Abstract
Background
Fungi are asexually and sexually reproducing organisms that can combine the evolutionary advantages of the two reproductive modes. However, for many fungi the sexual cycle has never been observed in the field or in vitro and it remains unclear whether sexual reproduction is absent or cryptic. Nevertheless, there are indirect approaches to assess the occurrence of sex in a species, such as population studies, expression analysis of genes involved in mating processes and analysis of their selective constraints. The members of the Phialocephala fortinii s. l. - Acephala applanata species complex (PAC) are ascomycetes and the predominant dark septate endophytes that colonize woody plant roots. Despite their abundance in many ecosystems of the northern hemisphere, no sexual state has been identified to date and little is known about their reproductive biology, and how it shaped their evolutionary history and contributes to their ecological role in forest ecosystems. We therefore aimed at assessing the importance of sexual reproduction by indirect approaches that included molecular analyses of the mating type (MAT) genes involved in reproductive processes.
Results
The study included 19 PAC species and > 3, 000 strains that represented populations from different hosts, continents and ecosystems. Whereas A. applanata had a homothallic (self-fertile) MAT locus structure, all other species were structurally heterothallic (self-sterile). Compatible mating types were observed to co-occur more frequently than expected by chance. Moreover, in > 80% of the populations a 1:1 mating type ratio and gametic equilibrium were found. MAT genes were shown to evolve under strong purifying selection.
Conclusions
The signature of sex was found in worldwide populations of PAC species and functionality of MAT genes is likely preserved by purifying selection. We hypothesize that cryptic sex regularely occurs in the PAC and that further field studies and in vitro crosses will lead to the discovery of the sexual state. Although structurally heterothallic species prevail, it cannot be excluded that homothallism represents the ancestral breeding system in the PAC.
Background
The origin and maintenance of sexual reproduction is a controversially discussed topic in evolutionary biology as reflected by the multitude of theories that have been proposed to explain why sexual reproduction, although highly costly, is widely occurring in nature [1-6]. Asexual organisms have a two-fold advantage over sexual conspecifics and can effectively disseminate [7-10]. In contrast, sexual reproduction efficiently eliminates deleterious mutations [11] and creates genetic variation that favors natural selection and accelerates adaptation to changing environments [1]. However, many species are capable of reproducing both sexually and asexually and illustrate how difficult it is to provide a general explanation on the evolutionary significance of sex. Fungi combine the advantages of the two reproductive modes. Several reasons were proposed for why the cost of sex compared to asex is lower in fungi than for animals and plants [12] because (i) fungi can be isogamous and thus the contribution of resources to the zygote by the gametes is limited and (ii) many fungi are also homothallic (self-fertile) and do not depend on finding a compatible mate which reduces the cost of sex, whereas others are heterothallic (self-sterile) and mating is regulated by mating type factors. (iii) Moreover, the majority of fungi can alternate between asexual and sexual reproduction and thus sexuality can be adjusted to when opportunity costs are low, for example at the end of the growing season of a host plant on which a fungus is dependent when adverse conditions are disadvantageous for somatic growth. Sex is also linked to essential processes such as the formation of resistant spores that are able to survive unfavourable conditions and enable new genotypes to be spread into new environments. Once the link between sex and such essential processes has evolved, selection against frequent sexual recombination might be less effective.
However, for many fungi, especially for filamentous ascomycetes, only part of their life cycle is known. These taxa are classified as Deuteromycota or "fungi imperfecti" due to the lack of sexual morphology [13], but it is unclear whether sexual reproduction is absent, rare or cryptic because sexual morphology is often difficult to observe in nature or in the laboratory [14,15]. Thus, the importance of sexual reproduction in natural populations of such species remains an open question which can be addressed by direct and indirect approaches [16].
The direct apporach consists in searching for the sexual state (teleomorph) in the field or in the laboratory. However, it is often difficult to induce the teleomorph in vitro as many factors (e.g. nutrient media, temperature, light exposure, selection of compatible mating types) need to be optimized for a successful induction [17,18]. Moreover, although sexual structures obtained in the laboratory indicate that the ability for sexual reproduction has not been lost, its importance in the field remains to be established by population studies and by monitoring the teleomorph in the field [16]. Indirect approaches comprise population studies that test the null hypothesis of random mating and include analysis of gametic disequilibrium, mating type ratios, genotypic diversity and phylogenetic analysis [19,20]. Moreover, functionality of genes involved in mating processes can be tested by expression analyses and provide further evidence for sexual reproduction [21]. Alternatively, estimates of selective pressures acting on such genes might indicate whether functionality is preserved [22-26].
In this study we aimed at elucidating the importance of sexual reproduction in the Phialocephala fortinii sensu lato - Acephala applanata species complex (PAC) that belongs to the dark septate endophytes, a polyphyletic group of ascomycetes with characteristic melanized, septate hyphae that commonly colonize roots of woody plant species [27]. The PAC is composed of more than 20 species, eigth of which were formaly described [28,29], and they dominate the endophytic assemblages in roots of conifers and members of the Ericaceae in the northern hemisphere from polar to subtropical regions [30-32]. PAC species form communities of up to ten species [32,33] and species abundance distributions within these communities ususally follow a hyperbolic distribution with a few abundant species and many "rare" species, consistent with the community structures of many other organismal groups [34]. Interestingly, no biogeographical pattern was found for PAC species and community compositions did neither correlate with composition of the host species nor with climate [32] supporting the the hypothesis that "everything is everywhere" [35]. Despite the broad geographical occurrence of this fungal species complex, the teleomorph has never been observed for any of its species.
In ascomycetes, the mating type (MAT) locus exercises key regulatory functions involved in mating processes and usually defines homothallism and heterothallism [36,37]. In heterothallic ascomycetes the MAT locus is characterized by two alternative forms, MAT1-1 and MAT1-2, called idiomorphs because they contain dissimilar DNA sequences at the same chromosomal location. These encode proteins with conserved DNA binding domains that are involved in transcription regulation leading to the attraction of compatible mating types in order to initiate the mating process [37] and are also involved in controlling the regulation of internuclear recognition in later steps of sexual development [38-40]. In contrast, in homothallic species, a single individual generally contains all of the MAT genes and is thus capable of selfing [41]. In filamentous ascomycetes the simpliest idiomorphs consist of MAT1-1 that carries one gene called MAT1-1-1 encoding a protein with an α-domain binding motif and MAT1-2 that contains one gene called MAT1-2-1 encoding a protein with a high mobility group (HMG)-binding domain [42,43]. However, additional MAT genes are regularely found in ascomycetous species [42]. For example, in the helotialean species Pyrenopeziza brassicae and Rhynchosporium secalis that are closely related to the PAC [44] up to two additional MAT genes have been identified in the MAT1-1 idiomorph. One is designated MAT1-1-3, encodes a protein with an HMG-binding domain and is present in both species, whereas MAT1-1-4 encodes a metallothionein-like protein and is only present in P. brassicae [45,46].
In a recent study we cloned the MAT genes of eight PAC species [44]. and expected the MAT idiomorphs to be similar to those of their helotialean relatives. Seven examined species showed a heterothallic organization of the MAT locus because strains either contained the MAT1-1 idiomorph carrying MAT1-1-1 and MAT1-1-3 or the MAT1-2 idiomorph carrying MAT1-2-1. In contrast, in A. applanata the MAT locus structure was indicative of homothallism because all of the three MAT genes were present in single strains. However, it was neither possible to induce sexual reproductive structures in crossing experiments nor to show that MAT genes were expressed under laboratory conditions [44]. In contrast, analysis of multi-locus gametic disequilibrium for a limited number of Swiss populations of four PAC species showed that in most populations the index of association IA [47] did not deviate significantly from zero which is indicative of recombination [33,48]. Because PAC species are successful root colonizers of a multitude of plant species in many ecosystems, we hypotesize that sexual reproduction occurrs as it confers the ability to successfully adapt to different environments worldwide.
In the present study we aimed at gaining a more complete picture about the importance of sexual reproduction in this species complex. In particular, we determined (i) the MAT locus structure for 11 additional PAC species, (ii) whether opposite mating types in populations of the PAC species deviated from the 1:1 ratio expected under random mating, (iii) the deviation from gametic equilibrium in the fungal populations, (iv) the spatial distribution of mating types at the collection sites, and (v) the selective pressures acting on MAT genes.
Results
Organization of the MAT locus in PAC species
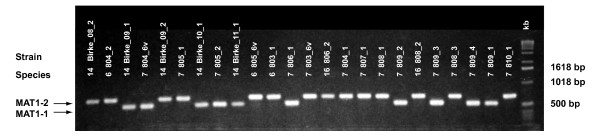
The multiplex PCR amplified either a MAT1-1 specific fragment of ~550 bp or a MAT1-2 specific fragment of ~750 bp in strains of any PAC species, except in the homothallic A. applanata for which no fragments were amplified, as expected due to its MAT locus organization [44] (Figure 1). During the screening of the collections, occasionally (in < 2.5% of the screened strains) both fragments were amplified in single strains, indicating potential homothallism, and in < 1.0% of the strains an amplification failure of the PCR product was recorded. However, re-analyzing newly prepared single-hyphal-tip cultures from a subset of eight of these strains, an amplification of single idiomorph-specific fragments was always obtained (see Additional file 1).
Figure 1.
Example of multiplex PCR. Multiplex PCR amplification of idiomorph specific bands in selected strains of different PAC species, using the primers Pf_HMG_R.03, Pf_HMG_F4 and Pf_MAT1-1F1c.
The MAT locus of 11 additional PAC species was sequenced and characterized for one MAT1-1 and MAT1-2 strain per species (Table 1), and its structure was found to be congruent with the MAT locus structure of the seven previously studied species [44]. The MAT1-1 idiomorphs contained the MAT1-1-1 and MAT1-1-3 gene whereas the MAT1-2 idiomorphs included the MAT1-2-1 gene.
Table 1.
Strains for which the complete mating type idiomorph was sequenced
| Species | Strain | Idiomorph | Geographic origin | Host/Substrate1 | Reference/Collector | GenBank accession No. |
|---|---|---|---|---|---|---|
| Phialocephala turicensis | 1_120_2 | MAT1-1 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347273] |
| 1_117_3 | MAT1-2 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347292] | |
| Phialocephala letzii | 2_120_3 | MAT1-1 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347274] |
| 2_116_2 | MAT1-2 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347293] | |
| Phialocephala europaea | 3_136_1 | MAT1-1 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347275] |
| 3_122_3 | MAT1-2 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347294] | |
| Phialocephala helvetica | 4_140_4 | MAT1-1 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347276] |
| 4_153_2 | MAT1-2 | Zürichberg, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347295] | |
| Phialocephala uotolensis | 5_265_3 | MAT1-1 | Uetliberg; Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347277] |
| 5_265_4 | MAT1-2 | Uetliberg; Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347296] | |
| Phialocephala subalpina | 6_30_4 | MAT1-1 | Bödmeren, Switzerland | root of Picea abies | Zaffarano et al. (2010) | [GenBank:HM347278] |
| 6_37_6v | MAT1-2 | Bödmeren, Switzerland | root of Vaccinium myrtillus | Zaffarano et al. (2010) | [GenBank:HM347297] | |
| Phialocephala fortinii s.s. | 7_K93_444a | MAT1-1 | Suonenjoki, Finland | roots of Pinus sylvestris | Zaffarano et al. (2010) | [GenBank:HM347279] |
| 7_6_7v | MAT1-2 | Bödmeren, Switzerland | root of Vaccinium myrtillus | Zaffarano et al. (2010) | [GenBank:HM347298] | |
| CSP8 | 8_L_05-7 | MAT1-1 | Sheep Creek, USA | n.a. | J.B. Zaerr | [GenBank:HM347280] |
| 8_L_02-5 | MAT1-2 | Sheep Creek, USA | n.a. | J.B. Zaerr | [GenBank:HM347299] | |
| CSP9 | 9_ST_43-1 | MAT1-1 | Walder Steinenberg, Switzerland | root of Picea abies | N. Brenn | [GenBank:HM347281] |
| 9_ST_02-3 | MAT1-2 | Walder Steinenberg Switzerland | root of Picea abies | N. Brenn | [GenBank:HM347300] | |
| CSP10 | 10_CS_15L_2 | MAT1-1 | Court, Switzerland | root of Picea abies | C.R. Grünig | [GenBank:HM347282] |
| 10_CS_9_1 | MAT1-2 | Court, Switzerland | root of Picea abies | C.R. Grünig | [GenBank:HM347301] | |
| CSP11 | 11_AF196-16 | MAT1-1 | Varena plantation, Lithuania | root of Picea abies | A. Menkis | [GenBank:HM347283] |
| 11_AF200-1 | MAT1-2 | Varena plantation, Lithuania | root of Picea abies | A. Menkis | [GenBank:HM347302] | |
| CSP12 | 12_405_1 | MAT1-1 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347284] |
| 12_418_1 | MAT1-2 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347303] | |
| CSP13 | 13_422_6v | MAT1-1 | Creux du Van, Switzerland | root of Vaccinium vitis-ideae | V. Queloz | [GenBank:HM347285] |
| 13_414_2 | MAT1-2 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347304] | |
| CSP14 | 14_426_4 | MAT1-1 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347286] |
| 14_444_6v | MAT1-2 | Creux du Van, Switzerland | root of Vaccinium myrtillus | V. Queloz | [GenBank:HM347305] | |
| CSP15 | 15_A_02-5 | MAT1-1 | Noonday, USA | n.a. | J. Hill | [GenBank:HM347287] |
| 15_A_08-4 | MAT1-2 | Noonday, USA | n.a. | J. Hill | [GenBank:HM347306] | |
| CSP16 | 16_428_6v | MAT1-1 | Creux du Van, Switzerland | root of Vaccinium vitis-ideae | V. Queloz | [GenBank:HM347288] |
| 16_404_2 | MAT1-2 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347307] | |
| CSP18 | 18_410_1 | MAT1-1 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347290] |
| 18_439_1 | MAT1-2 | Creux du Van, Switzerland | root of Picea abies | V. Queloz | [GenBank:HM347308] | |
| CSP19 | 19_1559-1 | MAT1-1 | Parco del Pollino, Italy | root of Abies alba | V. Queloz | [GenBank:HM347291] |
| 19_1545-1 | MAT1-2 | Parco del Pollino, Italy | root of Abies alba | V. Queloz | [GenBank:HM347309] |
1 n.a. = information not available
Mating type ratios and gametic disequilibrium in PAC populations
The vast majority of populations (> 80%) did not deviate significantly from the expected 1:1 ratio in mating types (Table 2). After applying Bonferroni correction to adjust for errors of multiple comparisons (type 1 error), no significant deviations remained. Moreover, even those populations with < 10 individuals contained both mating types. Only a single population was found that did not include both mating types. Similarly, only six out of 52 populations showed significant gametic disequilibrium and these were restricted to populations of P. subalpina (Table 2), Furthermore, in only two of these P. subalpina populations both unequal mating type frequencies and gametic disequilibria were recorded.
Table 2.
Mating type ratios for populations of species belonging to the Phialocephala fortinii s.l. - Acephala applanata species complex (PAC)
| Species | Study site | Country | Marker type1 | Number of strains | Mating type ratios | Gametic disequilibrium | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of MLH | MAT1-1 | MAT1-2 | Significance2 | Number of MLH3 | IA4 | |||||
| P. turicensis | Rothwald | Austria | M | 24 | 7 | 4 | 3 | n.a. | ||
| Etang de la Gruère | Switzerland | M | 203 | 12 | 9 | 3 | * | 12 | 0.28 | |
| Uetliberg | Switzerland | RFLP | 145 | 17 | 9 | 8 | 17 | 0.07 | ||
| Zürichberg | Switzerland | RFLP | 48 | 13 | 7 | 6 | 13 | 0.13 | ||
| P. letzii | Parabock | Switzerland | M | 58 | 11 | 6 | 5 | 11 | 0.69 | |
| Rothwald | Austria | M | 46 | 16 | 7 | 9 | 16 | -0.01 | ||
| Zürichberg | Switzerland | RFLP | 52 | 18 | 13 | 5 | * | 18 | 0.12 | |
| P. europaea | Redwater | Canada | M | 19 | 7 | 1 | 6 | n.a. | ||
| Alaska | USA | M | 19 | 12 | 7 | 5 | 13 | 0.41 | ||
| Catherine Creek | USA | M | 17 | 8 | 5 | 3 | n.a. | |||
| Sierra Nevada | USA | M | 10 | 9 | 7 | 2 | n.a. | |||
| Bödmeren | Switzerland | M | 28 | 19 | 7 | 12 | 17 | -0.18 | ||
| Scatlé | Switzerland | M | 36 | 31 | 12 | 19 | 25 | 0.02 | ||
| Rothwald | Austria | M | 86 | 40 | 23 | 17 | 38 | -0.12 | ||
| Alpes Maritimes | France | M | 77 | 43 | 24 | 19 | 34 | 0.05 | ||
| Creux du Van | Switzerland | M | 123 | 51 | 27 | 24 | 43 | -0.08 | ||
| Derberence | Switzerland | M | 179 | 64 | 28 | 36 | 48 | -0.05 | ||
| Runcaglia | Switzerland | M | 95 | 45 | 22 | 23 | 41 | 0.02 | ||
| Zürichberg | Switzerland | RFLP | 130 | 46 | 26 | 20 | 46 | 0.04 | ||
| P. helvetica | Yuba | USA | M | 25 | 5 | 3 | 2 | n.a. | ||
| Ala Archa | Kirgistan | M | 18 | 7 | 7 | 0 | n.a. | |||
| Baikalsee | Russia | M | 29 | 11 | 4 | 7 | 12 | 0.16 | ||
| Mansfield | Canada | M | 63 | 16 | 4 | 12 | * | 16 | 0.46 | |
| Sierra Nevada | USA | M | 47 | 30 | 17 | 13 | 30 | 0 | ||
| Zürichberg | Switzerland | RFLP | 40 | 15 | 11 | 4 | * | 15 | -0.03 | |
| Runcaglia | Switzerland | M | 98 | 36 | 20 | 16 | 36 | 0.02 | ||
| P. uotolensis | Alpes Maritimes | France | M | 17 | 6 | 4 | 2 | n.a. | ||
| P. subalpina | Creux du Van | Switzerland | M | 13 | 8 | 6 | 2 | n.a. | ||
| Etang de la Gruère | Switzerland | M | 15 | 14 | 8 | 6 | 14 | 0.05 | ||
| Massif Central/La Fage | France | M | 70 | 30 | 14 | 16 | 29 | -0.08 | ||
| Sheep Creek | USA | M | 12 | 7 | 2 | 5 | n.a. | |||
| Marsh Island | USA | M | 23 | 13 | 3 | 10 | * | 13 | 0.17 | |
| Pyrénées | France | M | 19 | 13 | 5 | 6 | 13 | 0.92 | ||
| Redwater | Canada | M | 12 | 11 | 3 | 8 | 10 | -0.06 | ||
| Baikalsee | Russia | M | 16 | 13 | 10 | 3 | * | 12 | 0.27 | |
| Bialowieza | Poland | M | 60 | 40 | 10 | 30 | ** | 40 | 2.12*** | |
| Noonday | USA | M | 22 | 15 | 8 | 7 | 14 | 0.11 | ||
| Grass Creek | USA | M | 28 | 22 | 11 | 11 | 21 | -0.22 | ||
| Rothwald | Austria | M | 43 | 23 | 12 | 11 | 23 | 0.70* | ||
| Derberence | Switzerland | M | 37 | 26 | 11 | 15 | 26 | 1.66*** | ||
| Valcartier | Canada | M | 35 | 30 | 17 | 13 | 28 | 0.48 | ||
| Bödmeren | Switzerland | M | 90 | 54 | 23 | 31 | 44 | 0.58 | ||
| Scatlé | Switzerland | M | 118 | 74 | 42 | 32 | 71 | 0.71** | ||
| Alpes Maritimes | France | M | 125 | 75 | 46 | 29 | * | 75 | 0.58* | |
| Campolino | Italia | M | 138 | 79 | 43 | 36 | 86 | 0.41 | ||
| Kevo | Finland | M | 138 | 87 | 44 | 43 | 72 | 0.52* | ||
| P. fortinii s.s. | Glentanar | Scottland | M | 10 | 5 | 3 | 2 | |||
| Bödmeren | Switzerland | M | 10 | 8 | 4 | 4 | ||||
| Marsh Island | USA | M | 17 | 10 | 5 | 5 | 10 | -0.17 | ||
| Noonday | USA | M | 11 | 7 | 2 | 5 | ||||
| Hill of Fare | Scottland | M | 23 | 15 | 9 | 6 | 13 | -0.07 | ||
| Valcartier | Canada | M | 23 | 17 | 8 | 9 | 15 | 0.37 | ||
| Etang de la Gruère | Switzerland | M | 24 | 18 | 10 | 8 | 23 | 0.29 | ||
| Bialowieza | Poland | M | 92 | 51 | 35 | 16 | * | 51 | 0.05 | |
| La Fage | France | M | 102 | 37 | 16 | 21 | 38 | 0.23 | ||
| Le Pirou | France | M | 13 | 11 | 4 | 7 | 11 | 0.22 | ||
| Tschornohora | Ukraine | M | 76 | 41 | 28 | 13 | * | 41 | 0.25 | |
| Kevo | Finland | M | 100 | 54 | 24 | 30 | 54 | 0.28 | ||
| CSP8 | Global | Global | M | 42 | 20 | 9 | 11 | |||
| CSP9 | Global | Global | M | 10 | 5 | 4 | 1 | n.a. | ||
| CSP10 | Global | Global | M | 30 | 5 | 4 | 1 | n.a. | ||
| CSP11 | Fuji | Japan | M | 20 | 11 | 7 | 4 | 11 | 0.59 | |
| Kevo | Finland | M | 12 | 7 | 4 | 3 | n.a. | |||
| CSP12 | Creux du Van | Switzerland | M | 15 | 7 | 5 | 2 | n.a. | ||
| Chychkan | Kirgistan | M | 9 | 7 | 6 | 1 | n.a. | |||
| CSP13 | Creux du Van | Switzerland | M | 43 | 23 | 9 | 12 | 23 | 0.66 | |
| Alaska | USA | M | 14 | 10 | 7 | 3 | 11 | 0.41 | ||
| CSP14 | Kevo | Finland | M | 16 | 14 | 4 | 10 | 14 | -0.18 | |
| Creux du Van | Switzerland | M | 21 | 8 | 6 | 2 | n.a. | |||
| Zinal_Baiting | Switzerland | M | 72 | 26 | 14 | 12 | 25 | -0.19 | ||
| CSP15 | Noonday | USA | M | 28 | 14 | 6 | 8 | 15 | -0.7 | |
| CSP16 | Creux du Van | Switzerland | M | 9 | 3 | 2 | 1 | n.a. | ||
| CSP18 | Global | Global | M | 20 | 4 | 1 | 3 | n.a. | ||
| CSP19 | Global | Global | M | 30 | 5 | 4 | 1 | n.a. | ||
1 Molecular markers used for clone-correction were 11 single-copy Restriction Fragment Length Polymorphisms (RFLP) or 12 microsatellite loci (M).
2 n.a. = not available; significance test was not performed due to small sample size; * = significant deviation from a 1:1 ratio.
3 MLH = Multilocus haplotypes inferred by RFLP or M; MLHs with missing data were excluded to calculate the index of association (IA).
4 * = p < 0.05; ** = p < 0.01; *** = p < 0.001
Spatial distribution of mating types in selected study sites
The spatial distribution of mating types was analyzed for four PAC species in ten Swiss populations. In each population, strains of opposite mating type were found distributed over the whole study site (see Additional file 2). Strains of different mating types were regularly isolated from the same grid points indicating that they can be found in close physical proximity (Table 3). In addition, measures of association [Q] between opposite mating types for significant spatial structures ranged from weakly negative to strongly positive (Table 3), meaning that co-occurrence of strains with different mating types is often more frequently observed than expected by chance.
Table 3.
Spatial distribution of mating types for selected PAC populations
| Species | Study site | Number of strains | Fraction of grid points with only one strain | Number of grid points with > 1 strain | Number of grid points and identified mating types | Association | chi-test | significance1 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| MAT1-1 | MAT1-2 | both | [Q] | |||||||
| P. europaea | Bödmeren | 28 | 0.65 | 6 | 7 | 8 | 2 | 0.33 | 18.75 | * |
| Creux du Van | 135 | 0.26 | 35 | 21 | 15 | 11 | -0.03 | 4.41 | * | |
| Zürichberg | 129 | 0.24 | 42 | 20 | 21 | 14 | -0.22 | 3.60 | n.s. | |
| Scatlé | 35 | 0.21 | 11 | 2 | 6 | 6 | 0.94 | 14.54 | * | |
| Derberence | 186 | 0.19 | 51 | 9 | 30 | 24 | -0.01 | 5.11 | * | |
| P. helvetica | Zürichberg | 40 | 0.64 | 10 | 20 | 6 | 2 | -0.13 | 12.16 | * |
| P. subalpina | Bödmeren | 80 | 0.46 | 22 | 5 | 25 | 11 | 0.49 | 4.92 | * |
| Derberence | 36 | 0.70 | 7 | 9 | 12 | 2 | -0.03 | 12.80 | * | |
| Scatlé | 115 | 0.30 | 30 | 17 | 11 | 15 | 0.43 | 3.61 | n.s. | |
| CSP13 | Creux du Van | 43 | 0.61 | 9 | 7 | 13 | 3 | 0.25 | 12.06 | * |
1 * = significant at p < 0.05; n.s. = not significant
Analysis of selective pressures based on sequence information
Overall values of ω ranged between 0.26 and 0.46 for the three MAT genes and proportions of amino acid sites under purifying selection (ω < 1) were larger than proportions of sites reported under neutral selection (ω = 1) or positive, diversifying selection (ω > 1). For all three MAT genes, models that allow codons to evolve under positive selection (M2a and M8) did not fit the data significantly better than models that do not permit positive selection (M1a and M7) (Table 4). Although for the genes MAT1-1-3 and MAT1-2-1 the log likelihood values of Model M2a and M8 were higher than the associated models M1a and M7, the p-values of the LRT statistic were > 0.05. Nevertheless, for MAT1-1-3 and MAT1-2-1 the LRT test of M0 versus M3 was significant, suggesting variable selection pressure among sites. For these two genes a proportion of sites with ω > 1 was found in models M2a and M8, i.e. 7.6% of the sites with ω = 2.72 for MAT1-1-3 and 3.1% of the sites with ω = 5.76 for MAT1-2-1. Up to seven amino acid sites were identified as sites of positive selection using the BEB analysis in MAT1-1-3 but only one site had strong support with BEB posterior probability > 95% in model M8 (see Additional file 3). In MAT1-2-1 five amino acid sites were reported under positive selection but had weak support with BEB posterior probabilities. In MAT1-1-1, no proportions of sites with ω > 1 were reported.
Table 4.
Likelihood ratio tests comparing models of molecular evolution of MAT genes
| Gene | Analysis | λ1 | df1 | 2Δλ1 | p-value | dN/dS | Parameters |
|---|---|---|---|---|---|---|---|
| MAT1-1-1 | M0: one ratio | -2205.4733 | 4 | 5.9586 | 0.2023 | 0.4280 | ω = 0.4280 |
| M3: discrete (κ = 3) | -2202.4940 | 0.4462 | p0 = 0.3732, ω0 = 0.7052; p1 = 0.3932, ω1 = 0.1117; p2 = 0.2336, ω2 = 0.7052 | ||||
| M1a: nearly neutral | -2202.6642 | 2 | 0.0000 | 1.0000 | 0.4599 | p0 = 0.6556, ω0 = 0.1761; p1 = 0.3440, ω1 = 1 | |
| M2a: positive selection | -2202.6642 | 0.4599 | p0 = 0.6556, ω0 = 0.1761; p1 = 0.2332, ω1 = 1; p2 = 0.1109, ω2 = 1 | ||||
| M7: β | -2202.5615 | 2 | 0.0000 | 1.0000 | 0.4507 | p0 = 0.4117; q = 0.5019 | |
| M8: β & ω > 1 | -2202.5615 | 0.4507 | p0 = 0.1000 p = 0.4117 q = 0.5019; (p1 = 0.00001) ω = 1.0000 | ||||
| MAT1-1-3 | M0: one ratio | -1706.3379 | 4 | 20.5175 | 0.0004* | 0.295 | ω = 0.2951 |
| M3: discrete (κ = 3) | -1696.0792 | 0.3103 | p0 = 0.0007, ω0 = 0.1116; p1 = 0.9232, ω1 = 0.1117; p2 = 0.0761, ω2 = 2.7212 | ||||
| M1a: nearly neutral | -1697.3663 | 2 | 2.5742 | 0.2761 | 0.2598 | p0 = 0.7402, ω0 = 0; p1 = 0.2598, ω1 = 1 | |
| M2a: positive selection | -1696.0792 | 0.3103 | p0 = 0.9239, ω0 = 0.1117; p1 = 0, ω1 = 1; p2 = 0.0761, ω2 = 2.7212 | ||||
| M7: β | -1697.6108 | 2 | 3.0594 | 0.2166 | 0.3000 | p = 0.0050; q = 0.0119 | |
| M8: β & ω > 1 | -1696.0811 | 0.3104 | p0 = 0.9244 p = 12.5791 q = 99.0000; (p1 = 0.0756) ω = 2.7290 | ||||
| MAT1-2-1 | M0: one ratio | -2133.8605 | 4 | 11.1677 | 0.0247* | 0.3714 | ω = 0.37135 |
| M3: discrete (κ = 3) | -2128.2767 | 0.4212 | p0 = 0.3531, ω0 = 0.2495; p1 = 0.6157, ω1 = 0.2496; p2 = 0.0312, ω2 = 5.7427 | ||||
| M1a: nearly neutral | -2129.6740 | 2 | 2.7948 | 0.2472 | 0.3528 | p0 = 0.6472, ω0 = 0; p1 = 0.3528, ω1 = 1 | |
| M2a: positive selection | -2128.2767 | 0.4212 | p0 = 0.9688, ω0 = 0.2496; p1 = 0, ω1 = 1; p2 = 0.0312, ω2 = 5.7428 | ||||
| M7: β | -2129.8169 | 2 | 3.0800 | 0.2144 | 0.3387 | p = 0.0170; q = 0.0321 | |
| M8: β & ω > 1 | -2128.2769 | 0.4212 | p0 = 0.9690 p = 33.0717 q = 99.0000; (p1 = 0.0310) ω = 5.7612 |
Models of neutral evolution (M1a and M7) are tested against models that incorporate positive selection (M2a and M8). Comparison of M0 vs. M3 tests variable dN/dS rate ratios among sites. Model M3 classifies sites in the sequence into discrete classes (κ = 3) with both the dN/dS rate ratio ω0, ω1, ω3 and the proportions p0, p2 and p3 estimated from the data. For models M1a and M2a, values of p0, p1, and p2, are the proportions of sites inferred to be evolving under purifying selection, neutral evolution, and positive selection, respectively, and ω0, ω1 and ω2 are their corresponding dN/dS rate ratios. For models M7 and M8, the β distribution, β (p, q), describes the distribution of the dN/dS rate ratio between zero and one, and p0 is the proportion of sites within this distribution. In these models, p1 is the proportion of sites inferred to be under positive selection, and ω is the dN/dS rate ratio for those sites.
Discussion
Members of the Phialocephala fortinii s.l. - Acephala applanata species complex (PAC) have no known sexual state. In this study, however, footprints of sexual reproduction were found in populations from large global samples, providing evidence that sexual reproduction may occur in these fungal species.
MAT locus structure and evolution of homo- and heterothallism in the PAC species
Homothallic fungi can fertilize themselves whereas heterothallic fungi depend on another compatible individual for sexual reproduction to occur. In filamentous ascomycetes the MAT locus is the key determinant of breeding system and it has been shown that conversions between heterothallism and homothallism can be achieved by manipulating the MAT locus [41,49]. For example, the heterothallic Neurospora crassa was capable of self-fertilization after a strain was made carrying both mating types [50,51]. In contrast, a strain of the homothallic Giberella zeae was converted to self-sterility after those MAT genes were deleted that are present on opposite mating types in other closely related heterothallic species [49,52].
The structure of the MAT locus was mapped for 11 additional PAC species for which the MAT locus had not yet been characterized. All of these species possessed either the MAT1-1 or MAT1-2 idiomorph consistent with a heterothallic organization structure. The identified MAT genes MAT1-1-1 and MAT1-3-1 or MAT1-2-1 in the respective idiomorphs were of consistent lengths and their arrangement within the MAT locus and orientation with respect to each other was the same as found for other PAC species [44] and the other closely related helotialean species Pyrenopeziza brassicae, Oculimacula yallundae and Rhynchosporium secalis [45,46,53]. The only PAC species containing a homothallic MAT locus structure remains A. applanata [44]. A controversial topic in fungal biology is whether heterothallism or homothallism represents the ancestral state [37,41,49,54,55]. The question has been addressed in particular for the ascomycete genera Cochliobolus [40], and references therein] and Aspergillus [56], and references therein] that comprise heterothallic and homothallic species. In the heterothallic Cochliobolus species the MAT locus structure is conserved as strains are either MAT1-1 or MAT1-2 and contain the genes MAT1-1-1 or MAT1-2-1 with the same gene orientation within idiomorphs. In contrast, in the homothallic species the MAT locus structure is unique because single individuals possess both MAT genes that are either fused into a single ORF, are closely linked or likely present on different chromosomes. Due to this variation in MAT locus structure and phylogenetic evidence of independent evolution of self-fertility, the likely derived state in this genus was proposed to be homothallism [49]. Recombination is normally suppressed in the MAT locus because the sequences of the two idiomorphs are significantly diverged [57,58]. However, small identity island of 8 and 9 nucleotides were identified within MAT genes of the heterothallic Cochliobolus heterostrophus and rare recombination events between these small identity islands were suggested to be the likely mechanism for the conversion of reproductive modes in ancestral lineages [49]. In contrast, the prevalence of homothallic species, phylogenetic analyses and comparisons of genome sequences suggest that heterothallism is the derived state in the genus Aspergillus [59] albeit additional characterizations of MAT loci of supposedly asexual Aspergillus species may alter this view [56].
For PAC species both evolutionary trajectories of reproductive modes from homothallism to heterothallism and vice versa are conceivable. Because MAT genes control reproductive processes the information gained by comparing their DNA sequences might allow reconstructing the evolutionary history of a species complex and indicate mechanisms that led to changes in reproductive modes [49]. The basal position of A. applanata in the MAT gene phylogenies suggests that the MAT genes of A. applanata are older than the MAT genes of the remaining PAC species which are grouped in clusters, and thus are more recently derived and share a common ancestor (see Additional file 4). In fact, this finding is congruent with population genetic data and other phylogenetic data based on DNA sequences of housekeeping genes and RFLP loci that showed that A. applanata is the most diverging species in the PAC [60]. It is possible that a switch in reproductive mode of a homothallic ancestor was the source of the strong divergence between A. applanata and other PAC species and that thus, A. applanata represents an ancient lineage within the PAC. In populations of an ancestral homothallic species a structural change in the MAT locus, e.g. after a deletion of a MAT gene could have provoked a switch to heterothallism and created barriers to self-mating. These genetic barriers could have increased and maintained genetic diversity, eventually leading to speciation and radiation that gave rise to the current heterothallic species. The likely mechanisms that have initiated such a switch in the PAC might be explained by re-arrangements among idiomorphs and the influence of transposable elements on recombination events in the MAT locus of A. applanata [44]. On the other hand the heterothallic PAC species possess a conserved MAT locus structure with congruent MAT gene orientation and position within the MAT locus. This organization is also consistent with the MAT locus structure found in closely related helotialean species that are heterothallic [45,46,53]. Thus, the prevalence of structurally heterothallic species in the PAC and their conserved MAT locus organization are arguments in favor of heterothallism as ancestral breeding system.
Mating type distribution and gametic equilibrium
If sexual reproduction plays an important role in the life cycle of a fungal species, natural populations would be expected to show following characteristics [16,19]: (i) 1:1 ratio of mating types because frequency-dependent selection acts on the rare mating type that has the higher chance for mating, and (ii) gametic equilibrium at unlinked loci because under random mating sexual recombination will reduce non-random association of alleles. Both criteria were met for the majority of PAC populations. The 6 out of 52 populations that displayed gametic disequilibrium were restricted to the species P. subalpina. Independently of the reproductive mode several factors can cause non-random association of alleles that result in gametic disequilibrium. These factors include high frequency of rare alleles, genetic drift, population admixture and selection [16,61]. We therefore re-analyzed these collections of P. subalpina after pooling rare alleles but the results did not change (data not shown). We then tested whether traces of population admixture could be identified in the two populations using the program STRUCTURE [62] and evidence for admixture was found in two populations. Moreover, the strains collected from Bialowieza could be allocated with high posterior probabilities to two subpopulations and the index of association IA did not significantly deviate from zero in both of these subpopulations (data not shown).
The spatial distribution of mating type within study sites is often neglected in studies aiming at assessing the reproductive biology of fungal species. In this study opposite mating types were often collected at the same grid points within study sites. Moreover, the measures of spatial association Q were predominantly neutral to strongly positive, meaning that for several populations the occurrence of both mating types at the same grid point was higher than expected by chance alone. Thus, the physical proximity of compatible mating types in the study sites meets another prerequisite for successful sexual reproduction in the PAC.
Selection acting on mating type genes in PAC
If MAT genes play an important role in the initiation and further steps of sexual reproduction [39,40] they are expected to be functionally conserved in sexual species. All PAC species possessed structurally identical idiomorphs and translation of the MAT genes suggested that they encode functional proteins. In A. applanata the insertion of a transposable element in the 3' end of MAT1-2-1 caused a truncation of 30 amino acids in the translated protein [44]. Moreover, insertions of 3 and 9 bp were found in MAT1-1-1 of A. applanata and in MAT1-2 deletions as long as 18 bp were found for cryptic species CSP12, CSP14, P. letzii, P. europaea and A. applanata. However these indels did not result in stop codons or frame shifts that could affect functionality.
The molecular analyses revealed that all three MAT genes appear to be under strong purifying selection because sites under purifying selection were predominant. The overall values of ω obtained ranged between 0.26-0.46, and the proportions of sites under purifying selection (ω < 1) were much larger than the fraction evolving neutrally or under positive selection accounting for at least 65% of the sites. In only few studies the selective pressures on MAT genes were analyzed and similar overall values of ω were found ranging between 0.14 and 0.49 in sexual or presumed sexual fungi [22,23,25,26]. The predicted effect of long-term asexuality is the decay of genes specific for sex and recombination due to the relaxation of selective constraints [63] but the results of our study suggest that MAT genes in PAC species are highly conserved and functional.
Rapid divergence of reproductive proteins, due to positive selection and adaptive evolution, is believed to be important in the formation of reproductive barriers leading to speciation [64-66]. Competition for gamete recognition between individuals might explain the rapid evolution of reproductive proteins [67]. High levels of interspecific polymorphism have been found in MAT genes [22,68]. In our study the amount of polymorphism between the structurally heterothallic PAC species was also high and ranged between 8.7 and 9.4%. This was in the range found for non-coding RFLP loci and approximately 2.5-fold more than in levels found in the housekeeping gene beta-tubulin [60]. However, statistical evidence of positive selection of MAT gene, i.e. high levels of non-synonymous substitutions leading to changes in MAT proteins, has only been shown in the genus Neurospora [25]. Our results showed that models of evolution that allow positive selection (ω > 1) were not more likely than those incorporating neutral evolution. Nevertheless, the comparison of model M0 vs. M3 tested significant for variable pressure among sites in MAT1-1-3 and MAT1-2-1 and up to seven sites evolving under positive selection were identified in these genes, although only one was significant in the BEB analysis. It might be assumed that these and many more sites evolve under positive selection in MAT genes but that, in a strong background of purifying selection, they remain uncovered by the currently available analysis methods. However, whether the putative protein alterations at such positively selected sites have a biological significance is unknown.
Have PAC species a sexual life cycle?
In the present study we combined different indirect approaches to analyze the importance of sexual reproduction in the PAC. The population analyses were in accordance with random mating and sexual recombination in abundant collections from different ecosystems. Moreover, the presence of a conserved MAT locus in all PAC species suggests that MAT genes are functional and are still involved in mating processes. Yet, although the data does not allow rejecting the hypothesis of sexual recombination, the confirmation of the teleomorph in vitro is pending. Our first attempts to induce apothecia formation of PAC species failed when we tried to cross opposite mating types under laboratory conditions [44]. However, in vitro induction of the teleomorph is often difficult and time consuming and some perseverance is required [17,18]. Nevertheless, we successfully induced teleomorph formation of Phaeomollisia piceae, the closest known relative of the PAC [69]. P. piceae forms small darkly pigmented apothecia of less than 800 μm diameter. Considering that PAC species might form similar apothecia in the soil or on roots it seems likely that the teleomorph has been overlooked in field studies.
Conclusions
In this study we evaluated the importance of sexual reproduction in the PAC, the dominant dark septate root endophytes of woody plant species, and showed that the signature of sex is present in all of these putatively asexual species. The MAT locus resembles those of closely related heterothallic ascomycetes, is conserved and purifying selection likely preserves functionality of its genes. The hypothesis of sex occurring in the PAC cannot be rejected based on the random association of alleles at multiple loci, the equal mating type frequencies and the spatial distribution of opposite mating types in populations from different ecological habitats, hosts and continents. We believe that further field studies and in vitro crosses will finally lead to the discovery of the sexual state, which is the missing link to better understand how reproductive biology shaped the evolutionary history and influences the ecological role of the PAC.
Methods
Collection of PAC species and mating type screening
A total of 3, 639 strains derived from 33 study sites were included in the mating type screening (Table 2). These strains represent 19 PAC [32] species eight of which are formally described [28,29]. Several classes of molecular markers were developed for PAC species assignment including PCR fingerprints, single-copy restriction fragment length polymorphisms (RFLP), multi-locus DNA sequences, and microsatellites. Each of these molecular markers supported the delineation of multiple species in this complex, with concordant cryptic species defined by all markers [60,70]. PAC species communities were either collected on grid nets (196 m2, 74 grid points) [33] or on transects (50 m, 11 grid points) [32] from Europe (number of study sites n = 19), North America (n = 10) and Asia (n = 4). Collections included both undisturbed and managed forests.
Multiplex PCR
A multiplex PCR method was developed to discriminate between isolates carrying either the MAT1-1 or the MAT1-2 idiomorph based on available sequence data of the MAT locus of seven structurally heterothallic PAC species [44]. The primer Pf_HMG_R.03 (5'-TCCTCGAACCGGTGCCACGAATGACTCCGA-3') was designed to match a flank of the idiomorphic region that is conserved between the two mating types of these species. The MAT1-1 specific primer (Pf_HMG_F4: 5'-AGCTGAGCTCGAGACCTCGTTCGC-3') and a MAT1-2 specific primer (Pf_MAT1-1F1c: 5'-CTTGTACGGTCCGGCAATCCACA-3') were designed to generate DNA fragments of distinct length in combination with Pf_HMG_R.03 that could be easily differentiated on agarose gels. All PCRs were performed on a Biometra T1 thermal cycler in a 10 μl reaction volume containing approximately 2 ng template DNA, 5 pmol of each primer, 50 mM KCl, 10 mM Tris-HCl, 1, 5 mM MgCl2, 200 μM dNTPs (Promega AG, Dübendorf) and 0.2 U Taq polymerase (Promega AG, Dübendorf). Cycling conditions during PCR were 2 min at 94°C followed by 29 cycles of denaturation for 20 s at 94°C, annealing for 30 s at 60°C and extension for 60 s at 72°C, followed by a final extension step of 5 min at 72°C.
Mating type frequencies in PAC populations
The number of strains from a single study site and belonging to the same PAC species is here defined as a population. Before assessing significance of deviation from a 1:1 mating type ratio in a population, each population was clone-corrected using 12 microsatellite loci [70] or 11 single-copy RFLP markers [33], meaning that only one representative of each multi-locus genotype was included. Clone-correction is necessary because the same fungal thallus is found on several grid points. Five species [cryptic species (CSP) 8, 9, 10, 18 and 19] occurred at exceptionally low frequencies in the collection sites and represented less than 3% of all collected strains. For these species, the distribution of mating types was assessed after pooling all strains of a species, irrespective of the study site from which they were isolated. Significance was tested using the exact binomial test. In addition, Bonferroni corrections were performed for multiple comparisons.
Because species abundance distributions within local PAC communities follow a hyperbolic distribution with a few very abundant species and many "rare" species [48], clone-corrected datasets for such rare species resulted in a low number of distinct genotypes (< 10). In these cases, only the frequency of mating types was recorded without testing for significance.
Analysis of gametic disequilibrium
Significant deviations from multi-locus gametic equilibrium in a population were tested using the index of association IA and its variance according to Maynard Smith et al. [47]. All calculations of IA were based on clone-corrected datasets and multi-locus genotypes with missing data were excluded from the analysis.
Spatial distribution of mating types in selected study sites
Spatial distribution of mating types was analyzed for five Swiss study sites. Only the two most abundant PAC species per community were included in this analysis. For each of these species, the number of grid points was recorded where either MAT1-1 or MAT1-2 or both mating types were found. A contingency table test for independence of the spatial distribution of the mating types was performed and a measure of association between MAT1-1 and MAT1-2 (Q) was calculated as described in Pielou [71].
Studying the presence of both mating types in single isolates
In few cases, no amplification product was obtained in some strains, or both the MAT1-1 and MAT1-2 specific fragments were amplified in single strains. In order to study the reason for these observations, ten new single-hyphal tip cultures were prepared from the original slants of the three strains having displayed both PCR products and of the five strains from which no PCR product was obtained. After harvesting and extracting DNA of mycelia from both the original slants and the newly prepared single-hyphal tip cultures, the mating types were re-determined using the multiplex PCR.
Sequencing the MAT locus
The MAT locus was sequenced for 11 PAC species and compared to the MAT locus structure of 8 previously sequenced PAC species [44]. For each of these species the MAT locus of both the idiomorph MAT1-1 and MAT1-2 was sequenced (Table 1). The detailed sequencing strategy and the sequencing primers used are given in the Additional files 5 &6, and the MAT locus was annotated as described in [44]. PCR fragments were directly purified using an "ExoSap" protocol [70]. Cycle sequencing was performed with the Big-Dye v3.1 kit (Applied Biosystems) in a 10 μl reaction volume using 1 μl purified PCR amplification product, 0.5 μl BigDye v3.1, 1.9 μl 5 × Buffer, 5.6 μl ddH2O and 1 μl of the sequencing primer (10 μM). Cycle sequencing reactions were performed on a Biometra T1 thermal cycler with the following running conditions: 60 s at 96°C followed by 55 cycles of 10 s at 95°C, 5 s at 50°C and 4 min at 60°C. Dye-labeled fragments were cleaned using BigDye Xterminator Purification kit following manufacturer's instructions (Applied Biosystems). Samples were run on an ABI 3130xl DNA Analyser (Applied Biosystems).
Analysis of selective pressures
In order to assess what kind of selective pressure is acting on any of the three MAT genes a maximum likelihood (ML) method implemented in the program CODEML in the PAML 4.1 software package [72] was used to compare the rate of non-synonymous substitutions with the rate of synonymous substitutions (dN/dS = ω). Under neutrality ω is expected to be 1. When amino acid changes are favored, indicative of positive selection, ω is expected to be > 1, whereas under purifying selection amino acid changes are prevented and ω is expected to be < 1. Positive selection of small number of codons in the MAT genes may be masked when the entire gene is strongly affected by purifying selection. Therefore, a codon by codon approach was used based on the ML method implemented in CODEML that allows site by site identification of particular codons that have been evolving under repeated and strong positive selection [72]. The three MAT genes were analyzed separately and a gene tree describing the phylogenetic relationships of all taxa studied (one strain per species) was constructed using the ML method implemented in the program treefinder [73] using the best-fitting mutation model. The inferred trees served as the basis for the implementation of the ML methods in the CODML (see Additional file 4).
Three sets of models, commonly applied to test hypotheses of selection, were used. Following the suggestions of Yang [72] the site model pairs that appear to be particularly useful for real data analysis, are the M1a (nearly neutral) versus M2a (selection) and M7 (neutral) versus M8 (selection). However, model M0 (one-ratio) was also compared versus M3 (discrete) in order to see if the selective pressure is variable among sites. Because the comparison of M7 versus M8 is less conservative, this set of models may indicate positive selection even when none is detected by the M1a vs. M2a comparison.
The strength of positive selection was calculated using the Likelihood Ratio Test (LRT) [72] implemented in PAML by comparing twice the log likelihood difference in a chi-square test. The degree of freedom for the test statistic is determined by the difference in estimated parameters between the models of selective pressures being compared, i.e. four [M0 (one-ratio) vs. M3 (discrete)] or two [M1a (nearly neutral) vs. M2a (selection), and M8 (selection) vs. M7 (neutral)] degrees of freedom [72]. Codons that are identified as having evolved under positive selection have high posterior probabilities (p > 0.95). Posterior probabilities for such sites were estimated based on Bayes Empirical Bayes (BEB) analysis [74].
The MAT gene sequence of the homothallic A. applanata species was not included in this analysis because it would have inflated the alignments with deletions. For example, the MAT1-2-1 sequence of A. applanata contained a large deletion of 90 bp at its 3' end, due to the insertion of a transposable element in the MAT locus of the species [44]. Gaps in alignments cannot be handled by CODEML and the program automatically removes such sites. Thus, in order to prevent the loss of informative sites in the sequence of the other species, the A. applanata sequences were removed from the alignment prior to the analyses.
List of abbreviations
MAT: mating type; PAC: Phialocephala fortinii s.l. - Acephala applanata species complex.
Authors' contributions
PZ participated in the design of the study, conducted the analysis of selective pressures, drafted and wrote the manuscript. VQ participated in the molecular work and conducted microsatellite analysis. AD carried out molecular work and participated in the sequence alignment. CG designed and coordinated the study, performed statistical analysis and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Testing the presence of both idiomorphs in selected PAC strains. Testing the presence of both idiomorphs, or the absence of MAT genes in selected PAC strains.
Spatial distribution of mating types. Example of the spatial distribution of mating types for a population of Phialocephala subalpina and Phialocephala turicensis.
Sites in MAT genes identified to evolve under positive selection. Sites under positive selection were identified using the Bayes Empirical Bayes analysis under different CODML site models in the PAML software package.
Phylogenies inferred from MAT genes. Phylogenies inferred for the three genes MAT1-1-1, MAT1-1-3, and MAT1-2-1. The sequence of Acephala applanata served as outgroup in all three analysis.
Sequencing strategy for MAT idiomorphs. Sequencing strategy applied to sequence the complete MAT idiomorphs for PAC species. Sequences of primers are given in the Additional file 5.
Sequencing primers. Sequences of primers used to sequence the complete MAT idiomoprh of PAC species.
Contributor Information
Pascal L Zaffarano, Email: pascal.zaffarano@env.ethz.ch.
Valentin Queloz, Email: valentin.queloz@env.ethz.ch.
Angelo Duò, Email: angelo.duo@env.ethz.ch.
Christoph R Grünig, Email: christoph.gruenig@microsynth.ch.
Acknowledgements
The Genetic Diversity Center (GDC, ETH Zurich) provided facilities for collecting the sequence data. This work was associated with the GEDIHAP project of the Competence Center Environment and Sustainability (CCES) of the ETH Domain, Zurich, Switzerland.
References
- Bell G. The Masterpiece of nature: the evolution and genetics of sexuality. London Canberra: Croom Helm; 1982. [Google Scholar]
- Kondrashov AS. Classification of hypothesis on the advantage of aphimixis. J Hered. 1993;84(5):372–387. doi: 10.1093/oxfordjournals.jhered.a111358. [DOI] [PubMed] [Google Scholar]
- West S, Lively CM, Read AF. A pluralist approach to sex and recombination. J Evolution Biol. 1999;12(6):1003–1012. doi: 10.1046/j.1420-9101.1999.00119.x. [DOI] [Google Scholar]
- Agrawal AF. Evolution of sex: Why do organisms shuffle their genotypes? Curr Biol. 2006;16(17):R696–R704. doi: 10.1016/j.cub.2006.07.063. [DOI] [PubMed] [Google Scholar]
- Otto SP. The Evolutionary Enigma of Sex. Amer Natur. 2009;174:S1–S14. doi: 10.1086/599084. [DOI] [PubMed] [Google Scholar]
- Meirmans S, Strand R. Why Are There So Many Theories for Sex, and What Do We Do with Them? J Hered. 2010;101:S3–S12. doi: 10.1093/jhered/esq021. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. Evolution in sexual and asexual populations. Amer Natur. 1965;99(909):439–450. doi: 10.1086/282389. [DOI] [Google Scholar]
- Smith JM. The evolution of sex. Cambridge: Cambridge University Press; 1978. [Google Scholar]
- Charlesworth B. The Cost of Sex in Relation to Mating System. J Theor Biol. 1980;84(4):655–671. doi: 10.1016/S0022-5193(80)80026-9. [DOI] [PubMed] [Google Scholar]
- Hoekstra RF, Vanloo EN. The Cost of Sex in Hermaphrodite Populations with Variation in Functional Sex. J Theor Biol. 1986;122(4):441–452. doi: 10.1016/S0022-5193(86)80184-9. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Deleterious Mutations and the Evolution of Sexual Reproduction. Nature. 1988;336(6198):435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- Aanen D, Hoekstra RF. In: Sex in Fungi Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. Why sex is good: On fungi and beyond; pp. 527–534. [Google Scholar]
- Kirk PM, Cannon PF, David JC, Stalpers JA, eds. Dictionary of the fungi. 9. Oxon, UK: CAB International; 2001. [Google Scholar]
- Dyer PS, Ingram DS, Johnstone K. The Control of Sexual Morphogenesis in the Ascomycotina. Biol Rev. 1992;67(4):421–458. doi: 10.1111/j.1469-185X.1992.tb01189.x. [DOI] [Google Scholar]
- Kück U, Poeggeler S. Cryptic sex in fungi. Fung Biol Rev. 2009;23:86–90. doi: 10.1016/j.fbr.2009.10.004. [DOI] [Google Scholar]
- Milgroom MG. Recombination and the multilocus structure of fungal populations. Annu Rev Phytopathol. 1996;34:457–477. doi: 10.1146/annurev.phyto.34.1.457. [DOI] [PubMed] [Google Scholar]
- Magee PT, Magee BB. Through a glass opaquely: the biological significance of mating in Candida albicans. Curr Opin Microbiol. 2004;7(6):661–665. doi: 10.1016/j.mib.2004.10.003. [DOI] [PubMed] [Google Scholar]
- O'Gorman CM, Fuller HT, Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457(7228):471–U475. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Xu J. Fundamentals of fungal molecular population genetic analyses. Curr Issues Mol Biol. 2006;8:75–89. [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Fisher MC. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- Paoletti M, Rydholm C, Schwier EU, Anderson MJ, Szakacs G, Lutzoni F, Debeaupuis JP, Latge JP, Denning DW, Dyer PS. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr Biol. 2005;15(13):1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fung Genet Biol. 2004;41(6):600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Devier B, Aguileta G, Hood ME, Giraud T. Ancient Trans-specific Polymorphism at Pheromone Receptor Genes in Basidiomycetes. Genetics. 2009;181(1):209–223. doi: 10.1534/genetics.108.093708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Nygren K, Johannesson H. The evolution of the pheromonal signal system and its potential role for reproductive isolation in heterothallic Neurospora. Mol Biol Evol. 2008;25(1):168–178. doi: 10.1093/molbev/msm253. [DOI] [PubMed] [Google Scholar]
- Wik L, Karlsson M, Johannesson H. The evolutionary trajectory of the mating-type (mat) genes in Neurospora relates to reproductive behavior of taxa. BMC Evol Biol. 2008;8 doi: 10.1186/1471-2148-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Villavicencio M, Aguileta G, Giraud T, de Vienne DM, Lacoste S, Couloux A, Dupont J. Sex in Penicillium: Combined phylogenetic and experimental approaches. Fung Genet Biol. 2010;47(8):693–706. doi: 10.1016/j.fgb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Grünig CR, Queloz V, Sieber TN, Holdenrieder O. Dark septate endophytes (DSE) of the Phialocephala fortinii s.l. - Acephala applanata species complex in tree roots - classification, population biology and ecology. Botany. 2008;86(12):1355–1369. doi: 10.1139/B08-108. [DOI] [Google Scholar]
- Grünig CR, Duò A, Sieber TN, Holdenrieder O. Assignment of species rank to six reproductively isolated cryptic species of the Phialocephala fortinii s.l.-Acephala applanata species complex. Mycologia. 2008;100(1):47–67. doi: 10.3852/mycologia.100.1.47. [DOI] [PubMed] [Google Scholar]
- Grünig CR, Sieber TN. Molecular and phenotypic description of the widespread root symbiont Acephala applanata gen. et sp. nov., formerly known as dark septate endophyte Type 1. Mycologia. 2005;97(3):628–640. doi: 10.3852/mycologia.97.3.628. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yin L, Dai S. Diversity of root-associated fungal endophytes in Rhododendron fortunei in subtropical forests of China. Mycorrhiza. 2009;19(6):417–423. doi: 10.1007/s00572-009-0246-1. [DOI] [PubMed] [Google Scholar]
- Piercey MM, Graham SW, Currah RS. Patterns of genetic variation in Phialocephala fortinii across a broad latitudinal transect in Canada. Mycol Res. 2004;108(8):955–964. doi: 10.1017/S0953756204000528. [DOI] [PubMed] [Google Scholar]
- Queloz V, Sieber TN, Holdenrieder, McDonald BA, Grünig CR. No biogeographical pattern for a root-associated fungal species complex. Global Ecol Biogeog. 2011;20(1):160–169. doi: 10.1111/j.1466-8238.2010.00589.x. [DOI] [Google Scholar]
- Grünig CR, McDonald BA, Sieber TN, Rogers SO, Holdenrieder O. Evidence for subdivision of the root-endophyte Phialocephala fortinii into cryptic species and recombination within species. Fung Genet Biol. 2004;41(7):676–687. doi: 10.1016/j.fgb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- McGill BJ, Etienne RS, Gray JS, Alonso D, Anderson MJ, Benecha HK, Dornelas M, Enquist BJ, Green JL, He FL. et al. Species abundance distributions: moving beyond single prediction theories to integration within an ecological framework. Ecol Letters. 2007;10(10):995–1015. doi: 10.1111/j.1461-0248.2007.01094.x. [DOI] [PubMed] [Google Scholar]
- de Wit R, Bouvier T. 'Everything is everywhere, but, the environment selects'; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8(4):755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- Heitman J, Kronstad JW, Taylor JW, Casselton LA, eds. Sex in Fungi. Molecular Determination and Evolutionary Implications. Washington, D.C.: ASM Press; 2007. [Google Scholar]
- Coppin E, Debuchy R, Arnaise S, Picard M. Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev. 1997;61(4):411–428. doi: 10.1128/mmbr.61.4.411-428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casselton LA. Mate recognition in fungi. Heredity. 2002;88:142–147. doi: 10.1038/sj.hdy.6800035. [DOI] [PubMed] [Google Scholar]
- Stanton BC, Hull CM. In: Sex in Fungi Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. Mating-type locus control of cell identity; pp. 59–73. [Google Scholar]
- Turgeon BG, Debuchy R. In: Sex in Fungi Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. Cochliobolus and Podospora: mechanisms of sex determination and the evolution of reproductive lifestyle; pp. 93–121. [Google Scholar]
- Lin X, Heitman J. In: Sex in Fungi Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. Mechanisms of homothallism in fungi and transitions between heterothallism and homothallism; pp. 35–57. [Google Scholar]
- Debuchy R, Turgeon BG. In: The Mycota: Growth, differentiation and sexuality. Kües U, Fischer R, editor. Vol. 1. Berlin Springer; 2006. Mating-type structure, evolution, and function in Euascomycetes; pp. 293–323. [DOI] [Google Scholar]
- Turgeon G, Yoder OC. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fung Genet Biol. 2000;31(1):1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- Zaffarano PL, Duò A, Grünig CR. Characterization of the mating type (MAT) locus in the Phialocephala fortinii s.l. -Acephala applanata species complex. Fungal Genet Biol. 2010;47(9):761–772. doi: 10.1016/j.fgb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Singh G, Ashby Alison M. Cloning of the mating type loci from Pyrenopeziza brassicae reveals the presence of a novel mating type gene within a discomycete MAT 1-2 locus encoding a putative metallothionein-like protein. Mol Microbiol. 1998;30(4):799–806. doi: 10.1046/j.1365-2958.1998.01112.x. [DOI] [PubMed] [Google Scholar]
- Foster SJ, Fitt BDL. Isolation and characterisation of the mating-type (MAT) locus from Rhynchosporium secalis. Curr Genetics. 2003;44(5):277–286. doi: 10.1007/s00294-003-0445-9. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, Orourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünig CR, Duò A, Sieber TN. Population genetic analysis of Phialocephala fortinii s.l. and Acephala applanata in two undisturbed forests in Switzerland and evidence for new cryptic species. Fung Genet Biol. 2006;43(6):410–421. doi: 10.1016/j.fgb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Yun S-H, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci USA. 1999;96(10):5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Grotelueschen J, Metzenberg RL. Neurospora crassa A mating-type region. Proc Natl Acad Sci USA. 1990;87(13):4912–4916. doi: 10.1073/pnas.87.13.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staben C, Yanofsky C. Neurospora crassa a mating-type region. Proc Natl Acad Sci USA. 1990;87(13):4917–4921. doi: 10.1073/pnas.87.13.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lee T, Lee YW, Yun SH, Turgeon BG. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol Microbiol. 2003;50(1):145–152. doi: 10.1046/j.1365-2958.2003.03694.x. [DOI] [PubMed] [Google Scholar]
- Singh G, Dyer Paul S, Ashby Alison M. Intra-specific and inter-specific conservation of mating-type genes from the discomycete plant-pathogenic fungi Pyrenopeziza brassicae and Tapesia yallundae. Curr Genet. 1999;36(5):290–300. doi: 10.1007/s002940050503. [DOI] [PubMed] [Google Scholar]
- Nauta MJ, Hoekstra RF. Evolution of reproductive systems in filamentous ascomycetes. ll. Evolution of hermaphroditism and other reproductive strategies. Heredity. 1992;68:537–546. doi: 10.1038/hdy.1992.76. [DOI] [PubMed] [Google Scholar]
- Butler G. In: Sex in fungi: molecular determination and evolutionary implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. The evolution of MAT: The ascomycetes; pp. 3–18. [Google Scholar]
- Dyer PS. In: Sex in Fungi Molecular Determination and Evolutionary Implications. Heitman J, Kronstad JW, Taylor JW, editor. Casselton LA. Washington, DC: ASM Press; 2007. Sexual reproduction and significance of MAT in Aspergilli; pp. 123–142. [Google Scholar]
- Menkis A, Jacobson DJ, Gustafsson T, Johannesson H. The mating-type chromosome in the filamentous ascomycete Neurospora tetrasperma represents a model for early evolution of sex chromosomes. PLoS Genetics. 2008;4(3):e1000030. doi: 10.1371/journal.pgen.1000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D. The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol. 2009;24(2):94–102. doi: 10.1016/j.tree.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J. et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438(7071):1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Grünig CR, Brunner PC, Duò A, Sieber TN. Suitability of methods for species recognition in the Phialocephala fortinii -Acephala applanata species complex using DNA analysis. Fung Genet Biol. 2007;44(8):773–788. doi: 10.1016/j.fgb.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Linde CC, Zala M, McDonald BA. Molecular evidence for recent founder populations and human-mediated migration in the barley scald pathogen Rhynchosporium secalis. Mol Phylogenet Evol. 2009;51(3):454–464. doi: 10.1016/j.ympev.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normark BB, Judson OP, Moran NA. Genomic signatures of ancient asexual lineages. Biol J Linnean Soc. 2003;79(1):69–84. doi: 10.1046/j.1095-8312.2003.00182.x. [DOI] [Google Scholar]
- Snook RR, Chapman T, Moore PJ, Wedell N, Crudgington HS. Interactions between the sexes: new perspectives on sexual selection and reproductive isolation. Evol Ecol. 2009;23(1):71–91. doi: 10.1007/s10682-007-9215-3. [DOI] [Google Scholar]
- Turner LM, Hoekstra HE. Causes and consequences of the evolution of reproductive proteins. Int J Dev Biol. 2008;52(5-6):769–780. doi: 10.1387/ijdb.082577lt. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131(1):11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Palumbi SR. Speciation and the evolution of gamete recognition genes: pattern and process. Heredity. 2009;102(1):66–76. doi: 10.1038/hdy.2008.104. [DOI] [PubMed] [Google Scholar]
- Poeggeler S. Phylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes. Curr Genet. 1999;36(4):222–231. doi: 10.1007/s002940050494. [DOI] [PubMed] [Google Scholar]
- Grünig CR, Queloz V, Duò A, Sieber TN. Phylogeny of Phaeomollisia piceae gen. sp. nov.: a dark-septate conifer-needle endophyte and its relationships to Phialocephala and Acephala. Mycol Res. 2009;113(2):207–221. doi: 10.1016/j.mycres.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Queloz V, Duo A, Sieber TN, Grünig CR. Microsatellite size homoplasies and null alleles do not affect species diagnosis and population genetic analysis in a fungal species complex. Mol Ecol Ressources. 2010. [DOI] [PubMed]
- Pielou EC. Mathematical Ecology. New York: John Wiley & Sons; 1977. Association between pairs of species I: Individuals in discrete habitable units; pp. 203–239. [Google Scholar]
- Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Jobb G. TREEFINDER version. 2010. http://www.treefinder.de/
- Yang ZH, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Testing the presence of both idiomorphs in selected PAC strains. Testing the presence of both idiomorphs, or the absence of MAT genes in selected PAC strains.
Spatial distribution of mating types. Example of the spatial distribution of mating types for a population of Phialocephala subalpina and Phialocephala turicensis.
Sites in MAT genes identified to evolve under positive selection. Sites under positive selection were identified using the Bayes Empirical Bayes analysis under different CODML site models in the PAML software package.
Phylogenies inferred from MAT genes. Phylogenies inferred for the three genes MAT1-1-1, MAT1-1-3, and MAT1-2-1. The sequence of Acephala applanata served as outgroup in all three analysis.
Sequencing strategy for MAT idiomorphs. Sequencing strategy applied to sequence the complete MAT idiomorphs for PAC species. Sequences of primers are given in the Additional file 5.
Sequencing primers. Sequences of primers used to sequence the complete MAT idiomoprh of PAC species.