Abstract
New site-specific protein labeling (SSPL) reactions for targeting specific, short peptides could be useful for the real time detection of proteins inside of living cells. One SSPL approach matches bioorthogonal reagents with complementary peptides. Here, hydrazide reactive peptides were selected from phage-displayed libraries using reaction-based selections. Selection conditions included washes of varying pH and treatment with NaCNBH3 in order to specifically select reactive carbonyl containing peptides. Selected peptides were fused to T4 lysozyme or synthesized on filter paper for colorimetric assays of the peptide-hydrazide interaction. A peptide-lysozyme protein fusion demonstrated specific, covalent labeling by the Hydrazide Reactive (HyRe) peptides in crude bacterial cell lysates, sufficient for the specific detection of an over-expressed protein fusion. Chemical synthesis of a short HyRe tag variant and subsequent reaction with two structurally distinct hydrazide probes produced covalent adducts observable by MALDI-TOF MS and MS/MS. Rather than isolating reactive carbonyl-containing peptides, we observed reaction with the N–terminal His of HyRe tag 114, amino acid sequence HKSNHSSKNRE, which attacks the hydrazide carbonyl at neutral pH. However, at the pH used during selection wash steps (<6.0), an alternative imine-containing product is formed that can be reduced with sodium cyanoborohydride. MSMS further reveals that this low pH product forms an adduct on Ser6. Further optimization of the novel bimolecular reaction described here could provide a useful tool for in vivo protein labeling and bioconjugate synthesis. The reported selection and screening methods could be widely applicable to the identification of peptides capable of other site-specific protein labeling reactions with bioorthogonal reagents.
INTRODUCTION
Site-specific protein labeling (SSPL) provides a powerful tool for the in vivo tracking of protein localization, dynamics, and concentration.1,2 SSPL enables visualization of specific proteins through conjugation to an appropriately functionalized probe. Genetically encoded fluorescent protein fusions provide a facile route to site-specific protein labeling,3 and are the current standard in labeling technology;4 however, the large size and limitation to a fluorescent signal has stimulated a search for smaller and chemically flexible labeling strategies. One route, enzymatic labeling of genetically encoded peptide substrates, is useful in forming fluorescent conjugates in vivo, and has been reviewed recently.5,6 Another powerful approach harnesses unique peptide sequences capable of forming covalent bonds with chemical probes. The latter approach does not require additional enzymes or reagents. Here, for example, we describe the selection, screening, and characterization of hydrazide reactive peptides (Figure 1).
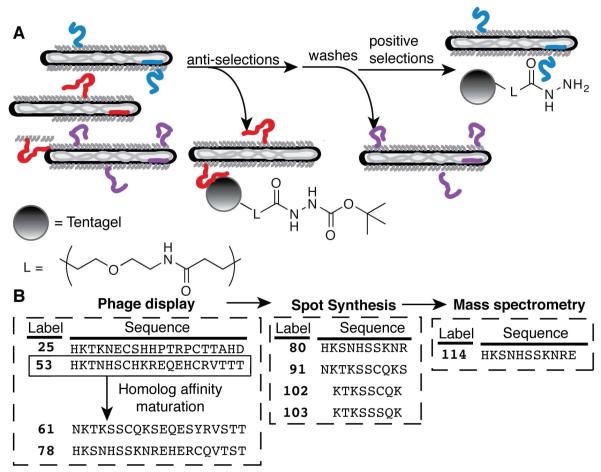
Figure 1.
(A) Selections for hydrazide binding peptides included anti-selections with Boc-protected hydrazide resin to remove non-specific peptides (e.g., the red peptide), positive selections with hydrazide-derivatized resin to capture hydrazide interacting peptides (e.g., the blue peptide), and extensive wash steps to remove non-binding peptides (e.g., the purple peptide). (B) The sequences of peptides discussed in the text from selections of phage-displayed peptide libraries, spot synthesis, and chemical synthesis followed by mass spectrometry analysis.
Non-enzymatic SSPL reactions have revolutionized protein purification and visualization. For example, short polyhistidine peptides (“His-tags”) selectively coordinate nickel (Ni2+), or cobalt (Co2+) chelated to a solid support. A genetically encoded His6-tag enabled purification of dihydrofolate reductase from crude, E. coli cell lysate under both native and denaturing conditions.7 Since used by innumerable laboratories for protein purification, His-tags also enable visualization via binding to a fluorescent, zinc-conjugated small molecule (HisZiFiT).8 Another standalone, probe-reactive peptide, uses the tetracysteine motif (amino acid sequence CCXXCC) to selectively react with fluorogenic bisarsenical probes in vitro and in vivo.9-11 Other short peptides capable of modification with metal-based fluorescent probes include labeling of tetraserine motifs by bis-boronic acids,12 polyaspartic acids by zinc fluorophores,13 and lanthanide binding peptides with lanthanide ions.14,15
Discovery of new SSPL peptide-probe pairs requires both a modestly reactive small molecule and a unique, complementary peptide. The small molecule must be stable to physiological conditions, yet reactive upon peptide binding. The partnering reactive peptide is ideally short enough to minimize misfolding and mislocalization of the fused protein, yet sufficiently long to ensure unique representation in the proteome. Suited for the identification of such complementary peptides, phage display enables the rapid sifting of vast libraries to identify peptides with affinity for essentially any target, including proteins16,17 small molecules,1 and inorganic materials.18 The filamentous bacteriophage used in phage display links the encoding DNA sequence to the displayed peptide, providing easy routes to both amplification and identification of the selected peptides.19,20
Two examples illustrate the utility of phage display to identify either non-covalent or covalent SSPL peptide-probe pairs. First, conventional affinity-based phage display identified tightly binding peptides that modulate the fluorescence of Texas red.21 After many rounds of optimization, a 38-residue peptide tethered via an intramolecular dimerization domain was developed that non-covalently bound a calcium-sensitive Texas red derivative with a KD of 80 pM.22 Secondly, reaction-based phage display targeted a diketone to isolate the 20-mer peptide rpf1368, which forms enaminones with various diketone-containing probes.23 The reported peptides from the initial rounds of selection for both of these examples proved functional yet inefficient. Further optimization of the peptides through appending additional libraries to previously selected sequences and performing more stringent selections ultimately yielded tighter binding or more reactive peptides.22,23
Other functionalities could be amenable to the discovery of bioorthogonal peptide-probe pairs using either affinity- or reaction-based phage display. Side chain modifying electrophiles, such as iodoacetamide and N-hydroxy succinamide are exceptional modifying agents for cysteine and lysine, respectively. These functionalities are insensitive to 3D sequence space, and can modify exposed side chains. Therefore, these reagents are restricted to use with purified proteins. Considering the nucleophilic nature of biopolymers, nucleophlic probes should be vastly more discriminating towards specific amino acid sequences. Thus, we focused on a nucleophilic functionality with known bioorthogonality towards proteins. Hydrazides do not react with unmodified biopolymers, but are used to label and isolate oxidized molecules with reactive carbonyls.24 Reactive carbonyl-containing proteins, for example, can be purified from biological samples using biotin hydrazide and surface-bound avidin for mass spectrometry identification.25,26
Reactive carbonyls can also be introduced into protein sequences through enzymatic labeling of peptide substrates. Ligation of a ketone-containing probe to an acceptor peptide with biotin ligase generates a reactive carbonyl for modification with hydrazide probes.27 Similarly, formylglycine-generating enzyme modification of the cysteine in the sequence LCTPSR also generates a reactive carbonyl for subsequent hydrazide labeling.28 These peptides are easily genetically incorporated as a fusion to proteins of interest for labeling proteins with either hydrazide- or aminooxy-derivatized compounds, after treatment with the appropriate enzyme.28,29 Unnatural amino acid incorporation can also introduce ketone-based amino acids.30,31
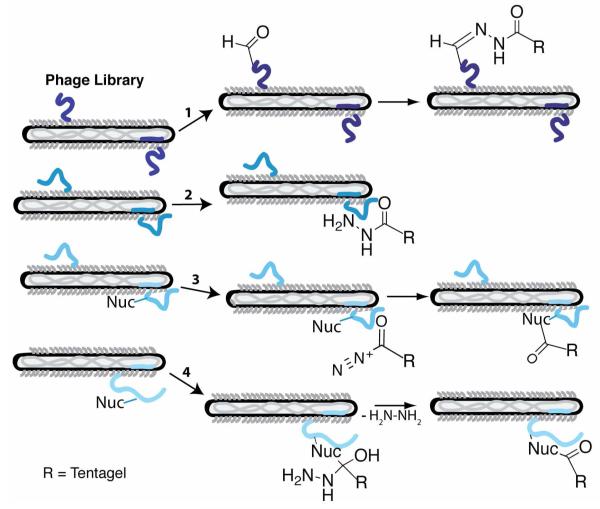
We sought to simplify protein labeling with hydrazide probes by identifying peptides with affinity for hydrazide, thus eliminating the need for the co-expression of a modifying enzyme. Hydrazides can interact with polypeptides in four different mechanisms (Figure 2). First, hydrazides would form hydrazones with reactive carbonyls present in oxidized peptides. A large number of spontaneous reactions introduce reactive carbonyls into protein sequences including conjugation to oxidized lipids and oxidation of susceptible amino acids (such as the side chains of arginine, histidine, lysine, or proline); both reactions occur during some age- and disease-related stresses.32,33 In addition, peptides from a phage-displayed library could act as substrates for carbonyl forming enzymes such as the formylglycine-generating enzyme, described above.34 Therefore, peptides susceptible to either enzymatic or atmospheric oxidation would react with hydrazide-based capture agents.
Figure 2.
Potential mechanisms for the selection of hydrazide interacting peptides from phage-displayed libraries include oxidation of susceptible peptides (1) for capture with hydrazide-derivatized Tentagel via hydrazone bond formation. Additionally, phage-displayed peptides could form high affinity non-covalent interactions with the hydrazide functionality (2). Alternatively, the hydrazide functional group could be oxidized and attacked by a peptide nucleophile liberating nitrogen gas (3). Lastly, a nucleophile from the displayed peptide could attack the carbonyl of the hydrazide functional group, displacing hydrazine (4).
Alternatively, peptides could non-covalently bind to the probe, as was discovered for the Texas red binding peptides. However, the small size of the hydrazide group might preclude high affinity binding. Contrary to peptide oxidation, the hydrazide functional group could be selectively oxidized, thereby presenting an electrophile for attack by a peptide nucleophile before displacement of nitrogen. The enzyme tyrosinase oxidizes acylhydrazide to an acyl diazene, to liberate nitrogen; and has been used as a chemical deprotection strategy.35 In the fourth mechanism, a peptide nucleophile could attack the hydrazide carbonyl, exhibiting similar reactivity as cysteine proteases such as cathepsin K inhibition by acylhydrazide inhibitors. In this example, the active site cysteine attacks the carbonyl of the acylhydrazide resulting in a stabilized tetrahedral intermediate.36
Thus, reaction-based phage selections for peptides interacting with the hydrazide functional group could explore a number of different reaction possibilities to expand the palette of bioorthogonal peptide-probe pairs (Figure 1). After five rounds of selection and DNA sequencing of the selected phage to determine trends of the peptide interactions with the hydrazide functionality, one sequence was chosen for further affinity maturation by homolog shotgun scanning. The selected peptides were screened for interaction with hydrazide probes through fusion to T4 lysozyme. After homolog affinity maturation, the interaction of all selected peptides with biotin hydrazide was compared through peptide spot synthesis and screening. The most promising short peptides identified by spot synthesis were chemically re-synthesized, treated with hydrazide-containing probes, and analyzed by mass spectrometry. Given the reactivity of hydrazides with ketones and aldehydes, we expected to isolate oxidation-susceptible peptides. Instead, the isolated 20-mer peptides are nucleophilic towards certain hydrazide derivatives.
EXPERIMENTAL PROCEDURES
Selections for hydrazide binding peptides
To remove background-binding peptides, an anti-selection was performed by incubating a previously described, phage-displayed peptide library37 with Boc-hydrazide Tentagel for 1 h at room temperature in a peptide reaction vessel. The unbound phage were next subjected to a positive selection through incubation with a new aliquot of deprotected, hydrazide Tentagel for 20-60 min with decreasing concentrations of resin in each round. The hydrazide-bound phage were washed three to six times with increasing volumes of PBS (from 10-50 mL) and times (from 2-20 min) for each subsequent round. Additional wash steps to enhance the stringency of selections included two washes with HCl (0.1 M, 1.5 mL) in rounds 1 and 2, and three to six washes of HCl (0.1 M, 50 mL) in rounds 4 and 5. The phage-resin mixture was then re-buffered with a wash of PBS (50 mL, 10 min) before being re-suspended in PBS (1.5 mL) for storage and propagation. Half of the phage-Tentagel suspension was incubated with 10 mL of XL-1 blue E. coli (OD600 = 0.5-1.0) for 20 min with shaking at 37 °C. A small aliquot of the infected cells were titered on LB-carbenicillin-agar plates, and the remaining culture was transferred to 2YT (250 mL) supplemented with M13-KO7 helper phage before overnight incubation at 37 °C with shaking. Individual colonies from the titer plate were sequenced following PCR using M13 primers (Supporting Information).
Bacterial subcloning and over-expression of HyRe-lysozyme fusion
The selected peptides were subcloned into a plasmid encoding each peptide fused through a GGGSG linker to the N-terminus of T4 lysozyme (Supporting Materials and Methods). Correctly ligated plasmids were transformed into heat shock competent BL21(DE3) E. coli. An overnight 37 °C culture (10 mL LB, 40 μg/mL kanamycin) was added to 1 L of LB media supplemented with kanamycin (40 μg/mL), and grown for an additional ≈2.5 h at 37 °C to an OD600 of 0.6. The addition of IPTG (1 mM) induced protein expression at 30 °C for 4 h. Following centrifugation and sonication, the crude supernatant was purified using cation exchange chromatography. Protein fusions were eluted with a gradient to 1 M NaCl and fractions containing the protein fusion were then combined and concentrated. Gel permeation chromatography further purified fusion proteins to >95% homogeneity (Figure S2 A in Supporting Information).
Biotin hydrazide and rhodamine B hydrazide binding assay to HyRe 53-T4 lysozyme fusion
Following purification, the HyRe peptide 53-T4 lysozyme fusion, the proteolyzed peptide 53-T4 lysozyme fusion, or wild-type T4 lysozyme were treated with either biotin hydrazide (1 mM in PBS for 1 h at room temperature) or an equivalent volume of buffer (negative controls) before the addition of SDS loading dye (20% volume). The solution was then incubated at 95 °C for 3 min to denature the proteins. The denatured protein samples were added to two 15% SDS-page gels before electrophoresis at a voltage of 130 V for 90 min. One protein gel was Coomassie-stained, and the other gel was transferred to a nitrocellulose membrane via electrophoresis in transfer buffer (25 mM Tris base; 192 mM glycine; 20% methanol; 0.02% SDS; pH 8.3) at 100 V for 100 min. This nitrocellulose membrane was blocked with 1.0% Tween 20 in PBS overnight at 4 °C. Biotinylation was assessed by incubating the membrane with SAV-AP at room temperature for 1 h. This membrane was washed five times with PT before visualization by treatment with 4-chloro-1-naphthol and 3,3′-diaminobenzidine, tetrahydrochloride (Pierce). Rhodamine B hydrazide (final concentration 1mM) was added to crude lysates for 1 h at room temperature, before SDS-PAGE and electrophoretic transfer to nitrocellulose as before. The blot was then visualized using a GE Typhoon scanner (532 nm laser, 580 bp 30 filter, 400 PMT voltage).
Spot synthesis and assay of HyRe peptide variants
Peptides were synthesized as C-terminal adducts to filter paper, and deprotected as outlined in reference 47. A background-binding assay was completed by incubating the sheets of spot synthesized peptides with SAV-HRP, before visualization with BCIP/NBT. The AP substrates, BCIP (135 mM in DMF) and NBT (61 mM in 70% DMF) were added to AP buffer (100 mM Tris, 100 mM NaCl, 2 mM MgCl2, 0.05% Tween-20, pH 9.5) to a final concentration of 445 μM and 403 μM respectively. This buffer was added to the spot-synthesized peptide cellulose sheets, and incubated for 5 min. After washing five times, images of the sheets were captured by digital photography under white light illumination. Peptides specifically binding to biotin hydrazide were then assayed by incubating biotin hydrazide (0.17 mM) in PBS for 2 h. The sheet was then washed five times (50 mL, 2 min each), before re-addition of SAV-HRP, re-visualization with BCIP/NBT, and re-imaging.
HyRe peptide synthesis and reaction with hydrazide derivatives
Peptides 103 and 114 were synthesized on a 0.2 mmol scale using standard solid phase peptide synthesis with Fmoc-protected amino acids (Aroz technologies) on Rink amide resin (Novabiochem). The synthesized peptides were cleaved, purified, and characterized, using standard conditions (Figures S7 in Supporting Information). The purified peptides were resuspended in water before dilution into PBS to a 1 mM final concentration. The hydrazides dissolved in DMSO were then added to each peptide (1 mM final concentration), and incubated at room temperature for 1-3 h before de-salting with C18 peptide desalting Zip-Tips (Varian; Palo Alto, CA) and characterized by MALDI-TOF MS.
RESULTS
Selections of hydrazide ligands from a phage library
The naïve, M13 phage-displayed peptide library included ≈2.5 × 1010 unique sequences fused to the major coat protein, P8. This collection of peptide sequences included 22 different configurations of disulfide-constrained peptide libraries and one linear peptide library, ranging in length from 8- to 20-mers37,38 (Table S1 in Supporting Information).
Molecular display selections can sometimes fail due to deleterious amplification of “background binding” peptides; such peptides are selected for binding to either the solid support or blocking agents employed during selection, and do not specifically bind to the target. Background binding peptides are removed from the phage library through anti-selections using similar selection conditions without the target. Thus, before each round of selection, an anti-selection with Boc-hydrazide Tentagel removed peptides with an affinity for Tentagel. The anti-selected phage library was added to a second aliquot of deprotected hydrazide Tentagel, and used to select phage-displayed peptides with affinity for hydrazide (Figure 1A). This water miscible solid support enables stringent wash steps to remove non-specific binding peptides during positive selections. Up to half a liter of different wash buffers was applied to 20 mg of resin during each round of selection. Additionally, in an effort to ensure capture of oxidized peptides, the phage library was subjected to treatment with sodium cyanoborohydride (NaCNBH3), and washes of varying pH. The reactive carbonyl-hydrazide reaction is reversible at low pH39, which in round 1 provided a route to selectively elute bound phage from the Tentagel. Sodium cyanoborohydride will reduce hydrazones, 40 permanently linking the selected phage to the Tentagel. After five rounds of increasingly stringent selection conditions (including shorter reaction times, less concentration of target resin, and increasing numbers of longer washes), the amino acid sequences of 74 phage-displayed, hydrazide interacting peptides were determined by DNA sequencing to yield 58 distinct sequences (Table S2 in Supporting Information).
In round one, 4.4×107 phage were isolated, but more stringent conditions in round 2 reduced the phage titers to 6.6×105 plaque forming units, and 2.2×106 phage colonies in round 3. The titers then equilibrated to 1×107 for the final two rounds of selection. More importantly, 30% of the first round of selection was identified as helper phage by sequencing, whereas 0% of the selectants in rounds 2-5 were helper phage. The reduced presence of helper phage in the selected pool demonstrates a successful selection for functional phage-displayed peptides.
The success of molecular display selections can be gauged by sequence homology and other trends in the sequences of selectants.41 In selections for binding to hydrazide Tentagel, hydrophilic amino acid side chains were enriched during selections, and hydrophobic amino acid side chains were almost entirely absent from the selected sequences. Furthermore, the sequences from selection rounds three through five were entirely derived from two of the 23 peptide scaffolds, X7CX4CX7 and X6CX7CX5 (where X = any of the 20 naturally occurring amino acids). Several peptides appeared in multiple rounds of selection, including peptide 25, identified in selection rounds two through five, and peptide 53, which accounted for ≈30% of the isolated sequences from the final two rounds of selection.
Next, peptides with potentially higher affinity for hydrazide were selected using homolog shotgun scanning. This method applies a protein-based, medicinal chemistry approach with a library composed of either the wild-type or closely homologous amino acid.42,43 This library strategy focuses peptide diversity space, and selects for subtle side chain substitutions that can collectively contribute large effects to binding affinity. Additionally, conserved residues during these selections can indicate functionally important side chains.
Chosen for its high frequency amongst the initial selectants, peptide 53 formed the basis for a homolog shotgun-scanning library. Unlike previous homolog shotgun scanning libraries, each amino acid position encoding His, Arg, or Lys was substituted with a library codon encoding His, Lys, Arg, Asn, Ser, Pro, or Gln. These additional amino acid residues were included due to their observed importance for hydrazide binding from the initial library selectants. Reaction-based selections analogous to those described above isolated an additional 23 hydrazide interacting peptides (Table S2 in Supporting Information). Peptide 61 was identified in homolog shotgun scanning rounds two through four, and peptide 78 dominated round 4, accounting for 60% of the selectants. Additionally, valine at position 17 and glutamic acid at positions 11 and 13 were highly conserved, suggesting the importance of those residues for hydrazide binding (Figure S1C in Supporting Information).
Notably, highly enriched peptides, 61 and 78, include only one cysteine residue. Thus, the intramolecular disulfide bonds engineered into the naïve libraries appeared unnecessary for hydrazide binding. This result contrasts with previous studies in which intramolecular disulfide bonds in peptides selected from phage-displayed peptide libraries contribute critical stability to peptide structure for target binding.44,45 This observation presented the opportunity, explored below, to eliminate cysteine residues in chemically synthesized versions of the selected peptides.
Typically, screens examining the functionality of individual phage-displayed selectants quantify the relative binding abilities of selectants from a library. Conventional screens for hydrazide binding with phage-displayed peptides proved problematic due to cross-reactivity between biotin hydrazide and contaminants remaining in the phage solutions, likely oxidized proteins. Two screening approaches explored the capabilities of the selected hydrazide interacting peptides. First, the gene for peptide 53 was fused to a test protein T4 lysozyme for colorimetric assays. Second, peptides and peptide variants from both the original selections and the homolog shotgun scanning selections were spot-synthesized on filter paper.
Hydrazide binding to peptide-T4 lysozyme fusions
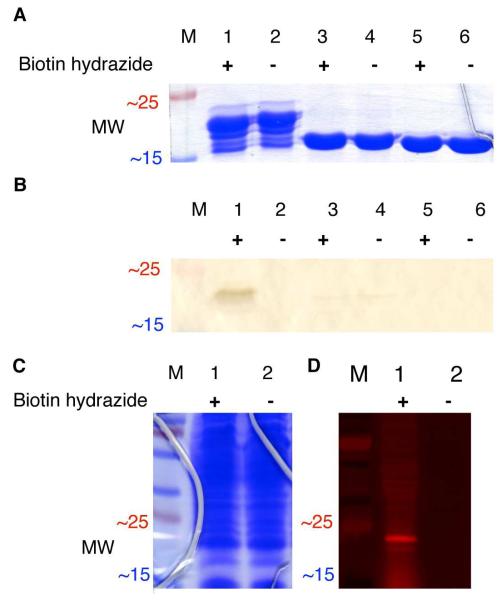
To test the reaction specificity of the selected peptides, the frequently selected peptide 53 was subcloned into an expression plasmid (pET-28c, Invitrogen) as a N-terminal fusion to the enzyme T4 lysozyme. The purified fusion protein was then treated with biotin hydrazide (1 mM) for two hours at room temperature in aqueous buffer (PBS at pH 7.2). SDS-PAGE and subsequent binding assay of the lysozyme fusions, either treated or untreated with biotin hydrazide, demonstrated that fusion to peptide 53 conferred biotin-hydrazide binding ability (Figure 3, lanes 1-2). Sustained affinity during the denaturing conditions required for SDS-PAGE (95 °C in SDS) can provide evidence for covalent labeling of peptide-probe interactions (e.g., the bis-arsenical tetracysteine peptide reaction).10 Therefore, additional mass spectrometry-based experiments were performed to identify the covalent adducts formed by Hydrazide Reactive (HyRe) peptide 53 and its variants reacting with hydrazide derivatives.
Figure 3.
(A) SDS-PAGE of the purified 53-lysozyme fusion protein (22.3 kDa in lanes 1-2), the fully proteolyzed 53-lysozyme fusion protein (19.2 kDa in lanes 3-4), and wild-type lysozyme (18.6 kDa in lanes 5-6). Each sample was either treated (+) or untreated (−) with biotin hydrazide (1 mM) for 1 h at room temperature before electrophoresis. (B) Western blot analysis for biotin hydrazide labeling the purified 53-lysozyme fusion protein. The protein gel and Western blot, excerpted for clarity here, are shown in full in Figure S2 in Supporting Information. (C) SDS-PAGE of the 53-lysozyme fusion protein in crude E. coli lysates, either treated (+) or untreated (−) with 1 mM rhodamine B hydrazide before separation by SDS-PAGE. (D) The protein lysate shown in C was then transferred electrophoretically to nitrocellulose, and imaged by fluorescence scanning. “M” indicates molecular weight standards.
As might be expected for fusions to a well-folded protein separated by a glycine linker (amino acid sequence of GGGSG), the peptide lysozyme fusions are susceptible to proteolysis (Figure 3, lanes 1-2). HyRe peptide 53 is completely removed from the N-terminus of lysozyme by proteolysis (Figure 3, lanes 3-4), as confirmed by MALDI-TOF mass spectrometry (Figure S2 C in the Supporting Information). Proteolytic removal of the HyRe tag prevents reaction with biotin hydrazide (Figure 3B, lane 3), and no signal is observed in the biotin-binding assay. Comparable results are observed for the negative control, treatment of wild-type lysozyme with biotin hydrazide (Figure 3B, lanes 5-6). Selective proteolysis from the N-terminus of lysozyme demonstrates that biotin hydrazide reacts site-specifically with the HyRe tag and not with the other amino acids of lysozyme.
Additionally, rhodamine B hydrazide reacts exclusively with the 53-lysozyme fusion protein in crude E. coli lysate (Figure 3D). Following SDS-PAGE and electrophoretic transfer to nitrocellulose of the crude lysate reaction mixture, the fluorescent probe labels only the fusion protein. Taken together, the experiments with lysozyme fusions show that two different hydrazide derivatives selectively label HyRe peptide 53. Therefore, further characterization of the interaction between HyRe peptide tags and hydrazide-containing probes focused on a covalent reaction.
Spot synthesis of hydrazide-interacting peptides
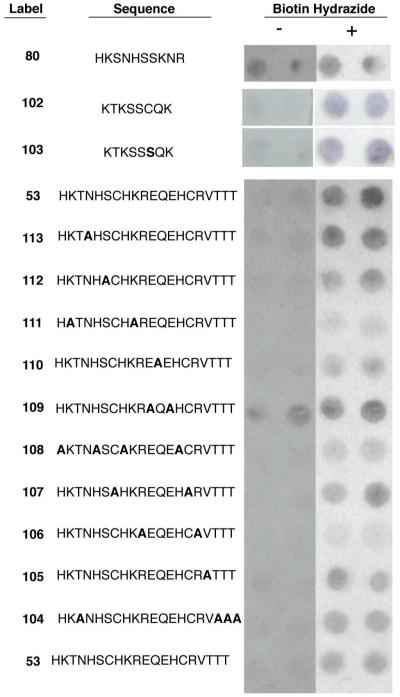
Peptides 61, 78, and 53, and variants of all three were chemically re-synthesized using spot synthesis. In this technique, each peptide is covalently attached as a C-terminal fusion to spatially segregated positions on a cellulose membrane.46,47 Spot synthesis allows for the simultaneous fabrication and screening of hundreds of different peptides in a format free from biological contaminants such as post-translational modifications by enzymes (e.g., formylglycine-generating enzyme)34 or reactive carbonyl-containing metabolites (e.g., pyridoxal phosphate).48,49 The synthesized analogs of peptides 61 and 78 included sequential 10-mers and single-site alanine substitutions. Scanning sequential 10-residue peptides could identify smaller active sequence of the HyRe tag. The spot-synthesized variants of peptide 53 included all 1, 2, 3, and 4 contiguous amino acid deletions. Additionally, the homolog shotgun scanning results suggested substitution of serine for cysteine in the HyRe peptide tags. Three different spot synthesis cellulose sheets containing over 350 different variants were synthesized and screened for binding to biotin hydrazide (Figures S3-S5 in Supporting Information).
Following deprotection of the spot-synthesized peptides, the relative binding affinity for biotin hydrazide was assayed by treatment with streptavidin conjugated to alkaline phosphatase (SAV-AP) and developed through addition of the AP substrates 5-bromo-4-chloro-3-indolyl phosphate (BCIP) and nitrotetrazolium blue (NBT). The levels of SAV-AP activity in each spot can provide an estimate of the efficiency of each peptide in the reaction with biotin hydrazide. However, many factors can influence the signal intensity at individual spots including peptide synthesis failure and purity.50 Additionally, identical sequences in different locations on the filter paper produced slightly variant levels of reactivity with hydrazide (see HyRe tag 53 in Figure 4). Thus, the qualitative analysis of spot synthesis reported here focuses on trends and requirements for hydrazide-peptide reactivity.
Figure 4.
Variants of selectant peptides were spot-synthesized and either treated (+) or untreated (−) with biotin hydrazide before binding to SAV-AP and visualization through addition of the AP substrate. The N-terminal 10-mer from peptide 78 produces a non-specific signal in the SAV-AP assay. An 8-mer from peptide 61 produces a specific signal for biotin hydrazide, as does the Cys7Ser substituted 8-mer, 103. Each type of amino acid in peptide 53 was substituted with alanine to identify key side chain functionalities required for the interaction with biotin hydrazide, while the other residues of peptide 53 remained unchanged. Bold letters indicate substitutions in the selected peptides.
Testing all possible contiguous 10-mers from both HyRe tags 61 (peptides 80-90) and 78 (peptides 91-101) revealed the importance of the N-terminal amino acids for reactivity with biotin hydrazide (Figure S6 in Supporting Information). The 10-residues at the N-terminus of peptide 78, labeled peptide 80, resulted in the highest overall signal intensity for reaction with biotin hydrazide on spot synthesis sheet 1, a result confirmed through duplicated synthesis and screens (Figure 4). However, peptide 80 also exhibited high background signal in the SAV-AP assay. This independent assay for background included all steps except treatment with biotin hydrazide, and measures peptide interaction with SAV-AP or the ability of the peptides to dephosphorylate BCIP.
The spot synthesis screens guided the synthesis of HyRe tags with both reactivity towards biotin hydrazide and reduced background binding. For example, HyRe peptide 91, the N-terminal 10 residues of HyRe tag 61, reacted efficiently with biotin hydrazide, but also had high background binding to SAV-AP. Further minimization via spot synthesis and screening identified HyRe peptide 102, an 8-mer peptide derived from HyRe tag 61, which resulted in the highest specific reactivity with biotin hydrazide relative to background levels. HyRe tag 102 includes one cysteine residue; therefore, the Cys7Ser substitution, peptide 103, was also spot-synthesized. This cysteine-free 8-mer peptide produced a signal equivalent to peptide 102, and also exhibited little background in the SAV-AP assay (Figure 4). Thus, as observed by homolog shotgun scanning, cysteine residues from the original selected HyRe tags can be omitted in homolog variants of peptide 53 without drastically reducing reactivity with biotin hydrazide.
Single point alanine substitutions of every residue in HyRe 61 and 78 had little effect on the qualitative reactivity of HyRe peptides with biotin hydrazide. Given the relative insensitivity for single point alanine substitutions, we next examined contributions made by each type of amino acid side chain to HyRe tag function by substituting all occurrences of each amino acid with alanine (Figure 4). For example, peptide 104 substituted all four threonines of peptide 53 with alanine. Subsequent reaction with biotin hydrazide and assay of the ten spot-synthesized peptides demonstrated the critical importance of arginine, histidine, and lysine side chains. Conversely, HyRe peptide 110 substituted Ala in place of both Glu residues, and resulted in increased background binding to SAV-AP. The importance of glutamic acid in position 11 also correlates to the conserved glutamic acids observed in homolog affinity selections (Figure S1 in Supporting Information). The spot-synthesis results guided the choice of peptides for large-scale synthesis.
Synthesis and mass spectrometry analysis of HyRe peptides
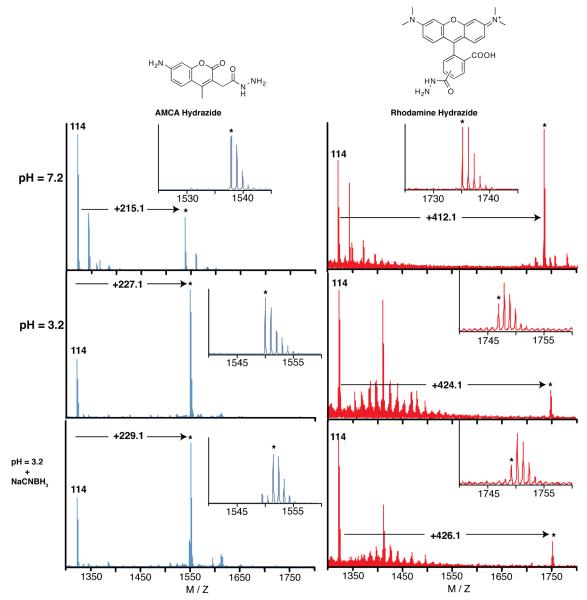
Two peptides identified initially from phage selections with hydrazide Tentagel and then minimized by screening spot-synthesized peptides were chosen for additional analysis: peptide 114, the N-terminal 11-residues from peptide 78 including the specificity determining Glu11, and peptide 103, residues 2 through 9 with the Cys7Ser substitution of HyRe peptide 61. After conventional solid-phase peptide synthesis, HPLC purification, and MALDI-TOF MS confirmation (Figure S7 in the Supporting Information), the synthesized HyRe peptides were treated with twelve different hydrazide-containing small molecules (structures shown in Figure S8) at equimolar concentrations (1 mM) in PBS, pH 7.2. The reactions were monitored by MALDI-TOF mass spectrometry (Figure 5). Interestingly, only AMCA hydrazide (aminomethylcoumarin acetate hydrazide) and rhodamine B hydrazide produced new mass adducts with either peptide.
Figure 5.
Treatment of peptide 114 with two hydrazide containing probes. Treatment with AMCA hydrazide at pH 7.2 produces a species with an additional mass of 215 mass units. However, treatment at pH 3.2 produces an adduct with an additional 227 mass units, which can be reduced with NaCNBH3. Treatment of 114 with rhodamine B hydrazide produces similar species at both pH 7.2 and 3.2, as well as upon reduction. The peaks marked with an asterisk (*) are the new adducts observed upon treatment with the hydrazide containing probes.
At pH 7.2, both HyRe peptides reacted with AMCA hydrazide to produce adducts with identical additional mass. The discussion here focuses on peptide 114, though similar results were observed for reactions of AMCA hydrazide with HyRe peptide 103 (Figures S7 in Supporting Information). Treatment of HyRe peptide 114 with AMCA hydrazide, followed by analysis with MALDI-TOF mass spectrometry, identified a new compound with an additional mass of 215 amu (Figure 5). Similarly, treatment of peptide 114 with rhodamine B hydrazide produced a new compound with an additional mass of 412. Both probes yield products with masses 32 amu less than the sums of the peptides and probes. The observation of new probe-specific higher molecular weight adducts is evidence for a single, covalent bond forming reaction between the selected peptide and hydrazide reagents. Furthermore, the difference in mass provides insight into the reaction mechanism.
MSMS analysis of the neutral pH peptide 114-AMCA hydrazide reaction reveals that the modification was restricted to daughter ions containing His1 and was absent from fragments lacking this residue. For example, a modified b-ion of the first residue (m/z = 481.2) was observed, while the y”-ion corresponding to residues 2-11 (lacking His1) possessed zero modifications (m/z = 1185.6). Additionally, both modified and unmodified b ions are observed for every residue, while no modified y” ions were observed. Therefore the modification resides on the N-terminal Histidine and the bond between AMCA hydrazide and the peptide is labile to MSMS conditions. This result was also observed in the analysis of the reaction product between 114 and rhodamine B hydrazide.
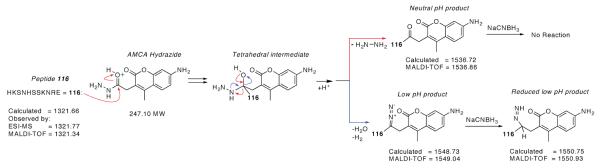
Addition of the hydrazide derivatives to HyRe peptide 114 followed by loss of hydrazine (32 amu) could account for the observed mass of the product (Scheme 1). In this proposed mechanism, the imidazole of the N-terminal His attacks the hydrazide carbonyl. Subsequent collapse of the tetrahedral intermediate displaces hydrazine. Additionally, since the leaving group hydrazine has not been directly observed, oxidation of the hydrazine, prior to displacement could contribute to the mechanism.
Scheme 1.
Proposed mechanism for the reaction between HyRe tag 114 and AMCA hydrazide. At pH = 7.2, loss of hydrazine justifies the observed loss of 32 mass units. At acidic pH (3.2 to 6.0), the loss of 20 mass units most likely results from a dehydration plus additional oxidation, to yield a product capable of reduction by NaCNBH3.
Notably, the HyRe peptide-hydrazide reaction takes place under mild conditions. The reaction occurs in aqueous buffer, at room temperature and physiological pH. No transition metal ions, organic solvents, and other proteins or co-factors are required. A peak corresponding to the adduct can be observed in the MALDI-TOF mass spectrum essentially immediately after the peptide and AMCA hydrazide are mixed, desalted, and applied to the mass spectrometer.
The reaction product could not be detected by reverse-phase HPLC and pH-neutral, ion-exchange chromatography. The reaction produces sufficient product for detection by fluorescence (peptide 53 fused to lysozyme in Figure 3), spot synthesis assays (Figure 4), and MS (peptide 114 in Figure 5). However, insufficient product is obtained for spectroscopic analysis, including HPLC detection. The MS analysis benefits from concentration by the de-salting Zip-Tips for sample preparation. Furthermore, the Zip-Tips appear to enrich for the product of the reaction with hydrazide, as the very hydrophilic HyRe peptide 114 is incompletely retained by C18 ZipTips (data not shown). However, after modification with either of the hydrophobic hydrazide fluorophores, the peptides could be preferentially retained on the Zip-Tips. Alternatively, the covalent products from the hydrazide reaction could be initiated by the MS conditions; however, MS is typically used to characterize covalent bonding. Despite these caveats, the interaction between the HyRe peptides and hydrazide derivatives is sufficiently strong for various assays.
The sensitivity of using MALDI-TOF mass spectrometry provided a method to investigate the basis for the selection of these peptides from the phage-displayed peptide libraries. As described above, the selections featured both washes with pH 5 buffer and the addition of NaCNBH3 in an attempt to select reactive carbonyl containing peptides. However, the electrophilic modification of the N-terminal His complicates interpretation of the selection conditions. Specifically, the imidazole side chain should be largely protonated at pH values <6, thus preventing modification on this side chain.
MS screening of conditions between pH 3 and 11 identified a new product formed at low pH values. For pH values ≤6, a different product resulted from the treatment of peptide 114 with AMCA and rhodamine B hydrazide. AMCA hydrazide added 227 and rhodamine B hydrazide added 424 amu (Figure 5). Both modifications result from the loss of 20 amu following addition of the probe to the peptide. Such conjugates could result from a condensation (−18 amu) accompanied by an additional oxidation (−2 amu). Additionally, these products from the low pH reaction, unlike the analogous neutral pH products, are reducible upon addition of NaCNBH3, producing new adducts with +2 additional mass units (Figure 5 and Scheme 1).
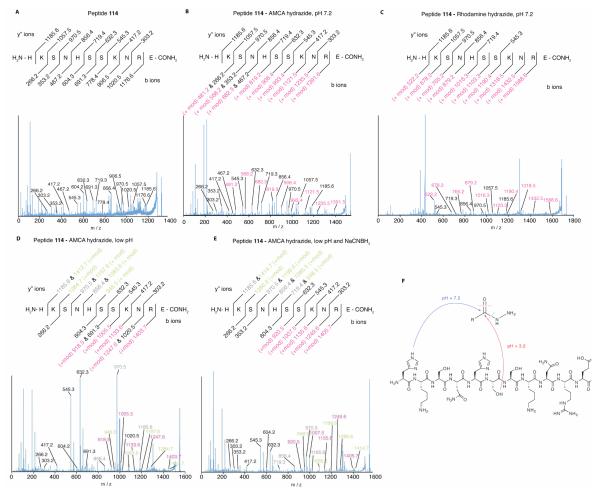
MSMS of the products from the low pH reaction of peptide 114 with AMCA hydrazide reveals that the fluorescent probe covalently attaches to a different amino acid, than was observed for neutral pH (Figure 6). MSMS analysis of the products from the AMCA hydrazide reaction demonstrates modification of the Ser6 side chain. Thus, the modification has switched residues from His1 at neutral pH to Ser6 at pH 5. This observation suggests a different mechanism for the selection of HyRe peptides from the phage-displayed peptide libraries. Consistent with the NaCNBH3 reduction, a diazo compound or other isomer could be formed.
Figure 6.
MSMS of products from the reaction of peptide 114 with AMCA and rhodamine B hydrazides. (A) MSMS of unmodified peptide 114 reveals all expected b and y” ions. (B) and (C) MSMS of both the AMCA- and rhodamine-modified peptides reveals an unmodified set of y” ions and a complete set of modified b ions indicating the covalent modification (+mod) occurs on the side chain of the N-terminal histidine. (D) MSMS of the reaction product at low pH and (E) following reduction with NaCNBH3 product indicates that the covalent modification occurs on Ser6. The MSMS chromatogram of peptide 114 modified with rhodamine B hydrazide at low pH was dominated by rhodamine cleavage adducts (data not shown). (F) A schematic diagram for the observed covalent modifications.
In conclusion, as shown by the results in Figures 3 through 5 the selected peptides can react with hydrazide derivatives at a range of different pH values and with sufficient efficiency for detection by MS, spot synthesis, and fluorescence blot assays.
DISCUSSION
The identification of short, standalone peptides capable of directing SSPL reactions remains a major challenge in modern chemical biology. The HyRe peptides offer an important step towards the goal of short peptides composed of naturally occurring amino acids that can react with hydrazide-based probes without requiring additional enzymes or reagents. The short sequence length of the peptides identified here and the availability of hydrazide-containing chemical probes suggest that HyRe tags could be used in a large number of applications with minimal perturbation, if the sequences are further optimized. As demonstrated here, HyRe peptide tags are capable of selectively detecting fusions in crude cell lysates on Western blots (Figure 3).
Selection and screens required careful planning to minimize false positives. To avoid the selection of streptavidin-binding peptides, streptavidin was omitted from the selection protocols. Thus, no sequences commonly associated with streptavidin binding were observed (e.g., HPQ-based sequences).41 Thus, any background binding in the spot synthesis results reflects non-specific interactions with SAV-AP or other components of the binding assay. Furthermore, the spot synthesis results clearly demonstrate the importance of 20-mer peptides for high specificity interactions between HyRe peptides and hydrazide derivatives. Shorter peptides resulted in higher background assay levels.
Though the HyRe peptides react with different hydrazide bearing probes, the results reaffirm the adage “you get what you screen [and select] for.”51 The selection conditions targeted hydrazide-derivatized Tentagel, which includes a linker between the PEG and the surface-exposed hydrazides. The resultant HyRe peptides also required an additional carbonyl at the γ-carbon of the hydrazide, found in both AMCA and rhodamine B hydrazide, for the covalent reaction observed by MS. Thus, the selection conditions isolated peptides requiring a functionality analogous to the linker of the targeted Tentagel. Interestingly, amide linkers have been found to participate in other examples of reaction discovery.52,53 Though designed to specifically capture reactive carbonyl containing peptides, the selection conditions identified peptides that react with hydrazide derivatives at neutral and low pH values, and subsequently with NaCNBH3.
Contrary to expectations, the selected peptides provide a nucleophile to react with the carbonyl of the hydrazide functionality. The HyRe tag-hydrazide reactivity reported here is analogous to acylhydrazide inhibitors of cathepsin K, in which an active site nucleophilic thiol forms a reversible covalent bond with the acylhydrazide carbonyl. Though this covalent bond could be observed by X-ray crystallography, the lability of the resultant adduct prevented product isolation by both dialysis and HPLC. Similarly, the efficiency of the hydrazide reaction by our selected peptides was insufficient for detection by HPLC.
In conclusion, this report demonstrates the reactivity of HyRe peptides composed of naturally occurring amino acids, which were selected from two generations of combinatorial libraries. As fusions to filter paper and lysozyme, the HyRe peptides react with both biotin hydrazide and rhodamine B hydrazide, respectively, to provide detectable adducts. Such reactions can provide an immediately usable method for labeling a single protein in complex mixtures, such as the cell lysate in Figure 3D. In the future, the reaction of the HyRe tags with hydrazide derivatives could provide a powerful new tool for bioconjugate synthesis and in vivo protein labeling.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge support from the National Cancer Institute of the NIH (R01 CA133592-01 to G.A.W.). We thank Drs. John Greaves and Ben Berhane at the UCI Mass Spectrometry Center for mass spectrometry expertise, Professor Tadhg Begley for a helpful suggestion, and Professor Jennifer Prescher for helpful comments during manuscript preparation.
Footnotes
SUPPORTING INFORMATION Supporting information including peptide sequences, spot synthesis assay results, and complete peptide characterizations available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- (1).Marks KM, Nolan GP. Chemical labeling strategies for cell biology. Nat. Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- (2).O’Hare HM, Johnsson K, Gautier A. Chemical probes shed light on protein function. Curr. Opin. Struct. Biol. 2007;17:488–494. doi: 10.1016/j.sbi.2007.07.005. [DOI] [PubMed] [Google Scholar]
- (3).Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- (4).Dobbie IM, Lowndes NF, Sullivan KF. Autofluorescent Proteins. Meth. Cell Biol. 2008;85:1–22. doi: 10.1016/S0091-679X(08)85001-7. [DOI] [PubMed] [Google Scholar]
- (5).Sletten E, Bertozzi C. Bioorthogonal Chemistry: Fishing for Selectivity in a Sea of Functionality. Angew. Chem., Int. Ed. 2009;48(38):6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Hackenberger CR, Schwarzer D. Chemoselective Ligation and Modification Strategies for Peptides and Proteins. Angew. Chem., Int. Ed. 2008;47(52):10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- (7).Hochuli E, Bannwarth W, Dobeli H, Gentz R, Stuber D. Genetic Approach to Facilitate Purification of Recombinant Proteins with a Novel Metal Chelate Adsorbent. Nat. Biotechnol. 1988;6:1321–1325. [Google Scholar]
- (8).Hauser CT, Tsien RY. A hexahistidine-Zn2+-dye label reveals STIM1 surface exposure. Proc. Natl. Acad. Sci. U.S.A. 2007;104(10):3693–3697. doi: 10.1073/pnas.0611713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Griffin BA, Adams SR, Tsien RY. Specific Covalent Labeling of Recombinant Protein Molecules Inside Live Cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- (10).Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. New Biarsenical Ligands and Tetracysteine Motifs for Protein Labeling in Vitro and in Vivo: Synthesis and Biological Applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- (11).Gaietta GM, Giepmans BNG, Deerinck TJ, Smith WB, Ngan L, Llopis J, Adams SR, Tsien RY, Ellisman MH. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 2006;103(47):17777–17782. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. Selective Recognition of Protein Tetraserine Motifs with a Cell-Permeable, Pro-fluorescent Bis-boronic Acid. J. Am. Chem. Soc. 2008;131:438–439. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ojida A, Honda K, Shinmi D, Kiyonaka S, Mori Y, Hamachi I. Oligo-Asp Tag/Zn(II) Complex Probe as a New Pair for Labeling and Fluorescence Imaging of Proteins. J. Am. Chem. Soc. 2006;128:10452–10459. doi: 10.1021/ja0618604. [DOI] [PubMed] [Google Scholar]
- (14).Franz KJ, Nitz M, Imperiali B. Lanthanide-Binding Tags as Versatile Protein Coexpression Probes. ChemBioChem. 2003;4:265–271. doi: 10.1002/cbic.200390046. [DOI] [PubMed] [Google Scholar]
- (15).Nitz M, Franz KJ, Maglathlin RL, Imperiali B. A Powerful Combinatorial Screen to Identify High-Affinity Terbium(III)-Binding Peptides. ChemBioChem. 2003;4:272–276. doi: 10.1002/cbic.200390047. [DOI] [PubMed] [Google Scholar]
- (16).Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem. Rev. 2005;105:4056–72. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- (17).Smith GP, Petrenko VA. Phage Display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- (18).Sarikaya M, Tamerler C, Schwartz DT, Baneyx F. Materials Assembly and Formation Using Engineered Polypeptides. Annu. Rev. Mater. Res. 2004;34:373–408. [Google Scholar]
- (19).Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- (20).Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- (21).Rozinov MN, Nolan GP. Evolution of peptides that modulate the spectral qualities of bound, small-molecule fluorophores. Chem. Biol. 1998;5:713–728. doi: 10.1016/s1074-5521(98)90664-0. [DOI] [PubMed] [Google Scholar]
- (22).Marks KM, Rosinov M, Nolan GP. In Vivo Targeting of Organic Calcium Sensors via Genetically Selected Peptides. Chem. Biol. 2004;11:347–356. doi: 10.1016/j.chembiol.2004.03.004. [DOI] [PubMed] [Google Scholar]
- (23).Tanaka F, Fuller R, Asawapornmongkol L, Warsinke A, Gobuty S. Development of a Small Peptide Tag for Covalent Labeling of Proteins. Bioconjugate Chem. 2007;18:1318–1324. doi: 10.1021/bc070080x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shaper JH, Stryer L. Accessibility of the carbohydrate moiety of membrane-bound rhodopsin to enzymatic and chemical modification. J. Supra. Struc. 1977;6:291–299. doi: 10.1002/jss.400060302. [DOI] [PubMed] [Google Scholar]
- (25).Mirzaei H, Regnier F. Creation of Allotypic Active Sites during Oxidative Stress. J. Proteome Res. 2006;5:2159–2168. doi: 10.1021/pr060021d. [DOI] [PubMed] [Google Scholar]
- (26).Madian AG, Regnier FE. Profiling Carbonylated Proteins in Human Plasma. J. Proteome Res. 9:1330–1343. doi: 10.1021/pr900890k. [DOI] [PubMed] [Google Scholar]
- (27).Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods. 2005;2:99–104. doi: 10.1038/nmeth735. [DOI] [PubMed] [Google Scholar]
- (28).Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR. Site-specific chemical modification of recombinant proteins produced in mammalian cells by using the genetically encoded aldehyde tag. Proc. Natl. Acad. Sci. U.S.A. 2009;106(9):3000–3005. doi: 10.1073/pnas.0807820106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat. Chem. Biol. 2007;3:321–322. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- (30).Wang L, Zhang Z, Brock A, Schultz PG. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2003;100(1):56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Datta D, Wang P, Carrico IS, Mayo SL, Tirrell DA. A Designed Phenylalanyl-tRNA Synthetase Variant Allows Efficient in Vivo Incorporation of Aryl Ketone Functionality into Proteins. J. Am. Chem. Soc. 2002;124:5652–5653. doi: 10.1021/ja0177096. [DOI] [PubMed] [Google Scholar]
- (32).Stadtman ER. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- (33).Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative Stress and Covalent Modification of Protein with Bioactive Aldehydes. J. Biol. Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schmidt B, Selmer T, Ingendoh A, von. Figurat K. A novel amino acid modification in sulfatases that is defective in multiple sulfatase deficiency. Cell. 1995;82:271–278. doi: 10.1016/0092-8674(95)90314-3. [DOI] [PubMed] [Google Scholar]
- (35).Muller GH, Waldmann H. The phenyl hydrazide as an enzyme-labile protecting group -- Oxidative cleavage with mushroom tyrosinase. Tetrah. Lett. 1999;40:3549–3552. [Google Scholar]
- (36).Thompson SK, Halbert SM, Bossard MJ, Tomaszek TA, Levy MA, Zhao B, Smith WW, Abdel-Meguid SS, Janson CA, D’Alessio KJ, McQueney MS, Amegadzie BY, Hanning CR, DesJarlais RL, Briand J, Sarkar SK, Huddleston MJ, Ijames CF, Carr SA, Garnes KT, Shu A, Heys JR, Bradbeer J, Zembryki D, Lee-Rykaczewski L, James IE, Lark MW, Drake FH, Gowen M, Gleason JG, Veber DF. Design of potent and selective human cathepsin K inhibitors that span the active site. Proc. Natl. Acad. Sci. U.S.A. 1997;94(26):14249–14254. doi: 10.1073/pnas.94.26.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Lamboy JA, Arter JA, Knopp KA, Der D, Overstreet CM, Palermo EF, Urakami H, Yu T-B, Tezgel O, Tew GN, Guan Z, Kuroda K, Weiss GA. Phage Wrapping with Cationic Polymers Eliminates Nonspecific Binding between M13 Phage and High pI Target Proteins. J. Am. Chem. Soc. 2009;131:16454–16460. doi: 10.1021/ja9050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lamboy JA, Tam PY, Lee LS, Jackson PJ, Avrantinis SK, Lee HJ, Corn RM, Weiss GA. Chemical and Genetic Wrappers for Improved Phage and RNA Display. ChemBioChem. 2008;9:2846–2852. doi: 10.1002/cbic.200800366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Jencks WP. Studies on the Mechanism of Oxime and Semicarbazone Formation. J. Am. Chem. Soc. 1959;81:475–481. [Google Scholar]
- (40).Borch RF, Bernstein MD, Durst HD. Cyanohydridoborate anion as a selective reducing agent. J. Am. Chem. Soc. 1971;93:2897–2904. [Google Scholar]
- (41).Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: a source of specific protein binding molecules. Science. 1990;249:404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- (42).Levin AM, Murase K, Jackson PJ, Flinspach ML, Poulos TL, Weiss GA. Double Barrel Shotgun Scanning of the Caveolin-1 Scaffolding Domain. ACS Chem. Biol. 2007;2:493–500. doi: 10.1021/cb700055t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Cochran JR, Kim Y-S, Lippow SM, Rao B, Wittrup KD. Improved mutants from directed evolution are biased to orthologous substitutions. Prot. Eng. Des. Sel. 2006;19(6):245–253. doi: 10.1093/protein/gzl006. [DOI] [PubMed] [Google Scholar]
- (44).Murase K, Morrison KL, Tam PY, Stafford RL, Jurnak F, Weiss GA. EF-Tu Binding Peptides Identified, Dissected, and Affinity Optimized by Phage Display. Chem. Biol. 2003;10:161–168. doi: 10.1016/s1074-5521(03)00025-5. [DOI] [PubMed] [Google Scholar]
- (45).Angelini A, Heinis C. Post-translational modification of genetically encoded polypeptide libraries. Curr. Opin. Chem. Biol. 2011;15:355–361. doi: 10.1016/j.cbpa.2011.03.009. [DOI] [PubMed] [Google Scholar]
- (46).Frank R. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron. 1992;48:9217–9232. [Google Scholar]
- (47).Hilpert K, Winkler DFH, Hancock REW. Peptide arrays on cellulose support: SPOT synthesis, a time and cost efficient method for synthesis of large numbers of peptides in a parallel and addressable fashion. Nat. Protoc. 2007;2:1333–1349. doi: 10.1038/nprot.2007.160. [DOI] [PubMed] [Google Scholar]
- (48).Scheck RA, Francis MB. Regioselective labeling of antibodies through N-terminal transamination. ACS Chem. Biol. 2007;2:247–51. doi: 10.1021/cb6003959. [DOI] [PubMed] [Google Scholar]
- (49).Witus LS, Moore T, Thuronyi BW, Esser-Kahn AP, Scheck R. a, Iavarone AT, Francis MB. Identification of highly reactive sequences for PLP-mediated bioconjugation using a combinatorial peptide library. J. Am. Chem. Soc. 2010;132:16812–16817. doi: 10.1021/ja105429n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Hilpert K, Volkmer-Engert R, Walter T, Hancock REW. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005:1008–1012. doi: 10.1038/nbt1113. [DOI] [PubMed] [Google Scholar]
- (51).You L, Arnold FH. Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Prot. Eng. Des. Sel. 1996;9(1):77–83. doi: 10.1093/protein/9.1.77. [DOI] [PubMed] [Google Scholar]
- (52).Momiyama N, Kanan MW, Liu DR. Synthesis of Acyclic α,β-Unsaturated Ketones via Pd(II)-Catalyzed Intermolecular Reaction of Alkynamides and Alkenes. J. Am. Chem. Soc. 2007;129:2230–2231. doi: 10.1021/ja068886f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature. 2004;431:545–549. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.