Abstract
Background
Genes in the major histocompatibility complex (MHC) have a critical role in both the innate and adaptive immune responses because of their involvement in presenting foreign peptides to T cells. However, the nature has remained largely unknown.
Results
We examined the genetic variation in MHC class IIB in half-smooth tongue sole (Cynoglossus semilaevis) after challenge with vibrio anguillarum. Two thousand and four hundred fry from 12 half-smooth tongue sole families were challenged with Vibrio anguillarum. To determine any association between alleles and resistance or susceptibility to V. anguillarum, 160 individuals from four high-resistance (HR, < 40.55% mortality) families and four low-resistance (LR, > 73.27% mortality) families were selected for MHC IIB exon2 gene sequence analysis. The MHC IIB exon2 genes of tongue sole displayed a high level of polymorphism and were discovered at least four loci. Meanwhile, the dN/dS [the ratio of non-synonymous (dN) substitutions to synonymous (dS) substitutions] in the peptide-binding region (PBR) was higher than that in the non-peptide-binding region (non-PBR). Eighty-eight alleles were discovered among 160 individuals, and 13 out of 88 alleles were used to analyze the distribution pattern between the resistant and susceptible families. Certain alleles presented in HR and LR with a different frequency, while other alleles were discovered in only the HR or LR families, not both. Five alleles, Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801, were found to be associated with susceptibility to V. anguillarum with a frequency of 1.25%, 1.25%, 1.25%, 1.25% and 2.5% in the HR families, and 35%, 33.75%, 27.5%, 16.25%, 15% in the LR families (p < 0.01, 0.01, 0.01, 0.01, 0.01), respectively. Four alleles, Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801 and Cyse-DBB*5901, were found to be associated with resistance to V. anguillarum, with a frequency of 13.75%, 11.25%, 11.25%, 8.75% in the HR families and 1.25%, 1.25%, 1.25%, 1.25% and 1.25% in the LR families (p < 0.01, 0.05, 0.05 and p = 0.064), respectively.
Conclusions
Elucidation of the role of MHC II B genes in half-smooth tongue sole should prove to be helpful to the in-depth development of marker-assisted selective breeding in half-smooth tongue sole.
Keywords: Cynoglossus semilaevis, Vibrio anguillarum, polymorphism, MHC IIB, susceptibility, resistance
Background
Major histocompatibility complex (MHC) molecules play a critical role in both innate and adaptive immunity by presenting foreign peptides to T cells in vertebrate organisms, and have been considered candidate molecular markers of an association between polymorphisms and resistance/susceptibility to diseases [1]. A combination of balanced and directional selection is thought to be responsible for allelic variation of MHC genes in vertebrate populations, because pathogen pressure varies at different times and locations [2]. Two classes of MHC are found in fish, MHC class I and class II molecules. The genes encode glycoproteins which bind peptides for the presentation of self and non-self peptides to T-cell receptors (TCR) [3].
The MHC class II molecules are symmetrical heterodimers, consisting of one alpha chain and one beta chain, with non-covalent contacts in which the alpha1 and beta1 domains form a peptide-binding region (PBR). In mammals, MHC class II genes are constitutively expressed in antigen-presenting cells such as macrophages, B cells, monocytes and dendritic cells, and have direct functional relevance in the immune response. Class I antigens are expressed in all somatic cells [1,4,5]. In teleosts, class I and class II genes were found to reside on different linkage groups [6-8]. Many MHC genes have been isolated, characterized expressed and analyzed in at least 30 different fish species over the last twenty years [9-14]. Multiple loci and a considerable number of alleles at each given locus were found in the classical MHC genes. The peptide-binding region (PBR) contains the highest level of polymorphisms in the MHC genes [15-29]. Certain MHC alleles of the class II genes linked to viral and bacterial diseases have been reported in some species [30-37]. The link between disease susceptibility/resistance and MHC polymorphism is crucial for detecting MHC alleles related to resistance in marine aquaculture species for molecular marker-assisted selective breeding programs [38].
Half-smooth tongue sole (Cynoglossus semilaevis) is widely cultured throughout the coastal areas of North China [39]. However, viral and bacterial diseases frequently occur in this cultured fish, and losses due to infectious disease limit the profitability and the extent of the development of the aquaculture [40,41]. One pathogen which is a significant threat to half-smooth tongue sole is Vibrio anguillarum [42]. Antibiotics have partially solved problem, but antibiotic residues in fish, environmental pollution and antibiotic resistance are questions about which grave concerns remain [43]. Therefore, the selective breeding of tongue sole with disease resistance, basing on molecular techniques which can enhance the resistance to specific pathogens, may be a good approach to solving these problems.
The half-smooth tongue sole MHC class IIB cDNA sequence and cDNA polymorphisms have been reported [40]. However, the polymorphisms at the DNA level and the link between specific alleles and resistance to V. anguillarum have not been elucidated yet. In the present study, we investigated the single nucleotide polymorphism (SNP) sites and polymorphisms in MHC II B exon2, and the association between certain alleles and disease resistance or susceptibility to Vibrio anguillarum, across 8 families of half-smooth tongue sole.
Methods
Fish and rearing
Eighteen full-sib families were established as reported [44], using a method for producing strains with a high growth rate and disease resistance. Male parents came from wild populations while female parents came from farming populations. Fertilized ova were hatched and reared at the breeding station at Minbo aquatic Co., Ltd. Located in Laizhou city, Shandong province, China. Each family was kept in a separate tank. The fry were fed a commercial diet using a standard feeding regimen [45].
Challenge test
For the challenge test, 200 individuals of each family (12 out of 18 families were large enough to be included), ten months old, were intraperitoneally injected with a 0.2 ml bacterial suspension of approximately 10,000,000 cells of V. anguillarum, while 16 individuals were injected with 0.9% saline as control [15]. Each fry weighed approximately 12-15 grams. The fry of each family were kept in a 1 m3 single tank with a fresh seawater supply at 23°C. This challenge experiment was performed twice and lasted for approximately two weeks. Mortality was recorded every day and the fin clips of all the fish were collected and preserved in absolute ethanol until use. The gross signs of fish mortality were based on a previous reporting method [42].
Sampling and DNA isolation
To identify whether MHC IIB exon2 alleles are associated with resistance or susceptibility to V. anguillarum, fin samples from each family of half-smooth tongue sole were collected and recorded from the first 20 to die and the last survivors at the time the bacterial challenge was terminated and preserved in absolute ethanol until use. High-resistance families (HR) with a survival rate (SR) > 59.45% and susceptible families or low-resistance families (LR) with a SR < 26.73% were selected from the challenge trials. The numbers fish which died or survived after the infection recorded for each family (Additional file 1).
Genomic DNA was isolated from the dorsal or caudal fin samples of 20 individuals per family (from the 4LR and 4HR families) using the phenol-chloroform method as described by Chen et al. [46]. The quality and concentration of DNA were assessed by agarose gel electrophoresis and then measured with a GENEQUANT Pro (Pharmacia Biotech Ltd.) RNA/DNA spectrophotometer. Finally, DNA was adjusted to 100 ng/μl and stored at -20°C.
Primer design and Polymerase Chain Reaction (PCR)
A pair of gene-specific primers was used for the PCR amplification of the MHC II B gene: hMPN12 (5'-CTCTCTTCTCTTCCTCCTCAC-3') and hMPC12 (5'-ACA CTCACCTGATTTAGCCA-3'). They were designed according to reported half-smooth tongue sole MHC II B cDNA sequences [40]. The primer pair was used to amplify part of exon1, and all of intron1 and exon2 from half-smooth tongue sole using a Polymerase Chain Reaction technique. A 25 μl PCR reaction mixture contained 1 μl of template DNA, 2.5 μl of 10×Taq polymerase buffer (TransGen Biotech), 1.5 mM MgCl2, 0.2 mM dNTP mix, 0.2 μM of the forward and reverse primers, and 1 unit of Taq polymerase (TransGen Biotech). The amplifications were performed on a Peltier Thermal Cycler (PTC-200). A Molecular Imager Gel Doc XR system (Bio-rad) was used to determine the PCR products by electrophoresis on a 1% agarose gel.
Cloning and sequencing
The PCR products were resolved by electrophoresis on 1.5% agarose gels. The fragments of interest were excised and purified with the QIAEX II gel extraction kit (Qiagen). The purified fragments were cloned into a PBS-T vector (Takara) according to the standard PBS-T vector protocol (Takara) and then transformed into TOP 10 Escherichia coli competent cells (TransGen Biotech). Forward and reverse M13 primers were used to screen for positive clones via PCR. Ten positive clones from the upper purified fragments were sequenced with an ABI 3730 automated sequencer using the M13+/- primer.
Genotyping, sequence analysis and statistical tests analysis
Sequence data were analyzed using DNASTAR 5.0 and DNAMAN software. The alignment was performed with MEGA4.0 [47]. The rate of synonymous substitution (dS) and non-synonymous substitution (dN) was calculated accord with an earlier report [47] using MEGA4.0 software. DAMBE and DnaSP5.0 software packages were used to analyze the polymorphisms [48]. Statistical analysis was carried out with SPSS13.0. Differences in the allelic frequency were verified using Fisher's exact test and the significance level [49] was determined for every individual (n = 160) and each family (n = 8).
The new alleles were designated Cyse-DBB*0101 to Cyse-DBB*6601 on the basis of the rules reported by Davies et al. [50]. Cyse refers to Cynoglossus semilaevis, D to class II, the first B to an uncharacterized family and the second B to β chain-encoding genes. In the first four digits after the asterisk, the first two digits refer to the major type (alleles that differ by at least five amino acid substitutions), while the last two digits refer to the subtype (alleles that differ by less than five amino acid substitutions within a single major type) [51,52].
Results
To analyze disease resistance among 12 half-smooth tongue sole families
The first specific mortality appeared after 16 h due to an ip injection of V. anguillarum, and the challenge test lasted two weeks, at which time the overall accumulated mortality reached 42.24%. The survival rate among the 12 test families ranged from 15% to79.25%, which was determined on the basis of each family. Here, we selected four high-resistance and four low-resistance families to ascertain whether MHC IIB exon2 alleles were associated with resistance to V. anguillarum among the 12 families of half-smooth tongue sole. The mean prevalence of survival of the four high-resistance families was 59.45%, while that of the four low-resistance families was considerably less at 26.73%.
To elucidate sequence polymorphism within exon2 of MHC IIB gene in 8 half-smooth tongue sole families
Eighty individuals from the four high-resistance families and eighty individuals from the four low-resistance families were used in the present study (Additional file 1). Nine to twelve positive clones per individual were sequenced and 1618 sequences were obtained. A fragment of 397 bp was obtained in reference to the complete half-smooth tongue sole MHC IIB cDNA sequence [40] and intron-exon boundary GT-AG rule. This fragment of 397 bp contains a part of exon1 (35 bp), the entire intron1 (84 bp, containing a 12 bp CA repeat sequence) and the entire exon2 of MHC IIB. A fragment of 270 bp containing the complete exon2 which encodes the β1 domain of the MHC IIB gene was also analyzed. The results indicated 88 different sequences, in which 88 novel alleles were designated (Table 1) belonging to 57 major allele types, following established allele nomenclature method [49,50].
Table 1.
Alleles and Genbank Accession Number of half-smooth tongue sole MHC class II exon2 gene
| Allele | GenBank Accession No. |
Allele | GenBank Accession No. |
Allele | GenBank Accession No. |
|---|---|---|---|---|---|
| Cyse-DBB*0101 | GU194838 | Cyse-DBB*2401 | GU194876 | Cyse-DBB*4601 | GU194918 |
| Cyse-DBB*0102 | GU194839 | Cyse-DBB*2501 | GU194877 | Cyse-DBB*4602 | GU194919 |
| Cyse-DBB*0201 | GU194840 | Cyse-DBB*2601 | GU194878 | Cyse-DBB*4701 | GU194921 |
| Cyse-DBB*0202 | GU194841 | Cyse-DBB*2602 | GU194879 | Cyse-DBB*4801 | GU194922 |
| Cyse-DBB*0301 | GU194842 | Cyse-DBB*2603 | GU194880 | Cyse-DBB*4802 | GU194923 |
| Cyse-DBB*0401 | GU194843 | Cyse-DBB*2801 | GU194882 | Cyse-DBB*4803 | GU194924 |
| Cyse-DBB*0701 | GU194847 | Cyse-DBB*2802 | GU194883 | Cyse-DBB*5002 | GU194927 |
| Cyse-DBB*0801 | GU194848 | Cyse-DBB*2803 | GU194884 | Cyse-DBB*5003 | GU194928 |
| Cyse-DBB*0901 | GU194850 | Cyse-DBB*2901 | GU194886 | Cyse-DBB*5101 | GU194929 |
| Cyse-DBB*1001 | GU194851 | Cyse-DBB*3002 | GU194888 | Cyse-DBB*5202 | GU194932 |
| Cyse-DBB*1002 | GU194852 | Cyse-DBB*3101 | GU194889 | Cyse-DBB*5401 | GU194934 |
| Cyse-DBB*1003 | GU194853 | Cyse-DBB*3102 | GU194890 | Cyse-DBB*5501 | GU194935 |
| Cyse-DBB*1201 | GU194855 | Cyse-DBB*3201 | GU194891 | Cyse-DBB*5601 | GU194936 |
| Cyse-DBB*1301 | GU194856 | Cyse-DBB*3301 | GU194892 | Cyse-DBB*5602 | GU194937 |
| Cyse-DBB*1402 | GU194858 | Cyse-DBB*3302 | GU194893 | Cyse-DBB*5604 | GU194939 |
| Cyse-DBB*1403 | GU194859 | Cyse-DBB*3401 | GU194896 | Cyse-DBB*5701 | GU194940 |
| Cyse-DBB*1501 | GU194860 | Cyse-DBB*3501 | GU194897 | Cyse-DBB*5801 | GU194941 |
| Cyse-DBB*1601 | GU194861 | Cyse-DBB*3701 | GU194902 | Cyse-DBB*5901 | GU194942 |
| Cyse-DBB*1602 | GU194862 | Cyse-DBB*3702 | GU194903 | Cyse-DBB*5902 | GU194943 |
| Cyse-DBB*1701 | GU194864 | Cyse-DBB*3901 | GU194905 | Cyse-DBB*6001 | GU194944 |
| Cyse-DBB*1702 | GU194865 | Cyse-DBB*4001 | GU194906 | Cyse-DBB*6002 | GU194945 |
| Cyse-DBB*1703 | GU194866 | Cyse-DBB*4002 | GU194907 | Cyse-DBB*6102 | GU194947 |
| Cyse-DBB*1801 | GU194867 | Cyse-DBB*4101 | GU194910 | Cyse-DBB*6201 | GU194948 |
| Cyse-DBB*2002 | GU194870 | Cyse-DBB*4201 | GU194911 | Cyse-DBB*6301 | GU194949 |
| Cyse-DBB*2101 | GU194871 | Cyse-DBB*4301 | GU194912 | Cyse-DBB*6401 | GU194950 |
| Cyse-DBB*2201 | GU194872 | Cyse-DBB*4302 | GU194913 | Cyse-DBB*6402 | GU194951 |
| Cyse-DBB*2202 | GU194873 | Cyse-DBB*4402 | GU194915 | Cyse-DBB*6403 | GU194952 |
| Cyse-DBB*2203 | GU194874 | Cyse-DBB*4501 | GU194916 | Cyse-DBB*6404 | GU194954 |
| Cyse-DBB*2301 | GU194875 | Cyse-DBB*4502 | GU194917 | Cyse-DBB*6501 | GU194955 |
| Cyse-DBB*6601 | GU194956 |
Gaps were not found in the full alignment of the 270 bp exon2 of the MHC IIB gene. A putative 90 amino acid peptide was based on a sequence alignment with the half-smooth tongue sole MHC II B cDNA sequence [40]. Among the 270 nucleotides, 72 regions and 121(44.8%) nucleotide positions were variable. The numbers of two-nucleotide mutation, three-nucleotide mutation and four-nucleotide mutation were 24, 11 and 1, respectively (Table 2). At the SNP sites, there were two kinds of nucleotide substitutions, i. e. transition (Table 2, Serial No. 1, 7, 11, 13, 18, 23, 28, 29, 32, 33, 35, 42, 43, 44, 46, 49, 52, 53, 54, 60 and 69) and transversion (Table 2, Serial No. 20, 21, 25, 59). Three kinds of mutation per site (Table 2, Serial No. 2, 4, 6, 9, 14, 15, 16, 22, 26, 30, 31, 36, 37, 41, 51, 56, 58, 61, 62, 63, 65, 67, 68 and 71) which revealed the mutation hotspots. 36 out of 72 mutation regions were multi-nucleotide co-mutations, ranging from two to five nucleotides per region. The SNP sites were located in a tight region from position 9 to 29 (Table 2), so this were most of the mutation hotspots of MHC exon2 herein must be located. The frequency ratio ranged from 0.989:0.011 (Table 2, Serial No.1, 23, 32, 49, 59 and 60) to 0.557:0.443 (Table 2, Serial No.7). No frame-shift mutation was observed in these sequences. The peptide binding regions in half-smooth tongue sole MHC II B were based on the corresponding peptide binding region identified in humans [53].
Table 2.
Distribution of SNP sites within exon2 of MHC IIB allelic sequences of half-smooth tongue sole
| Serial number |
Position | Base type |
Allele no. (n = 88) |
Frequency | Serial number |
Position | Base type |
Allele no. (n = 88) |
Frequency |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 6 | T | 87 | 0.989 | 39 | 104-106 | ATC | 51 | 0.580 |
| C | 1 | 0.011 | ATT | 1 | 0.011 | ||||
| 2 | 9-11 | CTA | 52 | 0.591 | ATG | 1 | 0.011 | ||
| GTA | 1 | 0.011 | CAG | 35 | 0.398 | ||||
| GAG | 35 | 0.398 | 40 | 109-111 | TCG | 84 | 0.955 | ||
| 3 | 12 | C | 17 | 0.193 | TCA | 2 | 0.023 | ||
| T | 34 | 0.386 | CCG | 1 | 0.011 | ||||
| A | 7 | 0.080 | TTG | 1 | 0.011 | ||||
| G | 30 | 0.341 | 41 | 124-126 | GGA | 49 | 0.557 | ||
| 4 | 13 | A | 82 | 0.932 | AGA | 12 | 0.136 | ||
| T | 5 | 0.057 | GAG | 27 | 0.307 | ||||
| G | 1 | 0.011 | 42 | 130 | A | 56 | 0.636 | ||
| 5 | 14-15 | AT | 30 | 0.341 | T | 32 | 0.364 | ||
| AC | 1 | 0.011 | 43 | 143 | C | 2 | 0.023 | ||
| TT | 55 | 0.625 | T | 86 | 0.977 | ||||
| CT | 2 | 0.023 | 44 | 148-149 | AT | 49 | 0.557 | ||
| 6 | 16 | C | 32 | 0.364 | TA | 8 | 0.091 | ||
| T | 20 | 0.227 | TT | 31 | 0.352 | ||||
| A | 36 | 0.409 | 45 | 156-157 | CC | 1 | 0.011 | ||
| 7 | 18 | G | 39 | 0.443 | TA | 24 | 0.273 | ||
| A | 49 | 0.557 | TC | 63 | 0.716 | ||||
| 8 | 19 | T | 33 | 0.375 | 46 | 163 | C | 87 | 0.989 |
| G | 20 | 0.227 | T | 1 | 0.011 | ||||
| C | 30 | 0.341 | 47 | 168-169 | AG | 71 | 0.807 | ||
| A | 5 | 0.057 | GC | 17 | 0.193 | ||||
| 9 | 20 | G | 46 | 0.523 | 48 | 170-172 | ATG | 75 | 0.852 |
| A | 34 | 0.386 | ATT | 9 | 0.102 | ||||
| C | 8 | 0.091 | TTG | 3 | 0.034 | ||||
| 10 | 21-23 | ACA | 53 | 0.602 | ACT | 1 | 0.011 | ||
| GTG | 35 | 0.398 | 49 | 174 | G | 1 | 0.011 | ||
| 11 | 24 | G | 74 | 0.841 | A | 87 | 0.989 | ||
| A | 14 | 0.159 | 50 | 177-178 | GA | 86 | 0.977 | ||
| 12 | 25 | A | 35 | 0.398 | AA | 1 | 0.011 | ||
| G | 31 | 0.352 | GG | 1 | 0.011 | ||||
| T | 19 | 0.216 | 51 | 181-183 | GTC | 80 | 0.909 | ||
| C | 3 | 0.034 | ATC | 7 | 0.080 | ||||
| 13 | 26 | T | 5 | 0.057 | GAA | 1 | 0.011 | ||
| C | 83 | 0.943 | 52 | 193 | C | 24 | 0.273 | ||
| 14 | 28-29 | CC | 52 | 0.591 | T | 64 | 0.727 | ||
| CA | 1 | 0.011 | 53 | 196 | A | 62 | 0.705 | ||
| GA | 35 | 0.398 | G | 26 | 0.295 | ||||
| 15 | 32-33 | CG | 35 | 0.398 | 54 | 198 | A | 69 | 0.784 |
| TG | 1 | 0.011 | G | 19 | 0.216 | ||||
| TC | 52 | 0.591 | 55 | 199-200 | GG | 40 | 0.455 | ||
| 16 | 38-39 | CA | 52 | 0.591 | GT | 18 | 0.205 | ||
| CG | 1 | 0.011 | TG | 8 | 0.091 | ||||
| TA | 35 | 0.398 | CG | 22 | 0.25 | ||||
| 17 | 40-41 | AC | 51 | 0.580 | 56 | 205 | A | 67 | 0.761 |
| GC | 1 | 0.011 | C | 20 | 0.227 | ||||
| AT | 35 | 0.398 | G | 1 | 0.011 | ||||
| CT | 1 | 0.011 | 57 | 207-208 | GA | 62 | 0.705 | ||
| 18 | 44 | C | 36 | 0.409 | AC | 3 | 0.034 | ||
| T | 52 | 0.591 | GG | 8 | 0.091 | ||||
| 19 | 47-49 | AAA | 52 | 0.591 | GC | 5 | 0.057 | ||
| AAG | 1 | 0.011 | 58 | 210-211 | AA | 86 | 0.977 | ||
| TAA | 19 | 0.216 | GA | 1 | 0.011 | ||||
| TGA | 16 | 0.182 | AT | 1 | 0.011 | ||||
| 20 | 51 | G | 53 | 0.602 | 59 | 218 | G | 87 | 0.989 |
| C | 35 | 0.398 | T | 1 | 0.011 | ||||
| 21 | 53 | G | 36 | 0.409 | 60 | 220 | A | 87 | 0.989 |
| C | 52 | 0.591 | G | 1 | 0.011 | ||||
| 22 | 55-56 | AC | 69 | 0.784 | 61 | 226-227 | TG | 47 | 0.534 |
| AT | 18 | 0.205 | TA | 40 | 0.455 | ||||
| GC | 1 | 0.011 | CG | 1 | 0.011 | ||||
| 23 | 58 | A | 87 | 0.989 | 62 | 228 | A | 50 | 0.568 |
| G | 1 | 0.011 | C | 29 | 0.330 | ||||
| 24 | 63-64 | GC | 51 | 0.580 | T | 9 | 0.102 | ||
| GT | 1 | 0.011 | 63 | 229 | A | 82 | 0.932 | ||
| GA | 1 | 0.011 | T | 2 | 0.023 | ||||
| CA | 35 | 0.398 | G | 4 | 0.045 | ||||
| 25 | 67 | A | 68 | 0.773 | 64 | 231-234 | AAC | 61 | 0.693 |
| T | 20 | 0.227 | ACT | 2 | 0.022 | ||||
| 26 | 72 | C | 13 | 0.148 | CAC | 20 | 0.227 | ||
| G | 40 | 0.455 | AGC | 5 | 0.057 | ||||
| T | 35 | 0.398 | 65 | 237-238 | GG | 79 | 0.898 | ||
| 27 | 74 | C | 40 | 0.455 | GA | 5 | 0.057 | ||
| G | 48 | 0.545 | AG | 4 | 0.045 | ||||
| 28 | 78 | C | 38 | 0.432 | 66 | 240-241 | AA | 10 | 0.114 |
| T | 50 | 0.568 | AT | 54 | 0.614 | ||||
| 29 | 80 | C | 35 | 0.398 | CT | 23 | 0.261 | ||
| T | 53 | 0.602 | GT | 1 | 0.011 | ||||
| 30 | 82-83 | AC | 52 | 0.591 | 67 | 242-243 | TG | 46 | 0.523 |
| AT | 35 | 0.398 | TT | 19 | 0.216 | ||||
| GT | 1 | 0.011 | GG | 23 | 0.261 | ||||
| 31 | 84-85 | TT | 30 | 0.341 | 68 | 245-246 | CT | 68 | 0.773 |
| CT | 23 | 0.261 | CA | 19 | 0.216 | ||||
| TA | 35 | 0.398 | AT | 1 | 0.011 | ||||
| 32 | 87 | A | 87 | 0.989 | 69 | 248 | T | 2 | 0.023 |
| G | 1 | 0.011 | C | 86 | 0.977 | ||||
| 33 | 90 | A | 86 | 0.977 | 70 | 250-253 | ACCA | 10 | 0.114 |
| G | 2 | 0.023 | ACGC | 64 | 0.727 | ||||
| 34 | 92-94 | ACT | 52 | 0.591 | AGCC | 1 | 0.011 | ||
| GAG | 36 | 0.409 | GGAC | 12 | 0.136 | ||||
| 35 | 96 | G | 35 | 0.398 | ACGG | 1 | 0.011 | ||
| A | 53 | 0.602 | 71 | 254-256 | TGC | 70 | 0.796 | ||
| 36 | 98-99 | GA | 42 | 0.477 | TGG | 12 | 0.136 | ||
| GT | 11 | 0.125 | GTT | 6 | 0.068 | ||||
| AT | 35 | 0.398 | 72 | 256-258 | TT | 12 | 0.136 | ||
| 37 | 100 | C | 1 | 0.011 | TC | 63 | 0.716 | ||
| A | 38 | 0.432 | CC | 2 | 0.023 | ||||
| T | 49 | 0.557 | TG | 11 | 0.125 | ||||
| 38 | 101-102 | CA | 34 | 0.386 | |||||
| CG | 16 | 0.182 | |||||||
| GA | 3 | 0.034 | |||||||
| TG | 31 | 0.352 | |||||||
| TA | 4 | 0.046 |
The variable positions of the PBR comprised 20 (87%) out of 23 and the polymorphic nucleotide PBR sites were 40 (57.97%) of 69. In the putative peptide-binding region, the ratio of non-synonymous (dN) substitution (0.261) was 1.7 times higher than that of synonymous (dS) substitution (0.153). The rates of dN and dS in the non-PBR were 0.087 and 0.159, respectively. All of the sequences were used to calculate these rates. The rate of ds in the non-PBR(0.159) was slightly higher than that of dS in the PBR(0.153), and dN in the PBR (0.261) occurred at a significantly higher rate than that in the non-PBR (0.087), but dS in the PBR (0.153) was a little lower than that in the non-PBR (0.159) (Table 3).
Table 3.
Synonymous (dS) and nonsynonymous (dN) substitution rate in the putative peptides binding region (PBR) and non-peptides binding region (non-PBR) among half-smooth tongue sole alleles
| Region | No. of codons |
dN(SE) | dS(SE) | dN/dS |
|---|---|---|---|---|
| PBR | 23 | 0.261 ± 0.033 | 0.153 ± 0.052 | 1.70 |
| Non-PBR | 67 | 0.087 ± 0.016 | 0.159 ± 0.034 | 0.547 |
| Total | 90 | 0.132 ± 0.017 | 0.157 ± 0.027 | 0.841 |
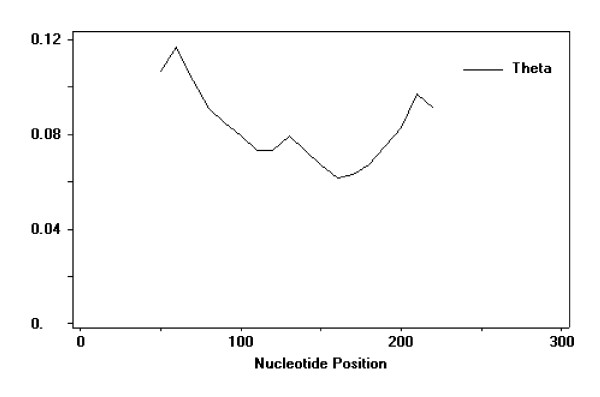
The per site nucleotide diversity Pi (p) was 0.13785, and per the site Theta-W value of the 88 sequences was 0.08876. Ninety-six out of the 121 variable sites were parsimony informative sites. The haplotype diversity (H) and the average number of nucleotide differences (k) were 1 and 37.220, respectively. DnaSP5.0 software was used to calculate these polymorphic values. The exon2 sequence of MHC IIB indicated high nucleotide diversity in the 8 families of tongue sole. Figure 1 shows the spatial distribution of the nucleotide diversity. Two peaks appeared at the downstream and upstream of exon2 of the MHC IIB sequences, respectively, while the Theta-W value in the middle region was lower.
Figure 1.
The nucleotide diversity within exon2 sequences of MHC IIB genes at the 88 alleles denoted by Theta-W. Sliding window length: 100; step size: 10.
To identify association between the MHC IIB alleles and disease resistance/susceptibility to V. anguillarum in half-smooth tongue sole
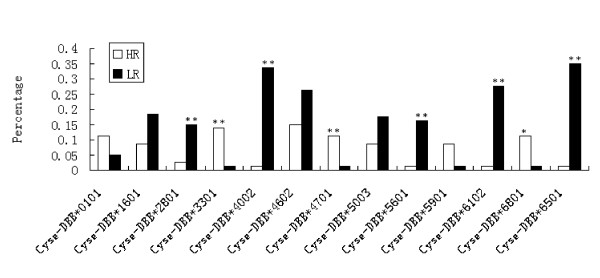
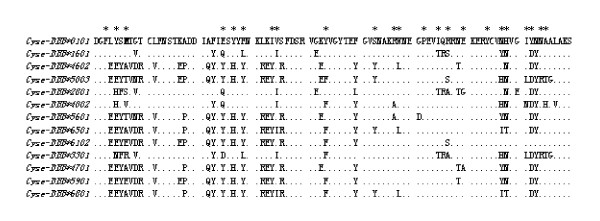
Additional file 2 shows the number of alleles per individual and the comparative individual number. An average ten clones per individual were sequenced, and 2 to 7 alleles per individual were discovered, which inferred the existence of at least seven alleles and four loci of the MHC IIB gene, in accordance with the reports of Xu et al. [40]. Among the 8 families examined, only 2.5% of the individuals were homozygous (all families were heterozygous) for exon2 of the MHC class IIB gene of tongue sole. Eighty-eight sequences resulted in eighty-eight different MHC IIB exon2 alleles deduced from 160 individuals. The distribution of the alleles was unequal. Certain alleles had a low frequency and were excluded from allele distribution analysis between the HR and LR families. Thirteen alleles were used for distribution analysis (Figure 2). The alleles Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801 and Cyse-DBB*5901 were more prevalent in individuals from the HR families (P = 0.005, 0.018, 0.018 and 0.064, respectively n = 160 individuals), while Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801 were more prevalent in individuals from low-resistance families, as shown by chi-square test (P < 0.01, 0.01, 0.01, 0.01, 0.01 respectively n = 160 individuals). Some alleles were not significantly different in the HR and LR families, such as Cyse-DBB*0101(P = 0.247), Cyse-DBB*1601(P = 0.107), Cyse-DBB*4602 (P = 0.117) and Cyse-DBB*5003 alleles (P = 0.159). Here we (deduced) show that Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801 and Cyse-DBB*5901 were associated with resistance, while Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801 were associated with susceptibility to V. anguillarum in half-smooth tongue sole. Alignment of the 13 deduced MHC IIB amino acid sequences (Figure 3) indicated that no specific single amino acid substitution was evidently involved in the resistance or susceptibility, as there was no specific amino acid substitution difference between the HR families and LR families.
Figure 2.
Sequence polymorphism analysis within exon 2 of MHCIIB gene. (Asterisks indicate the correlative amino acid that combines the antigen).
Figure 3.
Distribution of MHC class IIB alleles in high-resistance families individuals (white bars) and low-resistance families individuals (black bars) of half-smooth tongue sole. *Asterisks denote P < 0.05. ** denote P < 0.01.
4. Discussion
It is well known that MHC genes are vital components of both the innate and adaptive immune system. They present foreign peptides to T cells. Cloning and cDNA polymorphism of the MHC II B gene has been discussed [40]. In the present study, partial sequences of the MHC class IIB gene in different families of half-smooth tongue sole were isolated, then molecular polymorphisms as well as the link between alleles and resistance/susceptibility to V. anguillarum were analyzed.
Among the 72 mutated regions in the complete sequence of MHC IIB exon2, 36 regions were multi-nucleotide co-mutations, which indicate inter-allelic recombination took place in these regions. Moreover, no deletion, insertion or stop codon was observed, indicating that all of these alleles were functional genes. The frequency ratio of substituted nucleotides per mutation region was not equally distributed, which suggests that different regions might have different impact.
The rate of non-synonymous substitutions to synonymous substitutions (dN/dS) in the PBR and non-PBR of MHC IIB exon2 of half-smooth tongue sole was studied (Table 3). The dN/dS ratio was higher in the PBR than non-PBR, which corresponds with the results reported in other species [43,54-56]. The dN/dS ratio in exon2 was higher than 1. The location of the PBR sites in the MHC genes of fish was not yet defined, therefore PBR sites were identified using the model of Brown et al. [53] to define HLA-DRB, It was also in accordance with a previous application by Xu et al. [38] for half-smooth tongue sole. The 23 positions were used as PBR sites for in-depth study: 3, 5, 7, 25, 27, 29, 34, 35, 44, 53, 57, 58, 62, 65, 67, 71, 74, 77, 78, 82, 83, 85 and 86 (Figure 3).
It is possible that the PBR sites in fish do not exactly correspond to those in humans [57]. In mammals, MHC polymorphisms are maintained over long periods of time by balanced selection or positive selection at the non-synonymous sites specifying the PBR of the MHC [7]. The ratio between non-synonymous and synonymous substitutions in PBR sites of MHC IIB exon2 genes is greater than 1 (Table 3), as would be expected if the locus were evolving under a condition of balanced selection [58]. The number of alleles per individual ranged from 1 to 5, which showed that at least three loci existed per individual, a result is in accordance with previous studies [22,28,40]. Polymorphism of the 88 alleles in the 160 individuals was higher in half-smooth tongue sole than in Atlantic salmon [57,59] and cyprinid fish [54], and each family had 25-38 alleles. A few hypotheses have been put forward to interpret the abundant polymorphism of the MHC genes, including overdominant selection or heterozygous advantage [60], negative frequency-dependent selection [61,62] and balanced selection [24]. Pathogen-driven selection [26,60] is reported to be contributing to MHC gene diversity through both frequency-dependent selection and heterozygote advantage (over-dominance) [15]. In the present study, the high rate of dN/dS score and high levels of polymorphism which occurred in half-smooth tongue sole revealed that balanced selection is responsible for presence in the PBR domain of the MHC class IIB exon2 gene. This results in the high polymorphism levels in MHC IIB genes in half-smooth tongue sole. Due to the polymorphic nature of MHC genes, certain alleles/haplotypes may be associated with increased disease resistance. In the present study, the distinct distribution pattern of the alleles exhibited a relationship between MHC class IIB alleles and resistance/susceptibility to V. anguillarum in half-smooth tongue sole.
The Cyse-DBB*3301, Cyse-DBB*4701 and Cyse-DBB*6801 alleles which was found in three families, and the Cyse-DBB*5901 allele in two families, were markedly more frequent in HR families (13.75%, 11.25%, 11.25%, 8.75% respectively) than in LR families (1.25%, 1.25%, 1.25%, 1.25%, respectively). This suggests an association of the V. anguillarum disease resistance alleles in half-smooth tongue sole. The Cyse-DBB*6501, Cyse-DBB*4002 and Cyse-DBB*5601 alleles were found in two LR families (35%, 33.75% and 16.25% respectively) and one HR family (1.25%, 1.25% and 1.25%, respectively), while the Cyse-DBB*6102 allele was found in three LR families (27.5%) and one HR family (1.25%), Cyse-DBB*2801 was found in two LR families (15%) and two HR families (2.5%), which might be associated with susceptibility to V. anguillarum in half-smooth tongue sole. In the present study, statistical analysis was used to reveal the associations between the alleles and resistance or susceptibility to V. anguillarum in half-smooth tongue sole. The observed link between alleles Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801, Cyse-DBB*5901, Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801 and resistance/susceptibility to V. anguillarum supported the hypothesis that frequency-dependent selection is crucial for the maintenance of MHC variation [63]. This experimental result was in accord with reports in Atlantic salmon [64] and flounder [38]. However, it was not possible to identify a single allele which appeared in all HR families or all LR families. This might indicate the importance of multiple polymorphisms. One MHC haplotype has been reported to be significantly associated with resistance to Marek's disease in chickens [65], and MHC polymorphism was significantly associated with both juvenile survival and resistance to nematode parasites was also reported in Soay sheep [31].
A link between MHC polymorphism and resistance/susceptibility to disease in fish has been reported. Kjøglum et al. [5] demonstrated that fish with the genotypes UBA*0201/UBA*030 and DAA*0201/*0201 were the most resistant to infectious anaemia in Atlantic salmon, while fish with the genotypes UBA*0601/*080, DAA*0501/*0501 and UBA*0201/*030, DAA*0301/*0501 were the most susceptible, based on an analysis of the combined MHC class I and class II A genotypes. It is reported [15] that the allele combinations DAA*0201-*0201 and DAA*0301-*0301 displayed a significantly lower prevalence of death in homozygous fish than in Atlantic salmon containing one copy or no copy of the allele in Aeromonas salmonicida-challenged Atlantic salmon.
The Sasa-DAA-3'UTR 239 allele [36] was shown to be significantly associated with a decrease in the severity of amoebic gill disease in Atlantic salmon. It was also reported [66] that Sasa-B-04, at the non-classical class I locus, was highly associated with resistance to infectious hematopoietic necrosis in Atlantic salmon. The alleles Paol-DAB*4301 and Paol-DAB*1601 were shown to be associated with resistance and susceptibility to V. anguillarum in flounder [38].
In this study in half-smooth tongue sole, the alleles Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801 and Cyse-DBB*5901 were found to be associated with resistance while the Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801 alleles were associated with susceptibility to V. anguillarum. Associations of MHC with resistance or susceptibility to specific pathogens can also be derived through linkage disequilibrium with a resistance or susceptibility locus or gene, and may not be due to the MHC gene itself [55,67-69].
Conclusions
It can not ruled out that another linked gene, individual genetic background and different strains or populations may to some extent have caused the observed link, but here the Cyse-DBB*3301, Cyse-DBB*4701, Cyse-DBB*6801 and Cyse-DBB*5901 alleles were associated with resistance to V. anguillarum, while the Cyse-DBB*6501, Cyse-DBB*4002, Cyse-DBB*6102, Cyse-DBB*5601 and Cyse-DBB*2801 alleles were associated with susceptibility to V. anguillarum in half-smooth tongue sole. Further studies are needed to confirm the association between MHC class IIB exon2 gene with resistance to V. anguillarum in half-smooth tongue sole.
Authors' contributions
Professor SLC and MD designed of the study. MD carried out the molecular genetic studies, participated in the sequence alignment and wrote the final drafts of the manuscript. Professor SLC and YHL provided academic advising of this study. YL participated in the manuscript revision. MD and JFY were in charge of fish breeding. All authors read and approved the final manuscript.
Supplementary Material
Results of the infection with bacterial. Results of the infection with bacterial is presented. Numbers of high-resistance (HR, survivor rate(SR) > 59.45% when infected with the bacterium Vibrio anguillarum) and low-resistance (LR, SR < 26.73%)families of Cynoglossus semilaevis from which dead, surviving individuals were sampled.
The individual ID and corresponding number of allele. We presented the number of alleles per individual of half-smooth tongue sole and its corresponding individual number.
Contributor Information
Min Du, Email: du2005min@126.com.
Song-lin Chen, Email: chensl@ysfri.ac.cn.
Yan-hong Liu, Email: kidliu1968@126.com.
Yang Liu, Email: littlexueyang@163.com.
Jing-feng Yang, Email: yjfchina@gmail.com.
Acknowledgements
This work was supported by grants from 973 National Major Basic Research Program of China (2010CB126303) and Taishan scholar project Fund of Shan-dong province, China and Agriculture Science and Technology fund projects of China (2009GB23260436).
References
- Nikolich-Žugich J, Fremont DH, Miley MJ, Messaoudi I. The role of mhc polymorphism in anti-microbial resistance. Microbes and Infection. 2004;6:501–512. doi: 10.1016/j.micinf.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Edwards SW, Hedrick PW. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol Evol. 1998;13:305–311. doi: 10.1016/S0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- Srisapoome P, Ohira T, Hirono I, Aoki T. Cloning, characterization and expression of cDNA containing major histocompatibility complex classI, IIa and IIb genes of Japanese flounder. Paralichthys olivaceus. Fish Sci. 2004;70:264–276. doi: 10.1111/j.1444-2906.2003.00800.x. [DOI] [Google Scholar]
- Rothbard JB, Gefter ML. Interactions between immunogenetic peptides and Mhc proteins. Annu Rev Immunol. 1991;9:527–565. doi: 10.1146/annurev.iy.09.040191.002523. [DOI] [PubMed] [Google Scholar]
- Kjøglum S, Larsen S, Grimholt U, Bakke HG. How specic MHC classI andclass II combinations affect disease resistance against infectious salmon anaemia in Atlantic salmon(Salmo salar) Fish Shellfish Immunol. 2006;21:431–441. doi: 10.1016/j.fsi.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Hansen JD, Strassburger JC, Thorgaard GH, Young WP, DuPasquier L. Expression, linkage and polymorphism of MHC-related genes in rainbow trout, Oncorhynchus mykiss. J Immunol. 1999;163:774–786. [PubMed] [Google Scholar]
- Graser R, O'hUigin C, Vincek V, Meyer A, Klein J. Trans-specie polymorphism of class II Mhc loci in danio fishes. Immunogenetics. 1996;44:36–48. doi: 10.1007/BF02602655. [DOI] [PubMed] [Google Scholar]
- Sato A, Figueroa F, Murray BW, Malaga-Trillo E, Zaleska-Rutczynska Z, Sultmann H. Nonlinkage of major histocompatibility complex class I and class II loci in bony fishes. Immunogenetics. 2000;51:108–116. doi: 10.1007/s002510050019. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Nakanishi T, Kurosawa Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc Natl Acad Sci USA. 1990;87:6863–6867. doi: 10.1073/pnas.87.17.6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SL, Xu MY, Hu SN. Analysis of immune relevant genes expressed in red sea bream (Chrysophrys major) spleen Aquaculture. 2004;240:115–130. doi: 10.1016/j.aquaculture.2004.07.008. [DOI] [Google Scholar]
- Sato A, Figueroa F, O'Hugin C. Identification of major histocompatibility complex genes in the guppy,Poercilia reticulate. J Immunogenetics. 1995;43:38–49. doi: 10.1007/BF00186602. [DOI] [PubMed] [Google Scholar]
- Rodrigues PNS. Expression of major histocompatibility complex genes in carp (Cyprinus carpio L.) Dutch: Elsevier Science Ltd. and Academic Press Ltd; 1996. pp. 1–27. [Google Scholar]
- Reně JM Inference of structure and function of fish major histocompatibility complex (MHC), molecules from expressed genes Fish&Shellfish Immunology 19966305–318. 10.1006/fsim.1996.003121894570 [DOI] [Google Scholar]
- Godwin UB, Flores M, Quiniou S. MHC class II A gene in the channel catfish(Ictalurus punctatus) Dev Comp Immunol. 2000;24(6/7):609–622. doi: 10.1016/s0145-305x(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Larsen S, Nordmo R, Midtlying P, Kjoeglum S, Storest A. MHC polymorphism and disease resistance in Atlantic salmon(Salmo salar);facing pathogens with single expressed major histocompatibility class I and class II loci. Immunogenetics. 2003;55:210–219. doi: 10.1007/s00251-003-0567-8. [DOI] [PubMed] [Google Scholar]
- Shum BP, Azumi K, Zhang S. Unexpected beta microglobulin sequence diversity in individual rainbow trout. Proceedings of the National Academy of Sciences USA. 1996;93:2779–2784. doi: 10.1073/pnas.93.7.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein D, Ono H, O'hUighin C. Extensive MHC variability in cichlid fishes of Lake Malawi. Nature. 1993;6:330–334. doi: 10.1038/364330a0. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Duris T, Bront CR. High levels of MHC class II allelic diversity in lake trout from lake superior. J Heredity. 2000;91:359–363. doi: 10.1093/jhered/91.5.359. [DOI] [PubMed] [Google Scholar]
- Grimholt U, Olsaker I, Lindstrm DV. A study of variability in the MHC class II beta I and class I alpha2 domain exons of Atlantic salmon, Salmo salar L. Anim genet. 1994;25:147–153. doi: 10.1111/j.1365-2052.1994.tb00103.x. [DOI] [PubMed] [Google Scholar]
- Ono H, O'hUigin C, Tichy H. Major histocompatibility complex variation in two sepecies of cichlid fishes from Lake malawi. Mol Biol Evol. 1993;10:1060–1072. doi: 10.1093/oxfordjournals.molbev.a040055. [DOI] [PubMed] [Google Scholar]
- Figueroa F, Mayer WE, Sultmann H. Mhc class IIB gene evolution in East African cichlid fishes. Immunogenetics. 2000;51:556–575. doi: 10.1007/s002510000181. [DOI] [PubMed] [Google Scholar]
- Chen SL, Zhang YX, Xu MY. Molecular polymorphism and expression analysis of MHC class IIB gene from red sea bream (Chrysophrys major) Developmental and Comparative Immunology. 2006;30:407–418. doi: 10.1016/j.dci.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Prapansak S, Tsuyoshi O, Ikuo H. Cloning, characterization and expression of cDNA containing major histocompatibility complex classI, IIa and IIb genes of Japanese flounder Paralichthys olivaceus. Fisheries Science. 2004;70(2):264–276. doi: 10.1111/j.1444-2906.2003.00800.x. [DOI] [Google Scholar]
- Hardee JJ, Godwin U, Benedetto R. Major histocompatibility complex classA gene polymorphism in the striped bass. Immunogenetics. 1995;41:229–238. doi: 10.1007/BF00172064. [DOI] [PubMed] [Google Scholar]
- Walker RB, McConnell TJ, Walker RA. Polymorphism of the Mhc Mosa class IIb chain encoding gene in the striped bass (Morone saxatilis) Dev Comp Immunol. 1994;18(4):325–342. doi: 10.1016/S0145-305X(94)90358-1. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Vazquez M, Sato KEC. Evolution of the major histocompatibility complex: isolation of class II cDNA clones from the cartilaginous fish. Proc Natl Acad Sci USA. 1990;89:6688–6692. doi: 10.1073/pnas.89.15.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara MEC, Mckinney MF, Flajnik. The evolution origion of the major histocompatibility complex:polymorphism of class II cDNA clones from the cartilaginous fish. Eur J Immunol. 1993;23:2160–2165. doi: 10.1002/eji.1830230917. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Chen SL. Molecular identification, polymorphism and expression analysis of major histocompatibility complex class II A and B genes of turbot (Scophthalmus maximus) Mar Biotechnol. 2006;8:611–623. doi: 10.1007/s10126-005-6174-y. [DOI] [PubMed] [Google Scholar]
- Sültmann H, Mayer WE, Mayer. Zebrafish MHC class IIα chain encoding genes:polymorphism, expression, and function. Immunogenetics. 1993;38:408–420. doi: 10.1007/BF00184521. [DOI] [PubMed] [Google Scholar]
- Briles WE, Briles RW, Taffs RE, Stone HA. Resistance to a malignant lymphoma in chicken is mapped to subregion of major histocompatibility (B) complex. Science. 1983;219:977–979. doi: 10.1126/science.6823560. [DOI] [PubMed] [Google Scholar]
- Paterson S, Wilson K, Pemberton JM. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population. Proc Natl Acad Sci USA. 1998;95:3714–3719. doi: 10.1073/pnas.95.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AVS, Allsopp CEM, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA. Common West African HLA antigens associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- Flores-Villanueva PO, Hendel H, Caillat-Zucman S, Rappaport J, Burgos-Tiburcio A, Bertin-Maghit S. Associations of MHC ancestral haplotypes with resistance/susceptibility to AIDS disease development. J Immunology. 2003;170:1925–1929. doi: 10.4049/jimmunol.170.4.1925. [DOI] [PubMed] [Google Scholar]
- Gjedrem T, Salte R, Gjoen HM. Genetic variation in susceptibility of Atlantic salmon to furunculosis. Aquaculture. 1991;97:1–6. doi: 10.1016/0044-8486(91)90274-B. [DOI] [Google Scholar]
- Lohm J, Grahn M, Langefors A, Andersen O, Storset A, von Schantz T. Experimental evidence for major histocompatity complex-allele-specific resistance to a bacterial infection. Proc Biol Sci. 2002;269:2029–2033. doi: 10.1098/rspb.2002.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne JW, Cook MT, Nowak BF, Elliott NG. Major histocompatibility polymorphism associated with resistance towards amoebic gill disease in Atlantic salmon (Salmo salar L.) Fish Shellfish Immunol. 2007;22:707–717. doi: 10.1016/j.fsi.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Glover KA, Grimholt U, Bakke HG, Nilsen F, Storset A, Skaala. Major histocompatibility complex (MHC) variation and susceptibility to the sea louse Lepeophtheirus salmonis in Atlantic salmon Salmo salar. Dis Aquat Organ. 2007;76:57–65. doi: 10.3354/dao076057. [DOI] [PubMed] [Google Scholar]
- Xu TJ, Chen SL, Ji XS, Tian YS. MHC polymorphism and disease resistance to Vibrio anguillarum in 12 selective Japanese flounder (Paralichthys olivaceus) families. Fish & Shellfish Immunology. 2008;25(3):213–221. doi: 10.1016/j.fsi.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Chen SL, Tian YS, Yang JF, Shao CW, Ji XS, Zhai JM, Liao XL, Zhuang ZM, Su PZ, Xu JY, Sha ZX, Wu PF, Wang N. Artificial Gynogenesis and Sex Determination in Half-Smooth Tongue Sole (Cynoglossus semilaevis) Mar Biotechnol. 2009;11:243–251. doi: 10.1007/s10126-008-9139-0. [DOI] [PubMed] [Google Scholar]
- Xu Tj, Chen SL, Ji XS, Sha ZX. Molecular cloning, genomic structure, polymorphism and expression analysis of major histocompatibility complex class IIA and IIB genes of half-smooth tongue sole (Cynoglossus semilaevis) Fish & Shellfish Immunology. 2009;27(2):192–201. doi: 10.1016/j.fsi.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Liu YG, Bao BL, Liu LX, Wang L, Lin H. Isolation and characterization of polymorphic microsatellite loci from RAPD product in half-smooth tongue sole(Cynoglossus semilaevis) and a test of cross-species amplification. Molecular Ecology Resources. 2008;8:202–204. doi: 10.1111/j.1471-8286.2007.01923.x. [DOI] [PubMed] [Google Scholar]
- Tang XQ, Zhou L, Zhan WB. Isolation and Characterization of Pathogenic Listonella anguillarum of Diseased Half-Smooth Tongue Sole (Cynoglossus semilaevis Günther) J Ocean Univ Chin. 2008;7(3):343–351. doi: 10.1007/s11802-008-0343-3. [DOI] [Google Scholar]
- Zhang YX, Chen SL, Liu YG, Sha ZX, Liu ZJ. Major histocompatibilibility complex IIB allele polymorphism and its association with resistance/susceptibility to Vibrio anguillarum in Japanese Flounder (Paralichthys olivaceus) Mar Biotechnol. 2006;8:600–610. doi: 10.1007/s10126-005-6185-8. [DOI] [PubMed] [Google Scholar]
- Chen SL, Du M, Yang JF, Hu QM, Xu Y, Zhai JM. Development and characterization for growth rate and disease resistance of families in half-smooth tongue sole (Cynoglossus semilaevis) Journal of Fisheries of China. 2010;23(12):1789–1794. [Google Scholar]
- Lei JL. Marine Fish Culture Theory and Techniques [M] China Agriculture Press; 2004. [Google Scholar]
- Chen SL, Li J, Deng SP, Tian YS, Wang QY, Zhuang ZM, Sha ZX, Xu JY. Isolation of Female-Specific AFLP Markers and MolecularIdentification of Genetic Sex in Half-Smooth Tongue Sole(Cynoglossus semilaevis) Marine Biotechnology. 2007;9:173–280. doi: 10.1007/s10126-006-6081-x. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5:A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Rice WR. Analysis tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.2307/2409177. [DOI] [PubMed] [Google Scholar]
- Davies CJ, Andersson L, Ellis SA, Hensen EJ, Lewin HA, Mikko S. Nomenclature for factors of the BoLA system, report of the ISAG BoLA Nomenclature Committee. Anim Genet. 1997;28:159–168. doi: 10.1111/j.1365-2052.1997.00106.x. [DOI] [Google Scholar]
- Xu RF, Chen GH, Xu H, Qiang BZ, Li CC. Characterization of genetic polymorphism of novel MHCB-LBIIalleles in Chinese indigenous chickens. J Genet Genomics. 2007;34(2):109–118. doi: 10.1016/S1673-8527(07)60012-5. [DOI] [PubMed] [Google Scholar]
- Xu RF, Li K, Chen GH, Qiang YZ, Zhang YB, Lin L. Genetic variation within exon 2 of the MHC B-LB II gene inTibetan chicken. Acta Genet Sin. 2005;32(11):1136–1146. [PubMed] [Google Scholar]
- Li MH, Li K, Kantanen J, Feng Z, Fan B, Zhao SH. Allelic variations in exon2 of caprine MHC class II DRB3 gene in Chinese indigenous goats. Small Rumin Res. 2006;66:236–243. doi: 10.1016/j.smallrumres.2005.09.017. [DOI] [Google Scholar]
- Langefors Å, Lohm J, Grahn M, Andersen Ø, Schantz T. Association between major histocompatibility complex class II B alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc R Soc Lond B. 2000;268:479–485. doi: 10.1098/rspb.2000.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TJ, Parker KM, Hedrick PW. Major Histocompatibility Complex Differentiation in Sacramento River Chinook Salmon. Genetics. 1999;151:1115–1122. doi: 10.1093/genetics/151.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, Stern LJ, Urban RG. Strominger JL. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Ottová E, Simkova A, Martin JF, Bellocq JG, Gelnar M, Allienne JF. Evolution and trans-species polymorphism Of MHC class IIb genes in cyprinid fish. Fish & Shellfish Immunol. 2005;18:199–222. doi: 10.1016/j.fsi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Nei M. Nucleotide substitution at major histocompatibility complex class II loci:evidence for over-dominant selection. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stet RJM, Vries B, Mudde K, Hermsen T, Heerwaarden J, Shum BP. Unique haplotypes of co-segregating major histocompatibility class II A and class II B alleles in Atlantic salmon (Salmo salar) give rise to diverse class II genotypes. Immunogenetics. 2002;54:320–331. doi: 10.1007/s00251-002-0477-1. [DOI] [PubMed] [Google Scholar]
- Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade RW, McCallum HI. Overdominant vs frequency-dependent selection at MHC loci. Genetics. 1992;132:861–862. doi: 10.1093/genetics/132.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Ohta T. Population biology of antigen presentation by MHC class I molecules. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- Langefors Å, Lohm J, Grahn M, Andersen O, von Schantz T. Association between Mhc class II B alleles and resistance to Aeromonas salmonicida in Atlantic salmon. Proc R Soc Lond B. 2001;268:479–485. doi: 10.1098/rspb.2000.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon LD. Influence of the major histocompatibility complex on disease resistance and productivity. Poultry Science. 1987;66:802–811. doi: 10.3382/ps.0660802. [DOI] [PubMed] [Google Scholar]
- Miller KM, Winton JR, Schulze AD, Purcell MK, Ming TJ. Major histocompatibility complex loci are associated with susceptibility of Atlantic salmon to infectious hematopoietic necrosis virus. Environ Biol Fishes. 2004;69:307–316. [Google Scholar]
- Bengtsson BO, Thomson G. Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens. 1981;18:356–363. doi: 10.1111/j.1399-0039.1981.tb01404.x. [DOI] [PubMed] [Google Scholar]
- Kjøglum S, Grimholt U, Larsen S. Non-MHC genetic and tank effects influence disease challenge tests in Atlantic salmon (Salmo salar) Aquaculture. 2005;250:102–109. doi: 10.1016/j.aquaculture.2005.03.021. [DOI] [Google Scholar]
- Hedrick P, Kim T. In: Genetics of complex polymorphisms: parasites and maintenance of MHC variation. Singh R, Krimbas C, editor. Genetics, Evolution & Society. Harvard University Press: Cambridge; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results of the infection with bacterial. Results of the infection with bacterial is presented. Numbers of high-resistance (HR, survivor rate(SR) > 59.45% when infected with the bacterium Vibrio anguillarum) and low-resistance (LR, SR < 26.73%)families of Cynoglossus semilaevis from which dead, surviving individuals were sampled.
The individual ID and corresponding number of allele. We presented the number of alleles per individual of half-smooth tongue sole and its corresponding individual number.