of this cardiac glycoside on TRAIL-resistant U87 cells, we first performed PCR arrays on key components of the tumor necrosis factor (TNF)–signaling network 6 h after treatment of GBM cells with 1 µM lanatoside C. All 4 genes that showed >2-fold increase in the mRNA expression were members of the TNF superfamily. These genes were TNF (TNF superfamily, member 2; 4.04 ± 0.79), Fas L (TNF superfamily, member 6; 3.18 ± 0.32), lymphotoxin alpha (TNF superfamily, member 1; 2.665 ± 0.9), and CD40 (TNF receptor superfamily member 5; 2.995 ± 1.28; Supplementary Material, Fig. S2). We then determined the protein expression levels of caspase 3, 8, and 9 in U87 cells treated with lanatoside C alone or in combination with TRAIL. Although 1 µM of lanatoside C did kill these glioblastoma cells (∼50%), no cleavage of caspases was observed. On the other hand, the cleaved form of caspase 3 and 8 was detected after TRAIL treatment alone or in combination with lanatoside C (Fig. 4B).
Upon activation of cell surface death receptors, BID, a pro-apoptotic member of the Bcl2 family, is cleaved by caspase 8 to generate tBID.25 tBID translocates into the mitochondria to activate the mitochondrial cell death machinery. The processing of BID could only be observed in samples treated with TRAIL (Fig. 4B; tBID was not detected under our assay conditions). In addition, PARP cleavage, an indicator of apoptotic cell death,26 was also detected only in the presence of TRAIL but not in cells treated with lanatoside C alone (Fig. 4B). These results suggest that lanatoside C is killing GBM cells in a caspase-independent mechanism. The cleaved caspase 9 could not be detected in any of the treated samples, whereas procaspase 9 levels remained unchanged in all samples (Fig. 4B). To corroborate these findings, we used a bioluminescent assay that measures the activity of caspases 3, 7, and 8. Again, cells treated with lanatoside C alone did not show any activity of these caspases at any of the doses tested (0.2–1 µM; Supplementary Material, Fig. S3). TRAIL alone (without lanatoside C) showed a slight increase in the activity of caspases 3, 7, and 8. When TRAIL was combined with lanatoside C, a significant 2–3 fold increase in caspase 3, 7, and 8 activation was observed (P < .05) (Supplementary Material, Fig. S3).
Next, we sought to validate this caspase-independent cell killing triggered by lanatoside C in vivo. Subcutaneous U87 tumor xenografts received 4 consecutive injections of DMSO, lanatoside C, TRAIL, or both lanatoside C and TRAIL over a 48-h period. Tumor samples were collected at 60 h and lysed for protein analysis. Caspase 3/7 activity on tumor lysates was detected using the caspase bioluminescent assay. We did not detect any caspase activation upon treatment with lanatoside C or TRAIL alone, whereas a 4-fold-increase was observed in tumors treated with both lanatoside C and TRAIL (Fig. 4C). Total BID expression was lower only in the presense of TRAIL but not in the lanatoside C–treated tumors (Fig. 4D). These results all together confirm our in vitro findings that lanatoside C kills glioma cells using a caspase-independent cell death. The absence of caspase activity in TRAIL-treated tumors might be justified by the total absence of anti-tumor effect in mice that received TRAIL alone, as shown in Fig. 2.
We also tested this cardiac glycoside in combination with the pan-caspase inhibitor Z-VAD-FMK (zVAD; 100 µM). This drug failed to inhibit lanatoside C–induced cell death, with no effect on cell viability at 24 h after treatment with lanatoside C (Fig. 4E) and <10% protection after 48 h (data not shown). When testing lanatoside C combined with TRAIL, zVAD inhibited cell killing by up to 50% at 24 h (Fig. 4E). This is in accordance with the caspase activation observed during lanatoside C and TRAIL treatment, suggesting apoptotic cell death only when TRAIL is combined with lanatoside C, most likely due to the TRAIL-induced caspase activation. As a positive control, we used staurosporine (STS) a known inducer of apoptosis in human glioma cell lines, including U87.27 zVAD inhibited STS-induced apoptosis by >25% (Fig. 4E). Because zVAD can increase necrosis in some cells,11 we included another pan-caspase inhibitor, Q-VD-O-Ph (qVD). This caspase inhibitor also failed to protect against lanatoside C–induced cell death (Supplementary Material, Fig. S4).
To validate that lanatoside C–induced caspase-independent cell death is not exclusive to U87 cells, we also measured caspase 3/7 activity in 2 other glioma cell lines (U251 and Gli36), as well as primary GBM cells. In general, these cells had higher sensitivity to TRAIL, compared with U87. A significant increase in caspase 3/7 activity was observed only in the presence of TRAIL or both lanatoside C and TRAIL in all cells (Supplementary Material, Fig. S5A). Concomitant with the caspase activity, zVAD failed to protect from lanatoside C killing while significantly inhibiting TRAIL induced apoptosis in all cells (Supplementary Material, Fig. S5B).
Lanatoside C-Induced Cell Killing Can Overcome Apoptosis Resistance
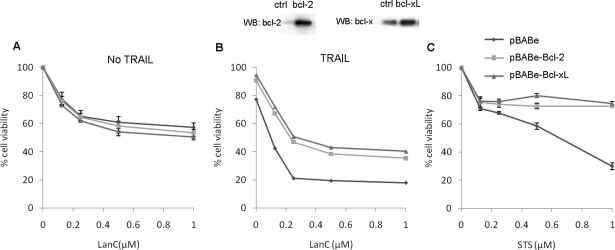
Because overexpression of anti-apoptotic proteins Bcl-2 and Bcl-xL conveys resistance to apoptosis,28 we generated U87 cell lines stably expressing these proteins. Cells overexpressing either Bcl-2 or Bcl-xL still died during lanatoside C treatment, suggesting that this cardiac glycoside mediates cell death independent of the Bcl-2 proteins (Fig. 5A). On the other hand, overexpression of either anti-apoptotic proteins conveyed 20%–30% protection of cell death induced when TRAIL was combined with lanatoside C (Fig. 5B). These results are in line with caspase activation, as well as with BID cleavage observed above, and they indicate an involvement of mitochondrial cell death when lanatoside C and TRAIL are combined. In these experiments, STS was used as a control to show the resistance of Bcl-2 or Bcl-xL–over-expressing cells to apoptosis (Fig. 5C). In summary, these results indicate that lanatoside C induces caspase-independent cell death in GBM cells, whereas a combination of lanatoside C and TRAIL leads to caspase activation most likely caused by TRAIL-induced apoptotic signaling.
Fig. 5.
Effect of Bcl-2 overexpression on lanatoside C–induced cell death. U87 cells stably expressing a control vector, Bcl-2, or Bcl-xL were treated with different concentrations of lanatoside C (A), lanatoside C and tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) (B) or staurosporine (STS; positive control) (C). Twenty-four hours later, cell viability was assayed using CellTiter-Glo. The upper panel shows western blot analysis for overexpression of Bcl2 or Bcl-xl in these cells.
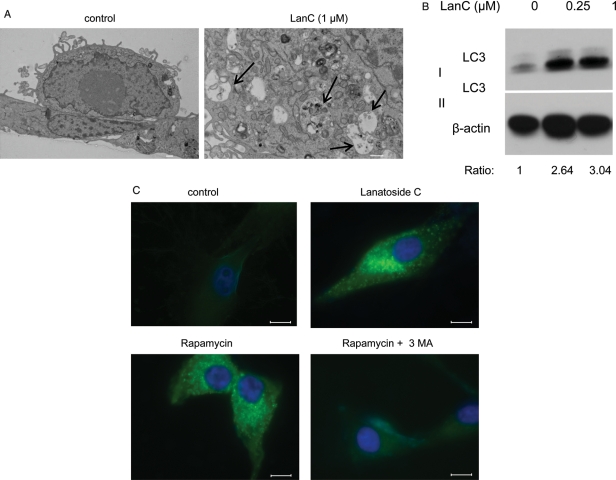
Lanatoside C Triggers Autophagy
Autophagy is characterized by the formation of double-membrane vesicles called autophagosomes.12 After exposure to lanatoside C for 6 h, autophagosomes were observed, as analyzed by electron microscopy (Fig. 6A). To confirm these results, we analyzed the expression of the microtubule-associated protein 1 light chain 3 (LC3-I), a marker of autophagy, and its LC3-II membrane-bound lipidated form expressed on the membrane of autophagosomes.29 Lanatoside C treatment induced an increase in LC3 II, confirming the induction of autophagy30 (Fig. 6B). We also treated U87 cells transfected with a plasmid expressing an LC3-GFP fusion protein with this drug. Twenty-four hours later, GFP punctuations were detected in the cytoplasm of the treated cells but not in the control samples (Fig. 6C). As a positive control, we treated cells with rapamycin, a chemical inducer of autophagy through mTOR signaling inhibition,31 which also showed GFP punctuations in the cytoplasm similar to those of lanatoside C-treated cells.
Fig. 6.
Lanatoside C induces autophagy. (A) Transmission electron micrograph of an untreated cell (original magnification, ×4000) or treated with 1 µM of lanatoside C (original magnification, ×8000). Arrows depict autophagosomes containing intracellular organelles (scale bar, 2 µm). (B) U87 cells treated for 16 h with lanatoside C (0.25 or 1 µM) were harvested and analyzed by western blotting using LC3 and β-actin antibodies. The ratio of LC3 II to β-actin is shown below the blot, with values normalized to the control sample. (C) U87 cells stably expressing LC3-GFP were treated with PBS, lanatoside C (1 µM), rapamycin (200 nM), or rapamycin (200 nM) in combination with 3MA (2 mM) for 24 h. Representative GFP fluorescent microscopic images of cells counterstained with DAPI (nuclei) are shown. Scale bar, 10 µm.
To investigate whether autophagy is the mechanism through which lanatoside C is killing the cells, we cotreated cells with this drug, as well as 2 different inhibitors of autophagy, 3-methyladenine32 and bafilomycin A1.33 Although these drugs efficiently abrogated the LC3-GFP punctuations in rapamycin-treated samples (Fig. 6C), suggesting an efficient inhibition of autophagy, they failed to protect against lanatoside C–mediated cell death (Supplementary Material, Fig. S6A). Furthermore, siRNA-mediated downregulation of the Beclin-1 gene, a key regulator of the mammalian autophagosome formation, failed to protect against lanatoside C–induced cell death (Supplementary Material, Fig. S6B). These data suggest that autophagy occurs as a downstream consequence of lanatoside C treatment but does not directly contribute to the cell death per se.
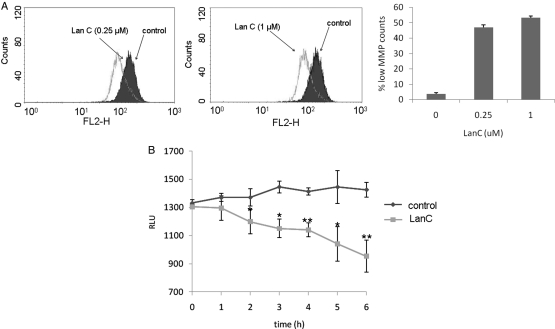
Lanatoside C Causes Loss of MMP and Early ATP Depletion Independently of RIPK1
Loss of MMP is another major determinant of cell death. When cells were treated with lanatoside C (0.25 or 1 µM), a significant decrease in MMP was detected, compared with nontreated cells (Fig. 7A). Because ATP depletion triggers autophagy,34 we evaluated the intracellular ATP levels during lanatoside C treatment. A decrease in ATP levels was detected as early as 2 h after treatment and continued over time. At 6 h, a 35% decrease in ATP levels was observed, compared with control samples (Fig. 7B). On the other hand, TRAIL-treated cells did not show any ATP depletion (data not shown).
Fig. 7.
Lanatoside C-induces mitochondrial damage and ATP depletion. (A) U87 cells were treated with 0.25 µM (left panel) or 1 µM (middle panel) lanatoside C. Four hours later, mitochondrial membrane potential (MMP) was measured after JC-1 staining, followed by flow cytometric analysis. The right panel represents quantitation analysis of (A) showing the percentage of cells with low MMP. (B) Time-course analysis of ATP levels following treatment of U87 cells with PBS or lanatoside C (1 µM) using CellTiter-Glo assay. All experiments were repeated at least 3 times to obtain statistical significance, *P≤ .05, by the Student's t test. **P≤ .001, by the Student's t test.
Although the cell death induced by lanatoside C is in line with necroptosis, a necrosis-programmed cell death,12 the specific inhibitor of this nonapoptotic cell death, necrostatin 1, did not show any protection when combined with lanatoside C (data not shown). Furthermore, the knock-down of RIP1, PARP2, and CYLD, reported to inhibit necroptosis,35 failed to protect against lanatoside C–induced cell death (data not shown). These data suggest that lanatoside C causes an alternative nonapoptotic cell death mechanism independent of the RIP1-mediated necroptotic pathway.
Discussion
In this study, we identified the cardiac glycoside lanatoside C as a potent sensitizer of GBM cells to TRAIL-induced apoptosis. Furthermore, this drug on its own showed a significant anti-glioma effect in culture as well as tumor xenograft in mice by activating an alternative cell-death pathway. To our knowledge, this is one of the first examples of use of caspase-independent cell-death inducers to trigger tumor regression in vivo. Recently, several studies suggested cardiac glycosides as potential therapies for different types of cancer,22 including GBM,36 although few have made it to the clinic because of their therapeutic index and/or their low antitumor activity, as well as their cardiotoxicity.37 When combined with a low dose of TRAIL, a low dose of lanatoside C provides a high anti-glioblastoma activity, both in cultured cells and in GBM xenografts in vivo. These results could present one leap toward testing of this drug combination for glioma therapy because TRAIL is currently being tested for the treatment of different tumors in humans. Also, cardiac glycosides have been approved by the US Food and Drug Administration and have been used for decades for the treatment of congestive heart failure.
A previous study showed a synergistic effect of cardiac glycosides with TRAIL in culture caused by upregulation of the death receptors (DR) on non–small cell lung cancer cells while reporting no toxicity of cardiac glycoside treatment without TRAIL.23 Another recent study of malignant epithelial cells had suggested that cardiac glycosides act as anoikis sensitizers while causing no cytotoxicity on adherent cells.38 Although it seems that the activity of cardiac glycosides is cell-type specific, their mechanism of action on cancer cells is not fully elucidated, and several possible mechanism have been suggested.22 A chemical genetic screen for small molecules with neuroprotective effect in ischemic stroke revealed 4 cardiac glycosides (neriifolin, ouabain, digitoxin, and digoxin).39 Neriifolin, being the most potent of the 4 hits, demonstrated a neuroprotective effect in hypoxia/ischemia animal models as well. This provides additional evidence of the use of cardiac glycosides in brain treatment and the lack of toxicity of these drugs on normal brain cells. Another major advantage of cardiac glycosides in GBM therapy may be their ability to cross the blood-brain barrier.40 Therefore, we decided to investigate the mechanism of action of lanatoside C and inspect a possible cross-talk between this cardiac glycoside and the TRAIL pathway. We observed that treatment of GBM cells with lanatoside C upregulated the mRNA expression of several genes that are members of the TNF superfamily. We also observed an upregulation of DR5 protein expression on the cell surface after lanatoside C treatment, suggesting that the sensitization of glioblastoma cells to TRAIL might be due to the cardiac glycoside-induced expression of this death receptor on the cell surface. A drug screen for inhibitors of polyglutamine-dependent activation of caspase-3 on human embryonic kidney cells (HEK293) by Piccioni et al41 identified 6 cardiac glycosides as validated drug hits. In the absence of caspase activation, a necrotic cell death mechanism induced through the DR can supervene.42 On the other hand, ATP depletion can trigger autophagy,34 and more interestingly, it can favor necrosis over apoptosis,43,44 partly because of the role of ATP in apoptosome formation and the activation of caspases.45,46 One possible explanation for lanatoside C–induced cell death is that this drug might indirectly inhibit caspase activity through ATP depletion while activating the death receptors, thereby inducing programmed necrosis.
A major remaining challenge is to translate the combined therapeutic strategy for glioma therapy, because of the inefficiency of TRAIL in crossing the blood-brain barrier.47 On the other hand, TRAIL has been infused locally into the brain by convection-enhanced delivery, which showed wide distribution in the brain without cytoxicity to normal cells and no clinical symptoms by hepatotoxicity evaluation, with the exception of modest inflammation immediately adjacent to the needle tract.48 TRAIL has also been delivered to brain tumors with adeno-associated viral vectors, as well as neural precursor cells expressing secreted TRAIL; both resulted in glioma regression in different models.49 These delivery strategies could potentially be tested in combination with systemic injection of lanatoside C.
Evading apoptosis is a hallmark of cancer.50 The apoptosis resistance mechanisms are made possible through a myriad of genetic and epigenetic processes. One example of these apoptosis-resistance mechanisms is the enhanced expression of Bcl-2 anti-apoptotic proteins.51 Here, we showed that lanatoside C can overcome this resistance by activating an alternative cell-death mechanism. Chemotherapeutic agents that induce nonapoptotic cell-death pathways might be of great benefit and could outwit tumor resistance. Moreover, combining drugs that induce different cell-death mechanisms, as seems to occur with lanatoside C (caspase-independent cell death) and TRAIL (apoptotic cell death) on GBM cells, may help achieve a maximal therapeutic benefit. A deeper understanding of programmed necrosis pathways and identification of markers for this caspase-independent mechanism are still needed. Use of drugs such as lanatoside C combined with known therapeutics, such as TRAIL, may be a useful strategy to counter resistance of cancer cells to apoptosis and provide new opportunities for cancer treatment in general and for GBM in particular.
Supplementary Material
Conflict of interest. The authors declare no conflict of interest.
Funding
This work was supported by grants from the National Institutes of Health (NIH)/National Cancer Institute 4R00CA126839, NIH/National Institute of Neurological Disorders 1R21NS061051, P30 NS045776 and 1R01NS064983 (B.A.T.) as well as Executive Committee on Research at Massachusetts General Hospital (C.B.) and the American Brain Tumor Association (T.W.).
Supplementary Material
Acknowledgments
We would like to thank Dr Robert Carter for providing the tumor tissue sections from patients; Dr Johan Skog for providing the primary glioblastoma cells; Dr Greg Cuny for providing us with necrostatin-1; Mrs Lisa Pike and Lee-Ann Tjon-Kon-Fat for technical help. We are thankful to Dr Esther Hulleman for Bcl-2 and Bcl-xL constructs, Drs Xandra Breakefield and Randall B. Murphy for providing significant advice on this work, and Drs Hang Lee and Patrick Sluss for statistical analysis guidance.
References
- 1.Sathornsumetee S, Rich JN. New approaches to primary brain tumor treatment. Anticancer Drugs. 2006;17(9):1003–1016. doi: 10.1097/01.cad.0000231473.00030.1f. doi:10.1097/01.cad.0000231473.00030.1f. [DOI] [PubMed] [Google Scholar]
- 2.Ashkenazi A, Pai RC, Fong S, et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104(2):155–162. doi: 10.1172/JCI6926. doi:10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan G, O'Rourke K, Chinnaiyan AM, et al. The receptor for the cytotoxic ligand TRAIL. Science. 1997;276(5309):111–113. doi: 10.1126/science.276.5309.111. doi:10.1126/science.276.5309.111. [DOI] [PubMed] [Google Scholar]
- 4.LeBlanc HN, Ashkenazi A. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 2003;10(1):66–75. doi: 10.1038/sj.cdd.4401187. doi:10.1038/sj.cdd.4401187. [DOI] [PubMed] [Google Scholar]
- 5.Fulda S, Wick W, Weller M, Debatin KM. Smac agonists sensitize for Apo2L/TRAIL- or anticancer drug-induced apoptosis and induce regression of malignant glioma in vivo. Nat Med. 2002;8(8):808–815. doi: 10.1038/nm735. [DOI] [PubMed] [Google Scholar]
- 6.Kasuga C, Ebata T, Kayagaki N, et al. Sensitization of human glioblastomas to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) by NF-kappaB inhibitors. Cancer Sci. 2004;95(10):840–844. doi: 10.1111/j.1349-7006.2004.tb02191.x. doi:10.1111/j.1349-7006.2004.tb02191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panner A, Parsa AT, Pieper RO. Use of APO2L/TRAIL with mTOR inhibitors in the treatment of glioblastoma multiforme. Expert Rev Anticancer Ther. 2006;6(9):1313–1322. doi: 10.1586/14737140.6.9.1313. doi:10.1586/14737140.6.9.1313. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S, Shen HM, Ong CN. Down-regulation of c-FLIP contributes to the sensitization effect of 3,3′-diindolylmethane on TRAIL-induced apoptosis in cancer cells. Mol Cancer Ther. 2005;4(12):1972–1981. doi: 10.1158/1535-7163.MCT-05-0249. doi:10.1158/1535-7163.MCT-05-0249. [DOI] [PubMed] [Google Scholar]
- 9.Kroemer G, El-Deiry WS, Golstein P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2005;12(Suppl 2):1463–1467. doi: 10.1038/sj.cdd.4401724. doi:10.1038/sj.cdd.4401724. [DOI] [PubMed] [Google Scholar]
- 10.Holler N, Zaru R, Micheau O, et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1(6):489–495. doi: 10.1038/82732. doi:10.1038/82732. [DOI] [PubMed] [Google Scholar]
- 11.Vercammen D, Beyaert R, Denecker G, et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187(9):1477–1485. doi: 10.1084/jem.187.9.1477. doi:10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degterev A, Huang Z, Boyce M, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1(2):112–119. doi: 10.1038/nchembio711. doi:10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 13.Badr CE, Hewett JW, Breakefield XO, Tannous BA. A highly sensitive assay for monitoring the secretory pathway and ER stress. PLoS ONE. 2007;2(6):e571. doi: 10.1371/journal.pone.0000571. doi:10.1371/journal.pone.0000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Badr CE, Niers JM, Tjon-Kon-Fat LA, Noske DP, Wurdinger T, Tannous BA. Real-time monitoring of nuclear factor kappaB activity in cultured cells and in animal models. Mol Imaging. 2009;8(5):278–290. [PMC free article] [PubMed] [Google Scholar]
- 15.Wurdinger T, Badr C, Pike L, et al. A secreted luciferase for ex vivo monitoring of in vivo processes. Nat Methods. 2008;5(2):171–173. doi: 10.1038/nmeth.1177. doi:10.1038/nmeth.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson WT, Giddings TH, Jr, Taylor MP, et al. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3(5):e156. doi: 10.1371/journal.pbio.0030156. doi:10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahana S, Finniss S, Cazacu S, et al. Proteasome inhibitors sensitize glioma cells and glioma stem cells to TRAIL-induced apoptosis by PKCε-dependent downregulation of AKT and XIAP expressions. Cell Signal. 2011;23(8):1348–1357. doi: 10.1016/j.cellsig.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Maguire CA, Deliolanis NC, Pike L, et al. Gaussia luciferase variant for high-throughput functional screening applications. Anal Chem. 2009;81(16):7102–7106. doi: 10.1021/ac901234r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tannous BA, Christensen AP, Pike L, et al. Mutant sodium channel for tumor therapy. Mol Ther. 2009;17(5):810–819. doi: 10.1038/mt.2009.33. doi:10.1038/mt.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badr CE, Wurdinger T, Tannous BA. Functional drug screening assay reveals potential glioma therapeutics. Assay Drug Dev Technol. 2010 doi: 10.1089/adt.2010.0324. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mekhail T, Kaur H, Ganapathi R, Budd GT, Elson P, Bukowski RM. Phase 1 trial of Anvirzel in patients with refractory solid tumors. Invest New Drugs. 2006;24(5):423–427. doi: 10.1007/s10637-006-7772-x. doi:10.1007/s10637-006-7772-x. [DOI] [PubMed] [Google Scholar]
- 22.Prassas I, Diamandis EP. Novel therapeutic applications of cardiac glycosides. Nat Rev Drug Discov. 2008;7(11):926–935. doi: 10.1038/nrd2682. doi:10.1038/nrd2682. [DOI] [PubMed] [Google Scholar]
- 23.Frese S, Frese-Schaper M, Andres AC, Miescher D, Zumkehr B, Schmid RA. Cardiac glycosides initiate Apo2L/TRAIL-induced apoptosis in non-small cell lung cancer cells by up-regulation of death receptors 4 and 5. Cancer Res. 2006;66(11):5867–5874. doi: 10.1158/0008-5472.CAN-05-3544. doi:10.1158/0008-5472.CAN-05-3544. [DOI] [PubMed] [Google Scholar]
- 24.Walczak H, Degli-Esposti MA, Johnson RS, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. Embo J. 1997;16(17):5386–5397. doi: 10.1093/emboj/16.17.5386. doi:10.1093/emboj/16.17.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94(4):481–490. doi: 10.1016/s0092-8674(00)81589-5. doi:10.1016/S0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 26.Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem Cell Biol. 1997;75(4):337–349. doi:10.1139/o97-043. [PubMed] [Google Scholar]
- 27.Harmalkar MN, Shirsat NV. Staurosporine-induced growth inhibition of glioma cells is accompanied by altered expression of cyclins, CDKs and CDK inhibitors. Neurochem Res. 2006;31(5):685–692. doi: 10.1007/s11064-006-9068-0. doi:10.1007/s11064-006-9068-0. [DOI] [PubMed] [Google Scholar]
- 28.Pommier Y, Sordet O, Antony S, Hayward RL, Kohn KW. Apoptosis defects and chemotherapy resistance: molecular interaction maps and networks. Oncogene. 2004;23(16):2934–2949. doi: 10.1038/sj.onc.1207515. doi:10.1038/sj.onc.1207515. [DOI] [PubMed] [Google Scholar]
- 29.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. doi:10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noda T, Fujita N, Yoshimori T. The late stages of autophagy: how does the end begin? Cell Death Differ. 2009;16(7):984–990. doi: 10.1038/cdd.2009.54. doi:10.1038/cdd.2009.54. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003;3(4):371–377. doi: 10.1016/s1471-4892(03)00071-7. doi:10.1016/S1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 32.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79(6):1889–1892. doi: 10.1073/pnas.79.6.1889. doi:10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manabe T, Yoshimori T, Henomatsu N, Tashiro Y. Inhibitors of vacuolar-type H(+)-ATPase suppresses proliferation of cultured cells. Journal of Cellular Physiology. 1993;157(3):445–452. doi: 10.1002/jcp.1041570303. doi:10.1002/jcp.1041570303. [DOI] [PubMed] [Google Scholar]
- 34.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6(6):439–448. doi: 10.1038/nrm1660. doi:10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 35.Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135(7):1311–1323. doi: 10.1016/j.cell.2008.10.044. doi:10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefranc F, Mijatovic T, Kondo Y, et al. Targeting the alpha 1 subunit of the sodium pump to combat glioblastoma cells. Neurosurgery. 2008;62(1):211–221. doi: 10.1227/01.NEU.0000311080.43024.0E. discussion 221–212 doi:10.1227/01.NEU.0000311080.43024.0E. [DOI] [PubMed] [Google Scholar]
- 37.Lefranc F, Kiss R. The sodium pump alpha1 subunit as a potential target to combat apoptosis-resistant glioblastomas. Neoplasia. 2008;10(3):198–206. doi: 10.1593/neo.07928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simpson CD, Mawji IA, Anyiwe K, et al. Inhibition of the sodium potassium adenosine triphosphatase pump sensitizes cancer cells to anoikis and prevents distant tumor formation. Cancer Res. 2009;69(7):2739–2747. doi: 10.1158/0008-5472.CAN-08-2530. doi:10.1158/0008-5472.CAN-08-2530. [DOI] [PubMed] [Google Scholar]
- 39.Wang JK, Portbury S, Thomas MB, et al. Cardiac glycosides provide neuroprotection against ischemic stroke: discovery by a brain slice-based compound screening platform. Proc Natl Acad Sci USA. 2006;103(27):10461–10466. doi: 10.1073/pnas.0600930103. doi:10.1073/pnas.0600930103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marx J, Pretorius E, Bornman MS. The neurotoxic effects of prenatal cardiac glycoside exposure: a hypothesis. Neurotoxicol Teratol. 2006;28(1):135–143. doi: 10.1016/j.ntt.2005.10.004. doi:10.1016/j.ntt.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 41.Piccioni F, Roman BR, Fischbeck KH, Taylor JP. A screen for drugs that protect against the cytotoxicity of polyglutamine-expanded androgen receptor. Hum Mol Genet. 2004;13(4):437–446. doi: 10.1093/hmg/ddh045. [DOI] [PubMed] [Google Scholar]
- 42.Jaattela M, Tschopp J. Caspase-independent cell death in T lymphocytes. Nat Immunol. 2003;4(5):416–423. doi: 10.1038/ni0503-416. doi:10.1038/ni0503-416. [DOI] [PubMed] [Google Scholar]
- 43.Eguchi Y, Shimizu S, Tsujimoto Y. Intracellular ATP levels determine cell death fate by apoptosis or necrosis. Cancer Res. 1997;57(10):1835–1840. [PubMed] [Google Scholar]
- 44.Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185(8):1481–1486. doi: 10.1084/jem.185.8.1481. doi:10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA. 2005;102(49):17545–17550. doi: 10.1073/pnas.0507900102. doi:10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES. Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J Biol Chem. 1999;274(25):17941–17945. doi: 10.1074/jbc.274.25.17941. doi:10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- 47.Xiang H, Nguyen CB, Kelley SK, Dybdal N, Escandon E. Tissue distribution, stability, and pharmacokinetics of Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand in human colon carcinoma COLO205 tumor-bearing nude mice. Drug Metab Dispos. 2004;32(11):1230–1238. doi: 10.1124/dmd.104.000323. doi:10.1124/dmd.104.000323. [DOI] [PubMed] [Google Scholar]
- 48.Saito R, Bringas JR, Panner A, et al. Convection-enhanced delivery of tumor necrosis factor-related apoptosis-inducing ligand with systemic administration of temozolomide prolongs survival in an intracranial glioblastoma xenograft model. Cancer Res. 2004;64(19):6858–6862. doi: 10.1158/0008-5472.CAN-04-1683. doi:10.1158/0008-5472.CAN-04-1683. [DOI] [PubMed] [Google Scholar]
- 49.Hingtgen S, Ren X, Terwilliger E, Classon M, Weissleder R, Shah K. Targeting multiple pathways in gliomas with stem cell and viral delivered S-TRAIL and Temozolomide. Mol Cancer Ther. 2008;7(11):3575–3585. doi: 10.1158/1535-7163.MCT-08-0640. doi:10.1158/1535-7163.MCT-08-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. doi:10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 51.Pavet V, Portal MM, Moulin JC, Herbrecht R, Gronemeyer H. Towards novel paradigms for cancer therapy. Oncogene. 2011;30(1):1–20. doi: 10.1038/onc.2010.460. doi:10.1038/onc.2010.460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.