Abstract
We previously reported that wild type (wt) hnRNP G exhibited tumor suppressive activity in human oral squamous cell carcinoma (HOSCC) cell lines lacking hnRNP G. Wt hnRNP G markedly inhibited the proliferation capacity, anchorage independency and in vivo tumorigencity of HOSCC cells and notably enhanced the DNA repair capabilities of these cells. In the present study, we studied the genetic and expression states of hnRNP G in normal, premalignant and malignant human oral tissues to further understand the relationship between the hnRNP G alterations and the development of human oral cancer. To correlate the cancer development and the level of hnRNP G expression, we performed an immunohistochemistry staining of hnRNP G in normal, premalignant and malignant human oral tissues. Moreover, we examined the entire coding regions of hnRNP G from selected samples to understand the cause of the alterations of the gene expression. The expression of hnRNP G was notably decreased or completely abolished in 80% of premalignant-dysplastic and malignant oral epithelial tissues, whereas 100% of normal and 90% of hyperplastic non-dysplastic epithelium showed high level of hnRNP G in the nucleus of the basal cell layers. Approximately 80% of HOSCC lacking the expression of hnRNP G showed genetic alteration in hnRNP G, i.e., point mutation and exonic deletion. This study suggest that genetic alterations and aberrant expression of hnRNP G occurring during oral carcinogenesis might be useful markers for the early detection of human oral cancer.
Keywords: hnRNP G, oral cancer, immunohistochemistry, genetic alteration
Introduction
Heterogeneous nuclear ribonucleoproteins (hnRNPs) are chromatin-associated RNA binding proteins that are initially described as a group of proteins that bound to RNA polymerase II transcripts and suggested to play a major role in RNA processing.1,2 About 30 hnRNP members are identified, and all of these proteins share a common feature of RNA binding domains such as the RBD/RRM/RNP-CS motif (RNA-binding domain/RNA-recognition motif/RNP-consensus motif), the RGG box, and the K-Homology (KH) motif.3 Besides its interaction with RNA, hnRNPs are shown to have protein-protein interaction in the absence of RNA,4 suggesting that hnRNPs may have a role other than RNA processing. In fact, a growing body of evidence supports a notion that hnRNPs are also involved in multiple cellular pathways including telomere biogenesis, chromatin remodeling, transcription, apoptosis, angiogenesis, and metastasis.5,6
Of 30 hnRNPs, hnRNP G (also known as RBMX) is one of the least characterized proteins for its biological functions. hnRNP G is a nuclear protein composed of 391 amino acid (a.a.) residues encompassing the RNA binding domain (a.a. 10–88) at the amino terminus of the protein.7 This structural feature suggests its ability to interact with RNA, which may be required for its role in RNA processing and metabolism.2 A recent study demonstrated that hnRNP G alters pre-mRNA splicing pattern by antagonizing the effects of Tra2β, a splicing activator 8 and a transcription factor.9 Therefore, the level of hnRNP G expression and its biological activities may have profound and multifaceted effects on the global gene expression profile in cells and maintenance of normal cellular homeostasis.
We recently identified two distinct biological functions of hnRNP G in the context of oral carcinogenesis. Ectopic expression of wt hnRNP G in cancer cells expressing negligible amount of hnRNP G resulted in the loss of malignant phenotypes of cells, indicating tumor suppressive functions of hnRNP G.10 Also, we found that hnRNP G is involved in DNA double strand break (DSB) repair by enhancing the fidelity of DNA end-joining activity in cells.11 Currently, we reported that hnRNP G elicits tumor-suppressive activity by transactivating tumor suppressor Txnip gene, suggesting its transcription factor.12 These observations suggest that the inactivation of hnRNP G plays a role in the development of human oral cancer.
In the present study, we performed an immunohistochemistry staining of hnRNP G in normal, premalignant and malignant human oral tissues to correlate the cancer development and the level of hnRNP G expression. Moreover, we examined the entire coding regions of hnRNP G from selected samples to understand the cause of the alterations of the gene expression. We found that The expression of hnRNP G was notably decreased or completely abolished in 80% of premalignant-dysplastic and malignant oral epithelial tissues, whereas 100% of normal and 90% of hyperplastic non-dysplastic epithelium showed high level of hnRNP G in the nucleus of the basal cell layers. Approximately 80% of HOSCC lacking the expression of hnRNP G showed mutations of hnRNP G. Thus, we conclude that genetic alterations and aberrant expression of hnRNP G occurring during oral carcinogenesis may be useful markers for the detection of human oral cancer.
Materials and Methods
Tissue preparation
Oral specimens that were previously collected for diagnostic purposes were obtained from the Oral Pathology Diagnostic Laboratory at the UCLA School of Dentistry. All tissue specimens were collected and processed according to the guidelines of the University of California at Los Angeles Institutional Review Board.
Immunohistochemistry
Immunohistochemical staining was performed as described previously.10 Briefly, four to five-micrometer tissue sections were cut from the block and deparaffinized in an oven at 60°C for 30 minutes followed by rehydration in xylenes and a graded series of ethanol. Antigen was retrieved using citrated buffer in a pressure cooker for 20 minutes. The endogenous peroxidase activity was quenched with 3% solution of H2O2. Tissues were blocked with 10% serum and incubated with hnRNP G antibody (G-17, Santa Cruz) at 1:50 for 90 minutes. The optimal concentration (1:50) of the primary antibody was first established using serially diluted primary antibody along with IgG as a negative control. The secondary antibody was incubated for 1 hour followed by 30 minutes incubation with HRP. Tissues were then developed with the 3,3’-diaminobenzidine (DAB) chromogen substrate for 3–4 minutes.
Evaluation of staining results
The immunohistochemical expression of hnRNP G protein was scored by two independent examiners including one pathologist (R.C.). The level of hnRNP G immunostaining was scored into four subgroups: (a) no staining (−, negative); (b) weak (+); (c) moderate (++); and (d) strong (+++). Strong staining corresponded to that of normal oral epithelium. Three dysplasia samples (#6, #9, and #12) from the previous study were included in this study.10
Genetic analysis of the hnRNP G gene
Intragenic mutation screening of the hnRNP G gene was performed by PCR-SSCP and subsequent sequencing of cloned PCR products. Each exon of hnRNP G gene was amplified by PCR using the primer sets (Table 1). The PCR reactions was performed in a volume of 25 µl consisting of 10 mM Tris HCl (pH 8.3), 50 mM KCl, 5 mM MgCl2, 0.5 units of recombinant Taq DNA polymerase, 0.5 µM of each primer, 1 µCi [alpha-32p]-deoxyadenosine 5'-triphosphate (Amersham), and 100ng of genomic DNA. The reaction was carried out using the following cyclic conditions: 95°C for 3 min of initial denaturation, 35 cycles of 95°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and 72°C for 5 min of final elongation. The PCR products were electrophoresed on a non-denaturing SSCP gel (6% polyacrylamide) for 16 hr at a constant 400 volts. After electrophoresis, the gel was transferred to 3M Whatman paper, dried, and subjected to autoradiography. PCR products showing abnormalities by PCR-SSCP analysis were cloned into pCRII using a TA™ Cloning Kit (Invitrogen) under the conditions recommended by the manufacturer. The nucleotide sequence of the cloned DNA was determined with a Taq dideoxy terminator cyclic sequencing kit on an ABI 377 automatic DNA sequencer (Perkin-Elmer; UCLA Sequencing Core Facility).
Table 1.
Primer sets for the coding exons of the hnRNP G gene.
| Exon | Length (bp) | Primer | Sequence (5’ to 3’) |
|---|---|---|---|
| 2 | 228 | E2F | GTGCGAAAGTAAAAGTGTCC |
| E2R | CTGTACTGCCAGACAACTCA | ||
| 3a | 267 | E3aF | AGTGGAAGGTGAGTTGTCTG |
| E3aR | CTTTCAAAGGTGACAAAAGC | ||
| 3b | 170 | E3bF | GAAACCAACAAATCAAGAGG |
| E3bR | GCTGTCCTAGACCAGACTTG | ||
| 4 | 304 | E4F | GCATTTTGAAGGACCAGTTA |
| E4R | AACTTATACACCAAACCCGA | ||
| 5 | 235 | E5F | TATTTTTGATGCAGATGACG |
| E5R | AGCATCATTCACCTCTCAAT | ||
| 6 | 298 | E6F | TAGGTGATCTAGCCTCCGTA |
| E6R | TGATAAGCTTTCCTTCAACC | ||
| 7 | 289 | E7F | GAGATGTTTATTTGTCCCCA |
| E7R | GTAAGCCACAGAAGCAAAGT | ||
| 8 | 241 | E8F | ACTTTGCTTCTGTGGCTTAC |
| E8R | ACACAGCCTAATAAACTGGC | ||
| 9a | 255 | E9aF | AGAGTTTAAAACCTCCCACC |
| E9aR | GGCTGCTTGAGTAACTGTCT | ||
| 9b | 258 | E9bF | AAGCAGTCGCTATGATGATT |
| E9bR | TAGTATCTGCTTCTGCCTCC | ||
| 9c | 228 | E9cF | ACGTGATTCCTACAGCAGTT |
| E9cR | GGGAACTTAACAGGGAATTT |
Statistical analysis
The numerical staining results were compared between groups with the Kruskal-Wallis test. The association between clinicalpathological factors and the hnRNP G staining was evaluated by ordinal regression models controlling for the type of tissue (normal, leukoplakia, dysplastic, SCC). Analyses were performed with Splus version 6 (Insightful Corp). P-values less than 0.05 were considered significant.
Results
Immunohistochemical expression of hnRNP G in normal, premalignant and malignant oral tissues
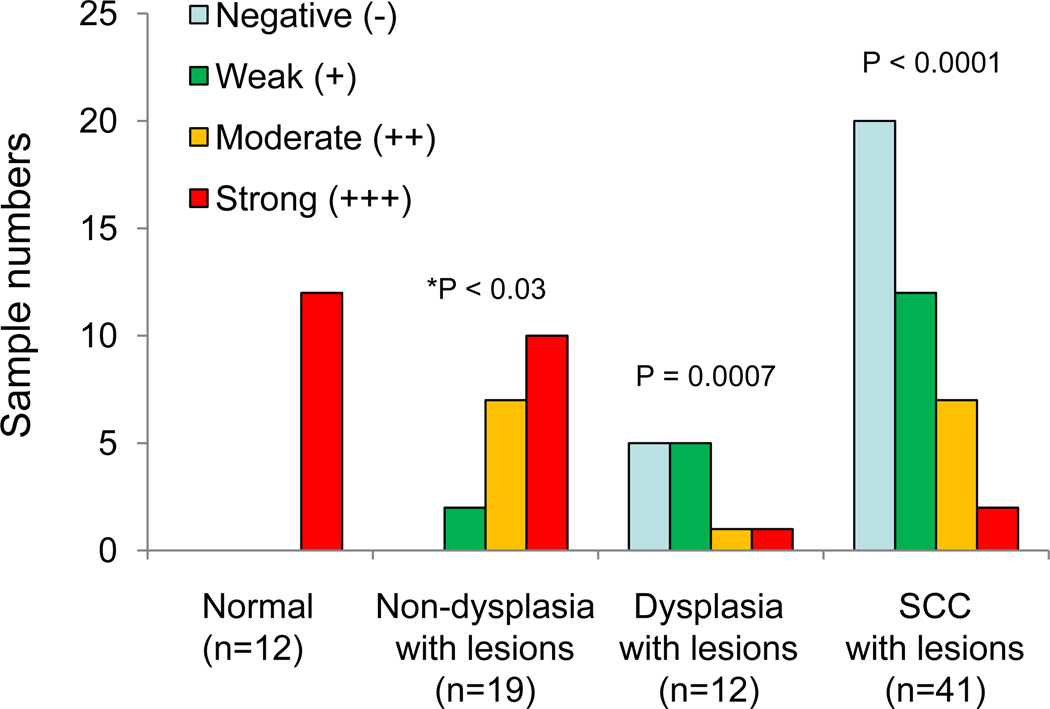
A typical hnRNP G immunohistochemical staining observation in normal, dysplaistic, and cancerous oral epithelium is shown in Figure 1, and the result is summarized in Figure 2. The hnRNP G staining was strong in all the examined normal oral epithelium (12/12 or 100%). All of these staining patterns were prominent in basal cell layer and were found in the nucleus, consistent with the previous findings that hnRNP G is a nuclear protein.11,13 In contrast, only 8% (1/12) of oral dysplasia cases showed strong hnRNP G staining, 8% (1/12) showing moderate staining, 42% (5/12) weak staining, and the rest 42% (5/12) no staining. Of the 41 HOSCC cases, 2 cases (5%) demonstrated strong hnRNP G staining, 7 (17%) with moderate staining, 12 (29%) with weak staining, and 20 (49%) with no staining. The intensity of hnRNP G expression was significantly different in cases with dysplasia (p=0.0007) and HOSCC (p<0.0001) when compared to that of normal, suggesting that the aberrant expression of hnRNP G may occur at the early stage of epithelial cell transformation during oral carcinogenesis. The intensity of staining was not different between subjects with dysplasia versus HOSCC (p=0.83).
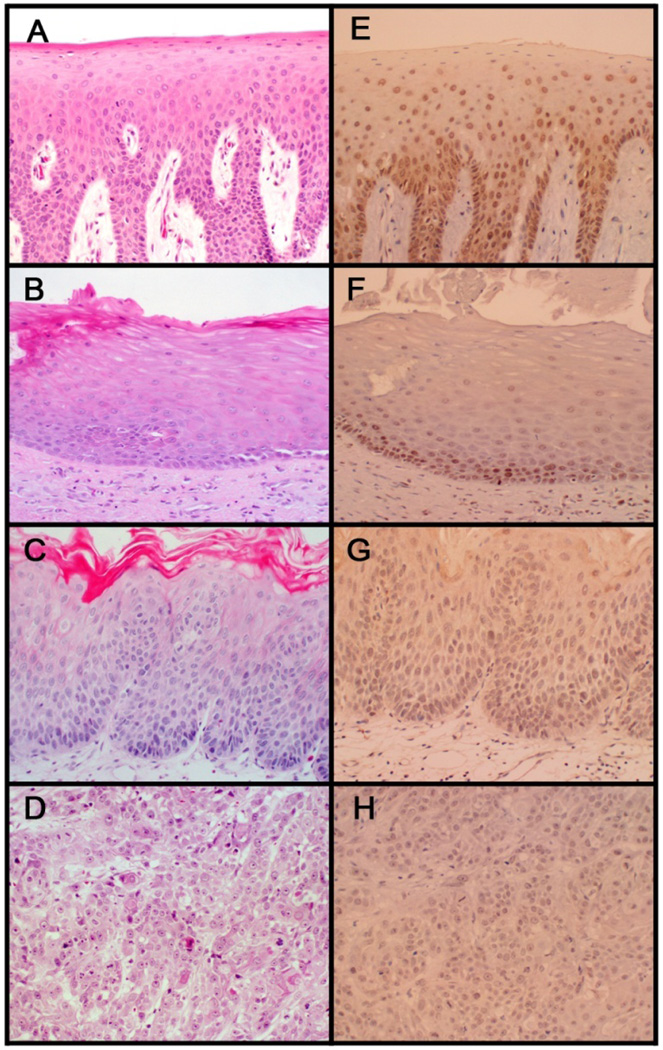
Figure 1. Representative example of hnRNP G immunohistochemical expression.
Strong nuclear staining was observed in normal oral epithelium (E), whereas majority of dysplastic (G) and cancerous (H) oral epithelium exhibited light or no hnRNP G staining. Note strong nuclear staining in keratotic white lesions that are not dysplastic or malignant (F). H&E staining of the respective tissue specimen is also shown (A, B, C, and D).
Figure 2. Correlation of hnRNP G expression with oral mucosal tissues representing the full spectrum of in situ oral carcinogenesis.
* Statistical analysis was performed compared to the staining intensity of normal oral epithelium.
Correlation between hnRNP G expression in oral cancers and the clinicopathological parameters of patients
The relation between clinicopathological parameters and the hnRNP G staining is summarized in Table 2. There was no statistically significant difference between the hnRNP G staining patterns and the clinicopathological parameters of patients (e.g., age, gender, clinical sites, and degree of differentiation).
Table 2.
Clinicopathological parameters and hnRNP G status in the specimens
| Pathologic category |
Age/sex | Degree of differentiation |
hnRNP G status | |
|---|---|---|---|---|
| Protein expressiona | Genetic alteration | |||
| Normal (n=12) | ||||
| 1 | M/39 | +++ | wtb | |
| 2 | M/57 | +++ | wt | |
| 3 | F/25 | +++ | ndc | |
| 4 | M/39 | +++ | nd | |
| 5 | F/65 | +++ | nd | |
| 6 | F/37 | +++ | wt | |
| 7 | F/44 | +++ | wt | |
| 8 | M/70 | +++ | wt | |
| 9 | F/65 | +++ | wt | |
| 10 | M/50 | +++ | wt | |
| 11 | M/65 | +++ | wt | |
| 12 | F/59 | +++ | wt | |
| Keratotic white lesion (n=19) | ||||
| 1 | M/73 | +++ | wt | |
| 2 | F/25 | +++ | nd | |
| 3 | F/76 | +++ | nd | |
| 4 | M/71 | + | wt | |
| 5 | M/70 | +++ | nd | |
| 6 | F/56 | + | nd | |
| 7 | M/66 | ++ | nd | |
| 8 | M/61 | +++ | wt | |
| 9 | M/29 | ++ | nd | |
| 10 | M/41 | ++ | nd | |
| 11 | F/55 | ++ | nd | |
| 12 | M/56 | ++ | nd | |
| 13 | M/54 | ++ | nd | |
| 14 | F/54 | +++ | nd | |
| 15 | F/67 | ++ | nd | |
| 16 | F/65 | +++ | nd | |
| 17 | F/46 | +++ | nd | |
| 18 | F/63 | +++ | wt | |
| 19 | F/67 | +++ | nd | |
| Dysplasia (n=12) | ||||
| 1 | F/83 | Mild | +++ | wt |
| 2 | M/53 | Moderate -severe | + | nd |
| 3 | M/72 | Mild | + | nd |
| 4 | M/53 | Mild | − | nd |
| 5 | M/53 | Mild | − | wt |
| 6 | M/77 | Mild | + | nd |
| 7 | M/54 | Mild | − | del.d exon 3, 4 |
| 8 | nae/na | Moderate -severe | + | nd |
| 9 | F/70 | Moderate -severe | − | nd |
| 10 | M/54 | Moderate -severe | ++ | wt |
| 11 | M/53 | Severe | − | wt |
| 12 | M/38 | Severe | + | wt |
| Squamous cell carcinomas (n=41) | ||||
| 1 | F/82 | Well | − | del. entire exon |
| 2 | M/75 | Well | − | del. exon 3,4,5,6,7,9 |
| 3 | M/71 | Well | − | del. entire exon |
| 4 | M/71 | Well | − | nd |
| 5 | F/nk | Moderate | ++ | missense mutation |
| 6 | F/76 | Moderate-well | + | wt |
| 7 | M/73 | Moderate | ++ | wt |
| 8 | F/79 | Moderate | − | del. exon 2,3,4 |
| 9 | M/70 | Moderate | − | nd |
| 10 | M/67 | Moderate | − | del. exon 7,8 |
| 11 | M/70 | Poor | − | del. exon 8,9 |
| 12 | M/81 | Well | + | wt |
| 13 | M/53 | Well | + | wt |
| 14 | M/71 | Moderate | − | del. exon 3,6,7 |
| 15 | F/53 | Moderate | − | del. entire exon |
| 16 | M/55 | Poor | − | del. exon 3,7,8 |
| 17 | F/78 | Moderate | − | del. entire exon |
| 18 | M/53 | Poor | ++ | wt |
| 19 | M/65 | Well | + | wt |
| 20 | M/66 | Poor | ++ | wt |
| 21 | M/62 | Well | + | del. exon 6,8 |
| 22 | F/52 | Well | − | del. exon 4,9 |
| 23 | F/84 | Moderate | − | del. exon 9 |
| 24 | M/73 | Moderate-poor | − | wt |
| 25 | M/63 | Well | − | wt |
| 26 | F/86 | Moderate | − | nd |
| 27 | F/61 | Moderate | + | del. exon 3,6,8 |
| 28 | M/72 | Well | + | del. exon 4,6, |
| 29 | M/70 | Well-moderate | ++ | wt |
| 30 | F/49 | Moderate | ++ | wt |
| 31 | M/69 | Well-moderate | + | wt |
| 32 | M/70 | Moderate | − | wt |
| 33 | F/53 | Moderate | + | del. exon 3,4,6,8 |
| 34 | M/88 | Moderate | − | del. entire exon |
| 35 | M/94 | Well | − | del. exon 2,3,4,5,6,7 |
| 36 | M/59 | Poor | +++ | wt |
| 37 | F/65 | Moderate | ++ | frameshift & missense |
| 38 | M/67 | Moderate | + | wt |
| 39 | M/50 | Moderate | + | wt |
| 40 | M/66 | Moderate | +++ | wt |
| 41 | M/43 | Moderate | + | frameshift & missense |
The level of hnRNP G immunostaining per each specimen was scored as negative (−), weak (+), moderate (++), or strong (+++) by two independent observations.
wild-type
not determined
deleted
not available
Immunohistochemical expression of hnRNP G in the basal cell layers keratotic white oral epithelium with no dysplastic features
Keratotic white lesions that cannot be scraped off with mechanical devices and cannot be characterized clinically or histologically as any other condition are generally termed leukoplakia. When these white lesions are present bilaterally, they are often clinically diagnosed as lichen planus. Oral lichen planus is an inflammatory condition that causes bilateral white striations, papules, or plaques on the buccal mucosa, tongue, and gingivae. Approximately 10% to 15% of these white lesions exhibit histological evidence of dysplasia or malignancy,14 although the evidence for lichen planus to further develop squamous cell carcinoma is speculative due to the lack of supporting data. However, chronic presence of these lesions may warrant clinicians to perform biopsy to rule out possible neoplasm. To examine hnRNP G expression in the keratotic white lesions, we stained for hnRNP G in 19 tissue samples that were clinically diagnosed as either leukoplakia or lichen planus but histologically non-dysplatic or non-malignant (Fig. 1 and 2). We found that 53% (10/19) had a strong staining intensity, 37% (7/19) moderate, 10% (1/19) weak, and 0% (0/19) no staining. Although there was a statistically significant difference in hnRNP G staining intensity between normal oral epithelium and the keratotic white lesions (p=0.03), the positivity of hnRNP G staining was very similar in these two groups (Fig. 2). For example, 90% of these keratotic white lesions exhibited positive staining, whereas 10% of these lesions exhibited no or weak staining. Also, the positive staining was localized in the nucleus of basal cell layers, similar observations that were seen in normal oral epithelium. These results suggest that hnRNP G expression may be confined to the normal histological architectures that do not exhibit any dysplastic or malignant features.
Genetic alteration of the hnRNP G gene in HOSCC
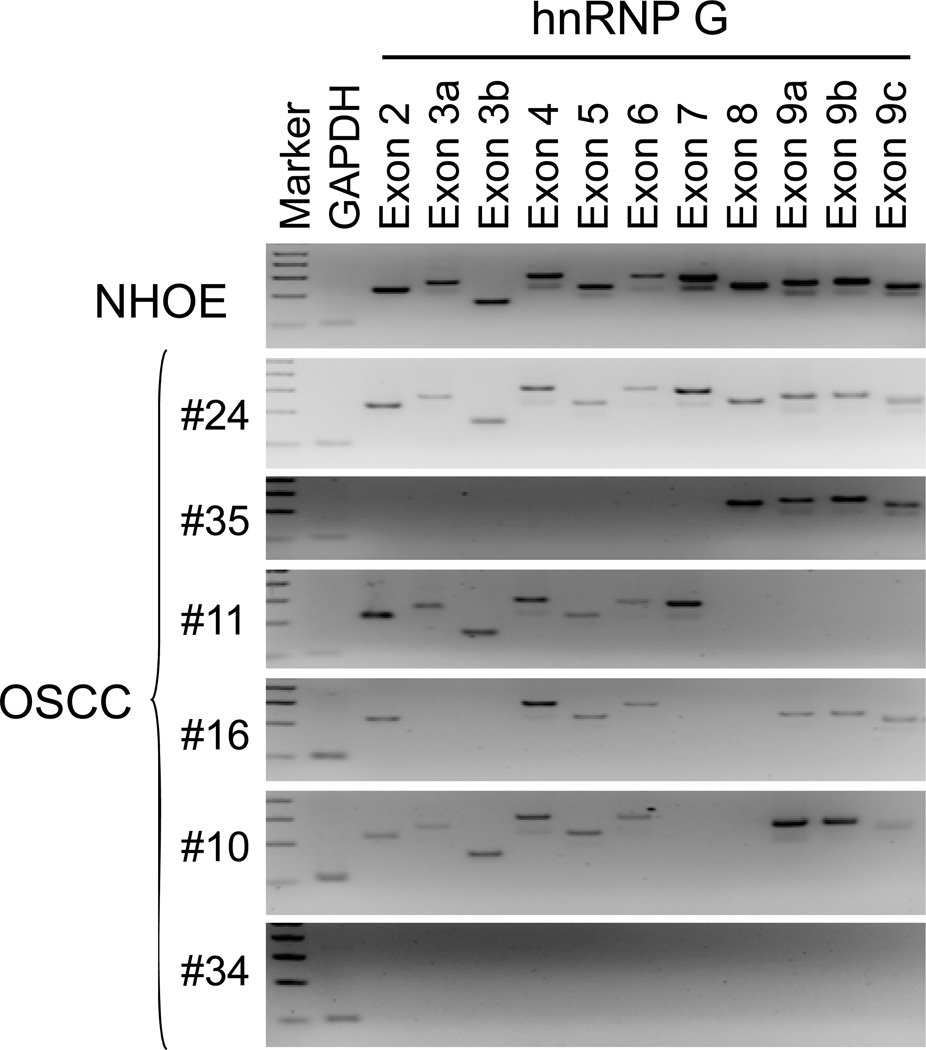
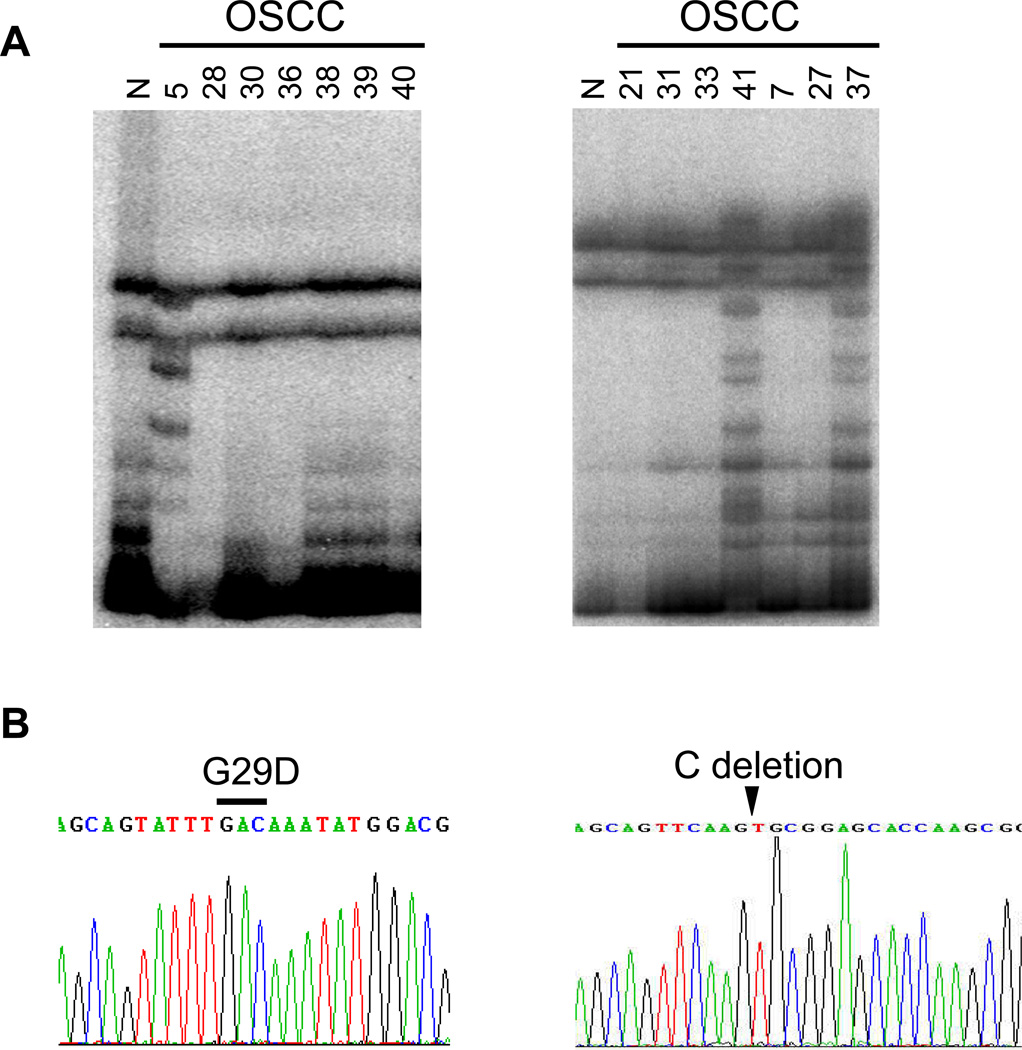
To further understand mechanism underlying the loss of hnRNP G expression, we examined the entire coding region (exon 2–9) of the gene in 17 hnRNP G-negative HOSCC by PCR-SSCP analysis. Abnormal band pattern was not observed in the tested HOSCC. However, we found various exonic deletion of the hnRNP G gene (Table 2 and Fig. 3). Fourteen of 17 showed either a partial or entire gene deletion of hnRNP G. Interestingly, one hnRNP G-negative dysplasia sample was found to have partial exonic deletion. We also examined genetic status of hnRNP G in 21 hnRNP G-positive HOSCC (Table 2). We found a missense mutation from Gly (GGC) to Asp (GAC) at codon 29 in exon 2 of case 5 (Fig. 4). Two HOSCC specimens, case 37 and 41 contain identical mutations, one C deletion at 368 and a base pair substitution at codon 369 from CGC to TGC (Fig. 4). The frameshift mutation abolishes the original stop codon of the hnRNP G gene, leading to complete loss of wt hnRNP G protein. Also, exonic deletions were detected from 4 hnRNP G-positive HOSCC but their expression level is extremely low (Table 2).
Figure 3. Representative example of hnRNP G genomic deletion in hnRNP G-negative HOSCC.
The entire coding region of hnRNP G (exon 2–9) were amplified from the genomic DNAs of normal human oral epithelium (NHOE) and HOSCC tissues. Amplification of GAPDH was served as a control for DNA quality.
Figure 4. hnRNP G mutations in hnRNP G-positive HOSCC.
A. PCR-SSCP analysis of exon 2 (left) and exon 9 (right) of the hnRNP G gene. Abnormal band patterns were noted in the exon 2 of case 5 and in the exon 9 of case 37 and 41. B. DNA sequencing analysis of the hnRNP G gene of case 5 and 37. A missense mutation from Gly (GGC) to Asp (GAC) at codon 29 in exon 2 was found in case 5. Identical mutations, one base pair C deletion at the 1st nucleotide in codon 368 and a base pair substitution from C to T at the 1st nucleotide in codon 369 were found in case 37. Identical C deletion was also detected from case 41 (data not shown).
Discussion
There is an increasing body of evidence that suggests the association of hnRNP family protein expression and oral cancer development. For example, hnRNP B1 was shown to be overexpressed in both oral and lung squamous cell carcinoma.15–17 Lee et al., observed the upregulation of hnRNP C in celecoxib-induced growth inhibition of human oral squamous cell carcinoma using proteomic approach.18 Also, hnRNP E2 was found to be significantly downregulated in human oral cancer tissues.19 Previously, we reported for the first time that hnRNP G has tumor suppressive effects in oral cancers. Here, archived oral mucosal tissues representing the full spectrum of in situ oral carcinogenesis were stained for hnRNP G protein expression by in situ immunohistochemistry. We further examined the level of hnRNP G expression in clinically-diagnosed leukoplakia/lichen planus specimens that are histologically non-dysplastic and non-malignant. We found that the expression of hnRNP G is closely associated with the non-dysplastic and non-malignant features of the tissues. Specifically, high expression of hnRNP G was observed in normal tissue samples, whereas reduced or no expression of hnRNP G is frequently observed in dysplastic and cancerous tissue samples. Since tumor suppressive effect of hnRNP G was demonstrated 10, it may be speculated that the loss of hnRNP G expression could be associated with oral carcinogenesis.
Interestingly, we also observed similar hnRNP G expression patterns in both normal epithelium and keratotic white lesions that exhibit no dysplastic features. Ninety percent of leukoplakia/lichen planus that exhibited non-dysplastic or non-malignant features showed strong staining patterns similar to that of normal tissues. The other 10% showed light or no staining. As 10–20% of keratotic white lesions become dysplasia or cancers, following-up with these cases in the future may help to elucidate possible correlation between hnRNP G expression and the progression of lesions into HOSCC. It may be possible that hnRNP G expression patterns hold a diagnostic value to evaluate possible progression of leukoplakia lesions to the dysplastic and/or malignant lesions.
hnRNP G mutation in human cancer has not been reported. The hnRNP G gene consists of 9 exons, and exon 1 of the gene is not translated. The functionally important domain of hnRNP G, a RNA-binding domain, is located within exon 2 and critical for the RNA-splicing function of hnRNP G.3,5 In the present study, we identified a missense mutation in exon 2. Although the biology effect of the mutant is not tested, we speculate that the mutant may loss its wild-type activities, including tumor suppression and DNA repair.10,11 We also detected an identical frameshift mutation in 2 different HOSCC tissues. This frameshift mutation results in loss of the original stop codon of the hnRNP G gene, suggesting a pathogenic mutation. Our study revealed that exonic deletion (partial and whole) is the most common type of genetic alteration in the hnRNP G gene. Among 22 samples with defective hnRNP G gene, 18 contain the exonic deletion type. Point mutation, such as substitution and single nucleotide deletion was identified only in 3 samples. These data indicate that unlike other cancer related genes, i.e., p53 and ras 20,21, hnRNP G gene has no mutational hot spots. Currently the reason is unclear, but we may expect possible mutational hotspots if more samples are examined.
Our genetic analysis revealed that genomic deletion of hnRNP G is a major responsible mechanism of the loss of hnRNP G expression in the hnRNP G-negative HOSCC. Eighty two % of hnRNP G-negative HOSCC lost either entire or a partial exon of the hnRNP G gene. Unexpectedly, we also observed hnRNP G genomic deletion in 4 hnRNP G-positive HOSCC. We are currently undertaking a study to understand this discrepancy. It is worthwhile to note that their levels of hnRNP G protein expression were extremely low compared to other hnRNP G-positive HOSCC. In addition to mutations and deletions, the loss of expression by promoter methylation plays an important part in the mechanism of gene inactivation. In our observation of series of oral cancers, three cases lacked hnRNP G expression, in the absence of a detectable mutation or genomic deletion. Therefore, methylation status in the promoter region of hnRNP G should be warranted to investigate.
In summary, our findings show a high frequency of hnRNP G protein reduction and loss of expression in precancerous and HOSCC tissue specimens, and suggest that reduction of this protein may play an important role in the early pathogenesis of oral squamous cell carcinomas. All these findings support the concept that hnRNP G gene and protein abnormalities could be used to develop new strategies for an early detection and molecular cancer therapy for a significant subset of human oral tumors.
Acknowledgments
Grant support: This work was supported by the grant DE14147 (K-H.S.) DE17121 (R.H.K.) and DE18295 (M.K.K.) from the National Institute of Dental and Craniofacial Research
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choi YD, Dreyfuss G. Isolation of the heterogeneous nuclear RNA-ribonucleoprotein complex (hnRNP): a unique supramolecular assembly. Proc Natl Acad Sci USA. 1984;81:7471–7475. doi: 10.1073/pnas.81.23.7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dreyfuss G, Matunis MJ, Pinol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 3.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 4.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. hnRNP A1 selectively interacts through its Gly-rich domain with different RNA-binding proteins. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 5.Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, Murray GI. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim Biophys Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Soulard M, Della Valle V, Siomi MC, Pinol-Roma S, Codogno P, Bauvy C, et al. hnRNP G: sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasim MT, Chernova TK, Chowdhury HM, Yue BG, Eperon IC. HnRNP G and Tra2beta: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet. 2003;12:1337–1348. doi: 10.1093/hmg/ddg136. [DOI] [PubMed] [Google Scholar]
- 9.Takemoto T, Nishio Y, Sekine O, Ikeuchi C, Nagai Y, Maeno Y, et al. RBMX is a novel hepatic transcriptional regulator of SREBP-1c gene response to high-fructose diet. FEBS Let. 2007;581:218–222. doi: 10.1016/j.febslet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 10.Shin KH, Kang MK, Kim RH, Christensen R, Park NH. Heterogeneous nuclear ribonucleoprotein G shows tumor suppressive effect against oral squamous cell carcinoma cells. Clin Cancer Res. 2006;12:3222–3228. doi: 10.1158/1078-0432.CCR-05-2656. [DOI] [PubMed] [Google Scholar]
- 11.Shin KH, Kim RH, Kang MK, Kim R, Kim S, Lim PK, et al. p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G. DNA Repair. 2007;6:830–840. doi: 10.1016/j.dnarep.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin KH, Kim RH, Kim R, Kang MK, Park NH. hnRNP G elicits tumor suppressive activity in part by up-regulating the expression of Txnip. Biochem Biophys Res Commun. 2008;372:880–885. doi: 10.1016/j.bbrc.2008.05.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soulard M, Barque JP, Della Valle V, Hernandez-Verdun D, Masson C, Danon F, Larsen CJ. A novel 43-kDa glycoprotein is detected in the nucleus of mammalian cells by autoantibodies from dogs with autoimmune disorders. Exp Cell Res. 1991;193:59–71. doi: 10.1016/0014-4827(91)90538-6. [DOI] [PubMed] [Google Scholar]
- 14.Sonis ST, Fazio RC, Fang L. Principles and Practices of Oral Medicine. 2nd Edition. W.B. Saunders Company; 1995. [Google Scholar]
- 15.Matsuyama S, Goto Y, Sueoka N, Ohkura Y, Tanaka Y, Nakachi K, Sueoka E. Heterogeneous nuclear ribonucleoprotein B1 expressed in esophageal squamous cell carcinomas as a new biomarker for diagnosis. Jpn J Cancer Res. 2000;91:658–663. doi: 10.1111/j.1349-7006.2000.tb00996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sueoka E, Sueoka N, Goto Y, Matsuyama S, Nishimura H, Sato M, et al. Heterogeneous nuclear ribonucleoprotein B1 as early cancer biomarker for occult cancer of human lungs and bronchial dysplasia. Cancer Res. 2001;61:1896–1902. [PubMed] [Google Scholar]
- 17.Wu S, Sato M, Endo C, Sakurada A, Dong B, Aikawa H, et al. hnRNP B1 protein may be a possible prognostic factor in squamous cell carcinoma of the lung. Lung Cancer. 2003;41:179–186. doi: 10.1016/s0169-5002(03)00226-5. [DOI] [PubMed] [Google Scholar]
- 18.Lee EJ, Kim SH, Kwark YE, Kim J. Heterogeneous nuclear ribonuclear protein C is increased in the celecoxib-induced growth inhibition of human oral squamous cell carcinoma. Exp Mol Med. 2006;38:203–209. doi: 10.1038/emm.2006.25. [DOI] [PubMed] [Google Scholar]
- 19.Roychoudhury P, Paul RR, Chowdhury R, Chaudhuri K. HnRNP E2 is downregulated in human oral cancer cells and the overexpression of hnRNP E2 induces apoptosis. Mol Carcinog. 2007;46:198–207. doi: 10.1002/mc.20265. [DOI] [PubMed] [Google Scholar]
- 20.Glazko GV, Koonin EV, Rogozin IB. Mutation hotspots in the p53 gene in tumors of different origin: correlation with evolutionary conservation and signs of positive selection. Biochim Biophys Acta. 2004;1679:95–106. doi: 10.1016/j.bbaexp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Feng Z, Hu W, Chen JX, Pao A, Li H, Rom W, et al. Preferential DNA damage and poor repair determine ras gene mutational hotspot in human cancer. J Natl Cancer Inst. 2002;94:1527–1536. doi: 10.1093/jnci/94.20.1527. [DOI] [PubMed] [Google Scholar]