Abstract
Thyroid hormone (T3) mediates diverse physiological functions including growth, differentiation, and energy homeostasis through the thyroid hormone receptors (TR). The TR bind DNA at specific recognition sequences in the promoter regions of their target genes known as the thyroid hormone response elements (TREs). Gene expression at TREs regulated by TRs is mediated by coregulator recruitment to the DNA bound receptor. This TR-coregulator interaction controls transcription of target genes by multiple mechanisms including covalent histone modifications and chromatin remodeling. Our previous studies identified a β-aminoketone as a potent inhibitor of the TR-coactivator interaction. We describe here the activity of one of these inhibitors in modulating effects of T3 signaling in comparison to an established ligand-competitive inhibitor of TR, NH-3. The β-aminoketone was found to reverse thyroid hormone induced gene expression by inhibiting coactivator recruitment at target gene promoters, thereby regulating downstream effects of thyroid hormone. While mimicking the downstream effects of a ligand competitive inhibitor at the molecular level, the β-aminoketone affects only a subset of the thyroid responsive signaling network. Thus antagonists directed to the coregulator binding site have distinct pharmacological properties relative to ligand based antagonists and may provide complementary activity in vivo.
INTRODUCTION
Nuclear receptors (NRs) have traditionally been targeted by drugs that competitively displace the physiological ligand with agonist or antagonist properties. More recently our and other groups have begun to focus on alternate strategies for targeting these receptors including the direct inhibition of nuclear receptor-coactivator interactions (1-8). We present here the first study comparing the mechanistic and cellular consequences of antagonizing NR function with a coactivator binding inhibitor or a ligand binding inhibitor.
The thyroid hormone receptors (TR) are nuclear receptors that respond to the thyroid hormone, 3,5,3'-triiodo-L-thyronine (T3), which is derived locally in responsive tissues from an endocrine hormone secreted by thyroid gland. There are 2 isoforms of the receptor: THRA (TRα) and THRB (TRβ) (9). TRα regulates cardiac rate and contractility whereas TRβ controls cholesterol homeostasis and TSH feedback suppression by T3. TR binds DNA as a heterodimer with another NR, the retinoid X receptor RXR, at TREs in the promoters of its target genes. T3 has profound effects on growth, development, metabolism and homeostasis (10). The signal from TR is carried forward by coactivator proteins that manipulate both chromatin structure and the composition of the transcriptional regulatory complex (11-14). Coactivator recruitment to TR is ligand dependent. In the unliganded state, corepressor proteins such as Nuclear Receptor Corepressor (NCoR) and Silencing mediator of Retinoic acid and Thyroid hormone receptor (SMRT) are bound to the thyroid receptors and suppress the expression of target genes. Upon T3 binding, the TR-ligand binding domain (TR-LBD) undergoes a conformational change resulting in repositioning of α helixes 3, 4, 5, and 12 of the protein to generate a hydrophobic groove for coactivator binding (15, 16). Coactivator molecules have a LXXLL (Leucine-X-X-Leucine-Leucine) domain in their structure (16-19), known as an NR box, which binds to this groove.
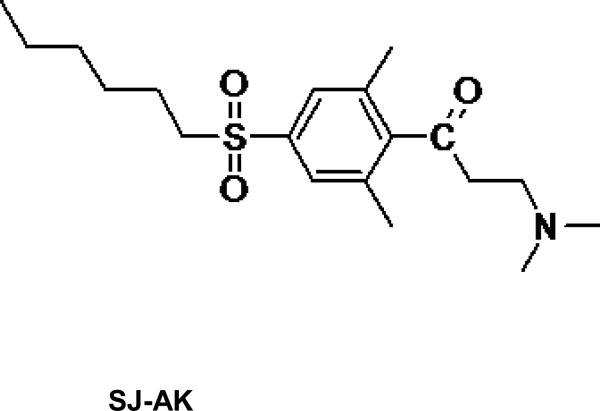
Previous work carried out in our laboratory identified β-aminoketones as inhibitors of the TR-coactivator interaction (8, 20). Chemical and pharmacological optimization of this series of inhibitors led to a second generation of more active and less toxic β-aminoketones including the compound SJ000311413, referred to as SJ-AK in this manuscript (Figure 1) (21, 22). The activity of these time dependent irreversible inhibitors was attributed to the generation of in-situ enones. These electrophilic enones selectively attack C298 in the coactivator binding site (AF-2) of the TR-LBD, forming a covalent adduct and inhibiting the TR-coactivator interaction (23). While SJ-AK was not the most fully optimized compound that was eventually produced, it was sufficiently potent and efficacious, non-toxic, and selective to justify undertaking the studies described herein in parallel to the further optimization that ultimately led to the most refined compounds (22).
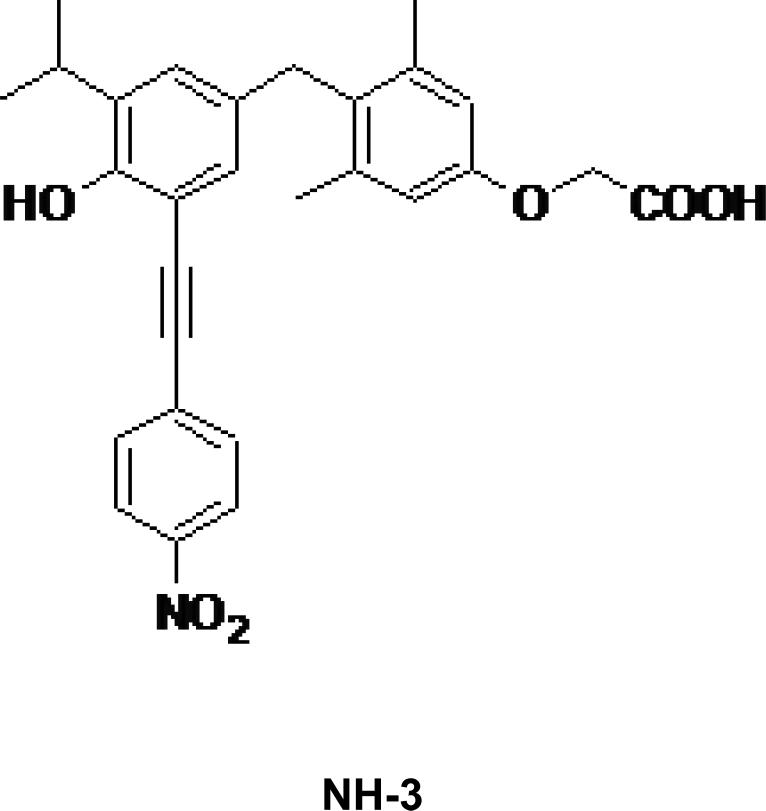
Figure 1.
Chemical structures of TR antagonists SJ-AK and NH-3
Below are described studies that compare the cellular mechanisms and effects of SJ-AK with those of NH-3, (24) a known ligand-competitive antagonist of TR (Figure 1). Inhibition of ligand binding and inhibition of receptor-coactivator interaction represent two major modalities for achieving antagonism of nuclear receptor function (Figure 2). Potentially a coactivator binding inhibitor may act simply as a replacement for ligand antagonists as coactivator binding is one step downstream of ligand binding in the T3 signaling network. However, there has not been a detailed systemic comparison of these types of antagonism, which may in practice give differing consequences in cells. This study compares the cellular effects of inhibition of ligand binding (achieved by NH-3) with those achieved by the inhibition of the receptor-coactivator interaction while maintaining ligand binding (achieved by SJ-AK).
Figure 2.
Graphical representation of the modes of actions of the 2 classes of nuclear receptor antagonists: SJ-AK (a thyroid receptor-coactivator interaction inhibitor) and NH-3 (a T3 competitive inhibitor). SJ-AK inhibits TR-coactivator (TR-CoA) interaction without effecting T3 binding and is thus not affected by T3 concentrations. NH-3 competes with T3 for binding to TR and secondarily effects coactivator binding. CoA-coactivator; CoR-co-repressor, RXR-retinoic × receptor.
RESULTS AND DISCUSSION
NH-3 and SJ-AK utilize two fundamentally differing modalities of antagonizing TR. NH-3 displaces T3 from its binding pocket in the receptor. Other examples of such NR targeted drugs include dexamethasone, an agonist of the glucocorticoid receptor, and bicalutamide, an antagonist of the androgen receptor. On the other hand, SJ-AK belongs to the new class of NR targeted compounds that directly inhibit the NR-coactivator interaction (22). The fundamental question here is how these two modes of modulating the same signaling cascade are similar in functional antagonism and how they are different.
Transcriptome effects of SJ-AK and NH-3 on T3 induced gene expression
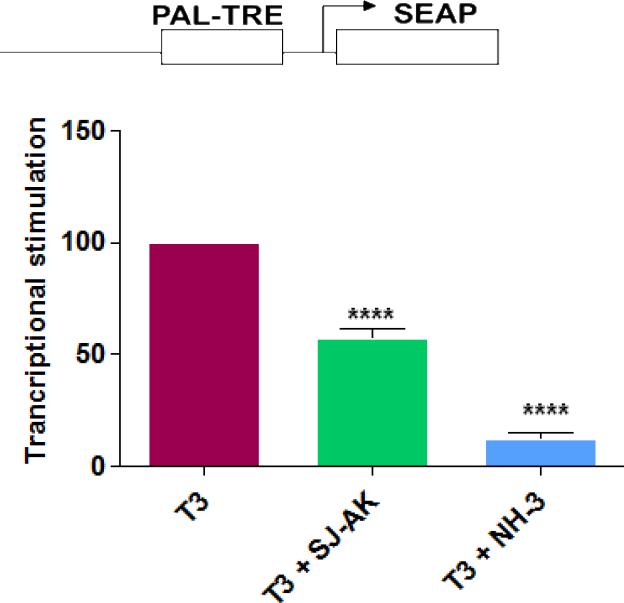
In order to establish the best concentrations of T3 and antagonist to use for genomic experiments both antagonists were characterized using reporter gene assay (Figure 3). We used a palindromic TRE (-AGGTCATGACCT-) construct driving the expression of a secreted alkaline phosphatase reporter (PAL-TRE-SEAP) to assay for T3 induced transcriptional activity. This reporter construct is routinely used in our lab as a screening tool to determine transcriptional inhibition potency of TR antagonists. Palindromic TRE is robustly induced by T3 in our transcription assays providing a sufficient range of effect to permit rank ordering of TR antagonists as per their activity in inhibiting T3 mediated transcription.
Figure 3.
Effect of SJ-AK and NH-3 on T3 induced gene expression. Transient transfections in HeK293 cells using palindromic TRE construct as the reporter. This reporter was transfected along with CMV-TRβ and β-Gal expression vectors. Cells were treated as indicated with T3 alone or in combination with SJ-AK or NH-3. Cells were harvested and SEAP activity assay was carried out. Data was normalized for β-gal activity. Transcriptional stimulation was expressed as a percentage of T3 alone transcriptional stimulation. **** indicates p < 0.001
Twenty four hours after compound treatment, the cell culture media was sampled and tested for secreted alkaline phosphatase activity. Transfected beta-galactosidase activity was used as internal control to normalize the SEAP expression levels. SJ-AK inhibited the T3 stimulation of this reporter by ~ 50% at 5 μM and NH-3 inhibited the T3 stimulation by 70% at 5 μM. This mirrors effects seen with other reporter constructs in prior studies (8, 24). Additionally the cytotoxicity of both antagonists was assessed and both were devoid of cytotoxicity or growth inhibition for most cell lines at these concentrations (Supplementary Table 1).
Liver is a key target tissue of thyroid hormone action. To assess the genomic effects of SJ-AK and NH-3 on T3 mediated gene expression, global transcript abundance studies were carried out in the hepatocellular carcinoma derived HepG2 cells, which maintain many aspects of hepatocyte function (25-29). These cells were treated for 24 hours with T3 alone and T3 in combination with either 5 μM SJ-AK or NH-3, saturating doses for each on simple TRE driven reporters. The mRNA from these treatments was analyzed using Affymetrix human U133 Plus 2.0 oligonucleotide gene chips to probe for changes in transcript abundance for 30,000 human genes. The experiments were carried out as three completely independent biologic replicates, each with internal triplicate replicates, for a total of 9 reads, and the data were pooled for analysis. Among the 54,613 probe sets on the microarray, 1403 (2.57%) had differential expression with FDR < 0.1 in response to T3. These profiles were additionally filtered for robustness by excluding probe sets called “Absent” across the entire data set, and by excluding probe sets with less than +/- 1.0 log2[signal] difference. Using these criteria, 193 probe sets were identified as “T3 target genes” (Figure 4A). Among these 163 were upregulated and 30 were downregulated by T3.
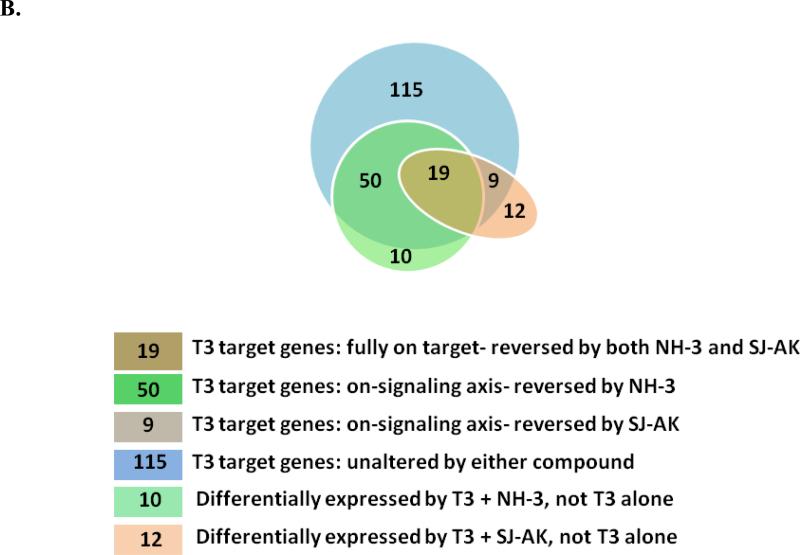
Figure 4.
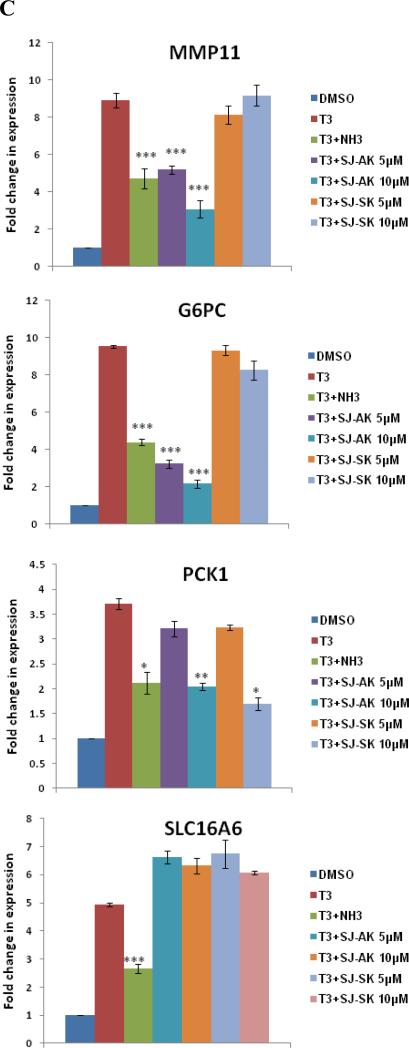
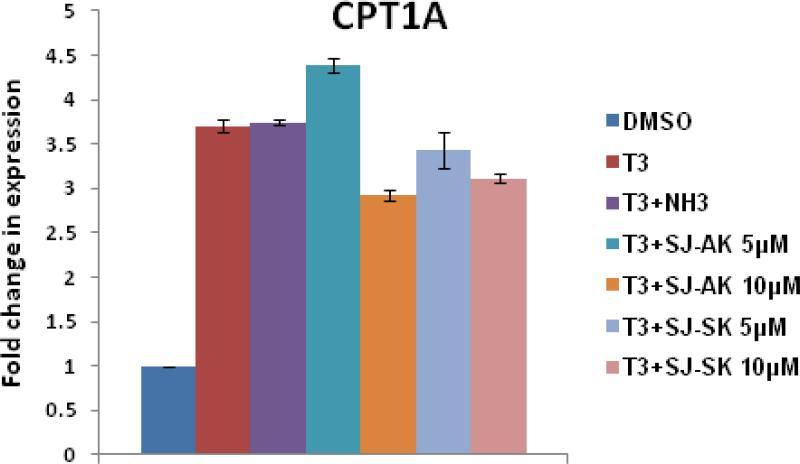
Microarray gene expression analysis in HepG2 cells. A, Representative heat map of the 78 T3 target probe sets whose differential expression by T3 is affected by the TR inhibitor compounds: SJ-AK and NH-3. Average signal log ratios vs. either DMSO or T3 are used to generate the heat map. B, Venn diagram illustrating the global gene expression effects of treatment of HepG2 cells with T3 alone or in combination with either NH-3 or SJ-AK. . C, qRT-PCR experiments on T3 target genes identified in the microarray experiment. T3 was added at a concentration of 100 nM alone or in combination with either SJ-AK or NH-3. qRT-PCR was carried out to amplify MMP11, PCK1, G6PC, SLC16A6, OSTβ, CYP24A1, CYP3A4 C8ORF4 and CPT1A genes. ΔΔCt method was used to calculate fold induction of expression. *** indicates p < 0.005, ** indicates p < 0.01 relative to T3 effect.
The observed T3 effects on gene expression (number of genes affected and their magnitude of change) in this study were consistent with other published microarray studies of T3 action in HepG2, as well as those using primary liver tissues of rat and mouse (30-33). The set of T3 target genes obtained from the microarray included a number of previously identified T3 target genes such as phosphoenolpyruvate carboxykinase (PCK1), glucose-6-phosphatase (G6PC), and matrix metalloproteinase 11 (MMP11) as well as many novel T3 target genes. Using the DAVID bioinformatics database (http://david.abcc.ncifcrf.gov), we performed gene ontology (GO) and pathway analysis to determine over-representation of specific biological pathways in T3 target genes. We found significant enrichment of genes involved in steroid metabolic processes, PPAR signaling, the complement pathway, and the coagulation pathway (Supplementary Table 2). Thus these studies of HepG2 cells are a reasonable surrogate for effects in a target tissue.
Co-treatment of HepG2 cells with T3 and either SJ-AK or NH-3 was performed to determine the extent and pattern of inhibitor effects on T3 agonized genes. Overall, both inhibitors reversed global expression changes induced by T3 treatment (Figure 4). Using +/- 0.5 log2[signal] as a measure of differential response to define reversal of T3 effect, we identified 69 probe sets that displayed reversal of T3 effect by NH-3 and 28 probe sets that displayed reversal by SJ-AK treatment. Nineteen probe sets had a concordant response greater than 0.5 log2 [signal] by both inhibitors. Among the 163 up-regulated genes, NH-3 reversed the T3 effect on 49 genes whereas among the 30 downregulated genes, NH-3 reversed the T3 effect on 20 genes. Among the 163 up-regulated genes, SJ-AK reversed T3 effect on 15 genes whereas among the 30 downregulated genes, SJ-AK reversed the T3 effect on 13 genes. There were 115 probe sets that were unaffected by either inhibitor. For one particular gene (unc5cl), SJ-AK exhibited agonist-like activity and actually enhanced the T3 effect of differential expression over DMSO (Supplementary Table 3). The profiles and gene annotations of the differentially expressed transcripts are provided in the supplementary information (Supplementary Table 3).
There were also instances in which T3 altered the expression of genes when administered in combination with either NH-3 or SJ-AK but not when administered alone). Both inhibitors demonstrate some activity against majority of T3 target genes but do not meet the set threshold for all genes. As shown in the Venn diagram in Figure 4B and Supplementary Table 4, there were 10 probesets that were differentially expressed by T3 in combination with NH-3 (T3 + NH-3 vs. DMSO, FDR <0.1, no absent call, +/- 1.0 log2[signal] difference) that were not altered by T3 alone (T3 + NH-3 vs. T3, > 0.5 log2[signal] difference). Using similar criteria, there were 12 probesets that were differentially expressed by T3 in combination with SJ-AK that were not altered by T3 alone.
In addition to co-treatment of T3 with the inhibitor compounds, we also carried out cell treatments using the inhibitor compounds alone to interrogate the possibility of inherent activities of these compounds in absence of exogenous T3 which could lead to off-target effects on downstream genes. Using the same analysis criteria as described above, we found NH-3 treatment caused the differential expression of only one gene whereas SJ-AK did not alter the expression of any gene on the array (Supplementary Table 5). Minimal effects of the inhibitor compounds on non-T3 target genes either alone (Supplementary Table 5) or in combination with T3 (Supplementary Table 4) illustrate the specificity of action of these thyroid receptor inhibitors. Furthermore, we also treated HepG2 cells with a compound named SJ-SK which is related to SJ-AK but unable to generate the active enone on the surface of TR protein. SJ-SK is inactive in inhibiting receptor-coactivator binding and T3 induction of TRE driven reporters (data not shown). Likewise, microarray analysis shows that SJ-SK is markedly less active than SJ-AK in reversing T3 induced gene expression changes in HepG2 cells (Supplementary Table 6). Raw microarray data can be accessed in the supplementary information (Supplementary Table 7).
We categorized the T3 target probesets as “fully on target” (reversed by both NH-3 and SJ-AK, 19 probesets), “on-signaling axis” (reversed by either NH-3 (50 probesets) or SJ-AK (9 probesets) but not both), “unique” (T3 induced only in presence of NH-3 or SJ-AK) and “unaltered” (not reversed by either) (Figure 4B). Since this categorization is based on an arbitrarily set threshold, it should not be interpreted as a rigid classification of activity of the inhibitors.
To validate the microarray observations, we performed qRT-PCR on a subset of putative T3 responsive genes identified by the microarray. Co-treatment with T3 (100 nM) and SJ-AK, NH-3, or SJ-SK at either 5 or 10 μM concentrations was carried out. Twenty four hours post treatment, RNA was isolated and qRT-PCR was carried out on cDNA made from the RNA samples. These experiments were carried out in two completely independent biological replicates, each with internal triplicate replicates for a total of 6 reads. We tested several different target genes identified in the microarray: matrix metallopeptidase 11 (MMP11); phosphoenolpyruvate carboxykinase (PCK1); glucose-6-phosphatase (G6PC); solute carrier family 16, member 6 (SLC16A6); organic solute transporter beta (OSTβ); cytochrome P450, family 24, subfamily A, polypeptide 1 (CYP24A1); cytochrome P450, family 3, subfamily A, polypeptide 4 (CYP3A4); chromosome 8 open reading frame 4 (C8ORF4) and carnitine palmitoyltransferase 1A (CPT1A) (Figure 4, panel C), some of which have been previously identified as T3 responsive by others (32, 34, 35). These genes belonged to all 3 categories of T3 targets identified my microarray: fully on-target (MMP11), on-signaling axis NH3 (G6PC, PCK1, SLC16A6, OSTβ, CYP24A1), on-signaling axis SJ-AK (CYP3A4, C8ORF4) and unaltered (CPT1A). There is good concordance between the microarray and real time PCR data except for G6PC gene where SJ-AK showed activity in qPCR but not in microarray experiment. NH-3 and SJ-AK activity ranged from significant inhibition of gene expression to no activity in agreement with the microarray data. NH-3 inhibits T3 mediated transcription of MMP11, G6PC, PCK1, SLC16A6, OSTβ, and CYP24A1 but not CYP3A4, C8ORF4 and CPT1A. SJ-AK inhibited the T3 mediated expression of MMP11, G6PC, PCK1 (at high concentration), CYP3A4 and C8ORF4 but not SLC16A6, OSTβ, CYP24A1 and CPT1A. At a concentration 5 μM, SJ-SK did not inhibit T3 induction of the genes we tested.
The genome wide transcriptional analysis shows that SJ-AK generally antagonizes TR signaling in a manner similar to NH-3 but targets a smaller and partially overlapping set of responsive genes. Most of the genes reversed by SJ-AK are a subset of the genes reversed by NH-3. These data validate the hypothesis that NR-coactivator interaction inhibitors will block nuclear receptor activated gene expression. Thus functionally, SJ-AK is a more restricted version of NH-3 in terms of its breadth of reversal of the T3 induced gene expression changes. However SJ-AK is a functional antagonist of T3 signaling that does not compete for T3 binding to the TR-LBD (8), thus eliminating the need to titrate against T3 in hyperthyroid conditions. As NH-3 has partial agonist activity at high concentrations as well as some cytotoxicity, its potential for use in hyperthyroidism is limited (36). In vivo studies on SJ-AK are required to determine if it is a better alternative to NH-3 in hyperthyroid conditions.
An unexpected finding in these studies was that there are a significant number of genes whose expression is not reversed by either inhibitor. While both inhibitors block ligand mediated coactivator recruitment to the activation function (AF-2) pocket in the C terminus of TR protein, these compounds do not target other coregulator binding surfaces that exist in the TR structure. For example, coactivators are also recruited to the activation function -1 (AF-1) pocket in the N terminus of TR in a ligand independent fashion (37). Therefore it is reasonable to expect that blocking just one site of coregulator interaction with these inhibitors may not result in complete loss of gene expression effects of T3 for at least some genes. Additionally, our data could also provide credence to the existence of alternate or non-TR dependent mechanisms of T3 action, such as those that have previously been suggested to be mediated via MAPK or PI3kinase-Akt (38, 39). It should be noted that at the twenty four hour time point, the T3 responsive gene list will include both the direct (TRE mediated) as well as indirect (non-TRE mediated) T3 target genes. NH-3 could potentially reverse more T3 responses if more aggressively titrated against a fixed dose of T3.
Effects of SJ-AK and NH-3 on Coregulators at Responding Genes
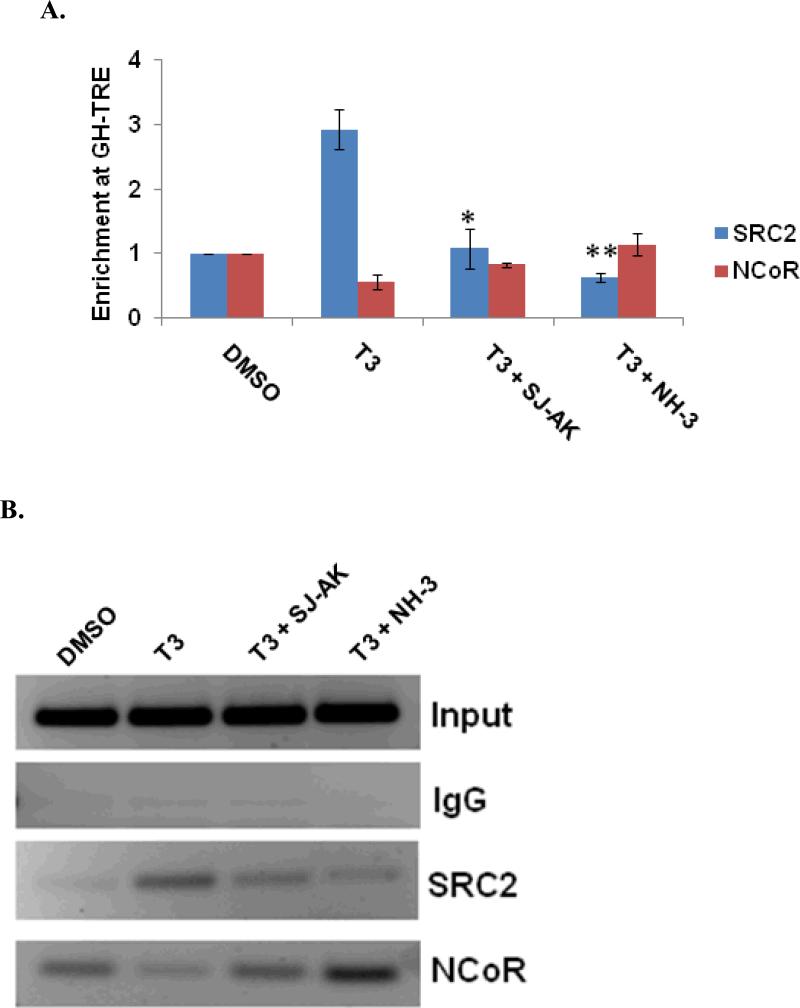
Having established the patterns of antagonism of T3 induced gene expression in cells by SJ-AK and NH-3 we further characterized the molecular mechanism of action of these compounds. To test whether these compounds inhibit the coactivator recruitment at the TRE sequences in the promoters of native T3 target genes, chromatin immunoprecipitation (ChIP) assays were performed in GH3 cells (Figure 5, panel A and B). The growth hormone gene is a classical, well-validated T3 target in anterior pituitary cells. The growth hormone TRE is an imperfect direct repeat located at -188 to -173 relative to transcription start site. TR binds this element as a heterodimer with RXR (40). Recruitment of cofactors to the growth hormone TRE was monitored 30 min after treatment with T3 in the presence or absence of either SJ-AK or NH-3. The coactivator SRC-2 was strongly recruited to the growth hormone gene TRE within 30 minutes of T3 addition. This recruitment was inhibited by both SJ-AK and NH-3. In the same experiment, we also determined the occupancy of NCoR (Nuclear receptor corepressor), a corepressor molecule, at the growth hormone TRE. NCoR shows greater promoter occupancy in the absence of T3. When T3 was present, NCoR left the growth hormone TRE. Co-treatment with T3 and either SJ-AK or NH-3 caused the reversal of the T3 effect, increasing NCoR binding at the Growth hormone TRE. Consistent with effects on gene transcription, NH-3 was more efficacious than SJ-AK in its effects on coregulator binding, when both were applied at concentrations higher than saturating. NH-3 has previously been reported to stimulate transient NCoR recruitment to the growth hormone TRE in GC pituitary cells (41). Thus both corepressor recruitment and coactivator loss most likely contribute to the action of SJ-AK and NH-3.
Figure 5.
Cofactor recruitment at growth hormone TRE. Chromatin Immunoprecipitation (ChIP) assay was carried out in GH3 cells. Cells were treated with T3 alone and T3 in concert with SJ-AK or NH-3. A, Enrichment at growth hormone TRE was determined using qRT-PCR analysis of the purified DNA. The PCR data was normalized for respective input samples and enrichment over DMSO control was determined using ΔΔCt method. B, Representative agarose gel image of the regular PCR analysis for the ChIP DNA. * indicates p < 0.05; ** indicates p < 0.01.
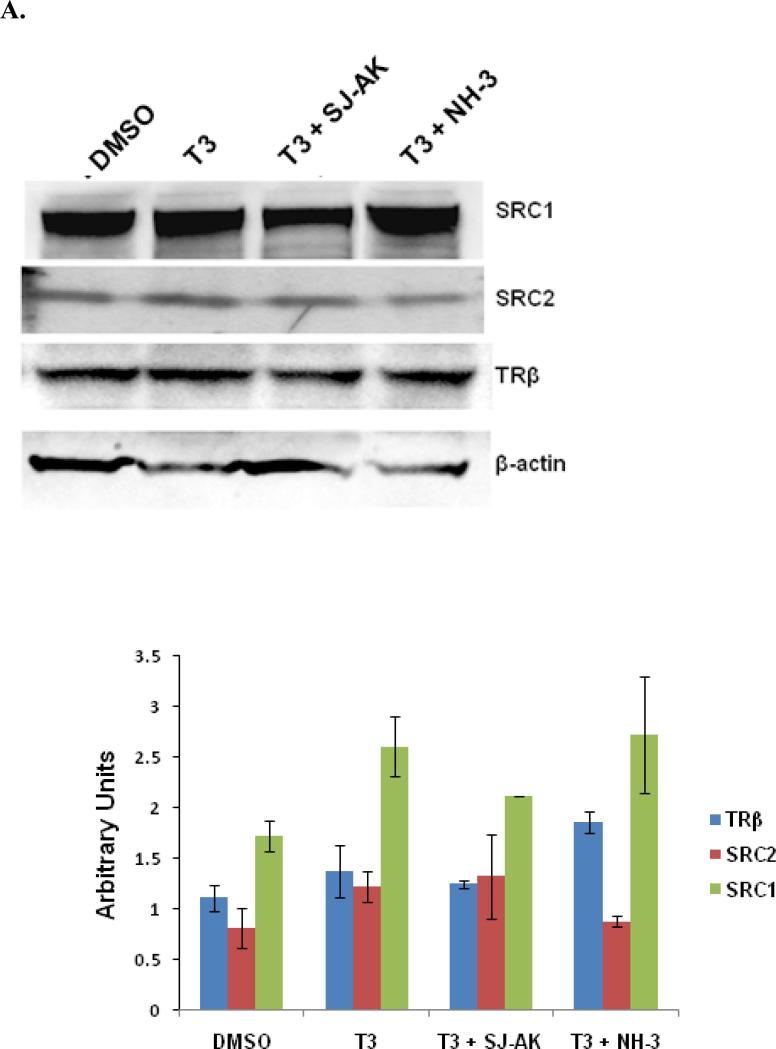
To establish that the observed effects actually represent changes in coregulator recruitment effects rather than changes in the basal expression, the steady state levels of coregulators and TRβ were monitored by western blotting (Figure 6, panel A). While some small changes in expression level are evident, grossly expression of the TRβ and cofactors is maintained under treatment with either inhibitor. Additionally, the nuclear/cytoplasmic partitioning of these proteins (Figure 6, panel B) was assessed. The nuclear enrichment of the cofactors was maintained under treatment by either inhibitor. Thus, changes in cofactor expression or cellular fractionation do not explain the activity of the inhibitor compounds. Hence, the genomic effects we observe are most likely directly related to inhibition of coactivator recruitment.
Figure 6.
Alternate mechanisms of SJ-AK and NH-3 action. A, GH3 cells were treated with T3 alone or in combination with SJ-AK or NH-3. Whole cell lysates were prepared from the cells and run out on SDS-PAGE gels. Western blotting was performed using indicated antibodies. Band quantitation is shown in the histogram plot. B, GH3 cells were fractionated into cytoplasmic and nuclear fractions after hormone and compound treatment. Equal amounts were run on SDS PAGE gels and immunoblotted for indicated proteins. Band quantitation is shown in the histogram plot.
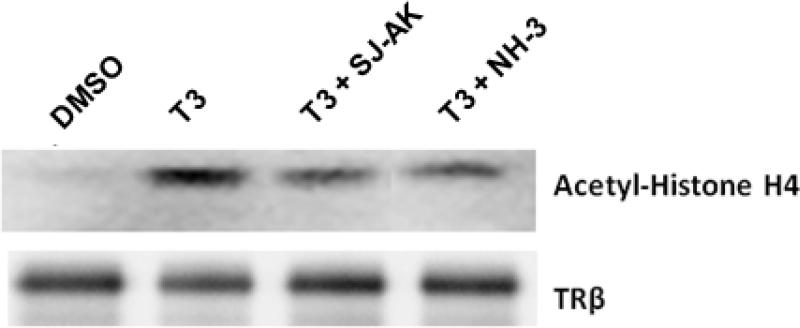
Coactivator proteins such as members of the p160 family and CBP/p300 homologues possess histone acetylation activity, which is responsible for unwinding the chromatin and facilitating transcription. An IP-HAT assay was used to investigate if SJ-AK and NH-3 alter the ability of TR to recruit functional HAT activity (Figure 7). TR and its bound coactivator proteins were immunoprecipitated using an anti- TRβ antibody and the resulting partially purified complexes where used as the HAT activity source in a histone acetylation reaction. Histone H4 acetylation was increased by immunocomplexes isolated from cells treated with T3 and this increase was absent in cells co-treated with T3 and either SJ-AK or NH-3. Taken together, these data show that SJ-AK and NH-3 functionally inhibits T3 signaling at responsive promoters by affecting the balance of co-regulatory proteins present at the responding promoter and therefore their function.
Figure 7.
IP-HAT assay. Nuclear extracts prepared from GH3 cells were immunoprecipitated using an anti-TRβ antibody. The TR immune-complexes were used to carry out acetylation of histone H4 protein. The resulting reaction mixtures were then separated by SDS–PAGE, transferred to membranes, and immunoblotted for acetyl-histone H4 and TRβ.
Functional effects of SJ-AK and NH-3 antagonism of thyroid hormone responses
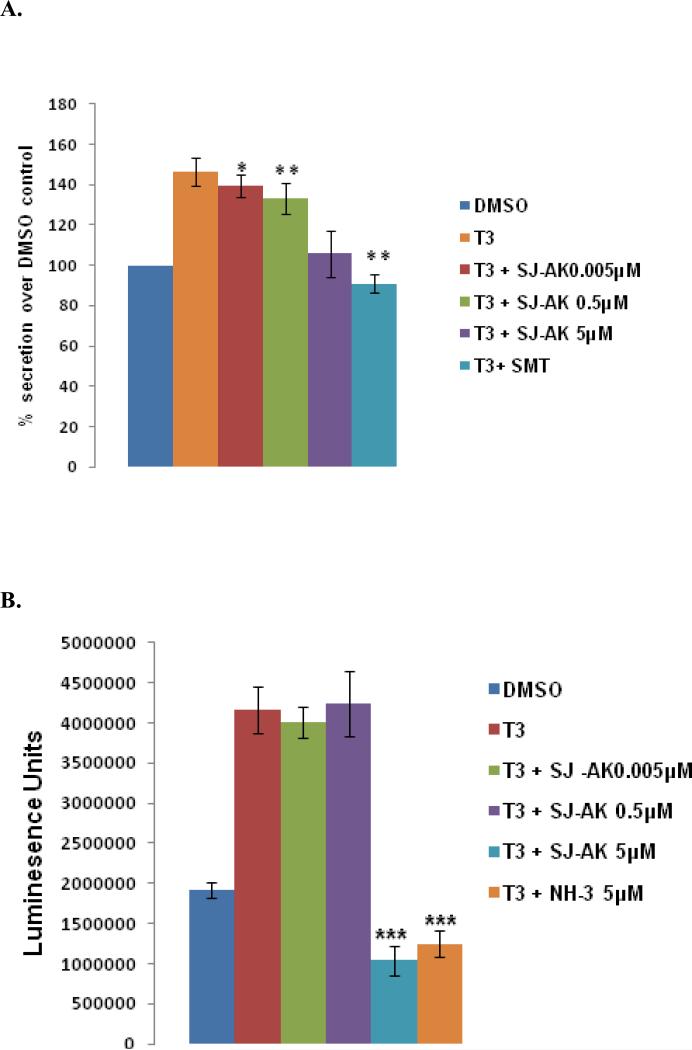
GH3 cells secrete growth hormone and prolactin (42). As growth hormone is a T3 target gene affected by both NH-3 and SJ-AK, we determined the levels of growth hormone in the culture media under different treatment conditions using a sandwich ELISA assay (Figure 7, panel A). T3 increased the secretion of growth hormone by 50% over a 24 hour period. SJ-AK partially antagonized growth hormone secretion, while somatostatin, the natural GH antagonist returned growth hormone secretion to basal levels. This shows that antagonistic effects of these small molecules on T3 induced gene expression are translated into functional consequences at the cellular level.
T3 also induces cell proliferation of GH3 and other pituitary derived cell types, by stimulating the secretion of an autocrine factor from these cells (43). This property of thyroid hormone can be used to identify inhibitors of thyroid hormone signaling in this cell line (44, 45). A cell proliferation assay was carried out to determine if these TR antagonists block the T3 effect (Figure 7, panel B). Treatment of proliferating GH3 cells with T3 alone or T3 in combination with either SJ-AK or NH-3 was followed by culture for 96 hours at which point the cell number was determined. T3 treatment caused a doubling of cell number in this time frame. This effect was fully antagonized by both SJ-AK (5μM) and NH-3 (5μM). At these concentrations, neither compound causes patent cytotoxicity to GH3 cells (data not shown).
Comparison of Two Modes of Thyroid Hormone Antagonism
The studies described herein address the antagonistic efficacy and molecular mechanisms underlying the cellular activity for two diverse TR antagonists: SJ-AK, which directly displaces coactivator proteins from the liganded TR LBD and NH-3, which competes with T3 for binding to the TR-LBD. Both compounds inhibit coactivator recruitment to the TR but use different means of achieving this effect. Multiple approaches were used to demonstrate that these compounds antagonize T3 signaling.
The antagonistic effects of both SJ-AK and NH-3 are evident only at a subset of T3 responsive genes. Neither NH-3 nor SJ-AK achieves a complete antagonism of T3 signaling in a genome-wide scale in HepG2 cells. This finding immediately implies that functional antagonism of thyroid hormone may be much more complex than previously thought and that careful in vivo study is warranted with both classes of inhibitors to elucidate the physiological consequences of partial changes to the T3 genomic responses. Many studies have shown that T3 can induce gene expression in an TR independent manner (46). Our studies suggest that there could likely be a significant number of T3 target genes for which these alternate mechanisms are relevant.
Additionally, the maximal efficacy of SJ-AK and NH-3 was often less than 100% of the T3 induction, even though both responses are saturable (22). Neither of the antagonists completely inhibited coactivator recruitment in the ChIP assay (Figure 5) with some residual SRC-2 binding being seen in the presence of both inhibitors. It is also possible that there are other coregulators whose binding is not inhibited by either NH-3 or SJ-AK. In addition, extent of inhibition of T3 response shows gene to gene variability as seen in the quantitative PCR experiments on the candidate genes we tested by real time PCR (Figure 4C). This suggests varying dependence on coactivator recruitment in the T3 induction of these genes. Meanwhile, the greater efficacy of NH-3 in inhibiting coactivator recruitment in the ChIP assay is reflected in its greater activity in inhibiting histone acetylation over SJ-AK. Taken together these findings imply that the formation of the coactivator containing transcriptional regulatory complex is more mutable than generally appreciated and that interaction mechanisms are likely to include both receptors at the promoter (SJ-AK and NH-3 do not affect RXR binding of coactivators).
Our studies show that SJ-AK has more restricted effects at the genomic level than the ligand antagonist NH-3. The narrower activity profile of SJ-AK potentially has greater utility in achieving selective antagonism of T3 effects rather than a broader antagonism that can be potentially achieved by NH-3. Directed functional studies focusing on specific pathways and physiological responses of T3 will elucidate any subtle and significant differences in SJ-AK and NH-3 action.
Modulation of nuclear receptor mediated transcription has very broad therapeutic utility. In the present study we have validated the nuclear receptor coactivator interaction interface as a target to modulate the activity and function of nuclear receptors in the cellular milieu and proven that this modulation should have functional consequences for the endocrine signaling of T3. Our study sets the ground for further development of such targeted compounds. β-aminoketones will provide enhanced tools to augment our understanding of thyroid hormone biology, provide pharmacological tools to modulate TR function, and help interrogate broadly the mechanism of action of TR and other NRs.
EXPERIMENTAL METHODS
Materials
HepG2 (cat. no. HB 8065) and GH3 (cat. no. CCL-82.1) cell lines were obtained from ATCC. Charcoal, dextran treated fetal bovine serum (cat. no. SH30068.03) was obtained from Hyclone laboratories; DMEM/F12 1:1 (cat. no. 16-405 CV) from Mediatech. RNA extraction kit (RNeasy Mini, cat. no. 74104) and Sybr green qPCR (Quantifast, cat. no. 204056) kit were obtained from Qiagen. Superscript cDNA synthesis kit (cat. no. 18080-051), Dynabeads protein G (cat. no. 100-04D), LDS sample buffer (cat. no. NP0008) were obtained from Invitrogen. Great escape SEAP chemiluminescence kit (cat. no. 631317) was obtained from Clontech. Wizard PCR clean up kit (cat. no. A9281) and Cell titer Glo (cat. no. G7573) was obtained from Promega. Protease Inhibitor cocktail (cat. no. P8340) and acetyl CoA (cat. no. A2056) were obtained from Sigma-Aldrich. Growth hormone Sandwich ELISA kit (cat. no. EZRMGH-45K) was obtained from Millipore; histone H4 (cat. no. M2504S) from New England Biolabs and proteinase K (cat. no. 03115887001) from Roche Normal Goat IgG, (cat. no. sc-2028), anti-SRC-2 (cat.no.sc-8996), anti-NCoR (cat.no.sc-1609) antibodies were obtained from Santa Cruz Biotechnology; normal rabbit IgG (cat. no. #2729), PGC-1 (cat. no. #2178) were obtained from Cell Signaling Technologies; SRC-1 antibody (cat.no.ab84) was from Abcam; anti-acetylated histone H4 (cat. no.06-866) was from Millipore and TRβ antibody (cat.no.MA1-216) was from Affinity Bioreagents.
Cell Culture, treatments and transfections
HepG2, Hek293T, and GH3 cells were cultured in DMEM containing 10% FBS at 37 °C, 100% humidity, and 5% CO2. Thyroid hormone and drug treatments were carried out in DMEM/F-12 (1:1 mixture) supplemented with 10% charcoal stripped FBS for indicated time points. T3 was used at a concentration of 100 nM in all assays. SJ-AK and NH-3 were added to the cells as DMSO solutions at concentrations of 5 μM or 10 μM, as indicated.
For transient transfections, Hek293T cells were plated at 6 × 106 cells dish-1 in 100 mm culture dishes in 10 mL of DMEM/F 12 (1:1 mixture) containing 2.5 mM L-glutamine and 10% heat inactivated CSS (FBS). After 10 h, the cells were at approximately 40-60 % confluence and were transfected by adding a transfection mixture containing CMV-TRβ (5 μg), palindromic TRE (-AGGTCATGACCT-) driven secreted alkaline phosphatase (PAL-SEAP, 10 μg), pSV-beta-galactosidase control vector (1.5 μg) and the transfection reagent Fugene-6 (1:3 w/v of DNA) and incubated overnight. After 18 h, the cells were trypsinized and plated in cell bind surface 96-well cell culture plates (Corning) at 40,000 cells well-1 in DMEM/F 12 medium (65 μL). Six hours after plating, compounds were added to the cell culture medium using a pin-tool (V&P Scientific) to the indicated concentrations.
Cytotoxicity Assay
Raji, HepG2, BJ, and HEK293 (ATCC) cells were grown to 80% confluence, collected, and plated at 700–1000 cells/well in 25 μl of medium/well in 384-well plates (Costar 3712). Compounds were diluted and transferred to cells as described above, and the plates were incubated for 72 h at 37 °C in 5% CO2. CellTiter-Glo (Promega) detection reagent was added following the manufacturer's instructions, and luminescence was measured using an EnVision (PerkinElmer Life Sciences) plate reader.
Cellular Drug Treatments and RNA extraction
HepG2 cells were split into 6 well plates at a density of 1 × 106 cells/well in DMEM/F-12 media with 10% CSS. 24 h later, the cells were treated with T3 or a combination of T3 with SJ-AK or NH-3. 24 h after treatment, cells were harvested for RNA using Qiagen RNeasy Mini (Qiagen), following the manufacturer's instructions. The resulting RNA was treated with DNase I (Invitrogen, Cat. No. 18068-015) to remove contaminating genomic DNA.
Microarray
RNA isolated as described above from HepG2 cells was used for microarray analysis. RNA quality was confirmed by analysis on the Agilent 2100 Bioanalyzer. Total RNA (100ng) was processed in the Hartwell Center microarray core according to the Affymetrix 3’ IVT Express target labeling protocol (https://www.affymetrix.com/support/downloads/manuals/3_ivt_express_kit_manual.pdf). Biotin-labeled cRNA (10μg) was hybridized overnight at 45 °C to the HG-U133Plus2 GeneChip array which interrogates more than 54,000 human transcripts. After staining and washing, arrays were scanned and expression values summarized using the MAS5 algorithm as implemented in the GCOS v1.4 software (Affymetrix, Santa Clara, CA). Detection calls (Present, Absent and Marginal) were determined using the default parameters of the software. Signals were normalized for each array by scaling to a 2% trimmed mean of 500. Prior to statistical analysis the MAS5 signals were variance stabilized by the started logarithm transformation (47). Analysis of differential expression was determined by ANOVA (Partek Genomics Suite v6.5, Partek Inc., St. Louis, MO).
Three completely independent replicates (different sets of cells, treated on different days, with independent variation of media, drug stocks, and all other variables) were carried out, each with internal triple replicates, for each condition. The data were then pooled for further work. The false discovery rate was estimated as described (48) and was controlled at a level of 0.1 or as otherwise stated. An additional filter was applied to select transcripts with robust expression by excluding probe sets called “Absent” across the entire dataset. Probe set annotations were obtained from the Affymetrix website (http://www.affymetrix.com/analysis/index.affx).
Real time PCR
RNA isolated as described above from HepG2 cells was used for microarray analysis. RNA quality was confirmed by analysis on the Agilent 2100 Bioanalyzer. Equal quantities of RNA were then reserve transcribed using Superscript III (Invitrogen), following the manufacturer's instructions. The resulting cDNA was diluted 1:50 in nuclease free water and used in real time PCR reactions with the Quantifast master mix (Qiagen) in an ABI 7900 HT. The following primers were used: 18S, forward: ATCCTCAGTGAGTTCTCCCG, reverse: CTTTGCCATCACTGCCATTA; PCK1, forward: ACGGATTCACCCTACGTGGT, reverse: CCCCACAGAATGGAGGCATTT, G6PC, forward: TCATCTTGGTGTCCGTGATCG, reverse: TTTATCAGGGGCACGGAAGTG and CPT1A, forward: GCCTCGTATGTGAGGCAAAA , reverse: TCATCAAGAAATGTCGCACG. MMP11 (cat. No. QT00024031), SLC16A6 (cat. no. QT00009338), OSTβ (cat. no. QT00239708), CYP24A1 (cat. no. QT00015428), CYP3A4 (cat. no. QT01672608), C8ORF4 (cat. no. QT00214165) real time PCR assays were ordered from Qiagen. The expression of target genes was normalized to the expression of the 18S subunit of the ribosome. The PCR quantitation was carried out using ΔΔCt method and data was expressed as fold change over DMSO treated controls.
The qPCR experiments were carried out completely independently from the microarray experiements. Two completely independent replicates (different sets of cells, treated on different days, with independent variation of media, drug stocks, and all other variables) were carried out, each with internal triple replicates, for each condition. The data were then pooled for further work.
SEAP assay
After transfection and 24 h treatment with T3 or a combination of T3 with SJ-AK o r N H-3, SEAP enzyme activity was measured using the Great Escape SEAP Chemiluminescence detection kit (Clontech), according to the manufacturer's instructions. Briefly, 10 μL of cell culture medium was transferred to each well of a 96 well plate, treated with 30 μL of assay buffer, and mixed gently using orbital shaker. The plate was sealed with aluminum foil, and incubated for 30 min in a 65 °C incubator. The plate was then placed on ice for 3 min, and then allowed to equilibrate to room temperature. 40 μL of CSPD substrate was added to each well, the plate incubated for 20 min at room temperature, and then luminescence signal read using an Envision plate reader (Perkin Elmer). The activity was determined by dividing beta-galactosidase signal as an internal control. Beta-galactosidase activity was determined using according to manufacturer's instructions (Thermo Scientific).
Chromatin Immunoprecipitation assay
GH3 cells, grown to 90% confluence in DMEM/F12 containing 10% charcoal stripped serum, were treated with T3 (100 nM) alone or T3 in combination with SJ-AK (5 μM) or NH-3 (5 μM). After 30 min., protein-DNA crosslinking was carried out using formaldehyde (1 %) for 10 min. at room temperature. The reaction was quenched with 125 mM glycine and the cells were washed twice with PBS and harvested in PBS containing protease inhibitor cocktail (Sigma). Cells were centrifuged (5 min, 450 g) and lysed into cell lysis buffer (50 mm Tris-HCl, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40) for 30 min. on ice. Cells were then centrifuged (1000 g, 5 min) and the resulting nuclear pellet resuspended in nuclear lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) with vortexing. The nuclear lysate was then sheared to 200-1000 bp segments using a Branson 450 sonicator (35% amplitude, 10sec on, 60 sec off, 6 pulses). Sheared DNA was then subjected to centrifugation (x g, 10 min) to pellet cellular debris.
An anti-SRC-2 antibody, anti-NCoR antibody, Normal rabbit IgG, and Normal goat IgG were adsorbed onto protein-G Dynabeads by incubating overnight at 4 °C. The sheared DNA was diluted in ChIP dilution buffer (16.7 mM Tris-HCl (pH 8), 167 mM NaCl, 1.1% Triton X-100, 1.2 mM EDTA, protease inhibitor cocktail) and pre-cleared with protein G Dynabeads (1 h; 4 ° C). Pre-cleared samples were then divided into equal aliquots and treated with antibody loaded Dynabeads for 2 hours. The beads were then washed with RIPA buffer (4 × 0.5 mL; 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 500 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100), LiCl buffer (4 × 0.5 mL, 20mM Tris, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate) and TE buffer (2 × 0.5 mL, 10 mM Tris-HCl pH8, 1 mM EDTA). Bound DNA was then eluted into elution buffer (0.15 mL, 20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 50 mM NaCl, 1% (wt vol-1) SDS, 50 mg ml–1 proteinase K) by vortexing (1300 rpm, 2 h, 68 °C). The eluted DNA was collected and elution was repeated a second time. Both elution fractions were combined and DNA was purified using Wizard PCR clean up kit. The purified DNA was eluted in 50 μL of elution buffer and was used in real time PCR and regular PCR to detect for rat Growth hormone TRE (GH-TRE). The primers used for GH-TRE were: forward: AAGGACACATTGGGTGGTCTCTGT and reverse: CACTGACAGCTTGTGCTGATGGAT. Regular PCR products were run out on agarose gels as shown. Real time PCR data was normalized to respective input for each sample and enrichment at GH-TRE was calculated for each sample over DMSO treated control. The fold enrichment at GH-TRE was plotted as shown.
Nuclear-cytoplasmic fractionation and western blot
GH3 cells were grown to confluence in 150 mm dishes in DMEM/F12 containing 10% CSS. After indicated treatments, cells were harvested into PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4) with protease inhibitor cocktail. The cells were swollen for 15 min on ice in a hypotonic buffer (20 mM HEPES, (pH 7.9), 10 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride). Cells were then lysed by adding 0.5% Nonidet P-40 and the lysates centrifuged (10,000 g, 4 °C, 30 s). The supernatant cytoplasmic fraction was collected and the residual nuclear pellet resuspended in a high salt buffer (20 mM HEPES, (pH 7.9), 0.42 mM NaCl, 1.5 mM MgCl2, 2.5% glycerol, 1 mM EDTA, 1 mm EGTA, 1 mM DTT) with shaking (1 h, 4 °C). The resulting resuspension was centrifuged (13,000 g, 4 °C, 15 min) and the nuclear extract was collected. The fractions were subjected to western blotting using standard procedures with anti-TRβ, SRC-2, SRC1, and PGC-1 antibodies.
IP-HAT assay
Nuclear extracts were prepared from GH3 cells as described above. 1 mg of nuclear extract protein was incubated with an anti-TRβ antibody overnight at 4 °C followed by mixing with Protein A/G agarose beads. The beads were washed 3 times with PBS (3 × 500 μL, 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.47 mM KH2PO4) and once with HAT assay buffer (1 × 500 μL, 50 mM Tris-HCl, (pH 8.0), 10% (v/v) glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM EDTA). The beads were resuspended in HAT assay buffer (30 μl), treated with acetyl CoA (final concentration 10 μM) and Histone H4 (1 μL) and incubated (30 °C, 30 minutes). After incubation, the samples were treated with LDS sample buffer and heated (95 °C, 10 min). Beads were removed by centrifugation and supernatant was subjected to SDS-PAGE (4-12% gel) and Western blotting using anti-acetylated histone H4 and TRβ primary antibodies.
Growth hormone assay
GH3 cells were split into 6-well plates and the cells were treated with thyroid hormone alone or in combination with SJ-AK at indicated concentrations. Somatostatin (SMT) at a concentration of 10 ng ml-1 was used as a positive control. The cell culture media (10 μL) was sampled at 24 h and assayed for growth hormone in a sandwich ELISA methodology (Millipore, Cat. No. EZRMGH-45K) as recommended by the manufacturer. The OD values measured using Spectramax reader (Molecular Devices) correlating with growth hormone quantity are plotted for different experimental treatments.
Cell growth assay
GH3 cells were seeded in 96-well plates (Costar, 3903) at a density of 5000 cells well-1 in DMEM/F12 media containing 10% charcoal stripped serum. The cells were allowed to continue in culture for 72 h. The cells were then treated with T3 or T3 in combination with SJ-AK or NH-3 at specified concentrations. The cells were kept in culture at 37 °C for 96 h. At that point the Cell-Titer Glo (Promega) measurements were carried out using EnVision plate reader (Perkin Elmer) to estimate cell number. The luminescence values were plotted for different treatment conditions.
Supplementary Material
Figure 8.
Functional effects of TR antagonism by SJ-AK and NH-3. A, Growth hormone secretion assay. GH3 cells were cultured with T3 or T3 in combination with SJ-AK or SMT for 24 hours. Cell culture media was sampled and assayed for the presence of growth hormone using a sandwich ELISA kit. Growth hormone secretion as a percentage of DMSO control is plotted. * indicates p < 0.05; ** indicates p < 0.01. B, Cell growth assay. GH3 cells seeded were treated as indicated and kept in culture for 96 hours. At that point, cell viability was assessed using a luminescent measure of ATP content. *** indicates p < 0.005.
Acknowledgements
This work was supported by NIH/NIDDK (R01 DK58080), St Jude Children's Research Hospital, and the American Lebanese Syrian Associated Charities (ALSAC). We acknowledge support from the Hartwell Center in carrying out the microarray studies.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Rodriguez AL, Tamrazi A, Collins ML, Katzenellenbogen JA. Design, synthesis, and in vitro biological evaluation of small molecule inhibitors of estrogen receptor alpha coactivator binding. J Med Chem. 2004;47:600–611. doi: 10.1021/jm030404c. [DOI] [PubMed] [Google Scholar]
- 2.Parent AA, Gunther JR, Katzenellenbogen JA. Blocking estrogen signaling after the hormone: pyrimidine-core inhibitors of estrogen receptor-coactivator binding. J Med Chem. 2008;51:6512–6530. doi: 10.1021/jm800698b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becerril J, Hamilton AD. Helix mimetics as inhibitors of the interaction of the estrogen receptor with coactivator peptides. Angew Chem Int Ed Engl. 2007;46:4471–4473. doi: 10.1002/anie.200700657. [DOI] [PubMed] [Google Scholar]
- 4.Gunther JR, Parent AA, Katzenellenbogen JA. Alternative inhibition of androgen receptor signaling: peptidomimetic pyrimidines as direct androgen receptor/coactivator disruptors. ACS Chem Biol. 2009;4:435–440. doi: 10.1021/cb900043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estebanez-Perpina E, Arnold LA, Nguyen P, Rodrigues ED, Mar E, Bateman R, Pallai P, Shokat KM, Baxter JD, Guy RK, Webb P, Fletterick RJ. A surface on the androgen receptor that allosterically regulates coactivator binding. Proc Natl Acad Sci U S A. 2007;104:16074–16079. doi: 10.1073/pnas.0708036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, Li H, Moore LB, Johnson MD, Maglich JM, Goodwin B, Ittoop OR, Wisely B, Creech K, Parks DJ, Collins JL, Willson TM, Kalpana GV, Venkatesh M, Xie W, Cho SY, Roboz J, Redinbo M, Moore JT, Mani S. The phytoestrogen coumestrol is a naturally occurring antagonist of the human pregnane X receptor. Mol Endocrinol. 2008;22:838–857. doi: 10.1210/me.2007-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H, Wang H, Sinz M, Zoeckler M, Staudinger J, Redinbo MR, Teotico DG, Locker J, Kalpana GV, Mani S. Inhibition of drug metabolism by blocking the activation of nuclear receptors by ketoconazole. Oncogene. 2007;26:258–268. doi: 10.1038/sj.onc.1209788. [DOI] [PubMed] [Google Scholar]
- 8.Arnold LA, Estebanez-Perpina E, Togashi M, Jouravel N, Shelat A, McReynolds AC, Mar E, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. Discovery of small molecule inhibitors of the interaction of the thyroid hormone receptor with transcriptional coregulators. J Biol Chem. 2005;280:43048–43055. doi: 10.1074/jbc.M506693200. [DOI] [PubMed] [Google Scholar]
- 9.Lazar MA. Thyroid hormone action: a binding contract. J Clin Invest. 2003;112:497–499. doi: 10.1172/JCI19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper DS, Greenspan FS, Ladenson PW. The Thyroid Gland. In: Shanahan J, editor. Greenspan's Basic and Clinical Endocrinology. 8th ed. McGraw Hill-Medical; USA: 2007. pp. 209–280. [Google Scholar]
- 11.Sadana P, Zhang Y, Song S, Cook GA, Elam MB, Park EA. Regulation of carnitine palmitoyltransferase I (CPT-Ialpha) gene expression by the peroxisome proliferator activated receptor gamma coactivator (PGC-1) isoforms. Mol Cell Endocrinol. 2007;267:6–16. doi: 10.1016/j.mce.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govindan M, Meng X, Denis CL, Webb P, Baxter JD, Walfish PG. Identification of CCR4 and other essential thyroid hormone receptor co-activators by modified yeast synthetic genetic array analysis. Proc Natl Acad Sci U S A. 2009;106:19854–19859. doi: 10.1073/pnas.0910134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Yang Q, Roeder RG. Dynamic interactions and cooperative functions of PGC-1alpha and MED1 in TRalpha-mediated activation of the brown-fat-specific UCP-1 gene. Mol Cell. 2009;35:755–768. doi: 10.1016/j.molcel.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 15.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 16.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore JM, Guy RK. Coregulator interactions with the thyroid hormone receptor. Mol Cell Proteomics. 2005;4:475–482. doi: 10.1074/mcp.R500001-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2004;2:E274. doi: 10.1371/journal.pbio.0020274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 20.Arnold LA, Estebanez-Perpina E, Togashi M, Shelat A, Ocasio CA, McReynolds AC, Nguyen P, Baxter JD, Fletterick RJ, Webb P, Guy RK. A high-throughput screening method to identify small molecule inhibitors of thyroid hormone receptor coactivator binding. Sci STKE. 2006;2006:l3. doi: 10.1126/stke.3412006pl3. [DOI] [PubMed] [Google Scholar]
- 21.Arnold LA, Kosinski A, Estebanez-Perpina E, Fletterick RJ, Guy RK. Inhibitors of the interaction of a thyroid hormone receptor and coactivators: preliminary structure-activity relationships. J Med Chem. 2007;50:5269–5280. doi: 10.1021/jm070556y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang JY, Arnold LA, Zhu F, Kosinski A, Mangano TJ, Setola V, Roth BL, Guy RK. Improvement of pharmacological properties of irreversible thyroid receptor coactivator binding inhibitors. J Med Chem. 2009;52:3892–3901. doi: 10.1021/jm9002704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estebanez-Perpina E, Arnold LA, Jouravel N, Togashi M, Blethrow J, Mar E, Nguyen P, Phillips KJ, Baxter JD, Webb P, Guy RK, Fletterick RJ. Structural insight into the mode of action of a direct inhibitor of coregulator binding to the thyroid hormone receptor. Mol Endocrinol. 2007;21:2919–2928. doi: 10.1210/me.2007-0174. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen NH, Apriletti JW, Cunha Lima ST, Webb P, Baxter JD, Scanlan TS. Rational design and synthesis of a novel thyroid hormone antagonist that blocks coactivator recruitment. J Med Chem. 2002;45:3310–3320. doi: 10.1021/jm0201013. [DOI] [PubMed] [Google Scholar]
- 25.Busch SJ, Barnhart RL, Martin GA, Flanagan MA, Jackson RL. Differential regulation of hepatic triglyceride lipase and 3-hydroxy-3-methylglutaryl-CoA reductase gene expression in a human hepatoma cell line, HepG2. J Biol Chem. 1990;265:22474–22479. [PubMed] [Google Scholar]
- 26.Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- 27.Darlington GJ, Kelly JH, Buffone GJ. Growth and hepatospecific gene expression of human hepatoma cells in a defined medium. In Vitro Cell Dev Biol. 1987;23:349–354. doi: 10.1007/BF02620991. [DOI] [PubMed] [Google Scholar]
- 28.Ostlund RE, Jr., Seemayer R, Gupta S, Kimmel R, Ostlund EL, Sherman WR. A stereospecific myo-inositol/D-chiro-inositol transporter in HepG2 liver cells. Identification with D-chiro-[3-3H]inositol. J Biol Chem. 1996;271:10073–10078. doi: 10.1074/jbc.271.17.10073. [DOI] [PubMed] [Google Scholar]
- 29.Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- 30.Yen PM, Feng X, Flamant F, Chen Y, Walker RL, Weiss RE, Chassande O, Samarut J, Refetoff S, Meltzer PS. Effects of ligand and thyroid hormone receptor isoforms on hepatic gene expression profiles of thyroid hormone receptor knockout mice. EMBO Rep. 2003;4:581–587. doi: 10.1038/sj.embor.embor862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores-Morales A, Gullberg H, Fernandez L, Stahlberg N, Lee NH, Vennstrom B, Norstedt G. Patterns of liver gene expression governed by TRbeta. Mol Endocrinol. 2002;16:1257–1268. doi: 10.1210/mend.16.6.0846. [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Jiang Y, Meltzer P, Yen PM. Thyroid hormone regulation of hepatic genes in vivo detected by complementary DNA microarray. Mol Endocrinol. 2000;14:947–955. doi: 10.1210/mend.14.7.0470. [DOI] [PubMed] [Google Scholar]
- 33.Chan IH, Privalsky ML. Isoform-specific transcriptional activity of overlapping target genes that respond to thyroid hormone receptors alpha1 and beta1. Mol Endocrinol. 2009;23:1758–1775. doi: 10.1210/me.2009-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loose DS, Cameron DK, Short HP, Hanson RW. Thyroid hormone regulates transcription of the gene for cytosolic phosphoenolpyruvate carboxykinase (GTP) in rat liver. Biochemistry. 1985;24:4509–4512. doi: 10.1021/bi00338a004. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig MG, Basset P, Anglard P. Multiple regulatory elements in the murine stromelysin-3 promoter. Evidence for direct control by CCAAT/enhancer-binding protein beta and thyroid and retinoid receptors. J Biol Chem. 2000;275:39981–39990. doi: 10.1074/jbc.M007529200. [DOI] [PubMed] [Google Scholar]
- 36.Grover GJ, Dunn C, Nguyen NH, Boulet J, Dong G, Domogauer J, Barbounis P, Scanlan TS. Pharmacological profile of the thyroid hormone receptor antagonist NH3 in rats. J Pharmacol Exp Ther. 2007;322:385–390. doi: 10.1124/jpet.106.116152. [DOI] [PubMed] [Google Scholar]
- 37.Oberste-Berghaus C, Zanger K, Hashimoto K, Cohen RN, Hollenberg AN, Wondisford FE. Thyroid hormone-independent interaction between the thyroid hormone receptor beta2 amino terminus and coactivators. J Biol Chem. 2000;275:1787–1792. doi: 10.1074/jbc.275.3.1787. [DOI] [PubMed] [Google Scholar]
- 38.Tang HY, Lin HY, Zhang S, Davis FB, Davis PJ. Thyroid hormone causes mitogen-activated protein kinase-dependent phosphorylation of the nuclear estrogen receptor. Endocrinology. 2004;145:3265–3272. doi: 10.1210/en.2004-0308. [DOI] [PubMed] [Google Scholar]
- 39.Hiroi Y, Kim HH, Ying H, Furuya F, Huang Z, Simoncini T, Noma K, Ueki K, Nguyen NH, Scanlan TS, Moskowitz MA, Cheng SY, Liao JK. Rapid nongenomic actions of thyroid hormone. Proc Natl Acad Sci U S A. 2006;103:14104–14109. doi: 10.1073/pnas.0601600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koenig RJ, Brent GA, Warne RL, Larsen PR, Moore DD. Thyroid hormone receptor binds to a site in the rat growth hormone promoter required for induction by thyroid hormone. Proc Natl Acad Sci U S A. 1987;84:5670–5674. doi: 10.1073/pnas.84.16.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shah V, Nguyen P, Nguyen NH, Togashi M, Scanlan TS, Baxter JD, Webb P. Complex actions of thyroid hormone receptor antagonist NH-3 on gene promoters in different cell lines. Mol Cell Endocrinol. 2008;296:69–77. doi: 10.1016/j.mce.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapp CA, Stachura ME, Tyler JM, Lee YS. GH3 cell secretion of growth hormone and prolactin increases spontaneously during perifusion. In Vitro Cell Dev Biol. 1987;23:686–690. doi: 10.1007/BF02620981. [DOI] [PubMed] [Google Scholar]
- 43.Miller MJ, Fels EC, Shapiro LE, Surks MI. L-triiodothyronine stimulates growth by means of an autocrine factor in a cultured growth-hormone-producing cell line. J Clin Invest. 1987;79:1773–1781. doi: 10.1172/JCI113018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kitamura S, Jinno N, Ohta S, Kuroki H, Fujimoto N. Thyroid hormonal activity of the flame retardants tetrabromobisphenol A and tetrachlorobisphenol A. Biochem Biophys Res Commun. 2002;293:554–559. doi: 10.1016/S0006-291X(02)00262-0. [DOI] [PubMed] [Google Scholar]
- 45.Schriks M, Vrabie CM, Gutleb AC, Faassen EJ, Rietjens IM, Murk AJ. T-screen to quantify functional potentiating, antagonistic and thyroid hormone-like activities of poly halogenated aromatic hydrocarbons (PHAHs) Toxicol In Vitro. 2006;20:490–498. doi: 10.1016/j.tiv.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Davis PJ, Davis FB, Cody V. Membrane receptors mediating thyroid hormone action. Trends Endocrinol Metab. 2005;16:429–435. doi: 10.1016/j.tem.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 47.Durbin BP, Hardin JS, Hawkins DM, Rocke DM. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics. 2002;18(Suppl 1):S105–110. doi: 10.1093/bioinformatics/18.suppl_1.s105. [DOI] [PubMed] [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.