Abstract
Various environmental and genetic factors affect the development and progression of skin cancers including melanoma. Melanoma development is initially triggered by environmental factors including ultraviolet (UV) light, and then genetic/epigenetic alterations occur in skin melanocytes. These first triggers alter the conditions of numerous genes and proteins, and they induce and/or reduce gene expression and activate and/or repress protein stability and activity, resulting in melanoma progression. Microphthalmia-associated transcription factor (MITF) is a master regulator gene of melanocyte development and differentiation and is also associated with melanoma development and progression. To find better approaches to molecular-based therapies for patients, understanding MITF function in skin melanoma development and progression is important. Here, we review the molecular networks associated with MITF in skin melanoma development and progression.
1. Introduction
Much evidence that environmental factors are correlated with various diseases has been accumulating. The environmental factors can be classified into physical [1–4], chemical [5–8], and biological [9, 10] factors. In addition to the environmental factors, genetic factors also have a great influence on the development and pathogenesis of various diseases [2, 11, 12]. Skin is a representative organ that directly suffers from environmental factors. There is much evidence showing that sunlight and ultraviolet light induce various skin cancers with modulation of the signaling of cell proliferation and DNA damage [13–16]. Therefore, roles of skin cancer-related molecules should be discussed with consideration of the effects of environmental factors. Moreover, the incidence of skin melanoma has recently been increasing at a greater rate than that of any other cancer [17]. In the USA, 68,130 cases of invasive melanoma and at least 48,000 cases of melanoma in-situ were diagnosed in a year [18]. Since melanoma is the most aggressive skin cancers [17, 18], we focus on skin melanoma in this paper.
Not only studies on humans including epidemiological research but also animal models can be useful for analyzing melanomagenesis [19–23]. For example, exposure of skin to oxygen might regulate development of benign melanocytic tumors with modulation of tumor immunity in animal models [19]. Ultraviolet (UV) light is correlated with malignant transformation from benign melanocytic tumors and melanoma [15, 24]. In addition to these environmental factors, various kinds of genetic factors have been reported as crucial factors of melanoma. For example, tyrosine kinases are important for the development and pathogenesis of melanoma in mice and humans [19, 21–23, 25–27]. Various membrane trafficking-associated molecules have also been reported to be involved in melanoma pathogenesis [28–30]. Moreover, some acidic glycosphingolipids have been reported to be expressed at high levels in melanomas and promote their malignant properties by activating cell growth and adhesion signals in melanoma cells [31–33].
Microphthalmia-associated transcription factor (MITF) is believed to be one of the master molecules to regulate melanomagenesis among the many previously reported melanoma-associated molecules. Therefore, we selected MITF as a cancer-associated molecule in melanoma and introduce recent findings regarding MITF in this paper.
2. Results
Melanocytes, melanin-producing cells that are widely distributed in several tissues from fungi to primates on the long evolutionary process, have multifunctionality for survival strategy [34–41]. Melanocytes are also present in skin surfaces and protect them from UV that damages DNA, thereby causing genotoxic mutations or skin cancers [42, 43], but once they transform, they can result in the development of one of the most serious cancers, melanoma.
2.1. Microphthalmia-Associated Transcription Factor
The MITF gene, encoding a basic-helix-loop-helix-leucine zipper transcription factor, is expressed in melanocytes, retinal pigmented epithelium, mast cells, osteoclasts, and melanoma [36, 37, 40–42, 44–49]. MITF protein forms dimers and binds to specific consensus DNA sequences in the promoter regions of various target genes to regulate several events including differentiation, proliferation, migration, invasion, and tumorigenesis (Figure 1) [50].
Figure 1.
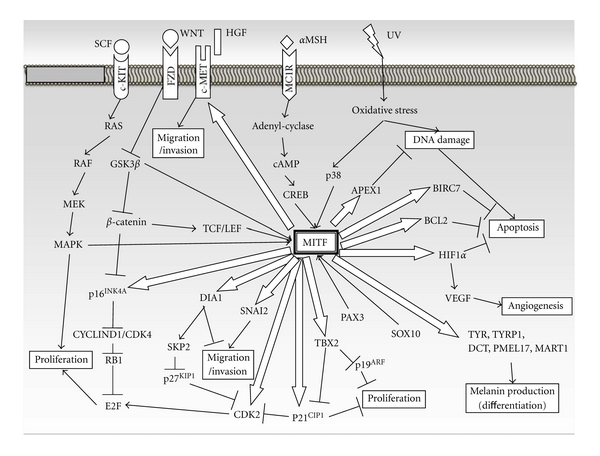
MITF-centered schematic scheme of cellular signaling on melanoma development and progression. Open arrows indicate direct transcriptional targets of MITF. The amount of transcripts of these targets is regulated by direct MITF binding to cis-elements in their promoter sequence. Black arrows or lines indicate signal cascades associated with melanoma development and progression. Arrows or lines toward MITF mean direct association by binding to the MITF promoter region or MITF protein. αMSH: melanocyte-stimulating hormone; BCL2: B-cell leukemia/lymphoma 2; BRN2: brain-2; CDK2/CDK4: cyclin-dependent kinase 2/4; CREB: cAMP-responsive element-binding protein; DCT: dopachrome tautomerase; DIA1: diaphanous homolog 1; FZD: frizzled; GSK3β: glycogen synthase kinase 3 beta; HGF: hepatocyte growth factor; HIF1α: hypoxia inducible factor 1, alpha subunit; MC1R: melanocortin 1 receptor; MDM2: transformed mouse 3T3 cell double minute 2; PMEL17: premelanosome protein; RB1: retinoblastoma 1; SCF: stem cell factor; SNAI2: snail homolog 2; SOX10: Sry-related HMG box 10; TYR: tyrosinase, WNT: wingless-related MMTV integration site.
2.2. Regulation of MITF Expression and Activity
Several transcription factors directly control MITF gene transcription to regulate melanocyte and melanoma development. Paired box 3 (PAX3) and Sry-related HMG box 10 (SOX10), highly correlated with melanocyte development and melanomagenesis [51–53], positively regulate MITF expression by directly binding to MITF promoter regions [54–57]. Activation of melanocortin 1 receptor (MC1R) by binding of alpha-melanocyte stimulating hormone (α-MSH) induces cAMP production via activation of adenylyl cyclase and phosphorylates cAMP response element-binding protein (CREB). Phosphorylated CREB directly binds to the MITF promoter region and stimulates MITF transcription [58, 59]. Wingless-type (WNT) signaling is often activated in human melanoma [60–63]. Activation of Frizzled receptors by binding of WNT molecules enhances interaction of β-catenin with TCF/LEF transcription factors, resulting in stimulation of MITF promoter activity [64–66].
Furthermore, MITF protein is modified by several factors after translation. Phosphorylation at Ser 301 of MITF is induced by UV through p38 stress-activated kinase [67], and Ser 298 of the protein is phosphorylated by GSK3β [65], resulting in stimulation of MITF transcriptional activity. The c-KIT receptor activated by stem cell factor (SCF, c-KIT ligand) phosphorylates Ser 73 and thereby increases MITF transcriptional activity followed by immediate degradation of MITF [59, 68], whereas sumoylation at Lys 182 and Lys 316 increases MITF transcriptional activity [69, 70].
2.3. Transcriptional Targets of MITF
MITF is associated with cellular senescence, apoptosis, proliferation, migration/invasion, and differentiation through regulating transcription of target genes.
Overcoming cellular senescence, acquisition of anti-apoptotic activity, and promotion of proliferation are critical cellular events for the initiation of tumorigenesis [50, 71–75]. CDKN2A and WAF1 genes encode senescence mediator proteins, p16 INK4A and p14 ARF , and p21 Cip1 , respectively, and are the well-known familial melanoma locus [71, 73, 76]. Copy number of CyclinD1 (CCND1) gene, a cell cycle mediator, is amplified in 25% of human melanomas [77]. MITF directly binds to the promoter regions of p16 Ink4a , p21 Cip1 , and CyclinD1 and positively regulates their transcription [78–80]. T-Box transcription factor 2 (TBX2) is highly expressed in melanoma cell lines and represses p19 ARF and p21 Cip1 , both of which are implicated as effectors of senescence, promotes proliferation, and suppresses senescence in melanoma [81, 82]. TBX2 has also been described as one of the MITF target genes [81]. These reports indicate that MITF is linked to melanoma development as a transcriptional activator of senescence-/proliferation-associated genes.
Antiapoptotic effect is a key process for melanoma development. B-cell leukemia/lymphoma 2 (BCL2) is an antiapoptotic gene and is widely expressed in human melanomas [83–85]. BCL2 is an MITF target gene and is activated at the transcription level [86]. Baculoviral IAP repeat containing 7/melanoma inhibitor of apoptosis (BIRC7/ML-IAP), which is an antiapoptotic regulator, is highly expressed in human melanomas [87] and provides resistance to apoptosis-based chemotherapeutic treatments [88]. BIRC7 transcription is also directly activated by MITF, and overexpression of BIRC7 rescued melanoma from apoptosis in MITF-depleted melanoma cells [87]. Antioxidative stress activity is important for melanocyte survival and melanoma development. Oxidative stress from environmental factors such as solar UV causes DNA damage and apoptosis. Recently, apurinic/apyrimidinic endonuclease1/redox factor-1 (APEX1/Ref1) has been identified as a MITF target gene and has been shown to be partially rescued from oxidative stress-induced apoptosis in MITF-depleted cells [89]. Hypoxia-inducible factor 1 α (HIF1α) has also been demonstrated to be activated at the transcription level by direct MITF binding to the HIF1α promoter region and acts as an antiapoptotic factor in melanoma cells [90]. Antiapoptotic activity and resistance to chemotherapy of melanoma are under the control of MITF activity.
Angiogenesis and invasion are critical steps for tumor progression, and these activities are enhanced in melanoma. MITF depletion in melanoma cells represses not only transcription of HIF1α but also that of vascular endothelial growth factor (VEGF), which is a target of HIF1α and has been demonstrated to be a major contributor to angiogenesis [90]. The c-MET protooncogene, which encodes hepatocyte growth factor receptor (HGFR), is highly expressed in human melanomas and linked to metastatic potential in melanomas. c-MET transcription is directly regulated by MITF [91]. SNAI2 has been reported to be a key player for epithelial-mesenchymal transition (EMT), which is a crucial phenomenon during invasion, metastasis of melanoma, by repressing E-cadherin transcription and stimulating fibronectin expression, and MITF directly activates the transcription level of SNAI2 [92–94]. On the other hand, MITF directly binds to the promoter region of diaphanous homolog 1 (DIAPH1, DIA1) gene and activates its transcription, resulting in inhibiting the invasiveness of melanoma by activation of actin polymerization [95].
MITF is also well known as a master regulator of melanin production. Melanin pigment is synthesized from tyrosine via an enzymatic process. This process is catalyzed by tyrosinase family proteins, tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and DCT (dopachrome tautomerase). After melanin production, melanin pigment is stored in melanosomes, which are organelles containing melanin, and is transported to the skin for UV protection. MART1 and PMEL17 are localized in melanosomes and contribute to melanosome maturation [96–99]. Direct regulation of melanin synthesis-associated genes at transcription levels by MITF stimulates melanin production. [100–103].
2.4. “Two-Faced” Function of Mitf in Melanoma Development and Progression
MITF is expressed in most human melanomas, and stability of its expression is essential for melanoma cell proliferation and survival [104]. In addition, amplification of the MITF locus was observed in human metastatic melanomas [105]. However, the expression level of MITF in melanomas is significantly lower than that in normal melanocytes, and higher expression level of MITF in melanoma represses cell proliferation even in the presence of oncogenic BRAF [106]. Most likely, MITF plays both cancer-promoting and cancer-inhibiting roles alternated by the expression level and/or activity. A low level of MITF expression promotes proliferation in melanoma, whereas a high level of MITF expression promotes differentiation through induction of cellular senescence and melanin production [107–109]. MITF has the ability to upregulate transcription of melanoma-promoting genes (e.g., CyclinD1, BCL2, c-MET) and also that of melanoma-repressing genes (e.g., p16 Ink4a , p21 Cip1 , DIAHP1). Taken together, MITF has two-faced functions in melanoma development and progression, and strict regulation of MITF in its expression and/or activity is likely to switch melanocytes to melanoma cells.
3. Concluding Remarks
Skin melanoma is an aggressive tumor of the skin, and patients have a poor prognosis. Analysis of the expression profile and function of MITF and identification of its target genes are important to better understand the complex system of melanoma development and progression. Expression patterns, functions, and many target genes of MITF have been reported by a number of groups, though the complicated functions of MITF in skin melanoma development and progression are still not well understood. Extensive analyses of MITF will lead to a better understanding of melanoma development and progression and to the establishment of more effective therapeutics.
References
- 1.Ida-Eto M, Ohgami N, Iida M, et al. Partial requirement of endothelin receptor B in spiral ganglion neurons for postnatal development of hearing. Journal of Biological Chemistry. 2011;286(34):29621–29626. doi: 10.1074/jbc.M111.236802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohgami N, Ida-Eto M, Sakashita N, et al. Partial impairment of c-Ret at tyrosine 1062 accelerates age-related hearing loss in mice. doi: 10.1016/j.neurobiolaging.2011.04.002. Neurobiology of Aging. In press. [DOI] [PubMed] [Google Scholar]
- 3.Wefstaedt P, Scheper V, Rieger H, Lenarz T, Stöver T. Neurotrophic factors of the GDNF family and their receptors are detectable in spiral ganglion cells of normal hearing as well as of deafened rats. Laryngo- Rhino- Otologie. 2006;85(11):802–808. doi: 10.1055/s-2006-925287. [DOI] [PubMed] [Google Scholar]
- 4.Ylikoski J, Pirvola U, Moshnyakov M, Palgi J, Arumae U, Saarma M. Expression patterns of neurotrophin and their receptor mRNAs in the rat inner ear. Hearing Research. 1993;65(1-2):69–78. doi: 10.1016/0378-5955(93)90202-c. [DOI] [PubMed] [Google Scholar]
- 5.Hossain K, Akhand AA, Kato M, et al. Arsenite induces apoptosis of murine T lymphocytes through membrane raft-linked signaling for activation of c-Jun amino-terminal kinase. Journal of Immunology. 2000;165(8):4290–4297. doi: 10.4049/jimmunol.165.8.4290. [DOI] [PubMed] [Google Scholar]
- 6.Kato M, Kumasaka MY, Takeda K, et al. L-cysteine as a regulator for arsenic-mediated cancer-promoting and anti-cancer effects. Toxicology in Vitro. 2011;25(3):623–629. doi: 10.1016/j.tiv.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Kato M, Onuma S, Kato Y, et al. Toxic elements in well water from Malaysia. Toxicological and Environmental Chemistry. 2010;92(9):1609–1612. [Google Scholar]
- 8.Kato M, Takeda K, Hossain K, et al. A redox-linked novel pathway for arsenic-mediated RET tyrosine kinase activation. Journal of Cellular Biochemistry. 2010;110(2):399–407. doi: 10.1002/jcb.22550. [DOI] [PubMed] [Google Scholar]
- 9.Kato M, Hattori T, Ikeda R, et al. Amount of pollen has an effect on the systemic and local levels of soluble ICAM-1 in patients with seasonal allergic rhinitis. Allergy. 1996;51(2):128–132. [PubMed] [Google Scholar]
- 10.Kato M, Hattori T, Kato Y, Matsumoto Y, Yamashita T, Nakashima I. Elevated soluble tumor necrosis factor receptor levels in seasonal allergic rhinitis patients. Allergy. 1999;54(3):278–282. doi: 10.1034/j.1398-9995.1999.00942.x. [DOI] [PubMed] [Google Scholar]
- 11.Kato M, Takeda K, Kawamoto Y, et al. Repair by Src kinase of function-impaired RET with multiple endocrine neoplasia type 2A mutation with substitutions of tyrosines in the COOH-terminal kinase domain for phenylalanine. Cancer Research. 2002;62(8):2414–2422. [PubMed] [Google Scholar]
- 12.Ohgami N, Ida-Eto M, Shimotake T, et al. c-Ret-mediated hearing loss in mice with Hirschsprung disease. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(29):13051–13056. doi: 10.1073/pnas.1004520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato M, Iida M, Goto Y, Kondo T, Yajima I. Sunlight exposure-mediated DNA damage in young adults. Cancer Epidemiology Biomarkers and Prevention. 2011;20(8):1622–1628. doi: 10.1158/1055-9965.EPI-11-0228. [DOI] [PubMed] [Google Scholar]
- 14.Kato M, Iwashita T, Akhand AA, et al. Molecular mechanism of activation and superactivation of Ret tyrosine kinases by ultraviolet light irradiation. Antioxidants and Redox Signaling. 2000;2(4):841–849. doi: 10.1089/ars.2000.2.4-841. [DOI] [PubMed] [Google Scholar]
- 15.Kato M, Iwashita T, Takeda K, et al. Ultraviolet light induces redox reaction-mediated dimerization and superactivation of oncogenic Ret tyrosine kinases. Molecular Biology of the Cell. 2000;11(1):93–101. doi: 10.1091/mbc.11.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato M, Ohgami N, Kawamoto Y, et al. Protective effect of hyperpigmented skin on UV-mediated cutaneous cancer development. Journal of Investigative Dermatology. 2007;127(5):1244–1249. doi: 10.1038/sj.jid.5700659. [DOI] [PubMed] [Google Scholar]
- 17.Hussein MR, Haemel AK, Sodilovsky O, Wood GS. Genomic instability in radial growth phase melanoma cell lines after ultraviolet irradiation. Journal of Clinical Pathology. 2005;58(4):389–396. doi: 10.1136/jcp.2004.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rigel DS. Epidemiology of melanoma. Seminars in Cutaneous Medicine and Surgery. 2010;29(4):204–209. doi: 10.1016/j.sder.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Kato M, Liu W, Akhand AA, et al. Linkage between melanocytic tumor development and early burst of Ret protein expression for tolerance induction in metallothionein-I/ret transgenic mouse lines. Oncogene. 1999;18(3):837–842. doi: 10.1038/sj.onc.1202329. [DOI] [PubMed] [Google Scholar]
- 20.Kato M, Liu W, Yi H, et al. The herbal medicine Sho-saiko-to inhibits growth and metastasis of malignant melanoma primarily developed in ret-transgenic mice. Journal of Investigative Dermatology. 1998;111(4):640–644. doi: 10.1046/j.1523-1747.1998.00341.x. [DOI] [PubMed] [Google Scholar]
- 21.Kato M, Takeda K, Kawamoto Y, et al. RET tyrosine kinase enhances hair growth in association with promotion of melanogenesis. Oncogene. 2001;20(51):7536–7541. doi: 10.1038/sj.onc.1204918. [DOI] [PubMed] [Google Scholar]
- 22.Kato M, Takeda K, Kawamoto Y, et al. c-kit-targeting immunotherapy for hereditary melanoma in a mouse model. Cancer Research. 2004;64(3):801–806. doi: 10.1158/0008-5472.can-03-2532. [DOI] [PubMed] [Google Scholar]
- 23.Kumasaka MY, Yajima I, Hossain K, et al. A novel mouse model for de novo melanoma. Cancer Research. 2010;70(1):24–29. doi: 10.1158/0008-5472.CAN-09-2838. [DOI] [PubMed] [Google Scholar]
- 24.Kato M, Liu W, Akhand AA, et al. Ultraviolet radiation induces both full activation of Ret kinase and malignant melanocytic tumor promotion in RFP-RET-transgenic mice. Journal of Investigative Dermatology. 2000;115(6):1157–1158. doi: 10.1046/j.1523-1747.2000.0202a-2.x. [DOI] [PubMed] [Google Scholar]
- 25.Iwamoto T, Takahashi M, Ito M, et al. Aberrant melanogenesis and melanocytic tumour development in transgenic mice that carry a metallothionein/ret fusion gene. EMBO Journal. 1991;10(11):3167–3175. doi: 10.1002/j.1460-2075.1991.tb04878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M, Takahashi M, Akhand AA, et al. Transgenic mouse model for skin malignant melanoma. Oncogene. 1998;17(14):1885–1888. doi: 10.1038/sj.onc.1202077. [DOI] [PubMed] [Google Scholar]
- 27.Ohshima Y, Yajima I, Takeda K, et al. c-RET molecule in malignant melanoma from oncogenic RET-carrying transgenic mice and human cell lines. PLoS ONE. 2010;5(4) doi: 10.1371/journal.pone.0010279. Article ID e10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. EMBO Journal. 2001;20(15):4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M, Wickner W. Vam10p defines a Sec18p-independent step of priming that allows yeast vacuole tethering. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6398–6403. doi: 10.1073/pnas.1132162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richmond A, Guo HF, Dhawan P, Yang J. How do chemokine/chemokine receptor activations affect tumorigenesis? Novartis Foundation Symposium. 2004;256:74–89. [PubMed] [Google Scholar]
- 31.Hamamura K, Furukawa K, Hayashi T, et al. Ganglioside GD3 promotes cell growth and invasion through p130Cas and paxillin in malignant melanoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):11041–11046. doi: 10.1073/pnas.0503658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohkawa Y, Miyazaki S, Hamamura K, et al. Ganglioside GD3 enhances adhesion signals and augments malignant properties of melanoma cells by recruiting integrins to glycolipid-enriched microdomains. Journal of Biological Chemistry. 2010;285(35):27213–27223. doi: 10.1074/jbc.M109.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamauchi Y, Furukawa K, Hamamura K, Furukawa K. Positive feedback loop between PI3K-Akt-mTORC1 signaling and the lipogenic pathway boosts Akt signaling: induction of the lipogenic pathway by a melanoma antigen. Cancer Research. 2011;71(14):4989–4997. doi: 10.1158/0008-5472.CAN-10-4108. [DOI] [PubMed] [Google Scholar]
- 34.Blois MS. Vitamin D, sunlight, and natural selection. Science. 1968;159(815):p. 652. [PubMed] [Google Scholar]
- 35.Kamei S, Yajima I, Yamamoto H, et al. Characterization of a novel member of the FGFR family, HrFGFR, in Halocynthia roretzi. Biochemical and Biophysical Research Communications. 2000;275(2):503–508. doi: 10.1006/bbrc.2000.3334. [DOI] [PubMed] [Google Scholar]
- 36.Kumasaka M, Sato S, Yajima I, Goding CR, Yamamoto H. Regulation of melanoblast and retinal pigment epithelium development by Xenopus laevis Mitf. Developmental Dynamics. 2005;234(3):523–534. doi: 10.1002/dvdy.20505. [DOI] [PubMed] [Google Scholar]
- 37.Kumasaka M, Sato S, Yajima I, Yamamoto H. Isolation and developmental expression of tyrosinase family genes in Xenopus laevis. Pigment Cell Research. 2003;16(5):455–462. doi: 10.1034/j.1600-0749.2003.00064.x. [DOI] [PubMed] [Google Scholar]
- 38.Puig I, Yajima I, Bonaventure J, Delmas V, Larue L. The tyrosinase promoter is active in a subset of vagal neural crest cells during early development in mice. Pigment Cell and Melanoma Research. 2009;22(3):331–334. doi: 10.1111/j.1755-148X.2009.00546.x. [DOI] [PubMed] [Google Scholar]
- 39.Sato S, Toyoda R, Katsuyama Y, et al. Structure and developmental expression of the ascidian TRP gene: insights into the evolution of pigment cell-specific gene expression. Developmental Dynamics. 1999;215(3):225–237. doi: 10.1002/(SICI)1097-0177(199907)215:3<225::AID-AJA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 40.Yajima I, Endo K, Sato S, et al. Cloning and functional analysis of ascidian Mitf in vivo: insights into the origin of vertebrate pigment cells. Mechanisms of Development. 2003;120(12):1489–1504. doi: 10.1016/j.mod.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Yajima I, Larue L. The location of heart melanocytes is specified and the level of pigmentation in the heart may correlate with coat color. Pigment Cell and Melanoma Research. 2008;21(4):471–476. doi: 10.1111/j.1755-148X.2008.00483.x. [DOI] [PubMed] [Google Scholar]
- 42.Goding CR. Melanocytes: the new black. International Journal of Biochemistry and Cell Biology. 2007;39(2):275–279. doi: 10.1016/j.biocel.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Robins A. Biological Perspectives on Human Pigmentation. New York, NY, USA: Cambridge University Press; 1991. [Google Scholar]
- 44.Hemesath TJ, Steingrimsson E, McGill G, et al. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes and Development. 1994;8(22):2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 45.Hodgkinson CA, Moore KJ, Nakayama A, et al. Mutations at the mouse Microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74(2):395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 46.Steingrímsson E, Copeland NG, Jenkins NA. Melanocytes and the Microphthalmia transcription factor network. Annual Review of Genetics. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- 47.Widlund HR, Fisher DE. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene. 2003;22(20):3035–3041. doi: 10.1038/sj.onc.1206443. [DOI] [PubMed] [Google Scholar]
- 48.Yajima I, Sato S, Kimura T, et al. An L1 element intronic insertion in the black-eyed white (Mitf(mi-bw)) gene: the loss of a single Mitf isoform responsible for the pigmentary defect and inner ear deafness. Human Molecular Genetics. 1999;8(8):1431–1441. doi: 10.1093/hmg/8.8.1431. [DOI] [PubMed] [Google Scholar]
- 49.Yasumoto KI, Amae S, Udono T, Fuse N, Takeda K, Shibahara S. A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work. Pigment Cell Research. 1998;11(6):329–336. doi: 10.1111/j.1600-0749.1998.tb00491.x. [DOI] [PubMed] [Google Scholar]
- 50.Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for Microphthalmia-associated transcription factor target genes. Pigment Cell and Melanoma Research. 2010;23(1):27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- 51.Khong HT, Rosenberg SA. The Waardenburg syndrome type 4 gene, SOX10, is a novel tumor-associated antigen identified in a patient with a dramatic response to immunotherapy. Cancer Research. 2002;62(11):3020–3023. [PMC free article] [PubMed] [Google Scholar]
- 52.Mascarenhas JB, Littlejohn EL, Wolsky RJ, et al. PAX3 and SOX10 activate MET receptor expression in melanoma. Pigment Cell and Melanoma Research. 2010;23(2):225–237. doi: 10.1111/j.1755-148X.2010.00667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plummer RS, Shea CR, Nelson M, et al. PAX3 expression in primary melanomas and nevi. Modern Pathology. 2008;21(5):525–530. doi: 10.1038/modpathol.3801019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bondurand N, Pingault V, Goerich DE, et al. Interaction among SOX10, PAX3 and MITF, three genes altered in Waardenburg syndrome. Human Molecular Genetics. 2000;9(13):1907–1917. doi: 10.1093/hmg/9.13.1907. [DOI] [PubMed] [Google Scholar]
- 55.Lee M, Goodall J, Verastegui C, Ballotti R, Goding CR. Direct regulation of the Microphthalmia promoter by Sox10 links Waardenburg-Shah syndrome (WS4)-associated hypopigmentation and deafness to WS2. Journal of Biological Chemistry. 2000;275(48):37978–37983. doi: 10.1074/jbc.M003816200. [DOI] [PubMed] [Google Scholar]
- 56.Potterf SB, Furumura M, Dunn KJ, Arnheiter H, Pavan WJ. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Human Genetics. 2000;107(1):1–6. doi: 10.1007/s004390000328. [DOI] [PubMed] [Google Scholar]
- 57.Verastegui C, Bille K, Ortonne JP, Ballotti R. Regulation of the Microphthalmia-associated transcription factor gene by the Waardenburg syndrome type 4 gene, SOX10. Journal of Biological Chemistry. 2000;275(40):30757–30760. doi: 10.1074/jbc.C000445200. [DOI] [PubMed] [Google Scholar]
- 58.Bertolotto C, Abbe P, Hemesath TJ, et al. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. Journal of Cell Biology. 1998;142(3):827–835. doi: 10.1083/jcb.142.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Price ER, Ding HF, Badalian T, et al. Lineage-specific signaling in melanocytes. C-Kit stimulation recruits p300/CBP to Microphthalmia. Journal of Biological Chemistry. 1998;273(29):17983–17986. doi: 10.1074/jbc.273.29.17983. [DOI] [PubMed] [Google Scholar]
- 60.Delmas V, Beermann F, Martinozzi S, et al. β-Catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes and Development. 2007;21(22):2923–2935. doi: 10.1101/gad.450107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochimica et Biophysica Acta. 2003;1653(1):1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 62.Omholt K, Platz A, Ringborg U, Hansson J. Cytoplasmic and nuclear accumulation of β-catenin is rarely caused by CTNNB1 exon 3 mutations in cutaneous malignant melanoma. International Journal of Cancer. 2001;92(6):839–842. doi: 10.1002/ijc.1270. [DOI] [PubMed] [Google Scholar]
- 63.Rimm DL, Caca K, Hu G, Harrison FB, Fearon ER. Frequent nuclear/cytoplasmic localization of β-catenin without exon 3 mutations in malignant melanoma. American Journal of Pathology. 1999;154(2):325–329. doi: 10.1016/s0002-9440(10)65278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorsky RI, Raible DW, Moon RT. Direct regulation of nacre, a zebrafish MITF homolog required for pigment cell formation, by the Wnt pathway. Genes and Development. 2000;14(2):158–162. [PMC free article] [PubMed] [Google Scholar]
- 65.Takeda K, Takemoto C, Kobayashi I, et al. Ser298 of MITF, a mutation site in Waardenburg syndrome type 2, is a phosphorylation site with functional significance. Human Molecular Genetics. 2000;9(1):125–132. doi: 10.1093/hmg/9.1.125. [DOI] [PubMed] [Google Scholar]
- 66.Widlund HR, Horstmann MA, Price ER, et al. β-Catenin-induced melanoma growth requires the downstream target Microphthalmia-associated transcription factor. Journal of Cell Biology. 2002;158(6):1079–1087. doi: 10.1083/jcb.200202049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mansky KC, Sankar U, Han J, Ostrowski MC. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-κB ligand signaling. Journal of Biological Chemistry. 2002;277(13):11077–11083. doi: 10.1074/jbc.M111696200. [DOI] [PubMed] [Google Scholar]
- 68.Wu M, Hemesath TJ, Takemoto CM, et al. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes and Development. 2000;14(3):301–312. [PMC free article] [PubMed] [Google Scholar]
- 69.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. Journal of Biological Chemistry. 2005;280(1):146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 70.Murakami H, Arnheiter H. Sumoylation modulates transcriptional activity of MITF in a promoter-specific manner. Pigment Cell Research. 2005;18(4):265–277. doi: 10.1111/j.1600-0749.2005.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bennett DC. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell and Melanoma Research. 2008;21(1):27–38. doi: 10.1111/j.1755-148X.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- 72.Collado M, Gil J, Efeyan A, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436(7051):p. 642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- 73.Gray-Schopfer VC, Cheong SC, Chong H, et al. Cellular senescence in naevi and immortalisation in melanoma: a role for p16? British Journal of Cancer. 2006;95(4):496–505. doi: 10.1038/sj.bjc.6603283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Michaloglou C, Vredeveld LCW, Soengas MS, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436(7051):720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 75.Mooi WJ, Peeper DS. Oncogene-induced cell senescence—halting on the road to cancer. The New England Journal of Medicine. 2006;355(10):1037–1046. doi: 10.1056/NEJMra062285. [DOI] [PubMed] [Google Scholar]
- 76.Vidal MJ, Loganzo F, Jr., de Oliveira AR, Hayward NK, Albino AP. Mutations and defective expression of the WAF1 p21 tumour-suppressor gene in malignant melanomas. Melanoma Research. 1995;5(4):243–250. doi: 10.1097/00008390-199508000-00006. [DOI] [PubMed] [Google Scholar]
- 77.Sauter ER, Yeo UC, von Stemm A, et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Research. 2002;62(11):3200–3206. [PubMed] [Google Scholar]
- 78.Carreira S, Goodall J, Aksan I, et al. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 2005;433(7027):764–769. doi: 10.1038/nature03269. [DOI] [PubMed] [Google Scholar]
- 79.Du J, Widlund HR, Horstmann MA, et al. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6(6):565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 80.Loercher AE, Tank EMH, Delston RB, Harbour JW. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. Journal of Cell Biology. 2005;168(1):35–40. doi: 10.1083/jcb.200410115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carreira S, Liu B, Goding CR. The gene encoding the T-box factor Tbx2 is a target for the Microphthalmia-associated transcription factor in melanocytes. Journal of Biological Chemistry. 2000;275(29):21920–21927. doi: 10.1074/jbc.M000035200. [DOI] [PubMed] [Google Scholar]
- 82.Vance KW, Carreira S, Brosch G, Goding CM. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Research. 2005;65(6):2260–2268. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 83.Cerroni L, Soyer HP, Kerl H. bcl-2 protein expression in cutaneous malignant melanoma and benign melanocytic nevi. American Journal of Dermatopathology. 1995;17(1):7–11. doi: 10.1097/00000372-199502000-00002. [DOI] [PubMed] [Google Scholar]
- 84.Ramsay JA, From L, Kahn HJ. bcl-2 protein expression in melanocytic neoplasms of the skin. Modern Pathology. 1995;8(2):150–154. [PubMed] [Google Scholar]
- 85.Selzer E, Schlagbauer-Wadl H, Okamoto I, Pehamberger H, Pötter R, Jansen B. Expression of Bcl-2 family members in human melanocytes, in melanoma metastases and in melanoma cell lines. Melanoma Research. 1998;8(3):197–203. doi: 10.1097/00008390-199806000-00001. [DOI] [PubMed] [Google Scholar]
- 86.McGill GG, Horstmann M, Widlund HR, et al. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 2002;109(6):707–718. doi: 10.1016/s0092-8674(02)00762-6. [DOI] [PubMed] [Google Scholar]
- 87.Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Current Biology. 2000;10(21):1359–1366. doi: 10.1016/s0960-9822(00)00781-8. [DOI] [PubMed] [Google Scholar]
- 88.Chang H, Schimmer AD. Livin/melanoma inhibitor of apoptosis protein as a potential therapeutic target for the treatment of malignancy. Molecular Cancer Therapeutics. 2007;6(1):24–30. doi: 10.1158/1535-7163.MCT-06-0443. [DOI] [PubMed] [Google Scholar]
- 89.Liu F, Fu Y, Meyskens FL. MiTF regulates cellular response to reactive oxygen species through transcriptional regulation of APE-1/Ref-1. Journal of Investigative Dermatology. 2009;129(2):422–431. doi: 10.1038/jid.2008.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Buscà R, Berra E, Gaggioli C, et al. Hypoxia-inducible factor 1α is a new target of Microphthalmia- associated transcription factor (MITF) in melanoma cells. Journal of Cell Biology. 2005;170(1):49–59. doi: 10.1083/jcb.200501067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McGill GG, Haq R, Nishimura EK, Fisher DE. c-Met expression is regulated by Mitf in the melanocyte lineage. Journal of Biological Chemistry. 2006;281(15):10365–10373. doi: 10.1074/jbc.M513094200. [DOI] [PubMed] [Google Scholar]
- 92.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nature Reviews Molecular Cell Biology. 2002;3(3):155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 94.Sánchez-Martín M, Rodríguez-García A, Pérez-Losada J, Sagrera A, Read AP, Sánchez-García I. SLUG (SNA12) deletions in patients with Waardenburg disease. Human Molecular Genetics. 2002;11(25):3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- 95.Carreira S, Goodall J, Denat L, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes and Development. 2006;20(24):3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Molecular Biology of the Cell. 2001;12(11):3451–3464. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoashi T, Watabe H, Muller J, Yamaguchi Y, Vieira WD, Hearing VJ. MART-1 is required for the function of the melanosomal matrix protein PMEL17/GP100 and the maturation of melanosomes. Journal of Biological Chemistry. 2005;280(14):14006–14016. doi: 10.1074/jbc.M413692200. [DOI] [PubMed] [Google Scholar]
- 98.Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. Journal of Cell Biology. 2001;152(4):809–824. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wasmeier C, Hume AN, Bolasco G, Seabra MC. Melanosomes at a glance. Journal of Cell Science. 2008;121, part 24:3995–3999. doi: 10.1242/jcs.040667. [DOI] [PubMed] [Google Scholar]
- 100.Bertolotto C, Buscà R, Abbe P, et al. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of Microphthalmia. Molecular and Cellular Biology. 1998;18(2):694–702. doi: 10.1128/mcb.18.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Du J, Miller AJ, Widlund HR, Horstmann MA, Ramaswamy S, Fisher DE. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. American Journal of Pathology. 2003;163(1):333–343. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yasumoto KI, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of Microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. Journal of Biological Chemistry. 1997;272(1):503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 103.Yavuzer U, Keenan E, Lowings P, Vachtenheim J, Currie G, Goding CR. The Microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10(1):123–134. [PubMed] [Google Scholar]
- 104.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends in Molecular Medicine. 2006;12(9):406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 105.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436(7047):117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 106.Wellbrock C, Marais R. Elevated expression of MITF counteracts B-RAF-stimulated melanocyte and melanoma cell proliferation. Journal of Cell Biology. 2005;170(5):703–708. doi: 10.1083/jcb.200505059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goding C, Meyskens FL., Jr. Microphthalmic-associated transcription factor integrates melanocyte biology and melanoma progression. Clinical Cancer Research. 2006;12(4):1069–1073. doi: 10.1158/1078-0432.CCR-05-2648. [DOI] [PubMed] [Google Scholar]
- 108.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445(7130):851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 109.Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell and Melanoma Research. 2010;23(6):746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]


