Abstract
Ameloblasts produce enamel matrix proteins such as amelogenin, ameloblastin, and amelotin during tooth development. The molecular mechanisms of ameloblast differentiation (amelogenesis) are currently not well understood. SP6 is a transcription factor of the Sp/KLF family that was recently found to regulate cell proliferation in a cell-type-specific manner. Sp6-deficient mice demonstrate characteristic tooth anomalies such as delayed eruption of the incisors and supernumerary teeth with disorganized amelogenesis. However, it remains unclear how Sp6 controls amelogenesis. In this study, we used SP6 high producer cells to identify SP6 target genes. Based on the observations that long-term culture of SP6 high producer cells reduced SP6 protein expression but not Sp6 mRNA expression, we found that SP6 is short lived and specifically degraded through a proteasome pathway. We established an in vitro inducible SP6 expression system coupled with siRNA knockdown and found a possible linkage between SP6 and amelogenesis through the regulation of amelotin and Rock1 gene expression by microarray analysis. Our findings suggest that the regulation of SP6 protein stability is one of the crucial steps in amelogenesis.
1. Introduction
Specificity Protein 6 (Sp6), also known as epiprofin, is a member of the SP/KLF transcription factor family [1–3]. Sp6 mRNA is expressed in dental epithelial cells during tooth development [3], and SP6 protein has been detected in the ameloblast layer of rat incisors accompanied by amelogenin expression [4]. The presence of two transcripts of Sp6 has been reported due to alternative promoter usage with tissue- and developmental stage-specific regulation, resulting in connections of two different first exons (epiprofin exon 1 and Sp6 exon 1) with the same second exon, which contains the coding region [3, 5]. However, we have recently reported that both transcripts are present in the same developmental stage in dental epithelial cells, demonstrating that the Sp6 gene comprises two first exons (exon 1a and exon 1b) as well as a second exon [6].
Two other research groups have developed Sp6-deficient mice to explore the function of SP6 in vivo, and they have reported several tooth abnormalities, including delayed eruption of incisors, supernumerary teeth, and enamel disorganization [7, 8]. We established a gain-of-function system in vitro and found that SP6 inhibits follistatin (Fst) gene expression in dental epithelial cells [4]. These findings show that Sp6 is indispensable for tooth development. However, the molecular basis for the biological functions of SP6 remains unclear.
In this study, we found that SP6 is a short lived protein that is specifically degraded via a proteasome pathway. Based on these findings, we established an in vitro SP6 induction system using a proteasome inhibitor, MG132, in combination with a small interfering RNA (siRNA) knockdown technique. The system sharply distinguished the presence or absence of SP6 protein in dental epithelial cells with the same genetic background. Taking advantage of this system, we screened SP6 target genes by DNA microarray analysis. Identification of amelotin (Amtn) and Rock1 (Rho-associated coiled-coil containing protein kinase 1) among the target genes suggested a specific role of SP6 in amelogenesis, providing a molecular basis for the direct link between SP6 function and tooth development.
2. Material and Methods
2.1. Sp6 High Producer C9 Cells
SP6 high producer cells, CHA9 cells, were previously established as a stable transformant clone [4], originating from the rat dental epithelial derived ameloblast-lineage clone, G5 [9]. G5 cells are a cloned cell derived from a rat enamel organ-derived dental epithelial sheet, expressed amelogenin mRNA, but not its protein [9]. CHA9 cells were recloned by serial limiting dilutions in the presence of G418 (400 μg/mL: Nakalai Tesque, Kyoto, Japan), and C9 cells, the clone with the highest production of SP6, were selected by Western blot analysis (WB). The cells were maintained in a combination of Dulbecco's modified Eagle's medium/Ham's F12 medium (D/F12, Nissui, Tokyo, Japan) supplemented with 10% foetal bovine serum (FBS, JRH Biosciences, Lenexa, Kan, USA).
2.2. Western Blot Analysis (WB)
Protein lysates were obtained by dissolving the cells in 2× sodium dodecyl sulfate (SDS) sample buffer, as described previously [4]. Protein concentration was measured using the BCA Protein Assay Kit (Pierce, Rockford, Ill, USA). Thirty μg of total protein were loaded on 10% SDS-polyacrylamide gels (SDS-PAGE). After electrophoresis, proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon; Millipore, Bedford, Mass, USA) and blocked with 5% skim milk in tris-buffered saline (TBS) (10 mM Tris-HCl [pH 8.0], 150 mM NaCl) with 0.05% Tween-20 for 3 h at room temperature (RT). The membranes were incubated with anti-HA antibody (1 : 5000; Santa Cruz Biotechnology, Santa Cruz, Calif, USA) overnight at 4°C, and immunodetection was performed using the same procedure described previously [4]. Quantification analysis was performed by densitometric scanning with “Quantity One” software (Bio-Rad laboratories, Hercules, CA, USA) according to the manufacturer's instructions. Lysates of Cos7 cells transfected with the CHA-Sp6 in pCIneo were used as a positive control if not specified.
2.3. Reverse Transcription-PCR (RT-PCR) and Quantitative PCR Analysis
Total RNA was purified using TRI Reagent (Molecular Research Center. Inc., Cincinnati, Ohio, USA) according to the manufacturer's instructions. After treatment with DNase I (Invitrogen, Carlsbad, Calif, USA), 500 ng of total RNA were converted into cDNA using an RNA PCR kit AMV Ver. 3.0 (Takara, Shiga, Japan) according to the manufacturer's instructions. Synthesized cDNA was subjected to either PCR analysis using Taq DNA polymerase (Promega, Madison, Wis, USA) or quantitative PCR (qPCR) using Thunderbird SYBR qPCR Mix (Toyobo Co. Ltd., Osaka, Japan). Gene-specific primer sets are shown in Table 1. qPCR was performed with a 7300 Real-Time PCR System (Applied Biosystems, Carlsbad, Calif, USA). The expression ratios were calculated from the threshold cycles and normalized to 18s rRNA results. All data were confirmed by triplicate experiments.
Table 1.
Gene-specific primers.
| Gene | Primer sequence |
|---|---|
| Amelotin | 5′-CCTCCTTATCCACCCCTTG-3′ |
| 5′-CCAACTGTGATGTGGTTTGC-3′ | |
| Car3 | 5′-CCTCTCTCTGGACCCTACC-3′ |
| 5′-TTTTGTCCAGGGCATCAAGG-3′ | |
| Follistatin | 5′-TTTTCTGTCCACCGGCAGCTCCAC-3′ |
| 5′-GCAAGATCCGGAGTGCTTCACT-3′ | |
| Osr2 | 5′-ATTCACGAGAGGACCCACAC-3′ |
| 5′-GTTTCGCCTGAACACTTTGC-3′ | |
| Pcm1 | 5′-CTTGGGTGGCATCAAACTCC-3′ |
| 5′-CCAAGTGAATCACGGTGTTG-3′ | |
| Rock1 | 5′-GGATGCTACCTGATCACCAG-3′ |
| 5′-CCGTAGGCAAACCCGCAAG-3′ | |
| Sp6 cds | 5′-CCGGCAATGCTAACCGCTGTCTGTG-3′ |
| 5′-GGCTCAGTTGGAGGACGCCGAGCTG-3′ | |
| Sp6 endogenous | 5′-GCGCCATCTTCAGACCCAC-3′ |
| 5′-CCACTTCGCAAGAGGATTTC-3′ | |
| Sp6 exogenous | 5′-GGCTAGAGTACTTAATACGACTCAC-3′ |
| 5′-CTCGAAGCATTAACCCTCACTAAAG-3′ | |
| 18s rRNA | 5′-TACCTGGTTGATCCTGCCAGTAGGAT-3′ |
| 5′-CCCGTCGGCATGTATTAGCTCTAGAA-3′ |
2.4. SP6 Protein Stability and Half-Life
For lysosomal analysis, C9 cells were treated with or without 1, 3, 10, and 20 mM of ammonium chloride (NH4Cl: Wako Pure Chemical Industries, Ltd. Osaka, Japan) at 7 and 24 h. For proteasomal analysis, Cbz-leu-leu-leucinal (MG132: Calbiochem, EMD Chemicals, Inc., San Diego, Calif, USA) or acetyl-leu-leu-norleucinal (ALLN: Calbiochem, EMD Biosciences Inc.) was applied to C9 cells. To investigate dose-dependent effects, C9 cells were treated with 0.3 and 10 μM of MG132 or 5, 10, and 15 μM of ALLN for 7 h. To analyze protein stability, cycloheximide (CHX; Wako Pure Chemical Industries, Ltd. Osaka, Japan) was used as described previously [10]. In brief, C9 cells were preincubated with 10 μM of MG132 for 8 h and washed with PBS(−) (137 mM NaCl, 8.1 mM Na2HPO4·12H2O, 2.68 mM KCl, 147 mM KH2PO4, [pH 7.4]) twice. The medium was then replaced with fresh medium, and 10 mM CHX was added either alone or together with MG132. The cells were harvested at the indicated time points, and the cell lysates were used for WB.
2.5. 20S Proteasome Activity
A 20S proteasome activity assay kit (Cayman Chemical Company, Ann Arbor, MI, USA) was used to analyze proteasome activity in accordance with the manufacturer's protocol. In brief, G5 and C9 cells at passage 7 (P7) and P28 were prepared with or without MG132 treatment for 8 h. Each cell preparation was transferred into individual tubes with 1 × 105 cells. The cells were then washed with 20S proteasome assay buffer. After centrifugation for 5 min at 500 g, the supernatant was aspirated. One hundred μL of 20S Proteasome Lysis buffer was added to the pellet and incubated at RT for 30 min. After centrifugation for 10 min at 1,000 g, 90 μL of the supernatant was transferred to a black 96-well detection plate. After 10 μL of buffer or control inhibitor solution was added to each well, 10 μL of substrate (Suc-LLVY-AMC) solution was added. The plate was then incubated for 1 h at 37°C. The fluorescent intensity of each well (excitation at 360 nm; emission at 480 nm) was analysed with Varioskan Flash (Thermo scientific, Waltham, Mass, USA) and SkanIt Software (Thermo scientific) according to the manufacturer's instructions. All experiments were performed in triplicate.
2.6. Immunocytochemistry (IC)
C9 cells were seeded in a 12-well plate. After treatment for 8 h with or without 10 μM MG132, the cells were washed with PBS(−) once. The cells were then fixed with 100% ethanol for 10 min at RT. After washing 3 times with PBS(−), the cells were permeabilized with 0.2% Triton X-100 in PBS(−) for 15 min at RT, then blocked with 4% BSA in PBS(−) for 30 min at RT. They were then incubated with HA-tagged antibody (1 : 250 dilution; Santa Cruz Biotechnology) overnight at 4°C. The next day, the cells were washed with PBS(−) 3 times and incubated with Alexa 594 anti-rabbit IgG (1 : 500 dilution; Molecular Probes, Eugene, Ore, USA) and 1 μg/mL Hoechst 33342 for 45 min at RT. After washing the cells 3 times with PBS(−), the wells were filled with PBS(−), and the plate was analysed by fluorescence microscopy (Table 2).
Table 2.
Antibody information.
| Antigen (clone) | Company | Code number | Dilution (WB) | Dilution (IC) |
|---|---|---|---|---|
| HA-tag (Y-11) | Santa Cruz biotechnology | sc-805 | 1 : 5000 | 1 : 250 |
| HA-tag (3F10) | Roche | #867423001 | 1 : 1000 | 1 : 250 |
| Ubiquitin | DAKO | Z0458 | 1 : 4500 | — |
| Sumo1 (FL-101) | Santa Cruz biotechnology | sc-9060 | 1 : 1000 | — |
| Sumo1 | Cell signalling | #4930 | 1 : 1000 | — |
| Sumo2/3 (18H8) | Cell signalling | #4971 | 1 : 1000 | — |
(Santa Cruz biotechnology, Inc., Santa Cruz, Calif, USA; DAKO, Glostrup, Denmark; Cell signalling, Danvers, Mass, USA).
2.7. Knockdown of Sp6 by siRNA Transfection
Twenty-five nM of Stealth RNAi siRNA for Sp6 (Sp6 siRNA; Invitrogen) or Stealth RNAi siRNA Negative Control Duplex Med GC #2 (GC; Invitrogen) was transfected into C9 cells at 30% confluency at P28 in a 12-well plate using Lipofectamine RNAiMAX (MAX; Invitrogen) for 24 or 48 h; the cells were then harvested. Ten μM of MG132 was added 8 h before harvesting the cells (i.e., the cells were subjected to 8 h of treatment with MG132).
2.8. Microarray Analysis
Total RNA was isolated from Sp6 siRNA-MG132- or GC-MG132-treated C9 samples using TRI Reagent. Purity was examined using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Calif, USA). Microarray analysis was performed using GeneChip Rat Gene 1.0 ST Arrays (Affymetrix, Santa Clara, Calif, USA). This microarray chip contains 27,342 oligonucleotides probes for known and unknown genes. First-strand cDNA was synthesized from 300 ng of total RNA using a WT Expression Kit (Affymetrix) according to the manufacturer's instructions. Ten μg of cRNA was added to the second-cycle cDNA reaction. cDNA was fragmented and end-labelled with a GeneChip WT Terminal Labeling Kit (Affymetrix). Approximately 5.5 μg of fragmented and labelled DNA target was hybridized to the array for 17 h at 45°C in a GeneChip Hybridization Oven 640 (Affymetrix) in accordance with the manufacturer's recommendations. Hybridized arrays were washed and stained on a GeneChip Fluidics Station 450 and scanned with a GeneChip Scanner 3000 7G (Affymetrix), and a CEL file was generated for each array. The data were analysed with GeneSpring GX11.0 (Silicon Genetics, Redwood City, Calif, USA). Expressed genes that showed fluorescence intensity greater than 100 were further analysed. Genes with more than a 2-fold difference between the Sp6 siRNA-treated samples and negative controls were selected.
2.9. Statistics
The immunoprecipitation, Western blot analyses, 20S proteasome activity, and qRT-PCR data presented are representative of three independent analyses conducted under the same experimental conditions. Determination of the half-life of the SP6 protein was conducted twice using individual analyses under the same experimental conditions. The data reported reflect the means ± S.D. of triplicate samples from the representative experiments. Student's t-test was utilized for proteasome activity by Microsoft Exel for Mac 2008 (Redmond, Wash, USA).
3. Results
3.1. SP6 Expression during Long-Term Culture
In a previous study, we established Sp6 stable transformants, CHA9 cells [4], using the dental epithelial clone, G5 cells [9]. However, we found that the level of SP6 was reduced in CHA9 cells during long-term culture. To investigate the molecular nature and function of SP6, we recloned a new SP6 high producer clone from CHA9 cells, C9 cells. Unexpectedly, by WB, we again found a significant reduction in the SP6 expression level in C9 cells at P28 compared with strong SP6 expression at P7 (Figure 1(a)). In contrast, as shown in Figure 1(b), the level of Sp6 mRNA cds, which detects the coding regions derived from both endogenous and exogenous Sp6 mRNA, was about 3.2-and 2.2-fold higher at P7 and P28, respectively, compared to that of the parent G5 cells. The polygonal epithelial cell-like morphology and growth rate were kept in C9 cells at P7 and P28. Based on these findings, we assumed that the stability of SP6 protein is involved in the regulation of SP6 expression.
Figure 1.

Downregulation of SP6 in C9 cells during long-term culture. (a) SP6 expression in C9 cells (P7 and P28) and in parent G5 cells. SP6 protein levels were analysed by WB using anti-HA antibody. Arrow: SP6; asterisk: nonspecific band and Posi: positive control. (b) Sp6 mRNA expression in C9 cells (P7 and P28) and G5 cells. Upper panel indicates schematic structure of the Sp6 gene and expression plasmid together with primer positions. Primers designed to detect three distinct types of Sp6 mRNA. Arrows indicate each primer's position and direction. Grey box: untranslated region; striped box: HA-tag; cds: coding region of Sp6; endo: endogenous Sp6; exo: exogenous Sp6. Middle panel shows the representative result of RT-PCR analysis. Lower panel shows densitometric analysis data of RT-PCR. Each Sp6 level was normalized by 18S rRNA level, and the relative ratio was calculated compared to that of G5 cells. The level of G5 cells was assigned as one.
3.2. Analysis of SP6 Stability
To explore the cause of the reduction of SP6 protein expression in C9 cells at P28, we examined two major pathways of protein degradation in the cells: the proteasome and lysosomal pathways [11, 12]. We first tested whether the lysosomal inhibitor NH4Cl had any effect on the inhibition of SP6 protein degradation. As shown in Figure 2(a), we could not detect any enhancement or induction of SP6 expression by NH4Cl at P7 or P28; rather we found an acceleration of its reduction due to cell toxicity at higher concentrations. Next we examined the effects of a proteasome inhibitor, MG132, on SP6 protein expression in C9 cells. In contrast to NH4Cl, C9 cells treated with 0.3 and 10 μM of MG132 for 7 h showed remarkably enhanced levels of SP6 protein expression at P7 and P28 in a dose-dependent manner (Figure 2(b), upper panel). C9 cells treated with another proteasome inhibitor, ALLN, also showed dose-dependent enhanced SP6 protein expression, although the optimal dose was different between P7 and P28 (Figure 2(b), lower panel). We further examined the effect of time course of MG132 treatment on SP6 expression. SP6 protein expression was gradually increased from 2 to 12 h after the addition of 10 μM MG132 to the culture, and more than 85% of maximal enhancement was obtained by 8 h of treatment, followed by a peak up to 68-fold increase at 12 h (Figure 2(c)). To prove the proteasome activity in dental epithelial cells, we measured the 20S proteasome activity in C9 cells at P7 and P28 as well as in the parent G5 cells. C9 at P28 and G5 cells had the similar levels of 20S proteasome activity (n = 3, P > 0.01). C9 at P28, proteasome activity became 67% of P7 (n = 3, P < 0.01). We also observed that G5 cells have statistically higher activity compared to that of Jurkat cells (n = 3, P < 0.01), which are a positive control with high 20S proteasome activity provided in a kit (Figure 2(d)), indicating a possible role of proteasome activity in dental epithelial function.
Figure 2.
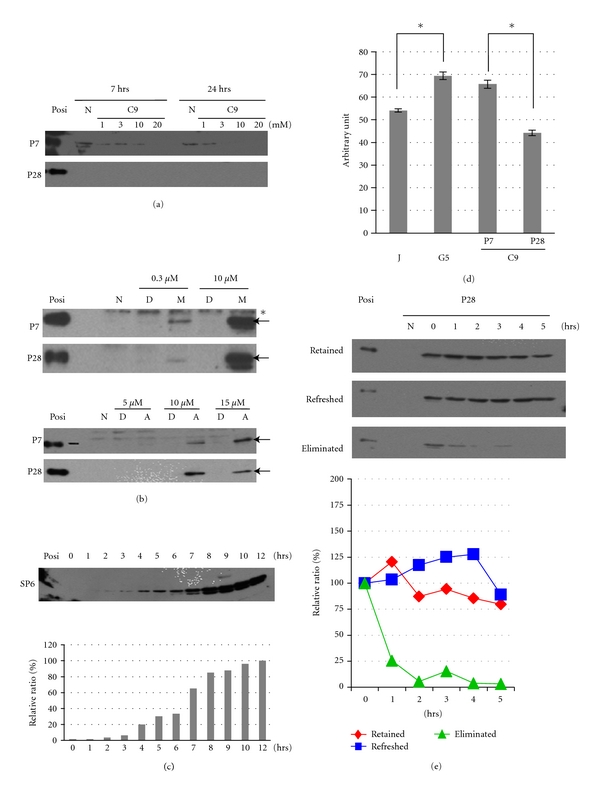
Effects of protease inhibitors on SP6 expression level in C9 cells. (a) Dose-dependent effects of the lysosomal inhibitor, NH4Cl, on SP6 expression in C9 cells. The SP6 expression level was evaluated by WB. Posi: positive control; N: no treatment. (b) Dose-dependent effects of MG132 on SP6 expression in C9 cells. Upper panel: the cells were treated with MG132 (M) or DMSO (D) as a control (vehicle alone) for 7 h. Lower panel: cells were treated with ALLN (A) or DMSO (D) for 7 h. Posi: positive control; N: no treatment; arrow: SP6; asterisk: nonspecific band. (c) Time course of the effects of MG132 treatment on SP6 induction in C9 cells. C9 cells at P28 were treated with 10 μM MG132 during the indicated time. Upper panel: SP6 expression analysed by WB; Posi: positive control. Lower panel: Relative levels of signal intensity of WB. Each SP6 level was shown as the relative level compared with the signal detected at 12 h (100%). (d) 20S proteasome activity in G5 and C9 cells (P7 and P28). J: Jurkat cells (positive control). The brackets show the significant difference demonstrated by Student's t-test (n = 3, *P < 0.01). (e) Half-life analysis of SP6 in C9 cells at P28 determined by blocking de novo protein synthesis. C9 cells were pretreated with MG132 for 8 h, and then 10 μg/mL CHX was added during the indicated time. Upper panel: SP6 expression detected by WB; Retained: no medium change (MG132 was retained) when CHX was added; Refreshed: new medium replaced the old medium, and MG132 was newly added to the culture with CHX; Eliminated: the medium was replaced without MG132, only CHX was added; Posi: positive control; N: untreated C9 cells. Upper panel: relative SP6 expression level ratios. Each signal level was normalized with that at time 0 (100%). Data were reproduced by two independent experiments.
We next analysed the stability of SP6 protein in C9 cells at P28 to determine whether SP6 is a short lived protein. C9 cells were cultured with 10 mM CHX in the presence or absence of MG132 as described in the experimental section. As shown in Figure 2(e), regardless of whether MG132 was retained continuously or refreshed, MG132 treatment maintained a similar SP6 level. When we changed the medium to wash out MG132, the level of SP6 was rapidly reduced to 25% of that of the peak within 1 h (Figure 2(e)). The half-life of the SP6 protein (t1/2) was assessed to be approximately 40 min. These results demonstrated that the SP6 protein is actually a short lived protein and is rapidly degraded through a proteasome pathway in dental epithelial cells.
3.3. In Vitro Regulation of SP6 Expression in Combination with MG132 Treatment and siRNA Knockdown
Although a proteasome inhibitor, MG132, could markedly induce and stabilize SP6 protein levels in C9 cells, it was not clear whether MG132-induced SP6 were able to function as a transcription factor. To address this question, we first examined the localization of the SP6 protein in MG132-treated C9 cells at P28 by immunocytochemical analysis. As shown in Figure 3(a), distinct SP6 signals was detected mainly in the nuclei of MG132-treated C9 cells and were detected hardly at all in untreated cells. Nuclear localization of SP6 was the same as native SP6 observed in vivo [4]. Next, to check whether MG132-induced SP6 could regulate the gene expression, we examined RT-PCR analysis of Fst mRNA since we previously demonstrated that it is specifically downregulated by SP6 (4). We confirmed the downregulation of Fst mRNA in MG132-treated C9 cells (data not shown), indicating that MG132-induced SP6 protein can act as a transcription factor. These findings prompted us to further analyze the SP6 target genes using an MG132-mediated SP6 induction system in C9 cells. This is great advantage to analyze SP6 functions in one cell line, in other words, with the same genetic background. Because when we simply compared transformant C9 cells with parental G5 cells, we cannot eliminate the influences upon gene expression due to the genomic insertion of transgene into C9 cells compared to parental G5 cells.
Figure 3.
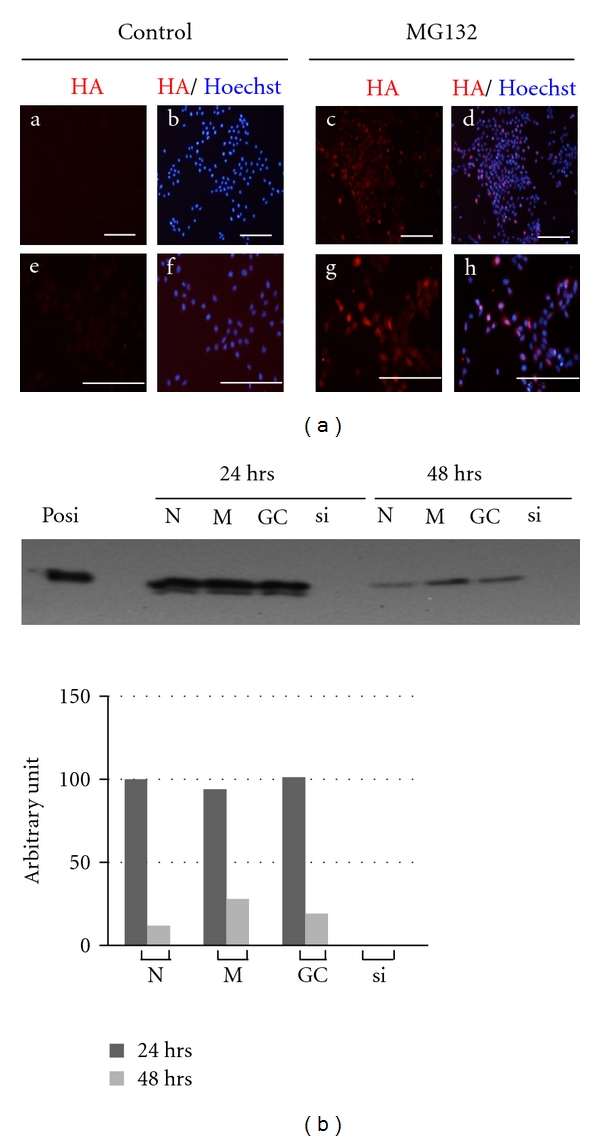
Specificity of SP6 induction by MG132 treatment. (a) Subcellular localization of SP6 in C9 cells treated with MG132. SP6 localization was analysed by immunofluorescence signals. Red colour indicates SP6 detected by anti-HA antibody, blue colour shows nuclei stained with Hoechst 33342. (a–d): low magnification; (e–h): high magnification. Scale bar indicates 200 μm. (b) Knockdown of SP6 in MG132-treated C9 cells by siRNA. Posi: positive control; N: no treatment; M: Lipofectamine RNAiMAX only; GC: negative control siRNA; si: siRNA against Sp6. Upper panel: SP6 expression was detected by WB. Lower panel: relative SP6 levels estimated by densitometric analysis of WB results.
To establish the specified SP6 induction system, we first checked the feasibility of in vitro MG132-mediated SP6 induction combined with siRNA knockdown to eliminate the dosage effect of Sp6 gene. As shown in Figure 3(b), SP6 expression was completely inhibited in C9 cells treated with Sp6-specific siRNA for both 24 h and 48 h compared with control siRNA (GC-) treated cells. In addition, we observed that C9 cells treated with siRNA for 48 h produced approximately 25% of the SP6 protein level at 24 h. Since the SP6 protein level in GC-treated cells was comparable with that in untreated cells (N) as well as that in the cells treated with transfection reagent (M) at 48 h, this reduction was not caused by reagent toxicity. These results confirmed that the in vitro MG132-mediated SP6 induction system combined with siRNA was a reliable experimental procedure for detecting transcriptional target genes.
3.4. Identification of Sp6 Target Genes by Microarray Analysis
To find out the tissue-specific function of SP6 as a transcription factor in dental epithelial cells, we performed microarray analysis using the in vitro MG132-mediated SP6 induction system to compare SP6 siRNA-treated samples and the control siRNA (GC) samples shown in Figure 3(b) at 24 h and 48 h. Among 27,342 probes on the array, 74 (known genes: 47; unknown genes: 27) and 310 genes (known genes: 190; unknown genes: 120) were regulated by SP6 presence or absence at 24 h and 48 h, respectively (Figure 4(a)). Twenty-one genes were commonly regulated among the 24 and 48 h-treated samples, 17 of which are known genes. Among these, we found several potential amelogenesis-related genes, including Amtn [13], Rock1 [14] and Car3 (carbonic anhydrase 3) [15] among all samples, Fst [4] in the 24 h group of samples, and Pcm1 (pericentriolar material 1) [16] and Osr2 (Odd-skipped 2) [17] in the 48 h group of samples. As shown in the right panel of Figure 4(a), the Amtn, Pcm1, and Rock1 genes were positively regulated, and the Fst, Car3, and Osr2 genes were negatively regulated, by the presence of SP6 protein. To validate the microarray data, we further analysed mRNA levels of these genes in C9 cells by qPCR analysis. The relative mRNA levels from the microarray and qPCR results correlated well (Figure 4(b)). Both the Amtn and Pcm1 genes were increased 8-9-fold at 48 h compared with 24 h, while the level of Rock1 mRNA was similar between the two time points. On the other hand, the Fst, Car3, and Osr2 genes were all reduced to 20%–50% of the original level at 48 h compared with 24 h. To further confirm the specificity of the relationship between SP6 protein expression and target gene response, we examined Amtn and Rock1 mRNA expression in C9 cells at P7 and P28. The expression of Amtn and Rock1 mRNAs was significantly downregulated at P28 to 4.5% and 30%, respectively, of that at P7 (Figure 4(c)). These data clearly demonstrated that the stabilization of the SP6 protein regulates the expression of the target genes in dental epithelial cells.
Figure 4.

Screening of Sp6 target genes by microarray. (a) Venn diagram showing a summary of microarray data; the number of genes that was changed more than 2-fold between the SP6 producing sample (GC: negative control of siRNA) and the SP6 knockdown sample (siRNA of SP6) is also indicated. Numbers with brackets are unknown genes and those without brackets are known genes. The right panel indicates that the expression levels of tooth-related genes were normalized using a control (GC) based on microarray data. (b) Confirmation of microarray data by qPCR. The fold changes were calculated and compared to that of 24 h control (GC 24; 100%). GC: negative control siRNA; si: siRNA against Sp6. All experiments were performed in triplicate. (c) The mRNA levels of the Amtn and Rock1 genes in C9 at P7 and P28. qPCR was performed with total RNA purified from C9 cells at P7 and P28 and normalized by the level of 18s rRNA. The levels of Amtn and Rock1 mRNA expression at P7 are indicated as 100%.
4. Discussion
Both loss- and gain-of-function studies have demonstrated that Sp6 is indispensable during development of teeth, skin, limbs, lungs, and so on. [4, 7, 8, 18]. However, little is known about the molecular basis for the biological role of Sp6 [19]. In this study, we obtained two important findings relating to SP6; one is its stability in nature, and the other is the potential target genes during tooth development.
The first finding was that the SP6 protein level is finely tuned by a ubiquitin-independent proteasome pathway in dental epithelial cells. SP6 protein is a short lived molecule (t1/2 = 40 min, Figure 2(e)) similar to the immediate-early gene product, c-Fos (t1/2 = 47 min during G0/G1 transition in Balb/C3T3 fibroblasts) [20]. Many transcription factors are short lived proteins and are degraded via a proteasome pathway [11], indicating that transcription factors are tightly regulated to maintain homeostasis at specific differentiation stages or tissue circumstances. Notably, the level of Sp6 mRNA was maintained at a similar level during long-term culture, although SP6 protein expression was transiently high during early cell passage and was maintained at a low level during later passages (Figures 1(a) and 1(b)). 20S proteasome activities in dental epithelial cells are constantly high (Figure 2(d)); however, the underlying mechanisms of transient SP6 expression during early passage remain unclear.
In general, proteasome degradation is mediated by ubiquitin-dependent or -independent pathways [21]. The ubiquitin-dependent pathway is known to be initiated by polyubiquitination or other posttranslational modifications, such as sumoylation and phosphorylation of the target proteins [22]. We found possible sites for these modifications in the rat SP6 sequence; however, we could not detect any modifications of SP6 (data not shown). It is quite interesting issue to see how SP6 was stabilized in C9 cells at P7 but not in C9 cells at P28 or in the parent G5 cells in relation to proteasome regulation. A unique structural feature of the SP6 protein among the Sp/KLF family is a proline-rich sequence at the N-terminal half of the SP6 protein [1, 2]. Since proline-rich sequences often form a protein-protein binding interface [23], the binding partner(s) of SP6 via this region could be involved in SP6 stability. Further investigations are required for better understanding of proteasome regulation of SP6 stability and its relationship to SP6 protein binding partners.
The second important finding was the identification of novel SP6 target genes linked with amelogenesis. We first developed an in vitro MG132-mediated SP6 induction system combined with siRNA knockdown technology (Figure 3). The most important feature of this in vitro system is that it enables direct investigation of the downstream target genes during SP6 functioning maintaining the same genetic background. Taking advantage of this system, we screened and identified potential target genes by microarray analysis. We found 6 tooth-related genes, Amtn, Pcm1, Rock1, Fst, Car3, and Osr2, which were regulated by the SP6 protein (Figure 4).
Among these, the Amtn gene was strikingly upregulated by the presence of SP6 protein. The Amtn gene was recently described as a novel ameloblast-specific gene [13, 24, 25], and it is highly conserved in mammals; the rat Amtn gene is located at chromosome 14p21 where it is quite close to other ameloblast-specific genes, including ameloblastin and enamelin at chromosome 14p22 [24, 25]. In addition, the AMTN protein has been detected in the enamel matrix at the postsecretory transition and maturation stage of ameloblasts during enamel development [13, 24, 26], since SP6 protein is expressed in the ameloblast layer from the presecretory stage to the secretory stage in the enamel-forming region [4], indicating a sequential expression pattern of SP6 and AMTN. Another SP6 target gene in amelogenesis is Rock1, a notable gene in amelogenesis. ROCK1 protein expression is localized in the inner enamel epithelium in presecretory through mature ameloblasts and the stratum intermedium in dental epithelial cells, regulating cell morphology, adhesion to control cell proliferation and cell polarity [14, 27]. Altogether, SP6 functions, at least in part, through Amtn or Rock1 regulation in amelogenesis. To the best of our knowledge, this is the first report to demonstrate a molecular connection between SP6 function and the late stage of amelogenesis related genes. In Figure 5, we summarize our current model of SP6 expression and its functional coupling with amelogenesis, incorporating our present study with previous observations [6, 18].
Figure 5.

Proposed model of Sp6 expression and function coupled with amelogenesis in dental epithelial cells. BMP and Wnt signals can enhance Sp6 promoter activity, resulting in the enhancement of Sp6 mRNA expression (6). The level of SP6 protein expression is regulated by proteasome activity, which would be physiologically controlled by a MG132-equivalent factor X. Stabilized SP6 protein is transferred into the nucleus and regulates the expression of amelotin and rock1 mRNA. BRE: BMP signal-responsive elements; WRE: Wnt signal-responsive elements.
In conclusions, we found that SP6 protein is short lived, and our in vitro SP6 induction system coupled with microarray analysis identified novel ameloblast-specific target genes. Our findings provide the molecular basis for the nature of the SP6 protein and its possible function as a transcription factor in dental epithelial cells. Further studies are required to analyze the tight regulation of both SP6 protein expression and stability during tooth development to clarify the biological roles of SP6. Specially we have to confirm direct binding of SP6 to the new target genes in this system and endogenous system in vitro. Furthermore, to link with in vivo function in amelogenesis, we need to establish ameloblast differentiation system to mimic in vivo status, for example, 3D culture or organ culture combined with siRNA technology and observe the differentiation markers. These issues may provide further hints in relation to the developmental regulatory system and cellular physiological homeostasis.
Acknowledgments
The authors would like to extend their special thanks to Mr. Hideaki Horikawa, Support Center for Advanced Medical Sciences, the University of Tokushima Graduate School, Institute of Health Biosciences, for his support with the microarray analyses. This work was partly supported by Grants-in-aid for Scientific Research (no. 21791805) and Cooperative Research Grant of the Institute for Enzyme Research, the University of Tokushima, as well as a Research Grant from KAO Health Science Research.
References
- 1.Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85(5):551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Scohy S, Gabant P, van Reeth T, et al. Identification of KLF13 and KLF14 (Sp6), novel members of the Sp6/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 3.Nakamura T, Unda F, de-Vega S, et al. The Kruppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. Journal of Biological Chemistry. 2004;279(1):626–634. doi: 10.1074/jbc.M307502200. [DOI] [PubMed] [Google Scholar]
- 4.Ruspita I, Miyoshi K, Muto T, Abe K, Horiguchi T, Noma T. Sp6 down regulation of follistatin gene expression in ameloblast. Journal of Medical Investigation. 2008;55(1-2):87–98. doi: 10.2152/jmi.55.87. [DOI] [PubMed] [Google Scholar]
- 5.Hertveldt V, De Mees C, Scohy S, Van Vooren P, Szpirer J, Szpirer C. The Sp6 locus uses several promoters and generate sense and antisense transcripts. Biochimie. 2007;89(11):1381–1387. doi: 10.1016/j.biochi.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Wahyudi IA, Horiguchi T, Miyoshi K, et al. Isolation and characterization of mouse specificity 6 promoter. The Indonesian Journal of Dental Research. 2010;1:21–34. [Google Scholar]
- 7.Nakamura T, De Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. Journal of Biological Chemistry. 2008;283(8):4825–4833. doi: 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- 8.Hertveldt V, Louryan S, Van Reeth T, et al. The development of several organs and appendages is impaired in mice lacking Sp6. Developmental Dynamics. 2008;237(4):883–892. doi: 10.1002/dvdy.21355. [DOI] [PubMed] [Google Scholar]
- 9.Abe K, Miyoshi K, Muto T, et al. Establishment and characterization of rat dental epithelial derived ameloblast-lineage clones. Journal of Bioscience and Bioengineering. 2007;103(5):479–485. doi: 10.1263/jbb.103.479. [DOI] [PubMed] [Google Scholar]
- 10.Azzu V, Mookerjee SA, Brand MD. Rapid turnover of mitochondrial uncoupling protein 3. Biochemical Journal. 2010;426(1):13–17. doi: 10.1042/BJ20091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee DH. Proteasome inhibitors: valuable new tools for cell biologists. Trends in Cell Biology. 1998;8(10):397–403. doi: 10.1016/s0962-8924(98)01346-4. [DOI] [PubMed] [Google Scholar]
- 12.Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation cellular topology and biochemistry of lysosomal lipid generation. Biochimica et Biophysica Acta. 2009;1793(4):674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Moffatt P, Smith CE, St.-Arnaud R. R, Simmons D, Wright JT, Nanci A. Cloning of rat amelotin and localization of the protein to the basal lamina of maturation stage ameloblasts and junctional epithelium. Biochemical Journal. 2006;399(1):37–46. doi: 10.1042/BJ20060662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otsu K, Kishigami R, Fujiwara N, Ishizeki K, Harada H. Functional role of Rho-kinase in ameloblast differentiation. Journal of Cellular Physiology. 2011;226(10):2527–2534. doi: 10.1002/jcp.22597. [DOI] [PubMed] [Google Scholar]
- 15.Lacruz RS, Hilvo M, Kurtz I, Paine ML. A survey of carbonic anhydrase mRNA expression in enamel cells. Biochemical and Biophysical Research Communications. 2010;393(4):883–887. doi: 10.1016/j.bbrc.2010.02.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balczon R, Simerly C, Takahashi D, Schatten G. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motility and the Cytoskeleton. 2002;52(3):183–192. doi: 10.1002/cm.10043. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323(5918):1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talamillo A, Delgado I, Nakamura T, et al. Role of epiprofin, a zinc-finger transcription factor, in limb development. Developmental Biology. 2010;337(2):363–374. doi: 10.1016/j.ydbio.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jimenez-Rojo L, Ibarretxe G, Aurrekoetxea M, et al. Epiprofin/Sp6: a new player in the regulation of tooth development. Histology and Histopathology. 2010;25:1621–1630. doi: 10.14670/HH-25.1621. [DOI] [PubMed] [Google Scholar]
- 20.Ferrara P, Andermarcher E, Bossis G, et al. The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene. 2003;22(10):1461–1474. doi: 10.1038/sj.onc.1206266. [DOI] [PubMed] [Google Scholar]
- 21.Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Moscow) 2009;74(13):1411–1442. doi: 10.1134/s000629790913001x. [DOI] [PubMed] [Google Scholar]
- 22.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nature Reviews Molecular Cell Biology. 2009;10(8):564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 23.Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259(5098):1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 24.Iwasaki K, Bajenova E, Somogyi-Ganss E, et al. Amelotin—a novel secreted, ameloblast-specific protein. Journal of Dental Research. 2005;84(12):1127–1132. doi: 10.1177/154405910508401207. [DOI] [PubMed] [Google Scholar]
- 25.Moffatt P, Smith CE, Sooknanan R, St-Arnaud R, Nanci A. Identification of secreted and membrane proteins in the rat incisor enamel organ using a signal-trap screening approach. European Journal of Oral Sciences. 2006;114(1):139–146. doi: 10.1111/j.1600-0722.2006.00318.x. [DOI] [PubMed] [Google Scholar]
- 26.Rauth RJ, Potter KS, Ngan AY, et al. Dental enamel: genes define biomechanics. Journal of the California Dental Association. 2009;37(12):863–868. [PMC free article] [PubMed] [Google Scholar]
- 27.Biz MT, Marques MR, Crema VO, Moriscot AS, Dos Santos MF. GTPases RhoA and Rac1 are important for amelogenin and DSPP expression during differentiation of ameloblast and odontoblas. Cell and Tissue Research. 2010;340(3):459–470. doi: 10.1007/s00441-010-0961-0. [DOI] [PubMed] [Google Scholar]


