Abstract
The proline-rich Akt substrate of 40 kilodaltons (PRAS40) was identified as a raptor-binding protein that is phosphorylated directly by mammalian target of rapamycin (mTOR) complex 1 (mTORC1) but not mTORC2 in vitro, predominantly at PRAS40 (Ser183). The binding of S6K1 and 4E-BP1 to raptor requires a TOR signaling (TOS) motif, which contains an essential Phe followed by four alternating acidic and small hydrophobic amino acids. PRAS40 binding to raptor was severely inhibited by mutation of PRAS40 (Phe129 to Ala). Immediately carboxyl-terminal to Phe129 are two small hydrophobic amino acid followed by two acidic residues. PRAS40 binding to raptor was also abolished by mutation of the major mTORC1 phosphorylation site, Ser183, to Asp. PRAS40 (Ser183) was phosphorylated in intact cells; this phosphorylation was inhibited by rapamycin, by 2-deoxyglucose, and by overexpression of the tuberous sclerosis complex heterodimer. PRAS40 (Ser183) phosphorylation was also inhibited reversibly by withdrawal of all or of only the branched chain amino acids; this inhibition was reversed by overexpression of the Rheb GTPase. Overex-pressed PRAS40 suppressed the phosphorylation of S6K1 and 4E-BP1 at their rapamycin-sensitive phosphorylation sites, and reciprocally, overexpression of S6K1 or 4E-BP1 suppressed phosphorylation of PRAS40 (Ser183) and its binding to raptor. RNA interference-induced depletion of PRAS40 enhanced the amino acid-stimulated phosphorylation of both S6K1 and 4E-BP1. These results establish PRAS40 as a physiological mTORC1 substrate that contains a variant TOS motif. Moreover, they indicate that the ability of raptor to bind endogenous substrates is limiting for the activity of mTORC1 in vivo and is therefore a potential locus of regulation.
The mammalian target of rapamycin (mTOR)3 is the founding member of the PI-3 kinase-related family of protein (Ser/Thr) kinases (PIKKs) and controls many important aspects of the cellular response to nutrient sufficiency and growth factors (1). The mTOR polypeptide is now known to function in two distinct, independently regulated hetero-oligomeric complexes, called mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Both complexes contain mTOR and the polypeptide mLST8/GβL; mTORC1 in addition contains raptor (an ortholog of Saccharomyces cerevisiae KOG1), which binds directly to the known mTORC1 substrates S6K1 and 4E-BP1 and is indispensable for their phosphorylation by mTOR in vivo and in vitro. mTORC2 lacks raptor but contains the polypeptides rictor (an ortholog to ScAVO3) and mSin1 (an ortholog of ScAVO1). mTORC2 is one of the activating kinases for Akt, previously called PDK2, and also regulates the actin cytoskeleton through as yet unidentified effectors. Although rapamycin, in complex with FKBP12, binds directly to mTOR in a segment just amino-terminal to the catalytic domain, only mTORC1 binds the FKBP12-rapamycin complex, and thus only mTORC1 is directly susceptible to inhibition by rapamycin.
Rapamycin is among the most selective kinase inhibitors known (2), and its longtime availability has enabled a detailed accounting of the cellular responses to inhibition of mTORC1. Perhaps the best appreciated and most general action of rapamycin in cell culture is its ability to inhibit cell growth (i.e. to suppress the accumulation of cellular mass) (3). This is accomplished by a selective inhibition of mRNA translation and by suppression of ribosomal biogenesis at both a transcriptional and translational level (1, 3). In addition, rapamycin activates autophagy and in some cells acts as a powerful inhibitor of proliferation (4, 5) and cell migration (6), actions that account for its current clinical applications as an immunosuppressant and in intravascular stents. Despite the considerable information as to the actions of rapamycin and thus mTORC1, both in cell culture and in vivo, very few direct mTORC1 substrates have been identified. S6K1 (7, 8) and 4E-BP1 (9, 10) were first identified as rapamycin-sensitive phosphoproteins in intact cells and were shown subsequently to be phosphorylated directly by mTOR in vitro at critical regulatory sites (11, 12). Schalm and Blenis (13) pointed out that the ability of these substrates to be regulated by mTORC1 in vivo depended on a short sequence (F(D/E)(F/I/L/M)(D/E)(L/I)) present in the noncatalytic amino-terminal flanking region of S6K1 and at the carboxyl terminus of 4E-BP1, which they named the TOR signaling (TOS) motif. Such a motif is also present in STAT3, another rapamycin-sensitive phosphoprotein (14). Subsequent work established that an intact TOS motif is required for the binding of S6K1 and 4E-BP1 to raptor (15–18), consistent with the view that raptor serves a necessary substrate-binding function in mTOR complex 1 (19). Although many potential TOS motifs are evident by BLAST analysis, we are unaware of validated mTORC1 substrates that have been identified thereby. Consequently, we sought novel candidate mTORC1 substrates by analyzing the cellular polypeptides that bound to recombinant raptor overexpressed in HEK293 cells. Herein, we describe the identification of PRAS40 (proline-rich Akt substrate of 40 kDa), previously identified as an Akt substrate and 14-3-3 binding partner (20), as a raptor-binding protein and a physiological substrate of mTORC1. During the preparation of this report, two papers (21, 22) appeared describing the ability of PRAS40 to bind raptor. Moreover, based on the ability of PRAS40 to antagonize the mTORC1-catalyzed phosphorylation of S6K1 and 4E-BP1, it was proposed that a primary function of PRAS40 is to inhibit mTORC1 signaling to its physiological substrates, a function that is ameliorated by Akt-catalyzed PRAS40 phosphorylation; the latter was claimed to reduce, in a 14-3-3-dependent manner, PRAS40 binding to raptor. In contrast, we demonstrate that PRAS40 is itself a physiological substrate of mTORC1 and, like other mTORC1 substrates, competes for a pool of raptor substrate-binding sites whose abundance is apparently limiting for mTORC1-catalyzed substrate phosphorylation in vivo. PRAS40 binding to raptor is abolished by mTORC1-catalyzed PRAS40 phosphorylation but is unaffected by mutation of the PRAS40 Akt phosphorylation site, Thr246. We propose that PRAS40 functions primarily to regulate an as yet poorly defined cellular process, under the joint control of Akt and mTORC1.
EXPERIMENTAL PROCEDURES
cDNAs and siRNAs
The expression vectors of FLAG-tagged raptor (pcDNA1-FLAG-raptor) (19), HA-tagged wild-type mTOR (pcDNA1-HA-mTOR), HA-tagged kinase-negative mTOR (pcDNA1-HA-mTOR NK), FLAG-tagged wild-type mTOR (pCMV5-FLAG-mTOR), FLAG-tagged rapamycin-resistant mTOR (pCMV5-FLAG-mTOR ST), FLAG-tagged rapamycin-resistant/kinase-negative mTOR (pCMV5-FLAG-mTOR ST/NK), FLAG-tagged 4E-BP1 (pCMV5-FLAG-4E-BP1) (23), HA-tagged S6K1 (pMT2-HAS6K1), HA-tagged kinase-negative S6K1 (pMT2-HA-S6K1 KM) (24), GST-fused S6K1 (pEBG2T-S6K1) (25), and GST-fused 4E-BP1 (pGEX4T1–4E-BP1) (19) were constructed as described. His6-tagged human PRAS40 (pReceiver-MO1-His6-PRAS40) was purchased from GeneCopoeia. The coding region was amplified by PCR with the addition of EcoRI and XhoI sites at 5′ and 3′ ends, respectively, and the PCR product was cloned into pcDNA3-Myc and pcDNA3-FLAG. The insert was mutated using a QuikChange™ site-directed mutagenesis kit (Stratagene). The mutants having Ala at Phe129 (F129A), Ser183 (S183A), and Thr246 (T246A) or Asp at Ser183 (S183D) and Thr246 (T246D) were generated in pcDNA3-Myc. The wild-type PRAS40 and the S183A mutant were also cloned into pGEX4T1 to produce GST fusion proteins in Escherichia coli. The human Rheb sequence was amplified by PCR from the QUICK-Clone™ cDNA of human brain (Clontech) with the additionofEcoRIandXhoIsitesat5′and3′ends,respectively,and the PCR product was cloned into pcDNA3-Myc. The expression vectors of FLAG-tagged TSC1 (pcDNA3-FLAG-TSC1) and FLAG-tagged TSC2 (pcDNA3-FLAG-TSC2) were kindly provided by Dr. David J. Kwiatkowski (Brigham and Women's Hospital, Boston).
Antibodies
The anti-mTOR antibody was produced as described previously (26). Rabbit polyclonal anti-peptide antibodies recognizing PRAS40, phosphorylated PRAS40 at Ser183, raptor, and rictor were produced against the following peptides by Immuno-Biological Laboratories: PRAS40 (P238), DLPRPRLNTSDFQKLKRKY (amino acids 238–256); phosphorylated PRAS40 (Ser(P)183), QYAKpSLPVS (amino acids 179–187) (in which pS represents phospho-Ser; raptor (R1), MESEMLQSPLLGLGEEDEA (amino acids 1–19); raptor (R984), IRKEREWRFLRNSRVRRQA (amino acids 984–1002); and rictor (R1291), GSSHTLPRRAQSLKA (amino acids 1291– 1305). In brief, rabbits were immunized with each synthetic peptide coupled with thyroglobulin, and the immunoglobulin G fractions against these peptide sequences were obtained from the immune sera using a column of antigen-coupled Activated Thiol Sepharose™ 4B (GE Healthcare Bio-Sciences). The following antibodies were purchased from commercial sources. Anti-FLAG (M2) was from Sigma; anti-Myc (9E10) and anti-HA (12CA5) were from Roche Applied Science; normal rabbit immunoglobulin, normal mouse immunoglobulin, anti-S6K1 (H-9), and anti-4E-BP1 (C-19) were from Santa Cruz Bio-technology, Inc. (Santa Cruz, CA); anti-phospho-AMPK (Thr(P)172) (40H9), anti-phospho-S6K1 (Thr(P)389) (1A5), anti-phospho-4E-BP1 (Ser(P)65), and anti-phospho-4E-BP1 (Thr(P)37/46) were from Cell Signaling Technology; anti-PRAS40 (73P21) and anti-phospho-PRAS40 (Thr(P)246) were from BioSource International; anti-GST was from Upstate Bio-technology; anti-β-actin was from Abcam.
Cell Culture and Treatment
HEK293 and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) containing 10% fetal bovine serum at 37 °C in a 5% CO2 incubator. HEK293 cells were transfected with each expression vector by the lipofection method using Lipofectamine reagent (Invitrogen) according to the manufacturer's protocol. For the small interfering RNA (siRNA) studies, HEK293 cells or HeLa cells were transfected with PRAS40 siRNA duplexes with 3′ dTdT overhangs corresponding to human PRAS40 mRNA (5′[notdef]GGGCAUUAGUGAUAAUGGA-3′) or (5′-GCGACUUCCAGAAGCUGAA-3′) (Qiagen) using Nucleofector 3 (AMAXA Biosystems). Scramble siRNA duplexes (iGENE Therapeutics) were employed as a control. For starvation of nutrients, cells were deprived of serum for 15 h, and the medium was replaced with fresh DMEM without serum for 1 h. For starvation of amino acids, the cells were washed twice with DMEM lacking amino acids, further incubated in the same medium for 2 h, and then stimulated with DMEM containing amino acids for 30 min in the presence or absence of 200 nm rapamycin. Where indicated, the amino acid mixture lacking branched-chain amino acids was employed instead of amino acids. For starvation of glucose, the cells were washed twice with DMEM lacking glucose (Sigma) and stimulated with DMEM lacking glucose in the presence or absence of 5.5 mm 2-deoxyglucose for 30 min. Where indicated, DMEM containing glucose was employed as a control.
Immunoprecipitation
This procedure was carried out essentially as described (19). All cells were lysed in ice-cold buffer A (20 mm Tris-HCl (pH 7.4), 120 mm NaCl, 1 mm EDTA, 5 mm EGTA, 50 mm β-glycerophosphate, 50 mm NaF, 0.3% CHAPS, 1 mm dithiothreitol, 4 μg/ml leupeptin, 4 μg/ml aprotinin). The immunoprecipitates were washed three times with buffer A and used for silver staining analysis or immunoblot. Where indicated, buffer A contained 1% Nonidet P-40 instead of 0.3% CHAPS. For the kinase assay, the immunoprecipitates were washed twice with buffer A and twice in buffer B (10 mm HEPES (pH 7.5), 50 mm β-glycerophosphate, 50 mm NaCl).
Immunoblot
The lysates and immunoprecipitates were separated by SDS-PAGE, and the proteins were transferred onto a polyvinylidene difluoride membrane. The membranes were blocked at room temperature with 5% skim milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) and were then incubated with each primary antibody, diluted in the 2% skim milk in TBST, for 1 h at room temperature or overnight at 4 °C. The commercial antibodies were used at dilutions of 1:500–1:1000, except for the anti-β-actin antibody employed at a dilution of 1:5000. After incubation with the horseradish peroxidase-conjugated secondary antibody, the proteins were visualized by the enhanced chemiluminescence method. When the same sample was analyzed with different antibodies, the membrane was stripped and employed for the subsequent immunoblot analysis. When the endogenous 4E-BP1 proteins were analyzed, the cell lysates were heated at 100 °C for 5 min and centrifuged at 18,000 × g for 30 min, and the heat-stable proteins were subjected to immunoblot (27).
Kinase Assay
The mTOR kinase assay was performed as previously described (19). GST and GST fusion proteins were prepared for substrates of the kinase assay, as previously described (17, 19). The purified MBP-PRAS40 was purchased from Bio-Source International. After the kinase reaction, the samples were separated by SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and analyzed by autoradiography using x-ray film or the Bioimaging Analyzer BAS2500 (Fujix). Then the membrane was immunoblotted with the appropriate antibody and visualized as described above.
Mass Spectrometry
The analysis was carried out as described previously (28, 29). Briefly, FLAG-raptor immunoprecipitates were separated by SDS-PAGE and visualized by silver staining. The band corresponding to p40 polypeptide was cut out and destained, and the proteins in gels were reduced and alkylated, followed by in-gel digestion with trypsin in 25 mm ammonium bicarbonate for 15 h at 37 °C. The resulting peptides were then subjected to the liquid chromatography electrospray ionization (ESI) mass spectrometry/mass spectrometry (MS/MS) by using a LCQ Advantage ion trap mass spectrometer (Thermo Finnigan). Protein identification according to product ion mass lists was performed by the product ion mass fingerprinting using MASCOT “MS/MS ion search.” For identification of in vitro phosphorylation sites in PRAS40, GST-PRAS40 phosphorylated by mTOR was separated by SDS-PAGE. The GST-PRAS40 visualized by reverse staining was cut out from a gel. Destained proteins were digested with trypsin in a gel as described above and subjected to nano-ESI mass spectrometry by using a Q-Tof2 quadrupole time-of-flight mass spectrometer (Micromass), as described previously.
Sequence Analysis
The amino acid sequence identity and similarity were analyzed by using the GENETYX program, version 8 (GENETYX).
RESULTS
Recombinant Raptor Binds Endogenous PRAS40 in an Amino Acid-dependent Manner
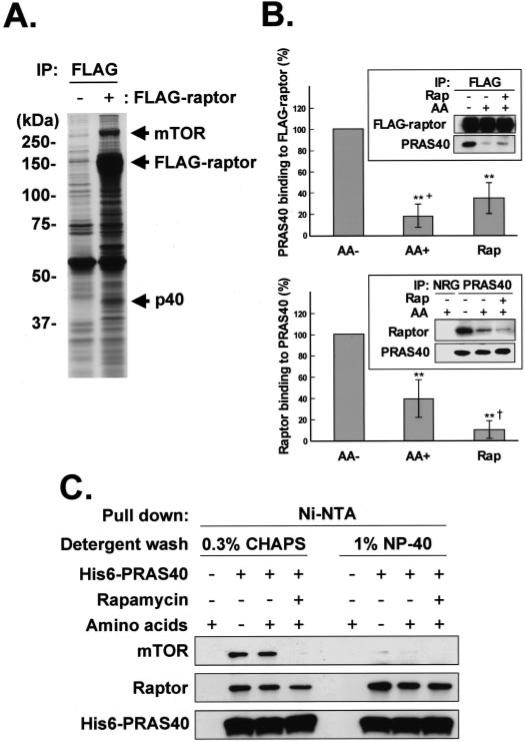
Using one-dimensional SDS-PAGE and silver staining, we compared the polypeptides retrieved in anti-FLAG immunoprecipitates prepared from HEK293 cells transfected with FLAG-raptor or with an empty vector. The most abundant cellular polypeptide recovered in the FLAG-raptor immmunoprecipitate was a protein of ~290 kDa (Fig. 1A), shown to be mTOR by immunoblot as well as by the peptide mass fingerprinting analysis (data not shown). Many of the other prominent polypeptide bands in the FLAG-raptor immunoprecipitate were identified by mass spectrometric analysis as raptor, presumably reflecting FLAG-raptor proteolysis. A polypeptide of ~40 kDa, however, contained several peptide fragments distinct from raptor, which were consistent with the amino acid sequence of PRAS40 (NCBI accession number NP_115751), a proline-rich protein first identified as an Akt substrate and 14-3-3 binding partner (20). To verify the ability of recombinant raptor to bind endogenous PRAS40, the FLAG-raptor immunoprecipitate was immunoblotted with an anti-PRAS40 antibody (Fig. 1B, top). A small amount of PRAS40 was detected in the FLAG-raptor immunoprecipitate prepared from serum-deprived cells, but the additional removal of ambient amino acids resulted in a large increase in FLAG-raptor-associated PRAS40. A similar pattern was observed if an immunoprecipitate of endogenous PRAS40 was probed for endogenous raptor (Fig. 1B, bottom). Whereas no raptor was coprecipitated with normal rabbit IgG, endogenous raptor was present in the anti-PRAS40 immunoprecipitate; moreover, amino acid withdrawal substantially increased the amount of raptor recovered with PRAS40, whereas rapamycin resulted in a modest decrease in PRAS40-associated raptor. To determine whether the association of PRAS40 with raptor required mTOR or mLST8, we expressed recombinant His6-PRAS40 in HEK293 cells, retrieved the His6-PRAS40 on Ni2+ -nitrilotriacetic acid beads, and washed the beads with buffer containing either 0.3% CHAPS or 1% Nonidet P-40; an immunoblot of the eluate from these beads was shown in Fig. 1C. Nonidet P-40, which causes the dissociation of an mTOR-mLST8 complex from raptor, did not diminish the recovery of raptor with His6-PRAS40. An amount of rapamycin sufficient to promote dissociation in vivo of raptor from mTORC1 modestly diminished the recovery of raptor with His6-PRAS40 (Fig. 1C). Thus, PRAS40 binds directly to raptor; this interaction occurs between the endogenous polypeptides, does not require mTOR, and is regulated by amino acid sufficiency.
FIGURE 1. Identification of PRAS40 as a raptor-binding protein.
A, isolation of the proteins interacting with raptor. HEK293 cells were transfected with FLAG-tagged raptor or the empty vector, and the cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. The immunoprecipitated (IP) proteins were applied to SDS-PAGE on 8% gel and visualized by silver staining. FLAG-tagged raptor, mTOR, and a 40-kDa protein (p40) are indicated by arrows. The positions of the molecular mass markers are shown in kDa. B, association of raptor and PRAS40 in the cells. HEK293 cells transfected with FLAG-tagged raptor (top) or without transfection (bottom) were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids (AA) in the presence or absence of rapamycin (Rap). The cell lysates were subjected to immunoprecipitation with the anti-FLAG (top) or anti-PRAS40 (bottom) antibodies. The immunoprecipitate by normal rabbit globulin (NRG) was employed as a control. Immunoblot was carried out with the anti-FLAG, anti-PRAS40, and anti-raptor antibodies, and the blots were scanned. A representative experiment is shown in the insets. In the upper experiments, the OD of the PRAS40 blot was divided by the OD of the corresponding FLAG-raptor blot, whereas in the lower experiments, the OD of the raptor blot was divided by the OD of the corresponding PRAS40 blot. In both sets of experiments, this ratio for the amino acid minus (AA–) condition was set to 100% and divided into the ratio for each of the other conditions. The bar graphs summarize the results of four (upper) or 12 (lower) experiments. **, a reduction as compared with AA–, p < 0.01; the plus symbol in the upper bar graph indicates that the AA+ values are less than those of the corresponding Rap values, p < 0.01; the dagger in the lower bar graph indicates that the Rap values are lower than the corresponding AA+ values, p < 0.01. C, HEK293 cells transfected with His6-tagged PRAS40 or mock-transfected were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids in the presence or absence of rapamycin. The cell lysates were subjected to His6 pull down with Ni2+-nitrilotriacetic acid beads. The proteins bound to the resin were washed with the buffer containing either 0.3% CHAPS or 1% Nonidet P-40. Immunoblot was carried out with the anti-mTOR, anti-raptor, and anti-PRAS40 antibodies.
PRAS40 (Phe129) Is Important for the Association with Raptor
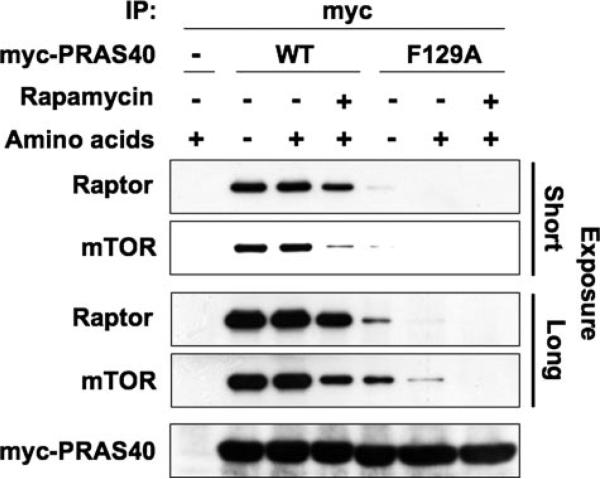
The binding of the mTORC1 substrates S6K1 and 4E-BP1 to raptor has been shown to require a specific five-amino acid sequence (F(D/E)(F/I/L/M)(D/E)(L/I)) called the TOS motif (13, 15–18). Human PRAS40 has six Phe residues, none of which are followed by a TOS motif consensus corresponding to those found in S6K1, 4E-BP1, or STAT3. The sequence in PRAS40 most similar to the TOS motif begins with Phe129, 129FVMDE133; this motif is conserved down through Xenopus tropicalis, slightly altered in Danio rerio (222FSMDE226) and lacking entirely in the putative Drosophila melanogaster ortholog, Lobe (supplemental Fig. 1). Nevertheless, we generated a PRAS40 mutant replacing Phe129 by Ala (F129A) and found that this mutant was greatly deficient in its ability to bind raptor as compared with wild-type PRAS40 (Fig. 2). Moreover, as seen in Figs. 1C and 2, the ability of PRAS40, when overex-pressed, to bind endogenous raptor was not noticeably enhanced by amino acid withdrawal, whereas rapamycin, at concentrations sufficient to promote dissociation of the raptor-mTOR complex, did decrease the amount of raptor recovered with recombinant PRAS40.
FIGURE 2. The involvement of a PRAS40 (Phe129), located in a TOS motif-like sequence, in PRAS40 binding to raptor.
HEK293 cells transfected with Myc-tagged wild-type PRAS40 (WT), Myc-tagged PRAS40 (F129A) mutant, or the empty vector (–) were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids in the presence or absence of rapamycin. The cell lysates were subjected to immunoprecipitation with the anti-Myc antibody. Immunoblot was carried out with the anti-mTOR, anti-raptor, and anti-Myc antibodies. The longer exposure reveals the persistence of a minor amount of raptor associated with the PRAS40(F129A) polypeptide, best shown by virtue of the slightly greater sensitivity of the anti-mTOR as compared with the anti-raptor antibody.
mTORC1 Phosphorylates PRAS40 in Vitro at Ser183
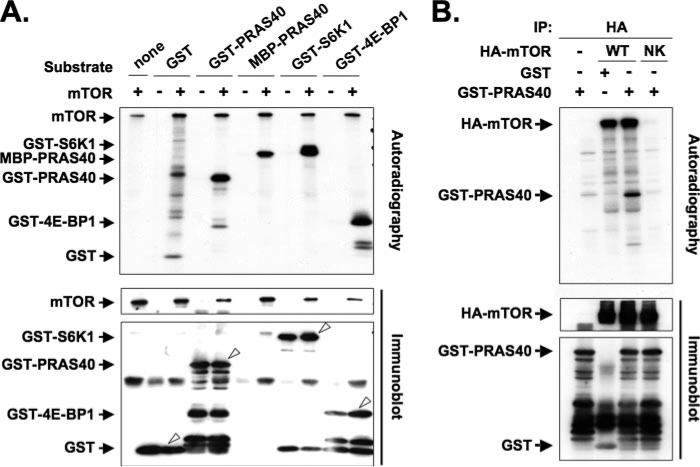
The ability of PRAS40 to associate with raptor through a TOS-like motif raised the possibility that PRAS40, like S6K1 and 4E-BP1, could serve as a substrate for the mTORC1 kinase. As seen in Fig. 3A, an immunoprecipitate of endogenous mTOR phosphorylated PRAS40 (as a prokaryotic recombinant GST or MBP fusion protein) in vitro to an extent comparable with GST-S6K1 or GST-4E-BP1 (the former purified from serum-deprived, rapamycin-treated HEK293 cells). This phosphorylation of PRAS40 was catalyzed by mTOR itself, inasmuch as only the active recombinant mTOR but not the kinase-negative mTOR (N2343K) mutant (23) catalyzed GST-PRAS40 phosphorylation in vitro (Fig. 3B). Tryptic digestion of PRAS40 followed by nano-ESI mass spectrometry was employed to identify the site(s) phosphorylated by mTOR in vitro. The experimental mass value (1395.75 Da) of the protonated peptide corresponding to the ion at m/z 698.38, which was observed only in the mass spectrum of GST-PRAS40 phosphorylated by mTOR (supplemental Fig. 2), was nearly identical to the calculated mass value (1395.71 Da) of a singly phosphorylated form of the PRAS40 tryptic peptide, 183SLPVSVPVWGFK194. Subjecting the m/z 698.38 ion to MS/MS established that Ser183, which is highly conserved (supplemental Fig. 1), is the phosphorylated residue (data not shown). In support of this identification, a PRAS40 (S183A) mutant was phosphorylated by mTOR in vitro to a greatly reduced extent as compared with wild-type PRAS40 (Fig. 4A), indicating that Ser183 is a major although not the sole site of mTOR-catalyzed PRAS40 phosphorylation in vitro.
FIGURE 3. Phosphorylation of PRAS40 by mTOR in vitro.
A, mTOR immunoprecipitated from HEK293 cells was used to phosphorylate GST, GST-PRAS40, MBP-PRAS40, GST-S6K1, and GST-4E-BP1 (1 μg each) as substrates. The immunoprecipitate by the normal mouse globulin (NMG) was employed as a control. The top panel shows autoradiography, and the lower two panels show immunoblot by using the anti-mTOR and anti-GST antibodies, respectively. The white arrowheads in the bottom panel indicate GST and GST fusion proteins. The stoichiometry of 32P incorporation (mol of P/mol of polypeptide) into the specific band was estimated as follows: GST, 0.01; GST-PRAS40, 0.14; MBP-PRAS40, 0.11; GST-S6K1, 0.20; GST-4E-BP1, 0.12. B, HEK293 cells were transfected with HA-tagged wild-type mTOR (WT), kinase-negative mTOR (N2343K) mutant (NK), or the empty vector (–). The immunoprecipitates were used to phosphorylate GST-PRAS40 and GST as substrates. The top panel shows autoradiography, and the lower two panels show immunoblot with anti-HA and anti-GST antibodies, respectively.
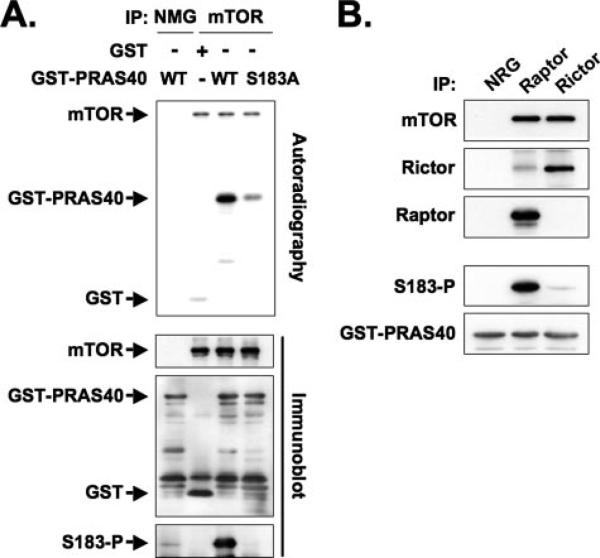
FIGURE 4. Identification of an in vitro mTORC1-catalyzed phosphorylation site in PRAS40.
A, mutation of PRAS40 Ser183 to Ala reduces PRAS40 phosphorylation by mTOR in vitro. mTOR immunoprecipitated from HEK293 cells was used to phosphorylate GST, GST-PRAS40, and GST-PRAS40 (S183A). The immunoprecipitate by the NMG was employed as a control. The top panel shows autoradiography, and the lower three panels show immunoblot by using the anti-mTOR, anti-GST, and phospho-specific anti-PRAS40 (Ser(P)183) antibodies, respectively. B, mTORC1-specific phosphorylation of PRAS40 in vitro. mTORC1 and mTORC2 were immunoprecipitated from HEK293 cells by the anti-raptor and anti-rictor antibodies, respectively, and the immunoprecipitates were used to phosphorylate GST-PRAS40 in vitro. The immunoprecipitate by the NRG was employed as a control. Immunoblot was carried out with the anti-mTOR, anti-rictor, anti-raptor, anti-PRAS40 (Ser(P)183), and anti-GST antibodies.
We generated a phospho-specific rabbit polyclonal antibody for PRAS40 (Ser(P)183); this antibody recognized PRAS40 after phosphorylation by mTOR in vitro but did not react with a PRAS40 (S183A) mutant (Fig. 4A, bottom). We first used this antibody to establish whether mTOR-catalyzed PRAS40 (Ser183) phosphorylation is catalyzed specifically by mTORC1. mTORC1 and mTORC2 were immunoprecipitated from HEK293 cells by the anti-raptor and the anti-rictor antibodies, respectively, and the immunoprecipitates were used to phosphorylate GST-PRAS40 in parallel in vitro kinase assays. mTORC1, which contains raptor, phosphorylated PRAS40 (Ser183), whereas mTORC2 did not (Fig. 4B). Consistent with its inability to phosphorylate GST-PRAS40 (Ser183), the anti-rictor immunoprecipitate did not contain PRAS40 (data not shown).
mTORC1 Phosphorylates PRAS40 at Ser183 in Vivo
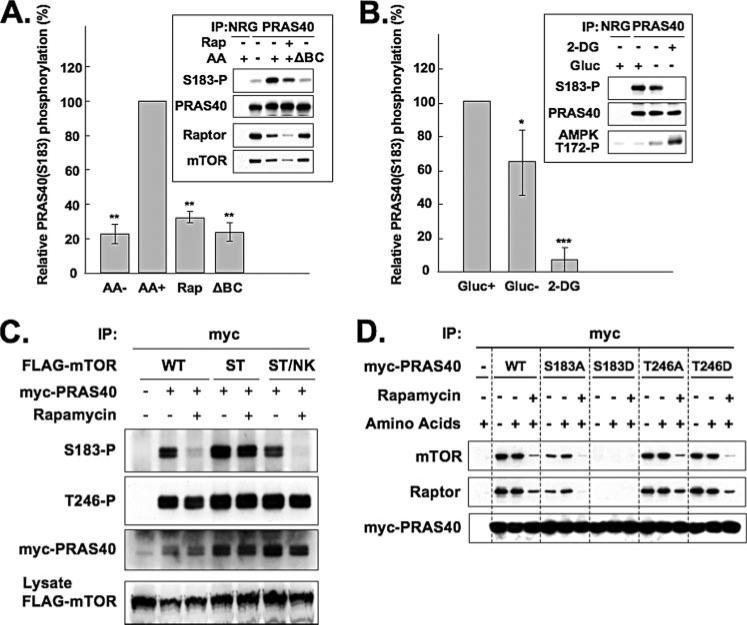
We employed the anti-PRAS40 (Ser(P)183) phosphospecific antibody to examine PRAS40 (Ser183) phosphorylation in intact cells. As seen in Fig. 5A, PRAS40 (Ser183) phosphorylation was evident in PRAS40 immunoprecipitated from HEK293 cells; this phosphorylation was inhibited by rapamycin as well as by withdrawal of all amino acids and equally by withdrawal of only the branched-chain amino acids, as observed previously for S6K1 and 4E-BP1 (30). Notably, the recovery of raptor in the PRAS40 immunoprecipitate was enhanced by withdrawal of all amino acids and equally by withdrawal of only the branched-chain amino acids; as before, rapamycin caused a modest decrease in the recovery of raptor with PRAS40. Amino acid stimulation, in addition to stimulating the phosphorylation of PRAS40 at Ser183, also induced the dissociation of raptor from PRAS40 (see also Fig. 1B). These results demonstrate that mTORC1 phosphorylates PRAS40 at Ser183 in vivo and suggest that PRAS40, once phosphorylated, dissociates from raptor.
FIGURE 5. Phosphorylation of PRAS40 at Ser183 in vivo.
A, effects of amino acids and rapamycin on phosphorylation of PRAS40 at Ser183. HEK293 cells were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids (AA) in the presence or absence of rapamycin (Rap). Where indicated, the amino acid mixture readded lacked branched-chain amino acids (ΔBC). The cell lysates were subjected to immunoprecipitation with the anti-PRAS40 antibody. The immunoprecipitate by the NRG was employed as a control. Immunoblot was carried out with the anti-PRAS40 (Ser(P)183), anti-PRAS40, anti-raptor, and anti-mTOR antibodies, and the blots were scanned. A representative experiment is shown in the inset. The OD of the PRAS40 (Ser(P)183) blot was divided by the OD of the corresponding PRAS40 blot. This ratio for the AA+ condition was set to 100% and divided into the ratio for each of the other conditions. The bar graphs summarize the results of three experiments. The double asterisks indicate a reduction as compared with AA+, p < 0.01. B, effect of ATP depletion on phosphorylation of PRAS40 at Ser183. HEK293 cells were deprived of serum and incubated in the absence of glucose (Gluc) alone or with added 2-deoxyglucose (2-DG). The cell lysates were subjected to immunoprecipitation with the anti-PRAS40 antibody. The immunoprecipitate by the NRG was employed as a control. Immunoblot was carried out with the anti-PRAS40 (Ser(P)183), anti-PRAS40, and anti-raptor antibodies. The aliquots of the cell lysates were subjected to immunoblot by the anti-AMPK (Thr(P)172) antibody. A representative experiment is shown in the insets. The OD of the PRAS40 (Ser(P)183) blot was divided by the OD of the corresponding PRAS40 blot. This ratio for the Gluc+ condition was set to 100% and divided into the ratio for each of the other conditions. The bar graphs summarize the results of four experiments. The double or single asterisks indicate a reduction as compared with Gluc+; p < 0.01 or p < 0.05, respectively. C, an active, rapamycin-resistant mTOR mutant overcomes rapamycin-induced dephosphorylation of PRAS40 (Ser183) in vivo. HEK293 cells co-transfected with Myc-tagged PRAS40 and either FLAG-tagged wild-type mTOR (WT), rapamycin-resistant mTOR (S2035T) mutant (ST), or rapamycin-resistant/kinase-negative mTOR (S2035T/N2343K) mutant (ST/NK) were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids in the presence or absence of rapamycin. The cell lysates were subjected to immunoprecipitation with the anti-Myc antibody. Immunoblot was carried out with the anti-PRAS40 (Ser(P)183), anti-PRAS40 (Thr(P)246), and anti-Myc antibodies. The aliquots of the cell lysates were subjected to immunoblot by the anti-FLAG antibody. D, mutation of PRAS40 (Ser183) to Asp abolishes association with raptor. HEK293 cells transfected with Myc-tagged PRAS40, its mutants, or the empty vector were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids in the presence or absence of rapamycin. The cell lysates were subjected to immunoprecipitation with the anti-Myc antibody. Immunoblot was carried out with the anti-mTOR, anti-raptor, and anti-Myc antibodies.
We next examined the effect of ATP depletion, which suppresses mTOR signaling through activation of the tuberous sclerosis complex (31), on PRAS40 (Ser183) phosphorylation (Fig. 5B). The activation of AMPK was monitored by the phosphorylation of its α subunit at Thr172. Withdrawal of glucose, which induces a modest AMPK activation, decreased PRAS40 (Ser183) phosphorylation; the nonmetabolizable glucose analog, 2-deoxyglucose, strongly activated AMPK and greatly suppressed PRAS40 (Ser183) phosphorylation. These results further support the conclusion that mTORC1 catalyzes PRAS40 (Ser183) phosphorylation.
Additional confirmation that mTORC1 is the rapamycin-sensitive kinase that phosphorylates PRAS40 (Ser183) in vivo was obtained by using the rapamycin-resistant mTOR (S2035T) mutant, which no longer binds the FKBP12-rapamycin complex (Fig. 5C). Overexpression of mTOR (S2035T), but not wild-type mTOR, is capable of rescuing S6K1 and 4E-BP1 from rapamycin-induced dephosphorylation (23). Similarly, mTOR (S2035T) rescued PRAS40 (Ser(P)183) from rapamycin-induced dephosphorylation, whereas neither wild-type mTOR nor a kinase-negative double mTOR mutant (S2035T/N2343K) was able to do so. Neither rapamycin nor the mTOR variants affected PRAS40 phosphorylation at the Akt site, Thr246 (Fig. 5C).
To evaluate independently the effect of Ser183 modification on PRAS40 binding to raptor, we created PRAS40 mutants wherein Ser183 was changed to Ala or Asp. In addition, inasmuch as PRAS40 is phosphorylated at Thr246 by Akt (20), the latter residue was also converted to Ala or Asp, and the ability of these PRAS40 mutants to bind endogenous raptor in amino acid-replete and -deficient cells and in the presence of rapamycin was examined (Fig. 5D). Mutation at Thr246, whether Ala or Asp, did not affect the binding of PRAS40 to raptor. The PRAS40 (S183A) mutant bound to raptor somewhat less than did wild-type PRAS40, whereas PRAS40 (S183D) exhibited no ability to bind to raptor. Rapamycin inhibited the association of raptor to all PRAS40 variants. These results are fully consistent with the view that phosphorylation of PRAS40 at Ser183 strongly inhibits PRAS40 binding to raptor, whereas modification of Thr246 does not alter PRAS40 binding to raptor. In addition, rapamycin interfered with the binding of PRAS40 to raptor through a mechanism independent of its ability to inhibit mTORC1-catalyzed PRAS40 (Ser183) phosphorylation; how this action of rapamycin relates to rapamycin's ability to interfere with the raptor/mTOR interaction is as yet unclear and is discussed below.
The TSC1/2 Complex and Rheb Regulate the Phosphorylation of PRAS40 at Ser183
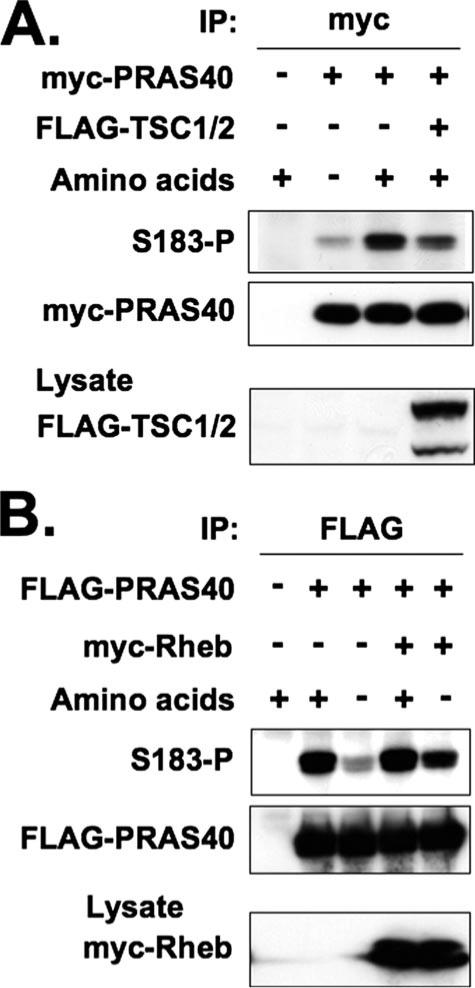
The tuberous sclerosis complex, a heterodimer of TSC1 and TSC2, is a negative regulator of mTORC1 signaling through its ability to activate the GTPase activity of Rheb (32). In turn, Rheb is a positive regulator of mTORC1, and overexpression of Rheb can restore mTORC1 phosphorylation of S6K1 and 4E-BP1 in cells deprived of amino acids (33). As seen in Fig. 6A, overexpression of TSC1/TSC2 inhibited the amino acid-induced phosphorylation of PRAS40 at Ser183. Reciprocally, overexpression of Rheb restored PRAS40 (Ser183) phosphorylation in the amino acid-starved cells (Fig. 6B). Thus, PRAS40 (Ser183) phosphorylation is regulated in a manner indistinguishable from that reported previously for other bona fide mTORC1 substrates.
FIGURE 6. Effects of TSC1/2 and Rheb on phosphorylation of PRAS40 at Ser183.
A, effect of TSC1/2 on phosphorylation of PRAS40. HEK293 cells cotransfected with Myc-tagged PRAS40 and either FLAG-tagged TSC1/2 or the empty vector were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids. The cells transfected with the empty vectors were employed as a control. The cell lysates were subjected to immunoprecipitation with the anti-Myc antibody. Immunoblot was carried out with the anti-PRAS40 (Ser(P)183) and anti-Myc antibodies. The aliquots of the cell lysates were subjected to immunoblot by the anti-FLAG antibody. B, effect of Rheb on phosphorylation of PRAS40. HEK293 cells cotransfected with FLAG-tagged PRAS40 and either Myc-tagged Rheb or the empty vector were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids. The cells transfected with the empty vectors were employed as a control. The cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. Immunoblot was carried out with the anti-PRAS40 (Ser(P)183) and anti-FLAG antibodies. The aliquots of the cell lysates were subjected to immunoblot by the anti-Myc antibody.
PRAS40 Competes with S6K1 and 4E-BP1 as a Substrate of mTOR
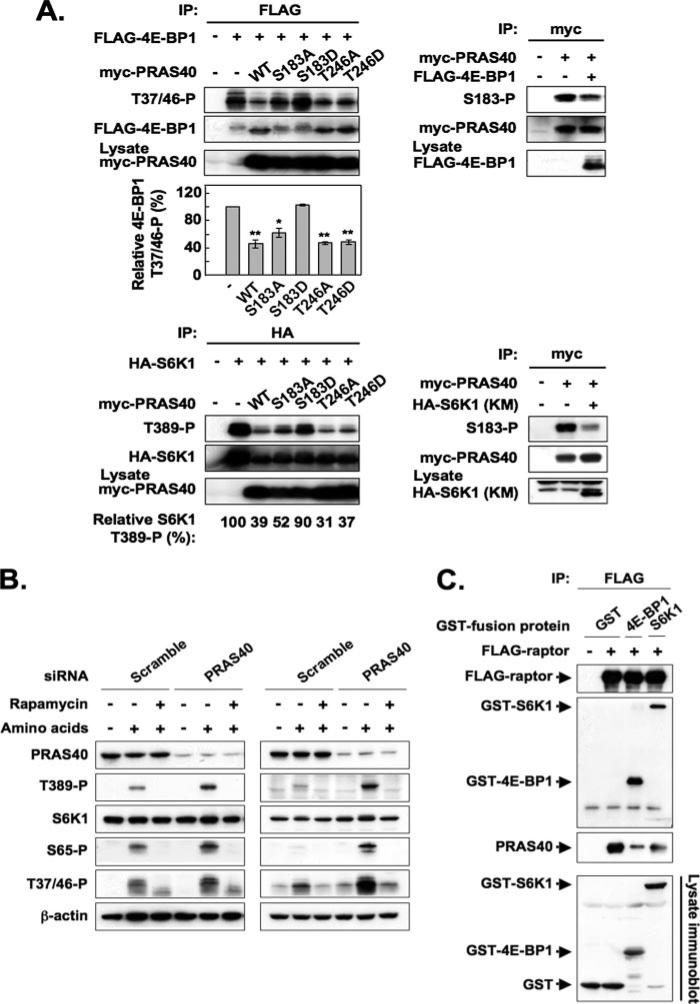
Although the site(s) on raptor responsible for binding S6K1 and 4E-BP1 have not been well defined, earlier work has shown that S6K1 and 4E-BP1 compete with each other as mTOR substrates (13, 34). We therefore examined the effect of PRAS40 overexpression on the phosphorylation of 4E-BP1 and S6K1 by mTORC1 in vivo and, conversely, the effect of overexpression of these bona fide mTORC1 substrates on PRAS40 (Ser183) phosphorylation in HEK293 cells (Fig. 7A). The overexpression of PRAS40 suppressed the phosphorylation of 4E-BP1 at Thr37/46 (Fig. 7A, upper left) and S6K1 at Thr389 (Fig. 7A, lower left), whereas the PRAS40 (S183D) mutant, which does not bind raptor, did not affect the phosphorylation of these two proteins. Notably, mutation of the PRAS40 Akt phosphorylation site, to either Ala or Asp, did not alter the ability of PRAS40 to suppress the phosphorylation of either 4E-BP1 (Fig. 7A, upper left) or S6K1 (Fig. 7A, lower left). Overexpression of 4E-BP1 (Fig. 7A, upper right) or kinase-negative S6K1 (Fig. 7A, lower right) each suppressed the phosphorylation of PRAS40 at Ser183. The impact of endogenous PRAS40 on mTORC1 phosphorylation of S6K1 and 4E-BP1 was examined by RNA interference depletion of endogenous PRAS40 (Fig. 7B). Partial depletion of PRAS40 increased the phosphorylation of S6K1 and 4E-BP1 observed in response to amino acid stimulation of HEK293 cells (Fig. 7B, left), and an even stronger stimulation was observed with PRAS40 depletion in HeLa cells (Fig. 7B, right). Accordingly, it appears that these three mTORC1 substrates can compete with one another for binding to a limiting pool of raptor. Direct evidence for competition among mTORC1 substrates for raptor is shown in Fig. 7C, wherein the co-expression of GST-S6K1 or GST-4E-BP1 with FLAG-raptor strongly inhibited the amount of endogenous PRAS40 recovered in the FLAG-raptor immunoprecipitate. These data indicate that diminishing the competition among mTORC1 substrates for raptor by diminishing the abundance of any substrate is sufficient to up-regulate phosphorylation of the remaining mTORC1 substrates, which presumably occurs without alteration of the intrinsic catalytic activity of mTOR. This response suggests that the abundance and/or affinity of raptor-substrate binding sites is limiting to the ability of mTORC1 to catalyze the phosphorylation of its substrates in vivo, even in serum/nutrient-replete cells. This in turn raises the possibility that the binding of substrates by raptor may be a locus at which mTORC1 activity is regulated, as previously suggested by Wang et al. (35).
FIGURE 7. Competitive effects among mTORC1 substrates for raptor binding and mTORC1 phosphorylation in vivo.
A, effect of overexpression of an mTORC1 substrate on phosphorylation of the other two substrates. In the experiments shown in the four panels on the upper left, HEK293 cells were co-transfected with FLAG-tagged 4E-BP1 and either empty vector, Myc-tagged PRAS40 wild type, or the PRAS40 mutants indicated. The cells were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids. The aliquots of the cell lysates were subjected to immunoblot by the anti-Myc antibody to verify comparable expression of the PRAS40 variants. An anti-FLAG immunoprecipitate was immunoblotted for 4E-BP1 (Thr(P)37/46) and FLAG. The upper three panels show the immunoblots from a representative experiment. The 4E-BP1 (Thr(P)37/46) blots were scanned, and the OD was divided by the OD of the corresponding FLAG-4E-BP1 blot. This ratio for the FLAG immunoprecipitate from cells that did not receive Myc-PRAS40 (vector only) was set to 100% and divided into the ratio obtained for each of the other FLAG immunoprecipitates. The bar graph summarizes the results from three experiments. The double or single asterisks indicate a reduction as compared with vector only; p < 0.01 or p <0.05, respectively. In the three panels shown at the lower left, HA-tagged S6K1 was co-expressed with either empty vector, Myc-tagged PRAS40 wild type, or the PRAS40 mutants indicated. The aliquots of the cell lysates were subjected to immunoblot by the anti-Myc antibody to verify comparable expression of the PRAS40 variants. An anti-HA immunoprecipitate was immunoblotted for S6K1 (Thr(P)389) and HA, and the blots were scanned. The OD of the S6K1 (Thr(P)389) blot was divided by the OD of the corresponding HA-S6K1 blot. This ratio for the HA immunoprecipitate from cells that did not receive Myc-PRAS40 (vectoronly) was set to 100% and divided into the ratio obtained for each of the other HA immunoprecipitates. These values of one experiment are shown below the bottom panel; a second experiment gave similar results. The panels on the right show the effects of 4E-BP1 (upper) and kinase-negative S6K1 (lower) co-expression on PRAS40 (Ser183) phosphorylation; cells were treated in a manner similar to the experiments on the left. B, effect of RNA interference-induced PRAS40 depletion on the phosphorylation of endogenous S6K1 and 4E-BP1. HEK293 cells (left) and HeLa cells (right) transfected with indicated siRNA were deprived of serum, further incubated without amino acids, and stimulated by the readdition of amino acids in the presence or absence of rapamycin. The cell lysates were subjected to immunoblot by the anti-PRAS40, anti-S6K1 (Thr(P)389), anti-S6K1, anti-4E-BP1 (Ser(P)65), anti-4E-BP1 (Thr(P)37/46), and anti-β-actin antibodies. C, effect of overexpression of 4E-BP1 and S6K1 on the association of endogenous PRAS40 with overexpressed recombinant raptor. HEK293 cells co-transfected with FLAG-tagged raptor and either GST, GST-fused 4E-BP1, or GST-fused S6K1 were deprived of serum and further incubated without amino acids. The cell lysates were subjected to immunoprecipitation with the anti-FLAG antibody. Immunoblot was carried out with the anti-FLAG, anti-GST, and anti-PRAS40 antibodies. The aliquots of the cell lysates were subjected to immunoblot by the anti-GST antibody.
DISCUSSION
The results presented above strongly support the conclusion that PRAS40 is a physiological substrate of mTORC1 kinase, whose phosphorylation at Ser183 is enabled by its binding to raptor. The interaction of PRAS40 with endogenous raptor is heavily dependent on PRAS40 (Phe129), inasmuch as mutation of this residue to Ala greatly reduces the binding of recombinant PRAS40 to raptor; whether, as with 4E-BP1, additional PRAS40 segments are necessary for optimal binding to raptor is not yet established. Vander Haar et al. (21) observed that Myc-PRAS40-(1–150) and -(150–234) are each unable to co-immunoprecipitate raptor, whereas PRAS40-(103–258) and -(1–234) are able to do so; taken together with the present data, this suggests that a PRAS40 segment between 150 and 234 is needed together with the region surrounding Phe129 for optimal binding to raptor. As with the TOS motif, the four PRAS40 residues carboxyl-terminal to Phe129 (i.e. 130VMDE133) consist of two hydrophobic (ϕ) and two acidic (Ac) amino acids; however, the arrangement of these residues in S6K1, 4E-BP1 (13), and STAT3 is Phe-Ac-ϕ-Ac-ϕ. Given the compelling evidence that PRAS40 is an endogenous substrate for mTORC1, we propose that the definition of a TOS motif be extended to include the motif Phe-ϕ-ϕ-Ac-Ac.
The binding of PRAS40 to raptor was also greatly inhibited by mutation of PRAS40 Ser183 to Asp, whereas mutation to Ala had a modest inhibitory effect on the interaction. We propose that the phosphorylation of Ser183, like its conversion from Ser to Asp, results in the dissociation of PRAS40 from raptor. A similar phenomenon was observed with 4E-BP1, i.e. mutation of the five mTORC1-catalyzed Ser/Thr phosphorylation sites to Glu abolished 4E-BP1 binding to raptor (19). We propose that the ability of amino acid withdrawal to inhibit PRAS40 (Ser183) phosphorylation in vivo may account for the ability of amino acid withdrawal to increase the binding of endogenous PRAS40 to raptor. By contrast, when PRAS40 was overexpressed, amino acid withdrawal caused little or no further increase in retrieval of endogenous raptor (Figs. 1C, 2, and 5D), perhaps because the cells are then expressing a large excess of PRAS40, much of which may not be phosphorylated at Ser183. This is also a likely explanation for why recombinant PRAS40 (S183A) does not exhibit increased binding to raptor as compared with recombinant wild-type PRAS40 (Fig. 5D).
The mechanism responsible for the ability of rapamycin to interfere with the association of endogenous PRAS40 with endogenous raptor (Figs. 1B (bottom) and 5A) is less obvious. Based on the ability of rapamycin to inhibit mTORC1-catalyzed phosphorylation of PRAS40 (Ser183) in vivo, rapamycin might be expected to increase rather than diminish the raptor-PRAS40 association. In fact, when raptor was overexpressed and presumably was present far in excess to endogenous mTOR, rapamycin did cause a small increase in the retrieval of endogenous PRAS40 by the recombinant raptor (Fig. 1B, top). Under those conditions, rapamycin, which (with FKBP12) binds only to mTOR, inhibited mTORC1 kinase and therefore diminished PRAS40 (Ser183) phosphorylation; we suggest that the excess raptor unassociated with mTOR is unaffected by the FKBP12-rapamycin complex, and the decreased phosphorylation of endogenous PRAS40 at Ser183 then results in increased binding of endogenous PRAS40 to the overexpressed recombinant raptor (Fig. 1B, top). In contrast, when the endogenous interactions were examined (Figs. 1B (bottom) and 5A) or when only PRAS40 was overexpressed (e.g. Figs. 1C, 2, and 5D), the inhibitory effect of rapamycin on the association of endogenous raptor with recombinant PRAS40 was highly pronounced. Under these conditions, most or all cellular raptor is in mTORC1 and thus susceptible to the inhibitory action of the FKBP12-rapamycin complex on the contiguous mTOR; inasmuch as the FKBP12-rapamycin complex binds only to mTOR, the effect of rapamycin on raptor must be exerted through mTOR. Although the concentrations of rapamycin employed in these experiments (200 nm) were sufficient to cause major dissociation of the mTOR-mLST8 heterodimer from raptor in vivo (Fig. 1C), it is unlikely that the loss of mTOR-mLST8 is itself the explanation for the loss of PRAS40 binding to raptor, inasmuch as complete dissociation of mTOR-mLST8 from raptor in vitro using Nonidet P-40 did not diminish the amount of PRAS40 bound to raptor (Fig. 1C). Thus, the ability of rapamycin (at 200 nm) to inhibit the binding of endogenous (Figs. 1B (bottom) and 5A) or recombinant PRAS40 (Figs. 1C, 2, and 5D) to endogenous raptor is not due to the loss of mTOR-mLST8 per se or to the dephosphorylation of PRAS40 (Ser183); rather, we propose that it reflects a decrease in the affinity of endogenous, mTORC1-associated raptor for PRAS40, which cannot be overcome by PRAS40 overexpression; generalizing this hypothesis, we propose that rapamycin may diminish the affinity of mTORC1-associated raptor for mTORC1 substrates in general. The mechanism for such an effect is unknown; although it is not due to the rapamycin-induced dissociation of raptor from mTOR-mLST8 per se (Fig. 1C), it may be that the rapamycin-induced dissociation of mTOR-mLST8 from raptor that occurs in vivo (Figs. 1C, 2, and 5D) eliminates some input from mTOR to raptor (e.g. phosphorylation) that is important for substrate binding. Clearly, much further work will be required to explore the validity of this hypothesis and the biochemical mechanisms responsible.
The identification of PRAS40 as a likely physiological substrate of mTORC1 raises the question of the function of mTORC1-catalyzed PRAS40 (Ser183) phosphorylation. Vander Haar et al. (21) and Sancak et al. (22) showed that PRAS40 overexpression reduces the size of a variety of cells, consistent with the ability of PRAS40 overexpression to suppress S6K1 and 4E-BP1 phosphorylation. This suggests (but does not establish) that endogenous PRAS40 is not a positive regulator of cell size. In our hands, neither transient nor stable overexpression or siRNA-mediated knockdown of PRAS40 affected the size of K562 cells (data not shown). Sancak et al. (22) showed that PRAS40 inhibits mTORC1-catalyzed phosphorylation of 4E-BP1 and S6K1 in vitro, and a PRAS40 phosphorylated by Akt was less inhibitory in vitro, whereas a PRAS40 (T246A) mutant was somewhat more inhibitory than wild-type PRAS40 during transient expression in vivo. Vander Haar et al. (21) both observed that sustained reduction of endogenous PRAS40 by lentivirus-encoded short hairpin RNA is accompanied by diminished levels of endogenous IRS1 polypeptide and diminished insulin activation of both Akt and S6K1. Consequently, both Vander Haar et al. (21) and Sancak et al. (22) propose that a primary function of endogenous PRAS40 is to suppress mTOR and up-regulate insulin signaling to Akt. The responses observed with sustained suppression of PRAS40, however, are those that would be expected from removal of a significant physiological mTORC1 substrate other than S6K1, especially one whose primary function is unrelated to the up-regulation of cell growth. As shown in the present studies, removal of PRAS40 would initially up-regulate the phosphorylation/activity of S6K1 (and other mTORC1 substrates), which in turn has been shown to result in the phosphorylation and enhanced degradation of IRS1 (36–38), reviewed in Ref. 39; these actions at the level of IRS1 would result in reduced insulin activation of Akt and S6K1 itself. Thus, although the present results do not eliminate a regulatory function for PRAS40 on mTORC1 or Akt signaling, they indicate strongly that the negative effects of PRAS40 depletion on Akt activation are likely to be largely or entirely attributable to the removal of an mTORC1 substrate rather than to a primary regulatory function of PRAS40.
The physiological function(s) of PRAS40 remain uncertain, and therefore the significance of the Akt-catalyzed phosphorylation of PRAS40 (Thr246), the subsequent binding of 14-3-3 (20, 40), and the mTORC1-catalyzed phosphorylation of Ser183 also remain obscure. It has been reported that amino acids are required for the binding of 14-3-3 to PRAS40 phosphorylated at Thr246 (40). It is unlikely that amino acid-stimulated phosphorylation at PRAS40 (Ser183) is a priming signal for the phosphorylation at Thr246 inasmuch as the insulin-stimulated phosphorylation at Thr246 is largely unaltered in rapamycin-treated cells (Fig. 5C) (20, 40). Alternatively, stable 14-3-3 binding to PRAS40 may require binding of the 14-3-3 dimer to PRAS40 at two independent sites (41, 42), one of which might be (Ser(P)183) itself.
PRAS40 has been implicated in the regulation of cell survival and apoptosis (43–45). The introduction of PRAS40 in mouse brain protects neuronal cells from apoptosis after ischemic injury, and the phosphorylation of PRAS40 at Thr246 by Akt in response to nerve growth factor is proposed to be important for the prevention of apoptosis (43). PRAS40 phosphorylated at Thr246 has been detected in mouse cortical neurons after ischemia and reperfusion (43), and the phosphorylation of PRAS40 (Thr246) and the subsequent binding of 14-3-3 are reported to be important for the survival of neuronal cells from apoptosis after ischemic injury in vivo (43, 45). Lobe, a putative D. melanogaster ortholog of PRAS40, is required for ventral growth of eye disc and cell survival during early eye development (46, 47). Therefore, PRAS40 seems to play an important role in cell survival conserved among different species. Phosphorylated PRAS40 (Thr246) is detected predominantly in the nuclei of H9c2-E2 cardiomyocytes and A14 fibroblasts after insulin stimulation (48). Thus, nuclear PRAS40, phosphorylated by mTORC1 and/or Akt may regulate the transcription levels of anti- and/or proapoptotic proteins. Moreover, the transcription of PRAS40 itself is enhanced in TSC1 knock-out mouse astrocytes in which the mTORC1 pathway is up-regulated (49), indicating that the expression level as well as the phosphorylation of PRAS40 is up-regulated by mTORC1 signaling. Thus, PRAS40 is a transcriptional and post-transcriptional target of mTORC1 regulation and thus likely to play an important, if as yet undefined role in mTORC1 action.
Acknowledgments
We express our appreciation to Shinji Kamada and Hidenori Matsuzaki for valuable discussions and to Masafumi Kamada and Satoshi Otsuka for technical advice. We are also grateful to Hiroe Miyamoto and Kaori Fujii for technical assistance and to Rie Kato, Yumiko Kawajiri, and Jeanette Prendable for secretarial assistance.
Footnotes
This work was supported in part by research grants from the Scientific Research Funds of the Ministry of Education, Culture, Sports, Science and Technology of Japan and CREST, Japan Science and Technology Agency. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
The abbreviations used are: mTOR, mammalian target of rapamycin; mTORC1 and 2, mTOR complex 1 and 2, respectively; TOS, TOR signaling; DMEM, Dulbecco's modified Eagle's medium; siRNA, small interfering RNA; ESI, electrospray ionization; MS, mass spectrometry; GST, glutathione S-transferase; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Wullschleger S, Loewith R, Hall MN. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Davies SP, Reddy H, Caivano M, Cohen P. Biochem. J. 2000;351:95–105. doi: 10.1042/0264-6021:3510095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fingar DC, Blenis J. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 4.Dumont FJ, Staruch MJ, Koprak SL, Melino MR, Sigal NH. J. Immunol. 1990;144:251–258. [PubMed] [Google Scholar]
- 5.Marx SO, Jayaraman T, Go LO, Marks AR. Circ. Res. 1995;76:412–417. doi: 10.1161/01.res.76.3.412. [DOI] [PubMed] [Google Scholar]
- 6.Poon M, Marx SO, Gallo R, Badimon JJ, Taubman MB, Marks AR. J. Clin. Investig. 1996;98:2277–2283. doi: 10.1172/JCI119038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J, Kuo CJ, Crabtree GR, Blenis J. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- 8.Price DJ, Grove JR, Calvo V, Avruch J, Bierer BE. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 9.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- 10.von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr., Abraham RT. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 12.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. Proc. Natl. Acad. Sci. U. S. A. 1998;95:1432–1437. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schalm SS, Blenis J. Curr. Biol. 2002;12:632–639. doi: 10.1016/s0960-9822(02)00762-5. [DOI] [PubMed] [Google Scholar]
- 14.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Curr. Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 15.Beugnet A, Wang X, Proud CG. J. Biol. Chem. 2003;278:40717–40722. doi: 10.1074/jbc.M308573200. [DOI] [PubMed] [Google Scholar]
- 16.Choi KM, McMahon LP, Lawrence JC., Jr. J. Biol. Chem. 2003;278:19667–19673. doi: 10.1074/jbc.M301142200. [DOI] [PubMed] [Google Scholar]
- 17.Nojima H, Tokunaga C, Eguchi S, Oshiro N, Hidayat S, Yoshino K, Hara K, Tanaka N, Avruch J, Yonezawa K. J. Biol. Chem. 2003;278:15461–15464. doi: 10.1074/jbc.C200665200. [DOI] [PubMed] [Google Scholar]
- 18.Schalm SS, Fingar DC, Sabatini DM, Blenis J. Curr. Biol. 2003;13:797–806. doi: 10.1016/s0960-9822(03)00329-4. [DOI] [PubMed] [Google Scholar]
- 19.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 20.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. J. Biol. Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 21.Vander Haar EV, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 22.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. J. Biol. Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 24.Weng QP, Andrabi K, Kozlowski MT, Grove JR, Avruch J. Mol. Cell. Biol. 1995;15:2333–2340. doi: 10.1128/mcb.15.5.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. Curr. Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 26.Nishiuma T, Hara K, Tsujishita Y, Kaneko K, Shii K, Yonezawa K. Biochem. Biophys. Res. Commun. 1998;252:440–444. doi: 10.1006/bbrc.1998.9671. [DOI] [PubMed] [Google Scholar]
- 27.Shigemitsu K, Tsujishita Y, Hara K, Nanahoshi M, Avruch J, Yonezawa K. J. Biol. Chem. 1999;274:1058–1065. doi: 10.1074/jbc.274.2.1058. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi Y, Shirai Y, Matsubara T, Sanse K, Kuriyama M, Oshiro N, Yoshino K, Yonezawa K, Ono Y, Saito N. J. Biol. Chem. 2006;281:31627–31637. doi: 10.1074/jbc.M606992200. [DOI] [PubMed] [Google Scholar]
- 29.Sakakibara K, Sato K, Yoshino K, Oshiro N, Hirahara S, Mahbub Hasan AK, Iwasaki T, Ueda Y, Iwao Y, Yonezawa K, Fukami Y. J. Biol. Chem. 2005;280:15029–15037. doi: 10.1074/jbc.M410538200. [DOI] [PubMed] [Google Scholar]
- 30.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 31.Inoki K, Zhu T, Guan KL. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Corradetti MN, Inoki K, Guan KL. Trends. Biochem. Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Curr. Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- 34.von Manteuffel SR, Dennis PB, Pullen N, Gingras AC, Sonenberg N, Thomas G. Mol. Cell. Biol. 1997;17:5426–5436. doi: 10.1128/mcb.17.9.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Rhodes CJ, Lawrence JC., Jr. J. Biol. Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- 36.Haruta T, Uno T, Kawahara J, Takano A, Egawa K, Sharma PM, Olefsky JM, Kobayashi M. Mol. Endocrinol. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 37.Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, Kobayashi M. Mol. Cell. Biol. 2001;21:5050–5062. doi: 10.1128/MCB.21.15.5050-5062.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF. J. Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah OJ, Hunter T. Cell Cycle. 2005;4:46–51. doi: 10.4161/cc.4.1.1343. [DOI] [PubMed] [Google Scholar]
- 40.Harthill JE, Pozuelo Rubio M, Milne FC, MacKintosh C. Biochem. J. 2002;368:565–572. doi: 10.1042/BJ20020838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzivion G, Avruch J. J. Biol. Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 42.Mackintosh C. Biochem. J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito A, Narasimhan P, Hayashi T, Okuno S, Ferrand-Drake M, Chan PH. J. Neurosci. 2004;24:1584–1593. doi: 10.1523/JNEUROSCI.5209-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Porter G. Acta Pharmacol. Sin. 2005;26:1253–1258. doi: 10.1111/j.1745-7254.2005.00184.x. [DOI] [PubMed] [Google Scholar]
- 45.Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Stroke. 2006;37:513–517. doi: 10.1161/01.STR.0000198826.56611.a2. [DOI] [PubMed] [Google Scholar]
- 46.Chern JJ, Choi KW. Development. 2002;129:4005–4013. doi: 10.1242/dev.129.17.4005. [DOI] [PubMed] [Google Scholar]
- 47.Singh A, Shi X, Choi KW. Development. 2006;133:4771–4781. doi: 10.1242/dev.02686. [DOI] [PubMed] [Google Scholar]
- 48.Nascimento EB, Fodor M, van der Zon GC, Jazet IM, Meinders AE, Voshol PJ, Vlasblom R, Baan B, Eckel J, Maassen JA, Diamant M, Ouwens DM. Diabetes. 2006;55:3221–3228. doi: 10.2337/db05-1390. [DOI] [PubMed] [Google Scholar]
- 49.Ess KC, Uhlmann EJ, Li W, Li H, Declue JE, Crino PB, Gutmann DH. Glia. 2004;46:28–40. doi: 10.1002/glia.10324. [DOI] [PubMed] [Google Scholar]