Abstract
An enzyme of unknown function within the amidohydrolase superfamily was discovered to catalyze the hydrolysis of N-6-substituted adenine derivatives, several of which are cytokinins. Cytokinins are a common type of plant hormone and N-6-substituted adenines are also found as modifications to tRNA. Patl2390, from Pseudoalteromonas atlantica T6c, was shown to hydrolytically deaminate N-6-isopentenyladenine to hypoxanthine and isopentenylamine with a kcat/Km of 1.2 × 107 M−1 s−1. Additional substrates include N-6-benzyl adenine, cis- and trans-zeatin, kinetin, O-6-methylguanine, N-6-butyladenine, N-6-methyladenine, N,N-dimethyladenine, 6-methoxypurine, 6-chloropurine, and 6-thiomethylpurine. This enzyme does not catalyze the deamination of adenine or adenosine. A comparative model of Patl2390 was computed using the three-dimensional crystal structure of Pa0148 (PDB code: 3PAO) as a structural template and docking was used to refine the model to accommodate experimentally identified substrates. This is the first identification of an enzyme that will hydrolyze an N-6 substituted side chain larger than methylamine from adenine.
With the rapidly increasing volume of protein sequence data available, a need has been created for a comprehensive strategy for analyzing and annotating newly sequenced genomes. Clusters of orthologous groups (COG) were created to aid in the assignment of function for related groups of proteins. Within the amidohydrolase superfamily (AHS) there are 24 clusters of orthologous groups as defined by NCBI. Members of the AHS may have either a mononuclear or binuclear metal center embedded within a (β/α)8-barrel structural fold with metal binding residues found at the C-terminal ends of β-strands 1, 4, 5, 6 and 8 (1). The metal center is responsible for activating a hydrolytic water molecule for nucleophilic attack on amino acids, sugars, nucleic acids and organophosphate esters. Three of these clusters of enzymes are known to catalyze aromatic deamination reactions: cog1001, cog0402 and cog1816. One of these clusters, cog1816, contains nearly 500 bacterial proteins that are currently annotated as adenosine deaminases (2). The prototypical adenine deaminase (ADE) and N-6-methyladenine deaminase (6-MAD) are members of cog1001 (3). Enzymes that deaminate guanine, cytosine, S-adenosylhomocysteine, thiomethyl adenosine, N-formimino-L-glutamate, and 8-oxoguanine are found in cog0402 (4).
A sequence similarity network for cog1816 at an E-value cutoff of 10−70 is presented in Figure 1 (5, 6). Enzymes with experimentally confirmed adenosine deaminase activity belong to Group 5 of cog1816, and enzymes recently identified as adenine deaminases belong to Group 3 (7). The authentic adenine and adenosine deaminases of cog1816, in addition to the other known deaminases from cog0402, share an invariant HxxE motif at the C-terminal end of β-strand 5. Eleven proteins at the periphery of Group 3 have an HxD motif instead of HxxE at this position in the protein sequence. These proteins (Patl2390 from Pseudoalteromonas atlantica, gi|109898705; Leum0809 from Leuconostoc mesenteroides, gi|116617920; Lsa0086 from Lactobacillus sakei, gi|81427699; Cja0578 from Cellvibrio japonicas, gi|192359911; MADE1015570 from Alteromonas macleodii, gi|332142506; Jden_1580 from Jonesia denitrificans DSM 20603, gi|256832804; Glaag_1976 from Glaciecola agarilytica 4H-3-7+YE-5, gi|332173667; ambt_10110 from Alteromonas sp. SN2, gi|332993493; Sros_2140 from Streptosporangium roseum DSM 43021, gi|271963671; Sked_12620 from Sanguibacter keddieii DSM 10542, gi|269794583; and LBU_0899 from Lactobacillus delbrueckii subsp. Bulgaricus 2038, gi|325125754) are highlighted in pink in Figure 1. These eleven proteins originate from organisms with an authentic adenine deaminase from either cog1001 or Group 3 of cog1816. Based on these observations, we predicted that the eleven proteins at the periphery of Group 3 from cog1816 will not deaminate adenine, but will deaminate an adenine-like substrate.
Figure 1.

Sequence similarity network created using Cytoscape(6) of cog1816 from the amidohydrolase superfamily. Each node in the network represents a single sequence, and each edge (depicted as lines) represents the pairwise connection between two sequences at a BLAST E-value of better than 1 × 10−70. Lengths of edges are not significant, except for tightly clustered groups which are more closely related than sequences with only a few connections. Group 5 contains the E. coli adenosine deaminase. The yellow dots in Group 3 represent proteins that will deaminate adenine. The proteins represented by the pink dots at the periphery of Group 3 are characterized in this paper.
Substrate Discovery
We established the substrate profile for one of the proteins at the periphery of Group 3 from cog1816; Patl2390 from P. atlantica T6c, an agarolytic bacterium (8). The gene for Patl2390 was cloned, expressed and the resultant protein purified to homogeneity. The purified protein contained 0.6 equivalents of Zn2+ per monomer. The substrate profile for Patl2390 was determined by monitoring the changes in absorbance after the addition of enzyme to a small library of modified purines, including adenosine, adenine, N-6-methyladenine, and N-6-isopentenyladenine. Based on the results of the initial screen, a larger collection of 6-substititued purines was assayed, including 6-chloropurine, 6-thiomethylpurine, N,N-dimethyladenine, trans-zeatin, kinetin, benzyladenine, 6-methoxypurine, N-butyladenine, O-6-methylguanine and cis-zeatin. The structures of the most active compounds are presented in Scheme 1. The enzyme exhibited a complete lack of activity (<0.005%) for the deamination of adenosine or adenine. Enzymatic conversion of substrate to product was verified by UV-vis spectroscopy and mass spectrometry. A coupled assay with alcohol dehydrogenase was used to detect the formation of methanol from 6-methoxypurine (9). The kinetic constants for the reactions of compounds 1–12 catalyzed by Patl2390 are presented in Table 1. The best substrate for Patl2390 is N-6-isopentenyladenine with a kcat/Km of 1.2 × 107 M−1s−1.
Scheme 1.
Table 1.
Kinetic constants for Patl2390
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1s−1) |
|---|---|---|---|
| 1 | 7.3 ± 0.1 | 0.6 ± 0.1 | 1.2 (0.2) × 107 |
| 2 | 8.5 ± 0.9 | 86 ± 7 | 9.9 (0.3) × 104 |
| 3 | 10.1 ± 0.5 | 9.2 ± 0.9 | 1.1 (0.1) × 106 |
| 4 | 10.2 ± 0.6 | 9 ± 2 | 1.1 (0.3) × 106 |
| 5 | 5.6 ± 0.5 | 9± 2 | 6.2 (0.2) × 105 |
| 6 | 6.5 ± 0.4 | 50 ± 6 | 1.3 (0.2) × 105 |
| 7 | 13.1 ± 0.6 | 10 ± 1 | 1.3 (0.1) × 106 |
| 8 | 11.4 ± 0.6 | 1.5 ± 0.3 | 8 (1) × 106 |
| 9 | 11.6 ± 0.6 | 7± 1 | 1.6 (0.3) × 106 |
| 10 | 13 ± 1 | 35 ± 5 | 3.6 (0.5) × 105 |
| 11 | _ | _ | 3.2 (0.3) × 104 |
| 12 | 13 ± 1 | 38 ± 5 | 3.3 (0.4) × 105 |
Structure of Patl2390
The determination of the three-dimensional structure of Patl2390 by X-ray crystallography has thus far failed. A comparative model was therefore constructed for Patl2390 using the most closely related structure in the PDB database as the template. This protein, Pa0148 from Pseudomonas aeruginosa PAO1 (PDB id: 3OU8) is an adenine deaminase from Group 3 of cog1816 and shares a 45% sequence identity to Patl2390. A protein sequence alignment of Patl2390 and Pa0148, along with four proteins predicted to have the same substrate profile as Patl2390, is presented in supplemental Figure S1. The crystal structure of Pa0148 with adenine bound (PDB id: 3PAO) was used as the template for comparative modeling by satisfaction of spatial restraints, as implemented in Modeller-9v8 (10). In the resulting model of Patl2390, most residues in the proximity of adenine are conserved relative to 3PAO, except for Asp203 and Phe257. The backbone and side chain conformations of segments near the two variable residues (Cys202 to Gln206, Pro252 to Ser261) were refined in the presence of adenine by Modeller for loops and by PLOP for side chains (11). The high-energy intermediate (HEI) (12, 13) library that contains ~22,500 different intermediate forms of 4207 KEGG (14, 15) molecules was docked to this refined model of Patl2390 by DOCK 3.5.54 (16, 17). Six known substrates that were included in the docking library (N-6-benzyladenine, cis-zeatin, kinetin, N-6-isopentenyladenine, trans-zeatin, N-6-methyladenine) were ranked 56, 60, 64, 88, 147, and 328, respectively. The docking poses of these six substrates are similar to the binding mode of adenine in 3PAO. For N-6-benzyladenine, the activated hydroxide attacks C-6 on the re-face of the purine ring, forming a tetrahedral intermediate with R-stereochemistry. Asp203 is hydrogen bonded to protonated N-1 of the purine ring.
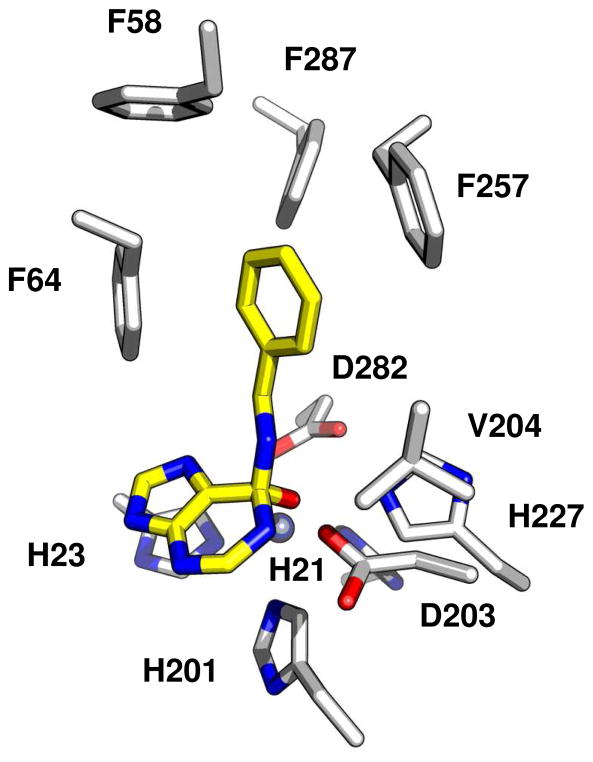
In the comparative model, residues that coordinate the mononuclear metal center (His21, His23, His201 and Asp282) and residues that apparently are involved in substrate recognition (Tyr68, Asp105, Ser174, and Asp283) superimpose with the aligned residues from the structural template. A deletion of two Patl2390 residues in the sequence 252PISNR--FV258 compared with the structural template, changes the second half of a two-turn helix in Pa0148 into a more flexible loop conformation containing residue Phe257. This deletion opens up a hydrophobic pocket that enables the binding of substrates with branched alkyl chains as well as aromatic substituents into the active site. Pi-stacking interactions are possible between the substituents attached to N-6 of the substrate with the side chains of Phe64 and Phe257.
Another region that is different in the structural model of Patl2390, compared to the template, is in the active site following β-strand 5. Instead of the HxxE motif found in Pa0148, Patl2390 has a two residue insertion with the sequence HxDxxQ at the C-terminal of β-strand 5. The invariant histidine in Patl2390 (His227) overlaps in the structural model with the corresponding histidine in Pa0148 (His196). The highly conserved glutamate in the HxxE motif is functionally quite critical since this residue has been shown to be essential for the delivery of a proton to N−1 during the deamination reaction in enzymes from the AHS that react with adenosine, adenine, and guanine (1, 18). The glutamine in the HxDxxQ motif in Patl2390 would be unable to function as a proton donor. The active site of the comparative model of Patl2390 with N-6-benzyladenine bound in the active site is presented in Figure 2.
Figure 2.
A structural model of the active site of Patl2390 with N-6-benzyladenine.
Since Patl2390 does not have an appropriately positioned glutamate after β-strand 5, residues that were likely to fill this function were identified from the initial comparative model. Asp203 from the HxDxxQ motif was mutated to an asparagine (D203N) and Gln206 was mutated to glutamate (Q206E). In addition to these two residues, a conserved glutamate after β-strand 4 that is in the vicinity of the active site was mutated to glutamine and alanine (E176Q and E176A). E176Q was insoluble but E176A could be purified to homogeneity. The kinetic constants for the E176A mutant using N-6-benzyladenine as a substrate were as follows: kcat = 0.013 s−1, Km = 4.9 μM and kcat/Km = 2600 M−1s−1. The value of kcat/Km has been reduced by nearly 4 orders of magnitude. The Q206E mutant exhibited a substantial reduction in the values of the kinetic constants relative to the wild type enzyme. The values of kcat, Km, and kcat/Km for this mutant are 0.11 s−1, 10 μM, and 1.1 × 104 M−1 s−1, respectively. We were unable to detect any activity (< 0.001% of wild-type enzyme) when Asp203 was mutated to an asparagine. Based on these results and the docking of known substrates into the active site of Patl2390, we conclude that Asp203 functions to deliver a proton to N-1 of the substrate during catalysis.
The substrates identified for Patl2390 are known as cytokinins. These compounds promote cell growth and division in plants (19). They are produced by nearly all plants, and have been isolated from red algae (20). Similar compounds are also found as modified adenosines in serine and phenylalanine tRNA (21–23). Naturally occurring cytokinins include N-6-benzyladenine, kinetin, cis-zeatin, trans-zeatin, and N-6-isopentenyladenine. N-6-isopentenyladenine, as well as cis- and trans-zeatin are believed to originate from dimethylallyl pyrophosphate transfer to AMP (21). The effects of cytokinins on plants have been well-studied but the metabolism of these compounds has received less attention (24). A cytokinin oxidase from Zea mays catalyzes the oxidative cleavage of N-6-isopentenyl or zeatin side chains of both the adenine and nucleoside forms (25).
The bacterium P. atlantica T6c was first isolated in association with marine red algae, which are known to contain agar in their cells walls and to produce cytokinins (20). P. atlantica T6c is able to degrade the cell walls of red algae through production of an extracellular agarase, which enables the bacteria to acquire nutrients from the algae (26). The enzyme identified here may function in salvaging of nutrients from algae through the conversion of cytokinins into hypoxanthine, which can then be incorporated into the purine salvage pathway. The reaction catalyzed by Patl2390 with isopentenyl adenine is presented in Scheme 2. The ability to remove a methoxy group from O-6-methylguanine may also indicate a role of this enzyme in DNA repair (27).
Scheme 2.
Strategy for Functional Annotation
Current estimates put the percent of unknown, uncertain or incorrect annotations of bacterial genes at more than 30% (28). For this investigation the NCBI database was used to construct a sequence similarity diagram for cog1816 from the amidohydrolase superfamily, which identified 11 proteins as having a high sequence identity to authentic adenine deaminases of Group 3, but were missing some key catalytic residues. The genes for these proteins are located in the vicinity of genes for other enzymes involved in purine metabolism, including the experimentally verified adenine deaminases. Using adenine as a structural scaffold, a small library of compounds was assembled to ultimately identify the substrate profile for Patl2390. We demonstrated that Patl2390 is capable of accepting cytokinins and several modified purines as substrates. No catalytic activity toward adenine or adenosine was observed. The construction of a comparative structural model for Patl2390 allowed for the identification of the structural rational for the preferential hydrolysis of large substituents attached to C-6 of the purine base.
MATERIALS and METHODS
Materials
All chemicals were purchased from Sigma unless otherwise stated. 6-methoxypurine was obtained from Tokyo Chemical Industry Co. N-6-methyladenine was procured from Spectrum. N-6-butyladenine was bought from Ryan Scientific. Cis-zeatin was ordered from MP Biomedicals and zeatin riboside was acquired from Carbosynth. O-6-methylguanine was obtained from LT Pharmatech.
Cloning and Purification of Patl2390 from Pseudoalteromonas atlantica T6c
Patl2390 was cloned from P. atlantica T6c genomic DNA (ATCC) using restriction sites for BamHI and EcoRI, digested and ligated into a pET-30a(+) vector. The cloned gene fragment was sequenced to verify fidelity of the PCR amplification. Patl2390 in a pET-30a(+) vector was overexpressed in BL21(DE3) cells (Novagen) and purified by affinity chromatography using a HISTrap column (GE Healthcare). Additional information is available in the Supplementary Information.
Activity Screens
Patl2390 (10 nM) was incubated for 16 hours with adenine, 2,6-diaminopurine, 6-mercaptopurine, 6-methylthiopurine, 6-chloropurine, 6-methylpurine, N-6-isopentyl adenine, isoguanine, N-6-methyladenine, zeatin, zeatin riboside, 4-aminopyrimidine, 4,6-diaminopyrimidine, 2,4-diaminopyrimidine, 7-methyladenine, toxopyrimidine, cytosine, 5-hydroxymethylcytosine, 2-chloroadenine and 2-dimethylaminoadenine. Substrate concentration was 80 μM for all screens. Activity was monitored by changes in absorbance between 240–300 nm on a SpectraMax384Plus spectrophotometer (Molecular Devices).
Measurement of Enzymatic Activity
Assays were conducted with 3–200 μM substrate. Dechlorination of 6-chloropurine was monitored by an increase in absorbance at 250 nm. Formation of hypoxanthine from N-6-isopentenyladenine, N-6-methyladenine, trans-zeatin, benzyladenine, 6-methoxypurine, and N-6-butyladenine was monitored at 270 nm. Decreases in absorbance at 275 nm were used to monitor formation of hypoxanthine from cis-zeatin and N,N-dimethyladenine. Conversion of 6-methylthiopurine and kinetin were monitored by decreases in absorbance at 290 nm and 274 nm, respectively. Formation of guanine from O-6-methylguanine was monitored at 255 nm. Difference extinction coefficients were calculated by subtracting the extinction coefficient of the product from the extinction coefficient for the substrate for 6-methylthiopurine (Δε290 = 16500 M−1 cm−1), N-6-methyladenine (Δε270 = 15000 M−1 cm−1), N-6-isopentenyladenine (Δε270 = 11700 M−1 cm−1), N,N-dimethyladenine (Δε275 = 18300 M−1 cm−1), trans-zeatin (Δε270 = 17700 M−1 cm−1), kinetin (Δε274 = 8900 M−1 cm−1), N-6-benzyladenine (Δε270 = 12300 M−1 cm−1), N-6-butyladenine (Δε270 = 12600 M−1 cm−1), cis-zeatin (Δε275 = 12000 M−1 cm−1). For 6-chloropurine (Δε250 = 9600 M−1 cm−1), O-6-methylguanine (Δε255 = 3200 M−1 cm−1), and 6-methoxypurine (Δε270 = 1937 M−1 cm−1), difference extinction coefficients were calculated by subtracting the extinction coefficient for the substrate from the extinction coefficient for the product. Production of the corresponding alkylamine side chains and hypoxanthine were confirmed by mass spectrometry for the reactions of N-6-benzyladenine, N-6-isopentenyladenine, N,N-dimethyladenine, N-6-methyladenine, N-6-butyladenine, cis- and trans-zeatin, 6-chloropurine, 6-methylthiopurine and kinetin. Production of methanol from 6-methoxypurine by 420 nM Patl2390 was detected using alcohol dehydrogenase (4 mg/mL) and monitoring the conversion of NAD+ to NADH at 340 nm (9). Removal of methanol from O-6-methylguanine was confirmed using a coupled assay with guanine deaminase to produce the demethylated and deaminated product, xanthine.
Mutation of E176Q, E176A, D203N and Q206E
Single-site mutations were constructed using the standard QuickChange PCR protocol according to the manufacturer’s instructions. The mutants were expressed and purified using the protocol used for wild-type Patl2390.
Metal Analysis
Metal content of the proteins was determined by ICP-MS (29). Protein samples for ICP-MS were digested with HNO3 by refluxing for ~45 minutes to prevent protein precipitation during the measurement. Protein concentration was adjusted to ~1.0 μM with 1% (v/v) HNO3.
Data Analysis
Sequence alignments were created using ClustalW at http://www.compbio.dundee.ac.uk/JalviewWS/services/ClustalWS. Steady-state kinetic data were analyzed using Softmax Pro version 5.4. Kinetic parameters were determined by fitting the data to Eq. 1 using the nonlinear least-squares fitting program in SigmaPlot 9.0, with A as the substrate concentration, Km is the Michaelis constant, v is the velocity of the reaction and kcat is the turnover number.
| (1) |
Model Building and Computational Docking
The sequence alignment between Patl2390 and Pa0148 was computed by MUSCLE (Multiple Sequence Comparison by Log-Expectation) (30). The original alignment was manually adjusted, so that the active site residue Asp203 in Patl2390 was aligned to Glu199 in Pa0148. The crystal structure of Pa0148 with adenine bound (PDB id: 3PAO) was used as the template. A total of 500 comparative models were generated with the standard “automodel” class in Modeller-9v8 (10). The model with the best DOPE (31) score was selected for refinement of backbone and side chain conformations of residues Cys202 to Gln206 and Pro252 to Ser261, using the “loopmodel” class in Modeller. Sidechains of residues in these two loops were subsequently refined using the “side chain prediction” protocol in PLOP (32), in the presence of adenine. This refinement resulted in two representative models of Patl2390, in which (1) residues Cys202 to Gln206 were refined with respect to the original model and (2) residues Cys202 to Gln206 and Pro252 to Ser261 were refined with respect to the original model. The high-energy intermediate (HEI) (12, 13) library of KEGG (14, 15) molecules was docked to both refined models of Patl2390 by DOCK 3.5.54 (16, 17). The second model yielding a higher enrichment for six known substrates (N-6-benzyladenine, cis-zeatin, kinetin, N-6-isopentenyladenine, trans-zeatin, N-6-methyladenine) was selected to represent the binding mode of N-6-benzyladenine in Patl2390 (Figure 2).
Supplementary Material
ABBREVIATIONS
- ADE
adenine deaminase
- AHS
amidohydrolase superfamily
- ICP-MS
inductively coupled plasma mass spectrometry
Footnotes
This work was supported in part by the NIH (GM 71790, GM 54762, and GM 93342) and the Robert A. Welch Foundation (A-840).
Supporting Information Available: A protein sequence alignment of Patl2390 with five other proteins (Figure S1) and additional information on the cloning and purification of Patl2390 is provided. This material is available free of charge via the Internet.
References
- 1.Seibert CM, Raushel FM. Structural and catalytic diversity within the amidohydrolase superfamily. Biochemistry. 2005;44:6383–6391. doi: 10.1021/bi047326v. [DOI] [PubMed] [Google Scholar]
- 2.Ho MC, Cassera MB, Madrid DC, Ting LM, Tyler PC, Kim K, Almo SC, Schramm VL. Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases. Biochemistry. 2009;48:9618–9626. doi: 10.1021/bi9012484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamat SS, Fan H, Sauder JM, Burley SK, Shoichet BK, Sali A, Raushel FM. Enzymatic deamination of the epigenetic base N-6-methyladenine. J Am Chem Soc. 2011;133:2080–2083. doi: 10.1021/ja110157u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall RS, Agarwal R, Hitchcock D, Sauder JM, Burley SK, Swaminathan S, Raushel FM. Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase. Biochemistry. 2010;49:4374–4382. doi: 10.1021/bi100252s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One. 2009;4:e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goble AM, Zhang Z, Sauder JM, Burley SK, Swaminathan S, Raushel FM. Pa0148 from Pseudomonas aeruginosa Catalyzes the Deamination of Adenine. Biochemistry. 2011;50:6589–6597. doi: 10.1021/bi200868u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yaphe W. The use of agarase from Pseudomonas atlantica in the identification of agar in marine algae (Rhodophyceae) Can J Microbiol. 1957;3:987–993. doi: 10.1139/m57-109. [DOI] [PubMed] [Google Scholar]
- 9.Bergmeyer HU, editor. Methods of Enzymatic Analysis. Academic Press; New York, NY: 1974. [Google Scholar]
- 10.Sali A, Blundell TL. Comparative Protein Modeling by Satisfaction of Spatial Restraints. Journal of Molecular Biology. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 11.Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, McGown G, Thorncroft M, Santibanez-Koref MF, Millington C, Arvai AS, Kroeger MD, Peterson LA, Williams DM, Fried MG, Margison GP, Pegg AE, Tainer JA. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature. 2009;459:808–813. doi: 10.1038/nature08076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hermann JC, Ghanem E, Li YC, Raushel FM, Irwin JJ, Shoichet BK. Predicting substrates by docking high-energy intermediates to enzyme structures. Journal of the American Chemical Society. 2006;128:15882–15891. doi: 10.1021/ja065860f. [DOI] [PubMed] [Google Scholar]
- 13.Hermann JC, Marti-Arbona R, Fedorov AA, Fedorov E, Almo SC, Shoichet BK, Raushel FM. Structure-based activity prediction for an enzyme of unknown function. Nature. 2007;448:775–U772. doi: 10.1038/nature05981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Research. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorber DM, Shoichet BK. Hierarchical docking of databases of multiple ligand conformations. Current Topics in Medicinal Chemistry. 2005;5:739–749. doi: 10.2174/1568026054637683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng EC, Shoichet BK, Kuntz ID. Automated Docking with Grid-Based Energy Evaluation. Journal of Computational Chemistry. 1992;13:505–524. [Google Scholar]
- 18.Mohamedali KA, Kurz LC, Rudolph FB. Site-directed mutagenesis of active site glutamate-217 in mouse adenosine deaminase. Biochemistry. 1996;35:1672–1680. doi: 10.1021/bi9514119. [DOI] [PubMed] [Google Scholar]
- 19.Skoog F, Hamzi HQ, Szweykow Am, Leonard NJ, Carraway KL, Fujii T, Helgeson JP, Loeppky RN. Cytokinins - Structure/Activity Relationships. Phytochemistry. 1967;6:1169. [Google Scholar]
- 20.Yokoya NS, Stirk WA, van Staden J, Novak O, Tureckova V, Pencik A, Strnad M. Endogenous Cytokinins, Auxins, and Abscisic Acid in Red Algae from Brazil. J Phycol. 2010;46:1198–1205. [Google Scholar]
- 21.Kakimoto T. Biosynthesis of cytokinins. J Plant Res. 2003;116:233–239. doi: 10.1007/s10265-003-0095-5. [DOI] [PubMed] [Google Scholar]
- 22.Uziel M, Gassen HG. Phenylalanine transfer ribonucleic acid from Escherichia coli B. Isolation and characterization of oligonucleotides from ribonuclease T-1 and ribonuclease A hydrolysates. Biochemistry. 1969;8:1643–1655. doi: 10.1021/bi00832a046. [DOI] [PubMed] [Google Scholar]
- 23.Nishimura S, Yamada Y, Ishikura H. The presence of 2-methylthio-N6-(delta-2- isopentenyl) adenosine in serine and phenylalanine transfer RNA’s from Escherichia coli. Biochim Biophys Acta. 1969;179:517–520. doi: 10.1016/0005-2787(69)90065-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Smith OC, McChesney J. Biosynthesis and cytokinin activity of 8-hydroxy and 2,8-dihydroxy derivatives of zeatin and N-6(increment-2-isopentenyl)adenine. Biochemistry. 1975;14:3088–3093. doi: 10.1021/bi00685a008. [DOI] [PubMed] [Google Scholar]
- 25.Whitty CD, Hall RH. A cytokinin oxidase in Zea mays. Can J Biochem. 1974;52:789–799. doi: 10.1139/o74-112. [DOI] [PubMed] [Google Scholar]
- 26.Holmstrom C, Kjelleberg S. Marine Pseudoalteromonas species are associated with higher organisms and produce biologically active extracellular agents. FEMS Microbiol Ecol. 1999;30:285–293. doi: 10.1111/j.1574-6941.1999.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 27.Morita R, Nakagawa N, Kuramitsu S, Masui R. An O6-methylguanine-DNA methyltransferase-like protein from Thermus thermophilus interacts with a nucleotide excision repair protein. J Biochem. 2008;144:267–277. doi: 10.1093/jb/mvn065. [DOI] [PubMed] [Google Scholar]
- 28.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall RS, Xiang DF, Xu C, Raushel FM. N-Acetyl-D-glucosamine-6-phosphate deacetylase: substrate activation via a single divalent metal ion. Biochemistry. 2007;46:7942–7952. doi: 10.1021/bi700543x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Science. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. Journal of Medicinal Chemistry. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.