Abstract
Posttranslational carbonylation of proteins by the covalent attachment of the lipid peroxidation product 4-hydroxy-2-nonenal (HNE) is a biomarker of oxidative stress. Tandem mass spectrometry (MS/MS) has become an essential tool for characterization of this modification. Chemical tagging methods have been used to facilitate the immunoaffinity-based enrichment or even quantification of HNE-modified peptides and proteins. With MS/MS spectra of the untagged modified peptides considered as references, a comparative evaluation is presented focusing on the impact of affinity-tagging with four carbonyl-specific reagents (2,4-dinitrophenyl hydrazine, biotin hydrazide, biotinamidohexanoic acid hydrazide and N’-aminooxymethylcarbonylhydrazino D-biotin) on collision-induced dissociation of the tagged HNE-carbonylated peptides. Our study has shown that chemical labeling may not be carried out successfully for all the peptides and with all the reagents. The attachment of a tag usually cannot circumvent the occurrence of strong neutral losses observed with untagged species and, in addition, fragmentation of the introduced tag may also be introduced. Chemical tagging of certain peptides may, nevertheless, afford more sequence ions upon MS/MS than the untagged carbonylated peptide, especially when Michael addition of the lipid peroxidation product occurs on cysteine residues. Therefore, tagging may increase the confidence of identifications of HNE-modified peptides by database searches.
1. Introduction
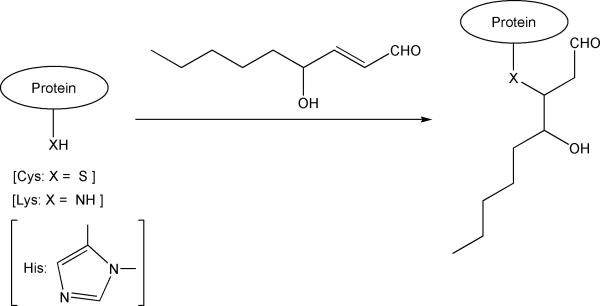
Protein carbonylation has been associated with various human diseases such as Alzheimer's disease, Parkinson's disease, chronic lung disease, chronic renal failure, diabetes, sepsis and sclerosis [1]. Generally, there are several types of amino acid oxidative modifications that can give rise to protein carbonyls [2–4]. Protein carbonyl derivatives can also be formed through reactions with reactive carbonyl compounds produced during oxidative conversion of various biomolecules such as lipids [5]. Among the reactive carbonyl compounds, 4-hydroxy-2-nonenal (HNE) has drawn particular attention and has been the most well studied lipid peroxidation end-product [6]. HNE is formed from polyunsaturated fatty acids present in biological membranes and it reacts readily with nucleophilic groups of protein amino acid side chains. Several studies have shown that covalent attachment of HNE to proteins lead to alteration in their structure and biological activity [7,8]. Modification by HNE occurs on nucleophilic side-chains of amino acid residues primarily via Michael addition or Schiff-base (imine) formation [9,10]. HNE modification through Michael addition involves reaction of the imidazole group of histidine (His), the ε-amino group of lysine (Lys), or the sulfhydryl group of cysteine (Cys) with the C=C double bond of HNE (Fig. 1). Schiff-base is also formed by the reaction of HNE with the ε-amino group of Lys. The reactivity of amino acids toward HNE has shown to be Cys>His>Lys [11]. Michael adducts generally represent 99% of HNE protein modifications, whereas Schiff-base adduct formation is less prevalent even in the presence of excess HNE and does not result in protein carbonylation [12,13].
Fig. 1.
Reaction of nucleophiles in amino acid side chains by Michael addition.
Protein targets of HNE-modification have been identified by 2-D polyacrylamide gel electrophoresis in which mass spectrometry is used merely for protein identification, mostly by peptide-mass fingerprinting and, thus, without seeking modification-specific information at the peptide level [14–16]. The availability of modern tandem mass spectrometers have prompted efforts to utilize them for the modification- and sequence-directed identification of carbonylation through the formation of covalent adducts with HNE. Due to the low abundance of this posttranslational modification, enrichment of HNE-modified peptides usually is required before mass spectrometric analyses [17]. Therefore, there has been much interest recently about development of methods to enrich carbonylated proteins and peptides for mass spectrometric analyses [18–22]. Solid-phase hydrazide chemistry has been employed for the enrichment of HNE-carbonylated peptides [17,20]. The feature of this method is that it recovers the modified species in its native, unlabeled form and may also allow for the use of sophisticated additional chemistry enabling partial 18O-labeling of reactive carbonyl modifications, which produces a unique isotope signature in mass spectra, to detect the modified peptides [23]. However, the solid-phase hydrazide reagent immobilized on controlled pore glass particles is not available commercially and, hence, has to be synthesized for the study, a task that some laboratories may not be prepared to perform. Recently, affinity columns have been made by immobilizing an antibody recognizing HNE-Michael adducts, and the use of these columns also yields samples of enriched untagged peptides [24]. However, the majority of the techniques rely on labeling (“tagging”) the carbonyl group for label-specific immunoaffinity enrichment.
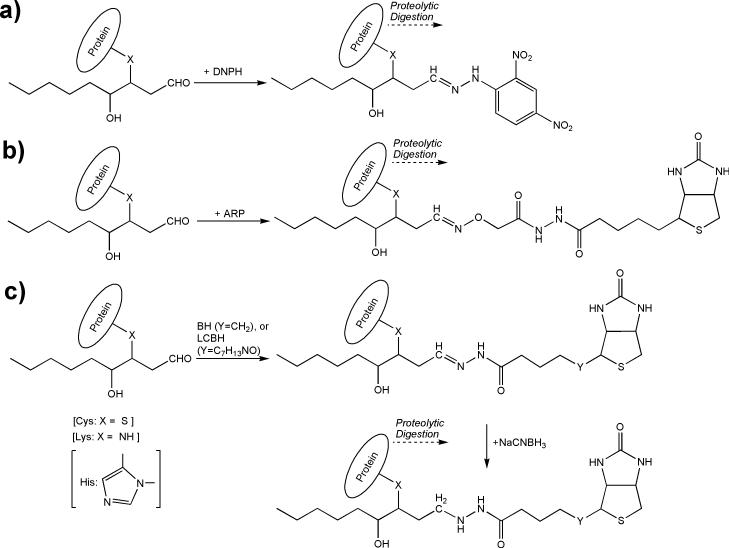
When gel electrophoresis is used for putative identification of carbonylated proteins [25,26], chemical derivatization of protein carbonyl groups are commonly carried out with 2,4-dinitrophenylhydrazine (DNPH, Fig. 2 (a)), a carbonyl-specific tag detected by anti-DNP antibodies (“oxyblot”) [27]. However, even this well-established procedure of redox proteomics have been scrutinized recently concerning methodological details [28]. Nevertheless, immunoaffinity purification of DNPH-derivatized malondialdehyde- and HNE-adducts of peptides has been developed to facilitate discovery-driven exploration of these posttranslational modifications by LC–MS/MS [29]. N’-aminooxymethylcarbonylhydrazino D-biotin, known as aldehyde-reactive probe (ARP, Fig. 2 (b)), has also been used to label HNE-carbonyls for biotin-avidin affinity enrichment and subsequent identification of protein targets in complex proteomes [18,30]. Additional biotin-based reagents such as biotin hydrazide (BH) [22] or biotinamidohexanoic acid hydrazide (a long-chain hydrazide-activated biotin, LCBH) [31] have also been applied for enrichment-enabling immunoaffinity tagging which requires an additional reduction step to prevent the loss of label in subsequent steps in the proteomics workflow, Fig. 2 (c) in studies directed to protein carbonylation and in conjunction with shotgun proteomic analyses. However, no evidence has been sought that tagging reactions perform uniformly for all peptides modified by HNE.
Fig. 2.
Labeling of HNE-carbonylated peptides with (a) DNPH, (b) ARP, and (c) BH and LCBH.
Collision-induced dissociation (CID) has been the prevalent method to obtain MS/MS spectra that potentially afford modification-directed identification at peptide level concerning HNE adducts. Most commonly, yet not exclusively [18,29], ion-trap instruments have been used for this purpose [17–31]. CID of HNE-modified peptides may, however, be confounded by the occurrence of strong neutral loss (NL) obscuring sequence ions necessary for peptide identification and localization of the site of modification [17–33]. Specifically, Michael adducts of HNE to peptides may produce NL of the lipid peroxidation product (–156 Da) upon CID, albeit the degree of this NL is apparently peptide-dependent [34]. Different chemical tagging methods have similarly revealed various extent of NLs of HNE-adducted tags [18,22,27]. ARP-labeled HNE carbonyls, like the relatively large non-cleavable isotope-coded affinity tag (ICAT) also containing a biotin moiety [35], display tag fragmentation upon CID [30] and, thus, may complicate MS/MS analysis of tagged peptides and subsequent database searching [36]. Overall, affinity tags for protein carbonylation were developed to permit selective enrichment, but their impact on MS/MS-based identification of HNE-modified peptides has not been addressed and/or evaluated.
Using selected tryptic peptides of proteins with biological importance as models [30], we compared labeling methods and CID features of untagged and chemically tagged HNE-carbonylated peptides. In particular, we focused on identification of this posttranslational modification by liquid chromatography–electrospray ionization tandem mass spectrometry (LC–ESI-MS/MS) on an ion trap instrument—the most common platform of discovery-driven proteomics in this field. Specifically, we evaluated four different HNE labeling methods (DNPH, ARP, BH and LCBH, as shown in Fig. 2) and the fragmentation of the resultant tagged species upon CID to survey the impact of tagging on the identification of HNE-modification by LC–ESI-MS/MS analyses in the context of shotgun-based redox proteomics.
2. Material and Methods
2.1. Chemicals and reagents
Peptides were custom-synthesized by Synthetic Biomolecules (San Diego, CA). HNE was purchased from Cayman Chemicals (Ann Arbor, MI). ARP was obtained from Dojindo Molecular Technologies (Rockville, MD). BH and LCBH were from Thermo Scientific Pierce (Rockford, IL). LC–MS grade water and acetonitrile were obtained from Fisher Scientific (Pittsburgh, PA). All other chemicals and reagents were purchased from Sigma Aldrich (St. Louis, MO).
2.2. HNE-modification and labeling of peptides
Peptides (1 mg/mL) in 100 mM phosphate buffer (pH 7.4) were incubated with 2 mM HNE in 37 °C for 2 h. Excess HNE was extracted by ethyl acetate. For ARP labeling [30], HNE-modified peptides were treated with 2.5 mM ARP in phosphate buffer (pH 7.4) at 37 °C for 3.5 h. DNPH labeling [27] was performed with 10 mM DNPH in 2 N HCl followed by 1-h incubation at room temperature. Excess DNPH was removed by ethyl acetate extraction. For BH or LCBH labeling [22], HNE-modified peptides (0.5 mg/mL) were mixed with 5 mM BH or 5 mM LCBH in 100 mM sodium acetate buffer (pH 5.5) followed by shaking for 2 h in vials wrapped into aluminum foil. The same volume of 15 mM sodium cyanoborohydride (NaCNBH3) in phosphate buffer was added to the above mixture on ice where the combined solution was kept for 40 min. Peptides after chemical labeling were cleaned up by C18-Ziptip (Millipore, Billerica, MA) and dried in a SpeedVac apparatus (Thermo Scientific Savant, San Jose, CA).
2.3. LC–ESI-MS/MS
An Eksigent nano-LC-2D system (Dublin, CA) with a 15 cm × 75 μm PepMap C18 (LC Packings, Dionex, San Francisco, CA) analytical nanoflow column was used for the analyses. Mobile phases containing solvent A [0.1% acetic acid and 99.9% water (v/v)] and solvent B [0.1% acetic acid and 99.9% acetonitrile (v/v)] were at a constant flow rate of 250 nL/min. Five μL of peptide mixture in 4.8% acetonitrile, 95.1% water and 0.1% acetic acid were automatically loaded onto the IntegraFrit™ sample trap (2.5 cm × 75 μm i.d.) (New Objective, Woburn, MA, USA) for sample concentration and desalting, at a flow rate of 1.5 μl/min in a loading solvent of 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in 94.9% (v/v) water. Separations were performed on the C18 column and equilibrated for 5 min at 4.8% solvent B, followed by a 45-min gradient to 40% solvent B. Solvent B was then held at 40% for 5 min, increasing up to 90% for the next 5 min and finally at 4.8% within 10 min. LC–ESI-MS/MS analysis was performed using a hybrid linear ion trap–Fourier transform ion cyclotron resonance (7-Tesla) mass spectrometer (LTQ-FT, Thermo Fisher, San Jose, CA) equipped with the manufacturer's nanoelectrospray ionization source and operated with the Xcalibur (version 2.0) and Tune Plus (version 2.2) data acquisition software.
For direct infusion analyses, HNE-modified peptides were prepared in methanol/water (50:50, v/v/) containing 1% acetic acid. The sample was electrosprayed at a flow rate of 3 μL/min. Precursor ions were isolated and, then, CID was performed in the linear ion trap. The isolation width was 4.0 Th. Normalized collision energy was set to 35%. MS/MS spectra were obtained by averaging 20-50 scans. For electron capture dissociation (ECD), precursor ions were isolated in the linear ion trap and, then, transferred into the ICR cell. The isolation width was 5.0 Th. ions trapped in the ICR cell were irradiated for 70 ms at –5 V cathode potential. ECD mass spectra were averaged over 40 scans.
Data-dependent mode of acquisition was utilized in which an accurate m/z survey scan was performed in the FT-ICR cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions. FT-ICR full-scan mass spectra were acquired at 50000 mass resolving power (m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping. CID was performed in the linear ion trap using a 4.0-Th isolation width and 35% normalized collision energy with helium as the collision gas. Singly charged precursors and unassigned charge states were excluded from the precursor selection. Also, the precursor ion that had been selected for CID was dynamically excluded from further MS/MS analysis for 60 s.
2.4. Data Analysis
Bioworks (version 3.3, Thermo) was used to generate peak lists for database search. Mascot (Matrix Science, London, UK; version 2.2) was used to identify peptides by searching the IPI human database (version 3.71, 86745 entries). Trypsin was selected as the digesting enzyme and one missed cleavage was allowed. X!Tandem (The GPM, thegpm.org; version 2007.01.01.1) was set up to search a subset of the ipi_HUMAN_v3.71 database. Mascot was searched with parent-ion and fragment-ion mass tolerances of 25 ppm and 0.80 Da respectively. Oxidation of methionine (15.9949 Da) and HNE modification (156.1150 Da), DNPH tagged modification (336.1434 Da), ARP tag (469.2359 Da), BH (398.2353 Da) or LCBH tag (511.3192 Da) on Cys/His/Lys residues were set as variable modifications in the Mascot search according to different labeling experiments. Scaffold (version Scaffold_3_00_01, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted at greater than 95% probability as determined by the Peptide Prophet algorithm [37]. Protein identifications were evaluated by the Protein Prophet algorithm [38]. In addition, MS/MS spectra were manually annotated using theoretical CID-MS/MS fragment ion masses obtained from the MS-Product program available through Protein Prospector (prospector.ucsf.edu). To discard potential false positives, validation was performed according to the flowchart in our earlier report [39].
3. Results and discussion
3.1. Tagging of HNE-carbonylated peptides and effect on CID
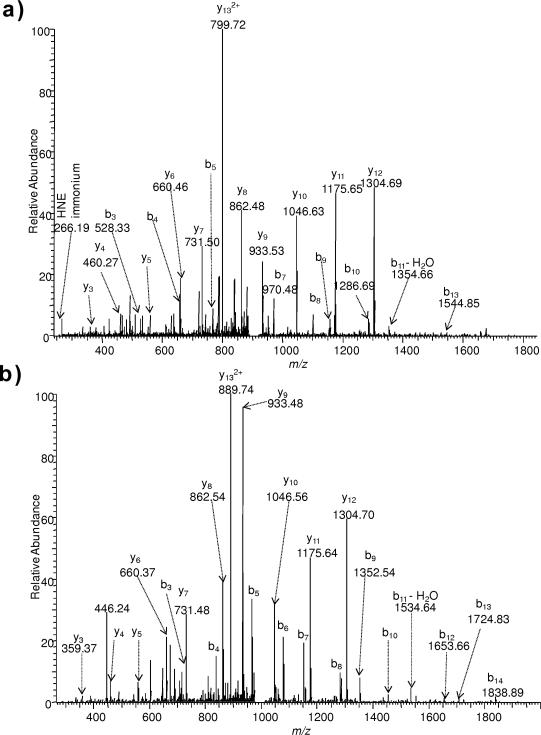
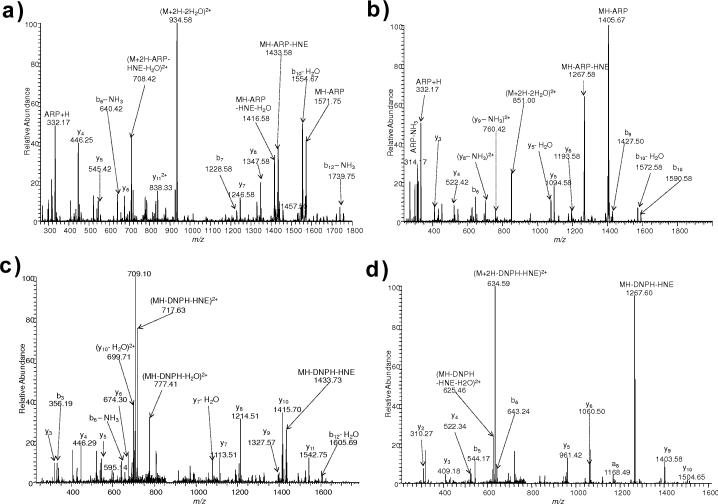
Four model peptides were selected from a previous report that identified protein targets of HNE-modification in human THP-1 monocytic cells after exposure to the lipid peroxidation end-product [30]. These peptides were identified to be carbonylated on His or Cys residues: FSH*EEIAMATVTALR represented a peptide with His, QVQSLTC*EVDALK and VTDDLVC*LVYK exemplified peptides with Cys as site of modification by the lipid peroxidation end-product (asterisks throughout the text indicate HNE-modification as a Michael adduct, Fig. 1), while PGHLQEGFGCVVTNR was chosen in which both His and Cys were susceptible to Michael addition of HNE. Fig. 3 displays the CID-MS/MS spectrum of the [M+2H]2+ ions for FSH*EEIAMATVTALR and DNPH-tagged FSH*EEIAMATVTALR demonstrating extensive fragmentation of the peptide backbone. No significant difference of fragmentation efficiency was found between HNE-modified and DNPH-tagged peptides (or ARP-tagged, BH-tagged and LCBH-tagged peptides, data not shown) in the among the MS/MS spectra; thus, extra steps of chemical labeling had no influence, because the covalently attached HNE was not prone to NL in this peptide.
Fig. 3.
ESI-MS/MS spectra for [M+2H]2+ ions of (a) FSH*EEIAMATVTALR (*represents HNE modification) (m/z 916.4846) and (b) DNPH-tagged FSH*EEIAMATVTALR (m/z 1006.4972).
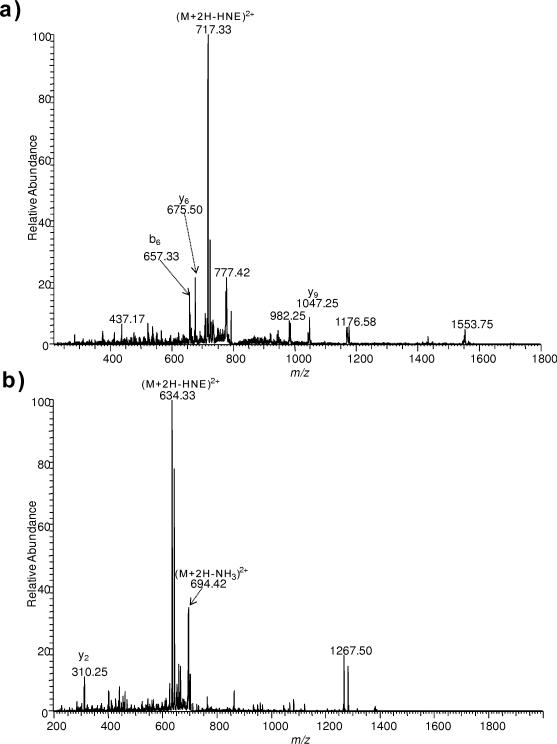
HNE-adducted QVQSLTC*EVDALK and VTDDLVC*LVYK, on the other hand, produced intense NL (–156 Da, corresponding to HNE loss) and this particular ion overwhelmed b- or y-type fragment ions; thus, very limited sequence information would be obtained through MS/MS, which may confound the identification of these HNE-modified peptides upon protein database search [32]. Compared to the MS/MS spectra of the untagged species (Fig. 4 (a, b)), more b- and y-ions of the sequence were detected in QVQSLTC*EVDALK and VTDDLVC*LVYK after ARP labeling (Fig. 5(a, b)). However, fragmentation of the ARP tag (loss of charged ARP and ARP-HNE) was observed, which may also hinder identification of peptides by lowering scores below the thresholds. Unlike NL that makes higher-order data-dependent methods possible [17,20, 32], tag fragmentation may be detrimental for the detection and identification of modified peptides, because the loss of ARP tag is also accompanied by a decrease in charge that would make another round of (MS3) impossible to program in a data-dependent manner. After DNPH labeling (Fig. 5(c, d)), more b- and y-type sequence ions of tagged peptides were observed in comparison with the untagged QVQSLTC*EVDALK and VTDDLVC*LVYK, although NL of DNPH tag (–336 Da) were apparently detected. On the other hand, these Cys-modified peptides could not be identified after BH or LCBH tagging. This might be explained by the prevalence of the cyclic hemiacetal form of the adduct under the condition of the corresponding tagging reactions [40,41]. Another possibility is that BH and LCBH have lower reactivity compared to DNPH or ARP, and it also requires two steps of chemical derivatization (formation of hydrazone linkage between biotin hydrazide and the aldehyde group followed stabilization through the reduction of the acid-labile C=N double bond, see Fig. 2 (c)), leading to diminished reaction yields. Peptides QVQSLTC*EVDALK and VTDDLVC*LVYK contained C-terminal Lys (K), but the latter residues were apparently not reactive to HNE to form stable Michael adducts under the experimental condition we employed because no modified Lys (K) was observed even after tagging with either ARP or DNPH (Table 1). Additionally, Lys was reported to preferentially form reversible Schiff-base with HNE [33] and the reactivity of Cys towards HNE is much higher than that of Lys [11].
Fig. 4.
ESI-MS/MS spectra for [M+2H]2+ ions of (a) QVQSLTC*EVDALK (m/z 795.3689) and (b) VTDDLVC*LVYK (m/z 712.3338).
Fig. 5.
ESI-MS/MS spectra for [M+2H]2+ ions of (a) ARP-tagged QVQSLTC*EVDALK (m/z 951.8689) and (b) ARP-tagged VTDDLVC*LVYK (m/z 868.8338), (c) DNPH-tagged QVQSLTC*EVDALK (m/z 885.4486) and (d) DNPH-tagged VTDDLVC*LVYK (m/z 802.4189).
Table 1.
Summary statistics of database searches (Mascot version 2.2, validated by X!Tandem and Peptide Prophet [37] in Scaffold version 3_00_01) for HNE-modified peptides and tagged HNE-modified peptides after LC-ESI-CID-MS/MS analyses.
| Peptides | Modifications | Charge | Mascot score (identity threshold) | Peptide probability [37] | Comment |
|---|---|---|---|---|---|
| FSH*EEIAMATVTALR | HNE (His) | +2 | 51.9 (36.2) | 95% | |
| +3 | 45.0 (36.2) | 95% | |||
| ARP (His) | +2 | 45.0 (36.5) | 95% | ||
| +3 | 53.5 (36.4) | 95% | |||
| DNPH (His) | +2 | 61.4 (36.7) | 95% | ||
| +3 | 43.2 (36.9) | 95% | |||
| BH (His) | +2 | 30.9(36.6) | 75% | ||
| +3 | 58.2(36.6) | 95% | |||
| LCBH (His) | +2 | 32.0(36.4) | 84% | ||
| +3 | 55.8(36.4) | 95% | |||
| QVQSLTC*EVDALK | HNE (Cys) | +2 | Not identified | – | Strong neutral lossa |
| ARP (Cys) | +2 | 27.1 (36.6) | 62% | Tag fragmentation | |
| DNPH (Cys) | +2 | 26.5 (36.7) | 64% | Neutral loss | |
| +3 | 16.8 (36.7) | 24% | Neutral loss | ||
| VTDDLVC*LVYK | HNE (Cys) | +2 | Not identified | – | Strong neutral lossa |
| ARP (Cys) | +2 | Not identified | – | Tag fragmentation | |
| +3 | Not identified | – | Tag fragmentation | ||
| DNPH (Cys) | +2 | 32.9 (36.6) | 89% | Neutral loss | |
| PGH*LQEGFGCVVTNR | HNE (His) | +2 | 72.8 (35.6) | 95% | Neutral loss |
| +3 | 50.3 (35.5) | 95% | Neutral loss | ||
| ARP (His) | +2, | 42.1 (35.7) | 95% | Tag fragmentation | |
| +3 | 44.9 (35.6) | 95% | Tag fragmentation | ||
| DNPH (His) | +3 | 27.7 (36.5) | 81% | Neutral loss | |
| BH | +3 | 47.0(35.8) | 95% | ||
| PGHLQEGFGC*VVTNR | HNE (Cys) | +2 | Not identified | – | |
| ARP (Cys) | +2 | 47.3 (36.0) | 95% | Neutral loss | |
| +3 | 42.7 (35.7) | 95% | Tag fragmentation | ||
| DNPH (Cys) | +2 | 41.2 (36.6) | 95% | Neutral loss | |
| PGH*LQEGFGC*VVTNR | HNE (Cys, His) | +2 | 29.1 (36.1) | 87% | Neutral lossb |
denotes HNE-modified amino acid residue, Michael adduct.
However, sequence information was obtained via NL-MS3 (see figure in the Supporting Information for details).
Preferential loss of HNE from the modified Cys (C*) was revealed by MS3 displayed in the Supporting Information (Fig. S3).
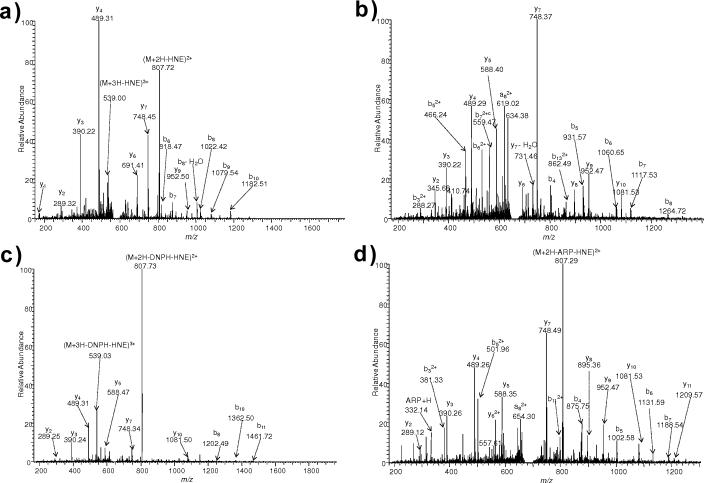
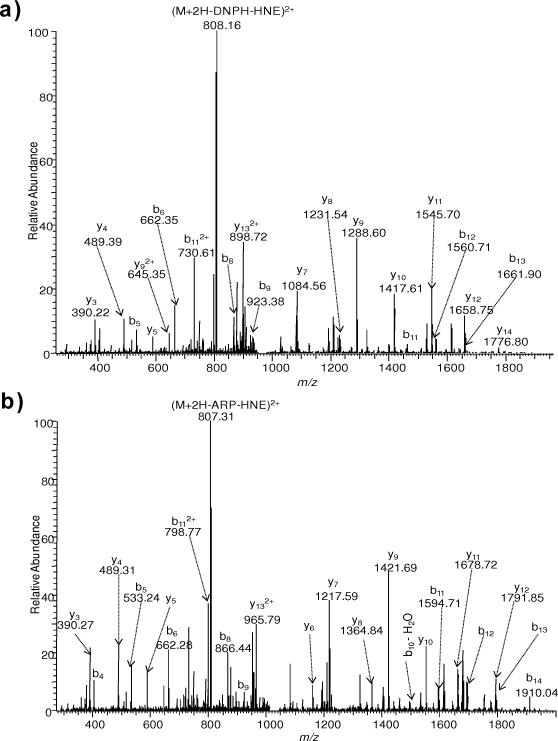
For peptide PGHLQEGFGCVVTNR, both His and Cys are possible sites for HNE modification. MS/MS spectrum of PGH*LQEGFGCVVTNR shows intense sequence ions with some HNE-NL observed at m/z 807.72 (Fig. 6 (a)). After BH labeling, similar fragmentation patterns were observed; however, no NLs were detected (Fig. 6 (b)). LCBH labeling did not afford any tagged HNE-modified peptide, possibly due to the hydrophobic long chain making tag conjugation to the modified peptide even more difficult to complete compared to BH. On the other hand, DNPH and ARP tagging of PGH*LQEGFGCVVTNR resulted in NLs and fragmentation of the tag, respectively (Fig. 6(c, d)). PGHLQEGFGC*VVTNR was not detected by LC-MS without tagging. However, intense b- and y-type sequence ions were observed, besides NL signals of ARP or DNPH tags, after ARP or DNPH labeling of PGHLQEGFGC*VVTNR (Fig. 7(a,b)). ARP or DNPH tags remove the carbonyl group of the Michael adduct through the introduction of a bulky substituent that also introduces steric hindrance to hamper NL. Unfortunately, neither BH nor LCBH tagged peptides were obtained from PGHLQEGFGC*VVTNR.
Fig. 6.
ESI-MS/MS spectra for [M+3H]3+ ions of (a) PGH*LQEGFGCVVTNR (m/z 590.6375) and (b) BH-tagged PGH*LQEGFGCVVTNR (m/z 671.3467), (c) DNPH-tagged PGH*LQEGFGCVVTNR (m/z 650.6541) and (d) ARP-tagged PGH*LQEGFGCVVTNR (m/z 695.3445).
Fig. 7.
ESI-MS/MS spectra for [M+2H]2+ ions of (a) DNPH-tagged PGHLQEGFGC*VVTNR (m/z 975.4825), (b) ARP-tagged PGHLQEGFGC*VVTNR (m/z 1042.0150).
In the untagged His- and Cys-modified peptide PGH*LQEGFGC*VVTNR, the MS/MS spectrum displayed intense NL signals (–156 Da and –312 Da for one and two HNE molecules, respectively) (Fig. 8). HNE-modification of His appeared to be relatively more stable towards CID compared to that of Cys. For peptides that contain more than one HNE labile modification site, NL signals may overwhelm the sequence ions, leading to ambiguous identification of the modification site. Nevertheless, few sequence ions from peptide backbone cleavages were clearly identifiable for PGH*LQEGFGC*VVTNR, which would implicate that co-modification of a His residue enabled the identification of modification on Cys even without tagging, while Cys-only modification (i.e., PGHLQEGFGC*VVTNR) was revealed only after tagging in this peptide sequence (Fig. 7(a,b)). However, additional observations are necessary to make this case. On the other hand, no tagging method was suitable to reveal the presence of this doubly-modified peptide by data-dependent LC–ESI-MS/MS. Therefore, already introduced tags may sterically hinder complete labeling in case of multiply modified peptides.
Fig. 8.
ESI-MS/MS spectra for [M+2H]2+ ion of PGH*LQEGFGC*VVTNR (m/z 963.5133).
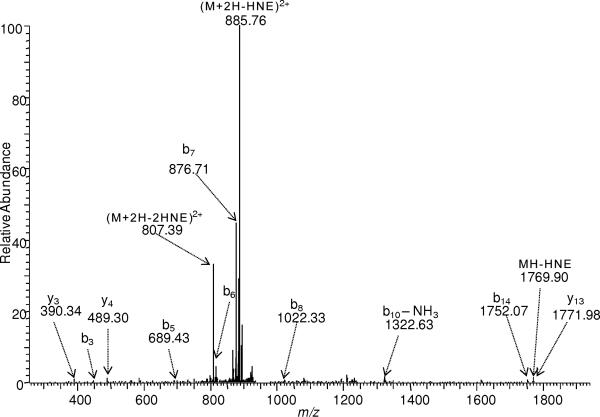
MS3 for the [M+2H-HNE]2+ NL-fragment ions of QVQSLTC*EVDALK, VTDDLVC*LVYK, and PGH*LQEGFGC*VVTNR have shown unambiguous peptide sequences (Fig. S1–S3). For peptide PGH*LQEGFGC*VVTNR, the MS3 spectrum also demonstrated that NL of HNE preferentially occurred from the modified Cys, when both HNE-His and HNE-Cys adducts were present in the peptide (Fig. S3). In addition to an earlier demonstration of ECD on untagged HNE-carbonylated peptides [17], the applicability of ECD to analyze tagged peptides has also been shown (Fig. S4). Although ECD generally retains the labile side-chain modifications including HNE adduction, some loss of HNE may be observed upon ECD [17]. Specifically, some neutral loss and tag fragmentation was also observed in case of the ARP-labeled VTDDLVC*LVYK (Fig. S4); however, it did not interfere significantly with the formation of sequence-revealing ECD fragment ions that also allowed for the localization of the modification.
3.2 Impact of tagging on the identification of site-specific HNE-modification by LC–ESI-MS/MS using CID
As summarized in Table 1, FSH*EEIAMATVTALR could be unequivocally identified as an HNE-modified peptide on His from fructose-bisphosphate aldolase A, an enzyme in the pathway of glycolysis, both in untagged and tagged form. For the HNE-modified Cys-containing peptides QVQSLTC*EVDALK from vimentin and VTDDLVC*LVYK from the signal recognition particle 9kD protein, no identification of modified peptide was made from the sample obtained without performing carbonyl labeling. Even widening the search criteria to allow for low peptide probability (20%), no modified peptide was matched either without carbonyl tagging. The ARP-tagged VTDDLVC*LVYK also displayed intense charged fragmentation of ARP tag and very few sequence ions (Fig. 5(b)); thus, no identification was made by database search. On the other hand, DNPH-tagged QVQSLTC*EVDALK and VTDDLVC*LVYK were identified with probability of 64% and 89%, respectively.
PGH*LQEGFGCVVTNR (from plasminogen activator inhibitor 1) was identified with 95% probability, although NL of HNE (–156 Da) was intense (Fig. 6(a)). ARP, BH, or LCBH labeling of PGH*LQEGFGCVVTNR produced prevalent sequence ions and His was identified as the HNE modification site (95%), while strong NL and charge-species loss of DNP-tagged HNE was found after DNPH labeling (see Fig. 6(c)); hence, corresponding peptide probability was 81%. Apparently because of the propensity of CID-induced NL, no HNE-Cys adducts were identified without carbonyl derivatization. On the other hand, ARP- or DNPH-tagging stabilized these adducts and made their identification and localization possible (with >95% peptide probability), although some NL of DNPH or ARP tags were nevertheless detected (Fig. 7(a, b)). Without tagging the modified peptide, HNE-carbonylated PGH*LQEGFGC*VVTNR was identified at 87% probability, although strong NL of one and two HNE molecules was observed (Fig. 8). On the other hand, database searches did not result in the identification of the doubly-modified peptide when DNPH, ARP, BH and LCBH were used for carbonyl labeling. NL-driven data-dependent MS3 acquisition [17,20,32], ECD [34] (or the related electron transfer dissociation [42], ETD) and NL-driven ECD [17] (or ETD) methods may be superior choices for revealing multiple HNE-modification sites within a peptide.
4. Conclusions
Our study was intended to provide a comparative evaluation and a guide to method selection to pursue redox proteomics with focus on posttranslational protein modifications by lipid peroxidation end-products, specifically by HNE. However, labeling efficiency, enrichment specificity and recovery should also be considered in addition to MS/MS behavior, when a method is selected for such a proteomic study. We have shown that immunoaffinity labeling of the carbonyl group on HNE-modified peptides may not be carried out successfully for all the peptides and with all the reagents under commonly employed reaction conditions. Even with a small set of carbonylated test peptides, BH and LCBH did not perform adequately in this regard, compared to ARP and DNPH. With CID performed in an ion trap to fragment multiply-charged ions produced by ESI, tag fragmentation was observed for ARP-labeled and DNPH-labeled peptides, which might impede the unambiguous identification of HNE modifications by LC–MS/MS followed by routine database search. On the other hand, strong NL of HNE was a liability that could prevent the identification of sequence from CID-MS/MS spectra of HNE-modified Cys-containing peptides without labeling the carbonyl group. ARP- or DNPH-tagging may also increase the probability of identifying HNE-adducted Cys in peptides by decreasing the propensity of NL. Data-dependent NL-based MS3 acquisition method [17,20,32] would also provide identification of Cys-modified peptides and this method may be especially beneficial for peptides containing two or more HNE modifications for which all tagging methods may have shortcomings due to chemical factors. CID-MS/MS generally performs adequately, when HNE-modification occurs on a single His of the peptide, although NL may occur and, hence, the use of NL-MS3 strategy usually provides additional information benefitting identification by protein database search. Ideally, a combination of analyses done both without and with appropriate tagging of HNE-carbonylated peptides would furnish overlapping and, also, complementary information on HNE-modifications of peptides by LC–ESI-MS/MS. The use of MALDI as an alternative method of ionization also is beneficial [30], and validation of search results should be performed even for high-score modified peptides to avoid false positives [24,39]. Immunoaffinity-labeling of HNE-containing peptides introduces, however, extra steps into sample preparation. Thus, it may diminish return on investment in costs and labor compared to the straightforward solid-phase hydrazide chemistry [17,20] or HNE Michael-adduct affinity columns [24] as enrichment methods for mass spectrometry-based proteomics focusing on this important posttranslational modification caused by oxidative stress.
Supplementary Material
Acknowledgments
The presented research was supported by the grant AG025384 from the National Institutes of Health (Bethesda, MD, USA) and by the Robert A. Welch Foundation (endowment number BK-0031).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dalle-Donne I, Giustarini D, Colombo R, Rossi R, Milzani A. Protein carbonylation in human diseases. Trends Mol Med. 2003;9:169–76. doi: 10.1016/s1471-4914(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann N Y Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 3.Hensley K, Floyd RA. Reactive oxygen species and protein oxidation in aging: A look back, a look ahead. Arch Biochem Biophys. 2002;397:377–83. doi: 10.1006/abbi.2001.2630. [DOI] [PubMed] [Google Scholar]
- 4.Uchida K. Histidine and lysine as targets of oxidative modification. Amino Acids. 2003;25:249–57. doi: 10.1007/s00726-003-0015-y. [DOI] [PubMed] [Google Scholar]
- 5.Negre-Salvayre A, Coatrieux C, Ingueneau C, Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins: potential role in diseases and therapeutic prospects for the inhibitors. Br J Pharmacol. 2008;153:6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-Hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Med Res Rev. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 7.Musatov A, Carroll CA, Liu YC, Henderson GI, Weintraub ST, Robinson NC. Identification of bovine heart cytochrome c oxidase subunits modified by the lipid peroxidation product 4-hydroxy-2-nonenal. Biochemistry. 2002;41:8212–20. doi: 10.1021/bi025896u. [DOI] [PubMed] [Google Scholar]
- 8.Bennaars-Eiden A, Higgins L, Hertzel AV, Kapphahn RJ, Ferrington DA, Bernlohr DA. Covalent modification of epithelial fatty acid-binding protein by 4-hydroxynonenal in vitro and in vivo. Evidence for a role in antioxidant biology. J Biol Chem. 2002;277:50693–702. doi: 10.1074/jbc.M209493200. [DOI] [PubMed] [Google Scholar]
- 9.Esterbauer H, Zollner H. Methods for determination of aldehydic lipid peroxidation products. Free Radic Biol Med. 1989;7:197–203. doi: 10.1016/0891-5849(89)90015-4. [DOI] [PubMed] [Google Scholar]
- 10.Poli G, Schaur RJ. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB Life. 2000;50:315–21. doi: 10.1080/713803726. [DOI] [PubMed] [Google Scholar]
- 11.Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143-144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- 12.Bruenner BA, Jones AD, German JB. Direct characterization of protein adducts of the lipid peroxidation product 4-hydroxy-2-nonenal using electrospray mass spectrometry. Chem Res Toxicol. 1995;8:552–9. doi: 10.1021/tx00046a009. [DOI] [PubMed] [Google Scholar]
- 13.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci U S A. 1992;89:4544–8. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perluigi M, Poon HF, Hensley K, Pierce WM, Klein JB, Calabrese V, De Marco C, Butterfield DA. Proteomic analysis of 4-hydroxy-2-nonenal-modified proteins in G93A-SOD1 transgenic mice - A model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;38:960–8. doi: 10.1016/j.freeradbiomed.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 15.Tanito M, Haniu H, Elliott MH, Singh AK, Matsumoto H, Anderson RE. Identification of 4-hydroxynonenal-modified retinal proteins induced by photooxidative stress prior to retinal degeneration. Free Radic Biol Med. 2006;41:1847–59. doi: 10.1016/j.freeradbiomed.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Perluigi M, Sultana R, Cenini G, Di Domenico F, Memo M, Pierce WM, Coccia R, Butterfield DA. Redox proteomics identification of 4-hydroxynonenal-modified brain proteins in Alzheimer's disease: Role of lipid peroxidation in Alzheimer's disease pathogenesis. Proteomics Clin Appl. 2009;3:682–93. doi: 10.1002/prca.200800161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal Chem. 2009;81:782–9. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chavez J, Wu J, Han B, Chung WG, Maier CS. New role for an old probe: affinity labeling of oxylipid protein conjugates by N'-aminooxymethylcarbonylhydrazino D-biotin. Anal Chem. 2006;78:6847–54. doi: 10.1021/ac0607257. [DOI] [PubMed] [Google Scholar]
- 19.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–63. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 20.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal Chem. 2007;79:3747–56. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 21.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79:3342–54. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 22.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–80. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roe MR, McGowan TF, Thomas F, Thompson LV, Griffin TJ. Targeted O-18-labeling for improved proteomic analysis of carbonylated peptides by mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1190–203. doi: 10.1016/j.jasms.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez D, Hernaez ML, Diez A, Puyet A, Bautista JM. Combined proteomic approaches for the identification of specific amino acid residues modified by 4-hydroxy-2-nonenal under physiological conditions. J Proteome Res. 2010;9:5770–81. doi: 10.1021/pr100555v. [DOI] [PubMed] [Google Scholar]
- 25.Sultana R, Newman SF, Huang Q, Butterfield DA. Detection of carbonylated proteins in two-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis separations. Methods Mol Biol. 2009;476:149–59. doi: 10.1007/978-1-59745-129-1_11. [DOI] [PubMed] [Google Scholar]
- 26.Prokai L, Yan LJ, Vera-Serrano JL, Stevens SM, Jr, Forster MJ. Mass spectrometry-based preliminary identification of mitochondrial proteins susceptible to age-related oxidative carbonylation in the rat brain. J Mass Spectrom. 2007;42:1583–89. doi: 10.1002/jms.1345. [DOI] [PubMed] [Google Scholar]
- 27.Levine RL, Williams JA, Stadtman EP, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–57. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 28.Linares M, Marin-Garcia P, Mendez D, Puyet A, Diez A, Bautista JM. Proteomic approaches to identifying carbonylated proteins in brain tissue. J Proteome Res. doi: 10.1021/pr101014e. E-Published 15 January 2011, DOI: 10.1021/pr101014e. [DOI] [PubMed] [Google Scholar]
- 29.Fenaille F, Tabet JC, Guy PA. Immunoaffinity purification and characterization of 4-hydroxy-2-nonenal- and malondialdehyde-modified peptides by electrospray ionization tandem mass spectrometry. Anal Chem. 2002;74:6298–304. doi: 10.1021/ac020443g. [DOI] [PubMed] [Google Scholar]
- 30.Chavez J, Chung WG, Miranda CL, Singhal M, Stevens JF, Maier CS. Site-specific protein adducts of 4-hydroxy-2(E)-nonenal in human THP-1 monocytic cells: protein carbonylation is diminished by ascorbic acid. Chem Res Toxicol. 2010;23:37–47. doi: 10.1021/tx9002462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimsrud PA, Picklo MJ, Griffin TJ, Bernlohr DA. Carbonylation of adipose proteins in obesity and insulin resistance - Identification of adipocyte fatty acid-binding protein as a cellular target of 4-hydroxynonenal. Mol Cell Proteomics. 2007;6:624–37. doi: 10.1074/mcp.M600120-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Stevens SM, Jr., Rauniyar N, Prokai L. Rapid characterization of covalent modifications to rat brain mitochondrial proteins after ex vivo exposure to 4-hydroxy-2-nonenal by liquid chromatography-tandem mass spectrometry using data-dependent and neutral loss-driven MS3 acquisition. J Mass Spectrom. 2007;42:1599–605. doi: 10.1002/jms.1349. [DOI] [PubMed] [Google Scholar]
- 33.Rauniyar N, Prokai L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics. 2009;9:5188–93. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rauniyar N, Prokai-Tatrai K, Prokai L. Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography-electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2007;45:398–410. doi: 10.1002/jms.1725. [DOI] [PubMed] [Google Scholar]
- 35.Borisov OV, Goshe MB, Conrads TP, Rakov VS, Veenstra TD, Smith RD. Low-energy collision-induced dissociation fragmentation analysis of cysteinyl-modified peptides. Anal Chem. 2002;74:2284–92. doi: 10.1021/ac010974p. [DOI] [PubMed] [Google Scholar]
- 36.Qiu Y, Sousa EA, Hewick RM, Wang JH. Acid-Labile Isotope-Coded Extractants: A class of reagents for quantitative mass spectrometric analysis of complex protein mixtures. Anal Chem. 2002;74:4969–79. doi: 10.1021/ac0256437. [DOI] [PubMed] [Google Scholar]
- 37.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 39.Stevens SM, Jr., Prokai-Tatrai K, Prokai L. Factors that contribute to the misidentification of tyrosine nitration by shotgun proteomics. Mol Cell Proteomics. 2008;7:2442–51. doi: 10.1074/mcp.M800065-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 41.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, et al. Stereochemical configuration of 4-hydroxy-2-nonenal-cysteine adducts and their stereoselective formation in a redox-regulated protein. J Biol Chem. 2009;284:28810–22. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–33. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.