Abstract
Many proteomic experiments require selective labeling of either N- or C-termini of proteins and recovery of terminal peptides. Although N-termini can be selectively labeled, selective labeling of protein C-termini has not been possible due to the difficulty in discriminating between the carboxyl group on the C-terminus versus that on aspartate and glutamate residues. Here we describe the first simple proteomic approach for positive selection of protein C-termini, Profiling Protein C-Termini by Enzymatic Labeling (ProC-TEL). ProC-TEL uses carboxypeptidase Y and other readily available reagents to selectively add an affinity tag to protein C-termini and to capture C-terminal peptides from complex cell lysates for mass spectrometry (MS) identification. Using ProC-TEL, we identify novel C-terminal processing and internal proteolytic cleavage events. These results indicate that ProC-TEL provides a straightforward approach for profiling C-terminal peptides and identifying protein processing in complex biological samples.
INTRODUCTION
Strategies for labeling protein termini are useful for a wide range of proteomic applications. After labeling proteins on the N-terminus, the corresponding N-terminal peptides can be selectively recovered and used for proteomic profiling. These profiling approaches can identify proteins, protein terminal modifications, and protein cleavage events in complex mixtures (1–3). For example, after a proteolytic cleavage, two products are generated, comprising one peptide with a new C-terminus (neo-C-terminus), and another peptide with a new N-terminus (neo-N-terminus). The neo-N- and neo-C-terminal peptides can be used to identify the specific site of proteolytic cleavage (1–5).
N-terminal labeling strategies have been an area of intense methodological development in proteomics (1–3). Reagents that react preferentially with N-terminal amines, as opposed to ε-amines on lysine residues, have been used to selectively recover N-terminal peptides (1, 2, 6). However, N-terminal labeling is not possible when the N-terminus is “blocked”, e.g. by acetylation, which occurs in as many as 85% eukaryotic cytosolic proteins (7). Furthermore, protein N-termini are highly susceptible to “trimming” by various aminopeptidases, resulting in proteins that have N-termini missing one, two, or three N-terminal amino acids (1, 3, 8). This can be problematic when attempting to identify proteolytic cleavage sites, since the N-terminal residue of a neo-N-terminal peptide may not reflect the initial cleavage, but may reflect subsequent trimming events.
These drawbacks of N-terminal labeling could be potentially overcome by using C-terminal labeling approaches because protein C-termini are rarely blocked compared to N-termini (9), and may not be as susceptible to C-terminal trimming. However, strategies for the selective labeling and capture of C-terminal peptides are lacking. One reason for the difficulty in labeling protein C-terminus is due to the lower reactivity of the carboxyl group relative to amino groups. Furthermore, selective labeling of the C-terminal carboxyl group is particularly challenging since the carboxyl group in aspartate and glutamate residues exhibits similar reactivity and is approximately 50 times more abundant in a typical protein (10).
Current approaches for C-terminal peptide enrichment rely on “negative selection,” in which non-C-terminal peptides are removed from a sample (11–13). One approach uses immobilized anhydrotrypsin, which binds tryptic peptides containing a C-terminal arginine or lysine (11). C-terminal peptides, which are unlikely to have a C-terminal arginine or lysine, remain in solution (11). Other negative enrichment strategies that rely on the differential charge or hydrophobicity between internal tryptic peptides and terminal peptides after chemical modification of carboxyl or amino groups have also been described (12, 13). However, since the C-terminal peptide is present at a low abundance relative to internal peptides, negative selection needs to be highly efficient in order to obtain samples that are enriched in C-terminal peptides. For this reason, “positive selection” approaches, in which the terminal peptide is selectively captured, are advantageous for C-terminal proteomics.
Because of the difficulty in selectively labeling protein C-termini using chemical approaches, we considered using an enzyme to selectively recognize and label protein C-termini. Carboxypeptidases are a class of enzymes that selectively recognize protein C-termini and hydrolyze C-terminal residues (14). This property has been utilized to liberate amino acids from protein C-terminus for C-terminal sequencing and identification (15). In some instances, proteases can exhibit both proteolysis activity as well as “transpeptidase” activity, in which an exogenous nucleophile, such as an amino acid, is added to a protein (16, 17). This activity arises after the catalytic serine attacks the scissile peptide bond in the substrate, forming an ester intermediate with the target protein. Although the target protein is usually released by subsequent hydrolysis of the ester with water, transpeptidation reactions occur when the ester is subjected to nucleophilic attack by an amino acid (16, 17).
Carboxypeptidase Y (CPY) could be used for C-terminal labeling since it exhibits transpeptidase activity, and has been used to replace C-terminal residues with labeled amino acids (16, 17). The transpeptidase activity is more prominent for proteins containing a carboxyl ester, which may bind more efficiently to CPY (17, 18). However, the usefulness of this reaction is limited since CPY-mediated transpeptidation reaction results in multiple products, comprising proteins in which variable numbers of C-terminal residues have been proteolyzed prior to the incorporation of a labeled residue (16, 17).
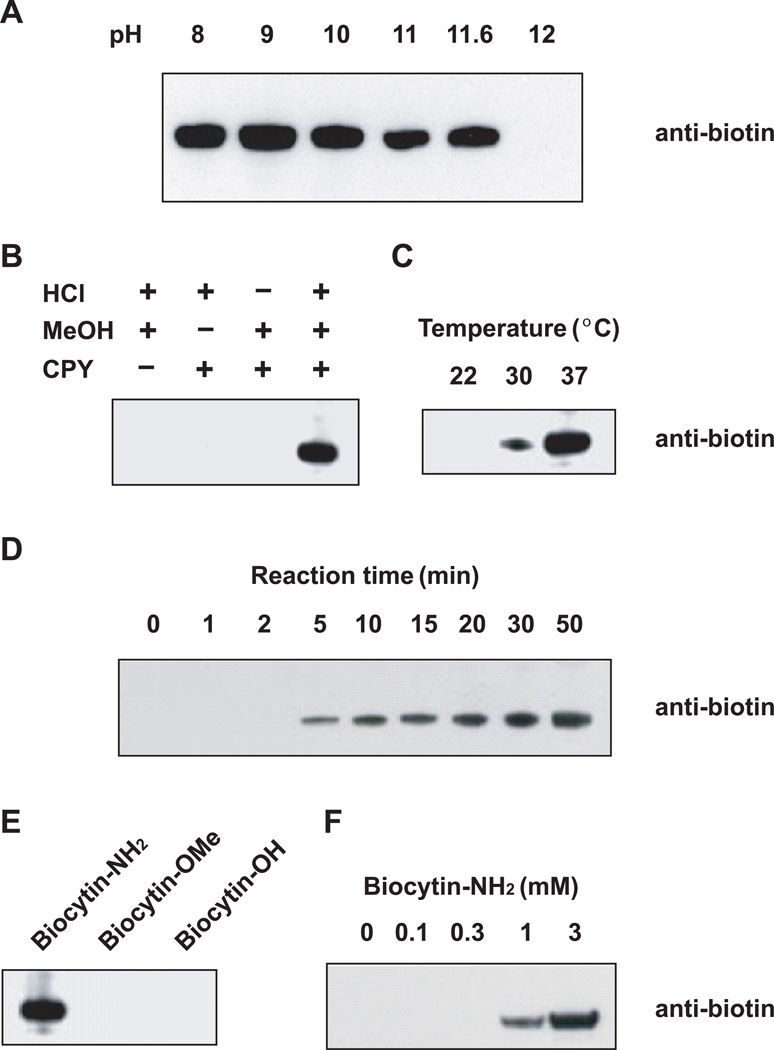
We considered the possibility that modifying the reaction conditions might favor transpeptidation over proteolysis. CPY-mediated proteolysis is most active at pH 5–7, with marked decreases occurring at pH > 9 (19). We therefore sought to identify pH conditions in which hydrolysis is minimized and transpeptidase activity is maintained. To measure CPY reaction products, we incubated CPY with biocytinamide, a biotinylated lysine derivative containing an α-amine as the nucleophile, and methyl esterified bovine serum albumin (BSA), prepared by incubation of BSA in methanolic HCl at 4°C for 48 h. This methyl esterification condition does not cause detectable protein degradation (Supplementary Figure 1). Labeling was performed over a range of pHs, and biocytinamide incorporation was determined by anti-biotin Western blotting. Biotinylated BSA was observed at pH 8, and reached a maximum level at pH 9, with decreasing levels observed until pH 11.6 (Figure 1, panel A). At pH 12, no biotinylated BSA was detected. Because proteolytic activity of CPY is suppressed by increasing pH (19), and pH 11.6 was the highest pH in which transpeptidation was observed, we used this pH for subsequent optimization experiments.
Figure 1. Identification of conditions leading to protein labeling by carboxypeptidase Y (CPY).
The results are evaluated by anti-biotin Western blotting analyses.
(A) CPY-catalyzed protein biotinylation takes place in a wide range of pHs. Methyl esterified BSA (1 mg/ml) was incubated with CPY and biocytinamide (3 mM) at the indicated pHs.
(B) Protein methyl esterification is required for CPY-catalyzed biotinylation.
(C) 37°C is an optimal temperature for CPY-catalyzed biotinylation.
(D) Time course of CPY-dependent biotinylation.
(E) Biocytinamide is the optimal nucleophile for CPY-catalyzed biotinylation. Methyl esterified BSA was incubated with CPY and 3 mM biocytinamide (biocytin-NH2), biocytin methyl ester (biocytin-OMe), or biocytin (biocytin-OH).
(F) Higher concentrations of biocytinamide lead to increased protein biotinylation.
We next optimized the labeling conditions for CPY-mediated biotinylation. We first addressed whether carboxyl esterification was required for CPY-mediated transpeptidation. Incubation of unmodified BSA with CPY at pH 11.6 resulted in no detectable biotinylation, while BSA incubated with methanolic HCl was sufficient for CPY-mediated biotinylation (Figure 1, panel B). Transpeptidation was optimal at 37°C (Figure 1, panel C), and increased over 50 min (Figure 1, panel D), with minimal increases observed with longer incubation (data not shown). The only biocytin derivative that was a substrate for transpeptidation was biocytinamide, while neither biocytin methyl ester or biocytin was suitable as a nucleophile (Figure 1, panel E). Biotinylation of BSA increased with increasing biocytinamide concentrations up to 3 mM, which was the maximum solubility achieved in the reaction buffer (Figure 1, panel F).
Transpeptidation was enhanced by the use of detergents, although some proteins were more efficiently labeled in the presence of 1% Triton X-100, while others were more efficiently labeled in the presence of 0.1% SDS (Supplementary Figure 2). In the case of insulin, the presence of both detergents provided the most efficient labeling. We therefore selected a reaction solution containing both detergents, although other buffer conditions could potentially facilitate the labeling of different proteins.
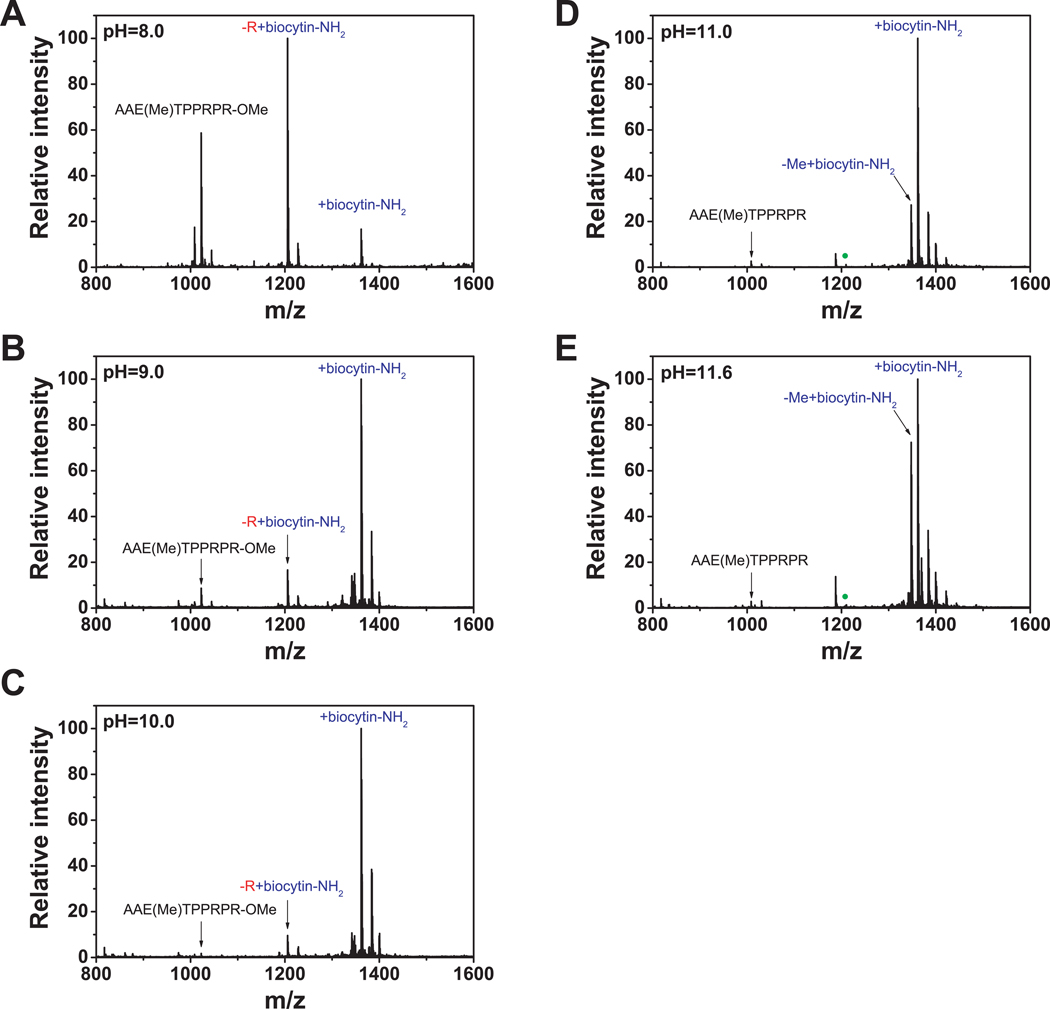
Previous protocols using CPY to label proteins at pH 6.5 (16) or pH 10 (17) resulted in the incorporation of amino acid labels at various positions in the C-terminus of proteins due to rounds of proteolysis prior to transpeptidase activity. To determine the reactions catalyzed by CPY at pH 11.6, we monitored the products using a methyl esterified model peptide, AAETPPRPR, by matrix-assisted laser desorption/ionization-time of flight-mass spectrometry (MALDI-TOF-MS). At pH 8.0, a mixture of products was observed, including a peptide in which a biocytinamide was added directly to the C-terminus, and a peptide in which the C-terminal arginine was hydrolyzed and replaced by biocytinamide (Figure 2, panel A). The identity of these peptides, as well as the position of biocytinamide incorporation, was confirmed by MS/MS (Supplementary Figures 3 and 4). Both products were also detected at pHs 9.0 and 10.0 (Figure 2, panels B and C). However, at higher pHs (Figure 2, panels D and E), only the C-terminally labeled peptide was detected, with no evidence for proteolysis. Similar results were seen for insulin (Supplementary Figure 5). Together, these experiments indicate that under high pH conditions, CPY exclusively catalyzes the addition of biocytinamide to the C-terminus of peptide methyl esters.
Figure 2. CPY-catalyzed C-terminal modifications are determined by the pH used during labeling.
(A, B, C) The products of CPY-mediated biotinylation were determined using a model peptide, AAETPPRPR, at pHs 8.0, 9.0 and 10.0, and the products were detected by MALDI-TOF-MS.
(D, E) CPY-mediated biotinylation at pHs 11.0 and 11.6 does not result in removal of the C-terminal amino acid. Negligible unmodified peptide was detected, indicating that C-terminal labeling with CPY is highly efficient under these reaction conditions. The green solid circles in (D) and (E) correspond to the position of the biocytinamide-modified peptide missing the C-terminal arginine, which was detected in (A), (B), and (C), but not in (D) and (E).
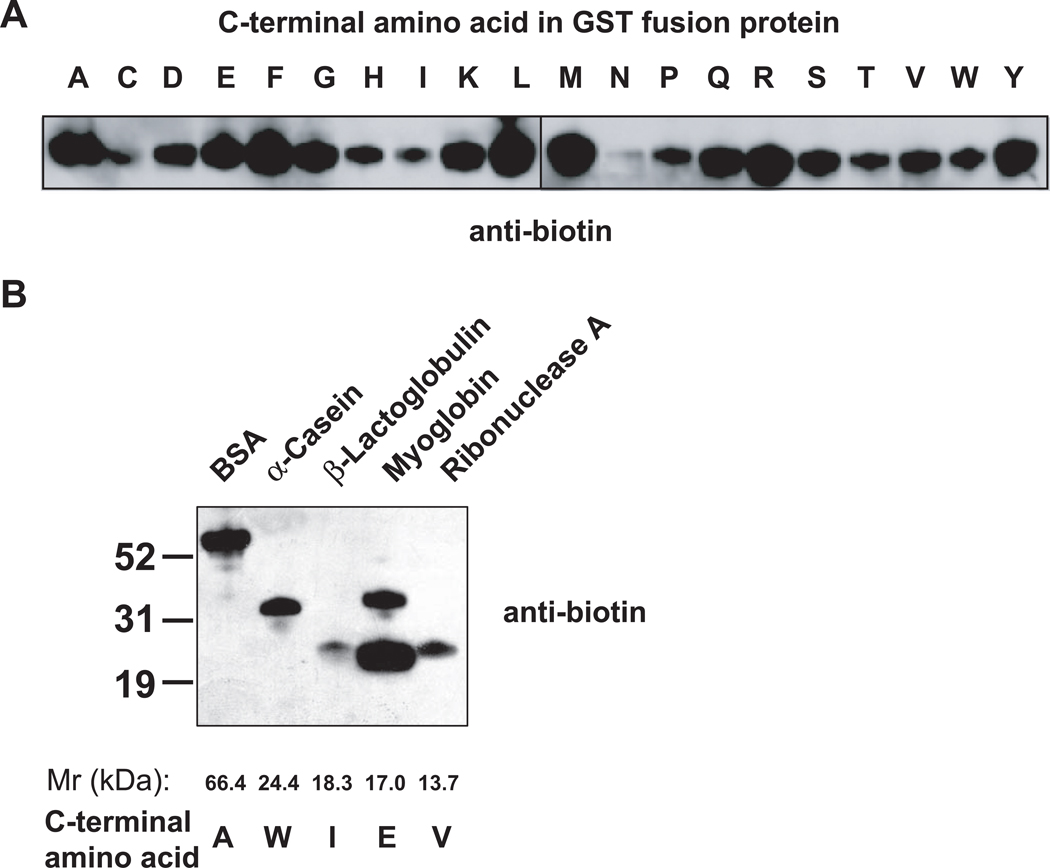
We next asked if CPY can label proteins containing diverse C-terminal residues since CPY-mediated amino acid hydrolysis exhibits broad tolerance for a range of C-terminal amino acids (20). Recombinant glutathione S-transferase (GST) containing each of the twenty amino acids as the C-terminal residue, were methyl esterified and biotinylated at pH 11.6. In each case, the GST fusion protein was labeled, although preference for different terminal amino acids was seen (Figure 3, panel A).
Figure 3. Effect of C-terminal amino acids on CPY-catalyzed protein biotinylation.
(A) GST fusion proteins containing each of twenty amino acids at the C-terminus can be biotinylated with CPY. Each GST protein (1 µg) was methyl esterified and incubated with CPY and biocytinamide at 37°C for 30 min. Because of the limited number of samples that can be loaded per gel, these experiments were conducted on two blots that were Western blotted with an anti-biotin antibody and exposed simultaneously.
(B) CPY-mediated biotinylation of naturally occurring proteins. The indicated proteins (1 µg) were methyl esterified and labeled with CPY and biocytinamide. The top band in the myoglobin lane is a contaminant.
We next assessed labeling of naturally occurring proteins. BSA, α-casein, β-lactoglobulin, myoglobin, and ribonuclease A were labeled by CPY-mediated transpeptidation (Figure 3, panel B) after methyl esterification. BSA and myoglobin were labeled most efficiently, with β-lactoglobulin labeled the least efficiently. The labeling of each of these proteins was consistent with the efficiency predicted by the labeling obtained with GST containing the different C-terminal residues. Since most proteomic applications involve comparing the abundance of the same protein derived from two different samples, the variability in labeling efficiency for different proteins should not prevent its application in quantitative proteomic experiments, if the labeling efficiency is reproducible. Indeed, the labeling efficiency is highly reproducible for the proteins tested in our experiments (Supplementary Figure 6).
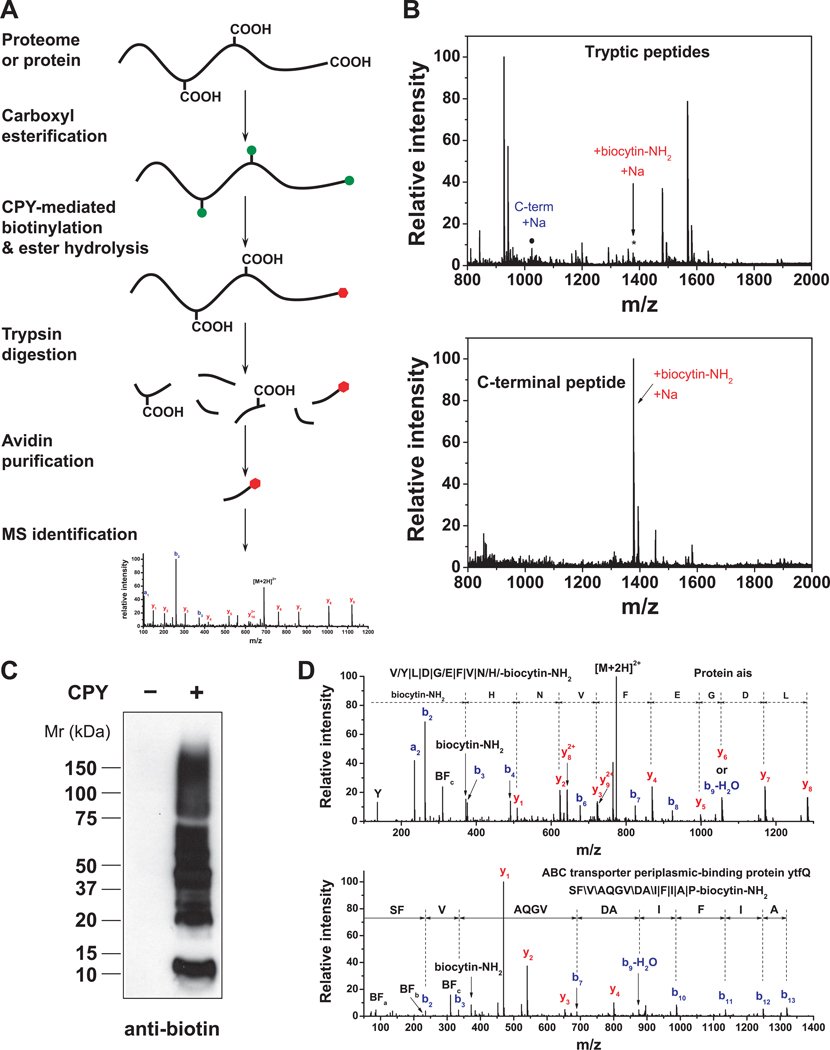
Based on the ability of CPY to selectively label protein C-termini, we devised ProC-TEL (Profiling Protein C-Termini by Enzymatic Labeling), in which protein mixtures, e.g. derived from cellular lysates, are methyl esterified, biotinylated with CPY, and then subjected to in-gel trypsinolysis. C-terminal peptides are then captured by monomeric avidin-affinity chromatography, and eluted for C-terminal peptide identification by MS (Figure 4, panel A).
Figure 4. Selective isolation of C-terminal peptides using ProC-TEL.
(A) ProC-TEL strategy for the isolation and identification of C-terminal peptides. Protein carboxyl groups (side chains and C-terminus) are first modified by methyl esterification (green circles). The C-termini are then biotinylated by CPY through the addition of a biotin-containing nucleophile at protein C-termini and methyl esters at the side chains of Glu and Asp are hydrolyzed at high pH at the same time. The biotinylated proteins are digested and the C-terminal peptides with affinity biotin tags (red hexagons) are isolated by avidin affinity chromatography for MS analysis.
(B) Isolation of the C-terminal peptide from a protein using ProC-TEL. After CPY labeling of BSA with biocytinamide and trypsinolysis, numerous tryptic peptides are detected by MALDI-TOF MS (top panel), including the biotinylated C-terminal peptide (asterisk). Following purification of peptides on monomeric avidin agarose, the biotinylated C-terminal peptide is readily detected, while the other unmodified peptides are almost completely removed (bottom panel).
(C) CPY-mediated labeling of proteins in E. coli lysates detected by an anti-biotin Western blotting.
(D) MS/MS spectra of representative C-terminal peptides identified from E. coli lysates using the ProC-TEL approach. The peptide sequence and modification is shown in the spectra and the b-ions and y-ions are labeled. The symbols, \, / and |, represent b-ions, y-ions, and both b-ions and y-ions, respectively. MS/MS spectra for the C-terminal peptide from Protein ais (top panel) and the neo-C-terminal peptide from the ABC transporter ytfQ (bottom panel) are shown. In the MS/MS spectra, three characteristic biotin fragment ions, BFa, BFb, BFc, and biocytinamide ion (biocytin-NH2) are detected for the C-terminal biotinylated peptides.
We first addressed whether C-terminal peptides generated by ProC-TEL would be sufficiently unique to identify parent proteins. The majority (62%) of ProC-TEL peptides prepared by virtual trypsin digestion of proteins listed in the Swiss-Prot E. coli protein database (v57.2) (21) are at least four amino acids long (Supplementary Figure 7, panels A and B), a length which may provide enough sequence information for protein identification in E. coli, depending on the MS instrumentation and identification algorithm (Supplementary Figure 7, panel C). Of these peptides, the majority match to a single protein in the database (Supplementary Figure 7, panel C). Thus, for ProC-TEL tryptic peptides, ~ 60% of the E. coli proteome could be unambiguously detected. However, since short peptides may be difficult to detect or identify, it is likely that a smaller fraction of the E. coli proteome can be detected. To increase the coverage of the proteome, the results from parallel experiments using additional proteases, such as Lys-C or Glu-C, could be merged with data obtained using trypsin.
To test whether ProC-TEL can be used to capture C-terminal peptides, we used BSA. After biotinylation and trypsinolysis, the modified C-terminal peptide was detected among numerous other tryptic peptides (Figure 4, panel B). However, after purification of peptides on monomeric avidin agarose, the trifluoroacetic acid eluate contained predominantly the C-terminal peptide, indicating that the labeled C-terminal peptide can be purified from tryptic digests. The identity of the C-terminal peptide, as well as the C-terminal biocytinamide was confirmed by MS/MS (Supplementary Figure 8).
We next determined whether ProC-TEL could recover C-terminal peptides from cellular lysates. Treatment of the esterified E. coli lysate with CPY resulted in extensive protein biotinylation (Figure 4, panel C). Following trypsinolysis and monomeric avidin purification, over 70 C-terminal peptides were identified (Supplementary Table 1). MS/MS analysis revealed readily detectable y- and b-ion series, as well as fragment ions characteristic of biotinylated peptides (Figure 4, panel D) (22). Although the majority of C-terminal peptides identified matched predicted C-terminal peptides, two classes of neo-C-terminal peptides were also identified (Supplementary Table 1). First, neo-C-terminal peptides consistent with trimming of protein C-termini accounted for ~5% of all C-terminal peptides. For example, C-terminal peptides from flagellar-hook protein flgE, glyceraldehyde-3-phosphate dehydrogenase, and peroxiredoxin osmC, were missing one or a few amino acids from the protein C-terminus. The second class of neo-C-terminal peptides was derived from internal proteolytic cleavages, accounting for ~26% of all C-terminal peptides. The majority of these have not been previously reported. Proteolytic cleavages were found in diverse proteins, such as the ABC-transporter ytfQ (Figure 4, panel D), glycerol-3-phosphate acyltransferase, osmotically-inducible protein Y, and tRNA-dihydrouridine synthase. From the secondary structures predicted by PSIPRED (23), we found that ~75% of the cleavage sites are located at, or close to unstructured regions. These data highlight the ability of ProC-TEL to reveal insights into protein processing.
Several modifications of the ProC-TEL procedure can readily be envisioned. Although we used biocytinamide, other affinity tags, such as amino acid amides containing azide or alkyne moieties, could be used to permit labeling using “click chemistry” approaches (24). Since ProC-TEL involves a lengthy esterification step, alternative methyl esterification reagents can be used, such as trimethylsilyldiazomethane (25), which allows rapid chemo-specific methylation of carboxyl groups under mild aqueous conditions. Additionally, labeling proteins with amino acid amides that contain stable isotopically labeled atoms could permit isotope coding of peptides from different samples for quantitative proteomic studies. Moreover, engineered CPY mutants (26) with increased transpeptidase activity could potentially improve the labeling efficiency. The yield of peptides can also be improved by using higher sensitivity MS instruments with a 2D-LC separation system, which can result in substantially more C-terminal peptide identifications.
Although terminal proteomics are commonly used to profile protein processing and to identify termini and neo-termini, C-terminal proteomics can be used to enhance the sensitivity of quantitative proteomics. The large number of peptides obtained following tryptic digestion can prevent thorough identification of a significant fraction of low abundant proteins in the sample due to instrument duty cycle, ion suppression, and dynamic range. Although this is partially alleviated using isotope-coded affinity tags in which only cysteine-containing peptides are analyzed (27), ProC-TEL can provide further simplification of a peptide mixture by providing a single C-terminal peptide per protein.
In summary, we have developed the first positive selection approach, ProC-TEL, to profile protein C-termini by enzymatic labeling. This strategy facilitates the isolation and identification of C-terminal peptides from complex biological samples. Recently, an approach for N-terminal labeling using a similar transpeptidation strategy has been described (2). This approach uses subtiligase, an engineered enzyme, to covalently link a cleavable glycolate ester-derivatized peptide tag to the N-termini of proteins, thereby facilitating the purification and subsequent protease-dependent release of N-terminal peptides. However, this approach is complex and requires specialized reagents that are not routinely available. An attractive feature of ProC-TEL is that it can be performed using CPY and biocytinamide, which are widely available and inexpensive laboratory reagents.
METHODS
CPY catalyzed-labeling of peptide and proteins
CPY catalyzed biotinylation was carried out under the conditions indicated in the figure legends. Briefly, methyl esterified peptide and proteins were incubated with CPY and biocytinamide at the indicated pH. The labeling was performed at 37°C for 30 min, and detected by an anti-biotin Western blotting unless otherwise indicated.
Sample preparation for MS analysis
Biotinylated proteins were separated by SDS-PAGE, subjected to in-gel trypsinolysis, and purified by monomeric avidin agarose prior to LC-MS/MS.
Supplementary Material
Acknowledgments
We thank A. Morrishow at Weill-Cornell Medical College (WCMC) MS Core Facility for assistance with MALDI-TOF-MS analysis, and J. Paige for assistance in initial sample preparation and LC-MS analysis. MS analyses were performed at the WCMC MS Core Facility using instrumentation supported by NIH RR19355 and RR22615. This work was supported by NIAID (AI068639) and the New York Speaker’s Fund for Biomedical Research (S.R.J.), and training grant T32CA062948 (S.Y.S. and G.X.).
Footnotes
Detailed Methods are available in the online Supporting Information.
Author contributions: S.R.J. and G.X. conceived and designed the experiments, S.Y.S. performed various pilot experiments, G.X. performed the majority of experiments, analyzed the data; and G.X. and S.R.J. wrote the paper.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, Eroshkin A, Smith JW, Tao WA, Salvesen GS. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu G, Shin SB, Jaffrey SR. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc Natl Acad Sci U S A. 2009;106:19310–19315. doi: 10.1073/pnas.0908958106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleifeld O, Doucet A, auf dem Keller U, Prudova A, Schilling O, Kainthan RK, Starr AE, Foster LJ, Kizhakkedathu JN, Overall CM. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol. 2010;28:281–288. doi: 10.1038/nbt.1611. [DOI] [PubMed] [Google Scholar]
- 5.Van Damme P, Martens L, Van Damme J, Hugelier K, Staes A, Vandekerckhove J, Gevaert K. Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat Methods. 2005;2:771–777. doi: 10.1038/nmeth792. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi M, Nakazawa T, Kuyama H, Obama T, Ando E, Okamura TA, Ueyama N, Norioka S. High-throughput method for N-terminal sequencing of proteins by MALDI mass spectrometry. Anal Chem. 2005;77:645–651. doi: 10.1021/ac048776w. [DOI] [PubMed] [Google Scholar]
- 7.Polevoda B, Sherman F. Nα-terminal acetylation of eukaryotic proteins. J Biol Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 8.Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 9.Nakazawa T, Yamaguchi M, Okamura TA, Ando E, Nishimura O, Tsunasawa S. Terminal proteomics: N- and C-terminal analyses for high-fidelity identification of proteins using MS. Proteomics. 2008;8:673–685. doi: 10.1002/pmic.200700084. [DOI] [PubMed] [Google Scholar]
- 10.Gerstein M. How representative are the known structures of the proteins in a complete genome? A comprehensive structural census. Fold Des. 1998;3:497–512. doi: 10.1016/S1359-0278(98)00066-2. [DOI] [PubMed] [Google Scholar]
- 11.Sechi S, Chait BT. A method to define the carboxyl terminal of proteins. Anal Chem. 2000;72:3374–3378. doi: 10.1021/ac000045i. [DOI] [PubMed] [Google Scholar]
- 12.Van Damme P, Staes A, Bronsoms S, Helsens K, Colaert N, Timmerman E, Aviles FX, Vandekerckhove J, Gevaert K. Complementary positional proteomics for screening substrates of endo- and exoproteases. Nat Methods. 2010;7:512–515. doi: 10.1038/nmeth.1469. [DOI] [PubMed] [Google Scholar]
- 13.Schilling O, Barre O, Huesgen PF, Overall CM. Proteome-wide analysis of protein carboxy termini: C terminomics. Nat Methods. 2010;7:508–511. doi: 10.1038/nmeth.1467. [DOI] [PubMed] [Google Scholar]
- 14.Skidgel RA, Erdos EG. Cellular carboxypeptidases. Immunol Rev. 1998;161:129–141. doi: 10.1111/j.1600-065x.1998.tb01577.x. [DOI] [PubMed] [Google Scholar]
- 15.Samyn B, Sergeant K, Castanheira P, Faro C, Van Beeumen J. A new method for C-terminal sequence analysis in the proteomic era. Nat Methods. 2005;2:193–200. doi: 10.1038/nmeth738. [DOI] [PubMed] [Google Scholar]
- 16.Berne PF, Schmitter JM, Blanquet S. Peptide and protein carboxyl-terminal labeling through carboxypeptidase Y-catalyzed transpeptidation. J Biol Chem. 1990;265:19551–19559. [PubMed] [Google Scholar]
- 17.Lin S, Lowe CR. C-Terminal labeling of immunoglobulin G with a cysteine derivative by carboxypeptidase Y catalyzed transpeptidation. Anal Biochem. 2000;285:127–134. doi: 10.1006/abio.2000.4723. [DOI] [PubMed] [Google Scholar]
- 18.Berne PF, Blanquet S, Schmitter JM. Carboxypeptidase Y-catalyzed transpeptidation of esterified oligo- and polypeptides and its use for the specific carboxyl-terminal labeling of proteins. J. Am. Chem. Soc. 1992;114:2603–2610. [Google Scholar]
- 19.Hayashi R, Bai Y, Hata T. Kinetic studies of carboxypeptidase Y. I. Kinetic parameters for the hydrolysis of synthetic substrates. J Biochem. 1975;77:69–79. [PubMed] [Google Scholar]
- 20.Hayashi R, Moore S, Stein WH. Carboxypeptidase from yeast. Large scale preparation and the application to COOH-terminal analysis of peptides and proteins. J Biol Chem. 1973;248:2296–2302. [PubMed] [Google Scholar]
- 21.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sioud S, Genestie B, Jahouh F, Martin P, Banoub J. Gasphase fragmentation study of biotin reagents using electrospray ionization tandem mass spectrometry on a quadrupole orthogonal time-of-flight hybrid instrument. Rapid Commun Mass Spectrom. 2009;23:1941–1956. doi: 10.1002/rcm.4091. [DOI] [PubMed] [Google Scholar]
- 23.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 24.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Leggio A, Liguori A, Perri F, Siciliano C, Viscomi MC. Methylation of alpha-amino acids and derivatives using trimethylsilyldiazomethane. Chem Biol Drug Des. 2009;73:287–291. doi: 10.1111/j.1747-0285.2009.00777.x. [DOI] [PubMed] [Google Scholar]
- 26.Stennicke HR, Olesen K, Sorensen SB, Breddam K. C-terminal incorporation of fluorogenic and affinity labels using wild-type and mutagenized carboxypeptidase Y. Anal Biochem. 1997;248:141–148. doi: 10.1006/abio.1997.9998. [DOI] [PubMed] [Google Scholar]
- 27.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.