Abstract
Mesenchymal condensation is critical for organogenesis, yet little is known about how this process is controlled. Here we show that Fgf8 and Sema3f produced by early dental epithelium respectively attract and repulse mesenchymal cells, which causes them to pack tightly together during mouse tooth development. Resulting mechanical compaction-induced changes in cell shape induce odontogenic transcription factors (Pax9, Msx1) and chemical cue (BMP4), and mechanical compression of mesenchyme is sufficient to induce tooth-specific cell fate switching. The inductive effects of cell compaction are mediated by suppression of the mechanical signaling molecule RhoA, and its over-expression prevents odontogenic induction. Thus, the mesenchymal condensation that drives tooth formation is induced by antagonistic epithelial morphogens that manifest their pattern-generating actions mechanically via changes in mesenchymal cell shape and altered mechanotransduction.
Keywords: Mesenchymal condensation, organogenesis, epithelial-mesenchymal interactions, mechanical forces, cell shape, Fgf8, Sema3f, Nrp2, Pax9, RhoA, tooth
Mesenchymal condensation, which is critical for the formation of many organs including tooth, cartilage, bone, muscle, tendon, kidney, and lung, occurs when previously dispersed mesenchymal cells gather together to differentiate into a single tissue type. It represents the earliest stage during organ formation when tissue-specific genes are activated, and development of this organogenetic capability played a key role in vertebrate evolution (Hall and Miyake, 1992; Smith and Hall, 1990). Various molecules and cellular mechanisms have been suggested to contribute to control of mesenchymal condensation, including changes in cell proliferation (Hall and Miyake, 2000), local deposition of extracellular matrix (ECM) components such as fibronectin and hyaluronate (Frenz et al., 1989; Knudson and Toole, 1985; Kulyk et al., 1989), centripetal haptotaxis driven by spatiotemporal patterns of ECM production (Downie and Newman, 1995; Newman SA, 1996), alterations of cell-cell or cell-ECM interactions that prevent cell movement away from a center (Frenz et al., 1989; Oberlender and Tuan, 1994) and inductive patterns of diffusible morphogens such as Fgfs, TGF-β and BMPs (Newman et al., 2008; Urban et al., 2006; Christley et al., 2007). However, the precise mechanism by which mesenchymal condensation is controlled at the level of tissue organization, as well as how this cell compaction process controls cell fate switching that drives subsequent organ development, still remains unknown.
In the condensed mesenchyme, cells change their shape and size dynamically, and this is accompanied by changes in cell fate that drive subsequent organ formation. The size and shape of the condensed cell mass also dictate the final three-dimensional (3D) form of the organ, and abnormal condensation can result in developmental defects (Hall and Miyake, 1992; Newman SA, 1996). Mechanical stimuli can modulate cell lineage commitment and control development of various tissues during embryogenesis (Carter and Wong, 1988; Farge, 2003; Mammoto and Ingber, 2010), and studies with cultured cells suggest that cell fate can be controlled mechanically by altering cell shape (Chen et al., 1997; McBeath et al., 2004). These observations raise the possibility that physical alterations in cells that result from cell compaction in the condensed mesenchyme also could play an active role in this differentiation process (Oster et al., 1983).
One reason that little is known about the mechanism by which epithelial and mesenchymal cells interact to initiate and control the mesenchymal compaction process, or how this process is linked to cell fate determination, is because of the lack of model systems to explore these mechanisms under controlled conditions in vitro. Here we studied one of the simplest models of whole organ formation – the embryonic tooth – to explore how mesenchymal condensation is controlled, and to determine whether physical forces contribute to the cell fate switching that drives organogenesis both in vivo and in vitro. Embryonic tooth has been previously used as an experimental system to identify multiple transcription factors (e.g. Msx1, Msx2, Egr-1, Pax9 and Runx2) and morphogens (e.g. Fgfs, BMPs, Wnt, Hh, TGF- β and activin) that are regulated in spatiotemporal manner by reciprocal epithelial-mesenchymal interactions, and that are critical for tooth organ formation during embryological development (Thesleff, 2003). Our results reveal that mesenchymal condensation in the tooth is driven by opposing stimulatory and repulsive migratory factors, and that compression-dependent changes in cell shape that occur secondary to physical compaction of cells within the tightly condensed mesenchyme are responsible for inducing expression of transcription factor genes that drive tooth organ formation.
RESULTS
Early dental epithelium induces mesenchymal condensation and Pax9 expression
In the mouse embryo, formation of the lower molar tooth is mediated by local thickening of the dental epithelium (DE) that causes placode formation at embryonic day 10 to 11 (E10–11). Budding of the DE into the underlying mesenchyme occurs at E12, and this is accompanied by a mesenchymal condensation as the connective tissue cells that are initially widely distributed compact together to form a dense cell mass directly beneath the invaginating DE by day E12–13 (Fig. 1a). Computerized image analysis confirmed that this condensed region at E13 spanned 83±3 µm from the epithelial-mesenchymal interface, and that mesenchymal cell packing densities in this condensed zone were more than twice than those observed in regions distant (>83 µm) from the interface after E13, whereas there was no significant difference in cell densities between these regions at earlier times (E10–E11) (Fig. 1b). Importantly, when tissues were labeled with BrdU, dividing cells were found to be distributed diffusely throughout the mesenchyme as previously shown by others (Shigemura et al., 1999), and there was no significant difference in cell growth rates between these adjacent and distant regions at the stage of E10–E13 (Fig. 1c). Hence, local variations in cell proliferation do not appear to drive mesenchymal condensation at these stages in the tooth.
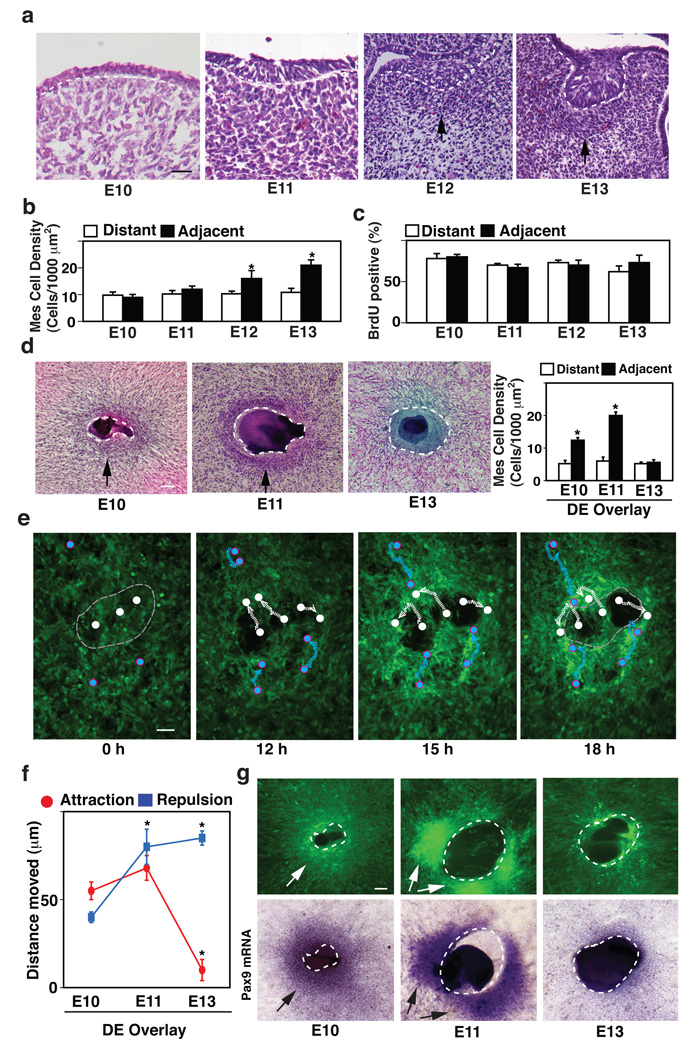
Figure 1. Induction of Pax9 expression and tooth formation by early dental epithelium (DE) in vitro and in vivo.
a, Hematoxylin and eosin (H/E) stained histological sections showing development of the lower molar tooth in the mouse embryo. Tips of black arrows abut on the lower edge of the condensed mesenchyme. b, Graph shows mesenchymal cell densities measured in regions adjacent to (<83 µm) or distant (>83 µm) from the epithelial-mesenchymal interface (dashed lines). c, Graph showing the percent of BrdU-positive mesenchymal cells measured in regions adjacent to (<83 µm) or distant (>83 µm) from the epithelial-mesenchymal interface at each stage. d, H/E-stained cell cultures containing a retracted ball of intact DE isolated from E10, 11 or 13 embryos (dashed lines) overlaid atop a monolayer of cells isolated from the dental mesenchyme (DM) of E10 embryos (black arrows indicate regions of mesenchymal condensation). Graph at right shows mesenchymal cell densities measured in the regions adjacent to (<81 µm) or distant from (>81 µm) the epithelial-mesenchymal interface in vitro. e, Fluorescence micrographs showing images of GFP-labeled mesenchymal cells overlaid with E11 DE and cultured for 0, 12, 15 or 18 h. Note that the E11 DE (which is not visible in this view) repulsed underlying mesenchymal cells and caused them to move peripherally (migration paths of representative mesenchymal cells are indicated by white circles and dashed lines), while it simultaneously attracted other cells (indicated by blue circles and dashed lines) that moved closer to the epithelial-mesenchymal interface (light gray dashed lines) from distant regions of the mesenchymal cell monolayer (see Fig. S1b and Supplementary Movie M1). f, Graph shows results of quantifying total migration distances of mesenchymal cells that were attracted or repulsed in experiments using overlays containing DE isolated from E10, 11 or 13 embryos. g, Fluorescence microscopic (top) and in situ hybridization (ISH) (bottom) views showing the cell compaction and Pax9 mRNA localization, respectively, in the mesenchymal condensation induced when confluent GFP-mesenchymal cell monolayers are overlaid with DE isolated from E10, 11 or 13 embryos and cultured for 2 days in vitro (dashed lines indicate epithelial-mesenchymal interface) (see also Fig.S1f–h). In all figures, scale bars = 50 µm; *, p < 0.01.
To explore how mesenchymal condensation is induced during tooth development, we developed a new in vitro model that recapitulates this condensation response. We isolated dental epithelium (DE) from E10–E11 placode regions or E13 mandibles from mouse tooth germs and overlaid the DEs on top of monolayers of cultured dental mesenchymal (DM) cells that we had previously isolated from E10 mouse tooth germs. The DEs formed spherical tissue masses that adhered to the underlying mesenchymal cell monolayer and remained viable for days. Interestingly, co-culture of early DE (E11) with DM for 2 days under these conditions induced the underlying mesenchymal cells to condense around it (Fig. 1d, left) with cell packing densities in this dense zone increasing by more than 2-fold compared with more distant regions (Fig. 1d, right). The condensed region was located within 81±2 µm of the epithelial-mesenchymal interface as defined by linear deposition of type IV collagen-containing basement membrane (BM) (Fig. S1a), thus closely mimicking the mesenchymal condensation response that this DE produces over a similar 2 day time course (E11 to E13) in vivo (Fig. 1a). Moreover, DE isolated at E13 failed to stimulate mesenchymal condensation in this in vitro model (Fig. 1d), which is again consistent with past work that showed E13 DE loses its inductive ability for producing odontogenesis in vivo (Mina and Kollar, 1987).
One mechanistic model for mesenchymal condensation in limb bud suggests that cell compaction results from ECM-driven translocation and directional migration of cells from the surrounding loose mesenchyme to the condensed region (Newman SA, 1996). We therefore used time lapse microscopy to analyze the contribution of cell movement to this in vitro mesenchymal condensation process using cultured DM cells labeled with green fluorescent protein (GFP). These studies revealed that the overlying E11 DE attracted some cells to migrate from distant regions of the dish to the epithelial-mesenchymal interface; however, it also actively repulsed mesenchymal cells in the underlying monolayer causing them to move peripherally, thereby clearing the region of the substrate directly beneath the DE cell mass (Fig. 1e, Fig. S1b and Supplementary Movie M1). These opposing migratory movements resulted in a major increase in cell packing density at the edge of the cleared zone (Fig. S1b). Quantification of the movements of individual cells confirmed that E10 and E11 DE attracted surrounding mesenchymal cells to a much greater degree than E13 DE, whereas E11 and E13 DE exhibited the greatest repulsing activities and thus, the net cell clearing activity progressively increased over time (Fig. 1f). DM cells without DE moved randomly at a slower velocity (2.5±1 µm/h) (Fig. S1c and Supplementary Movie M2) compared to DM cells combined with E11 DE that moved faster (8±2 µm/h for attraction and 7±3 µm/h for repulsion) and in a more oriented manner (Fig. 1e and Supplementary Movie M1). Again, we did not detect any significant local change in mesenchymal cell growth in these same regions during mesenchymal condensation in vitro (data not shown), just as we observed in vivo (Fig. 1c). Thus, these data suggest that E11 DE induces mesenchymal condensation by simultaneously attracting and repulsing mesenchymal cells, which causes a condensed cell mass to form in a dense border along the epithelial periphery (Fig. S1b).
Next we explored whether mesenchymal condensation induced via this mechanisms of antagonistic migratory movements contributes to cell fate switching during tooth differentiation. We began by isolating DM and DE from E10 to E13 embryos, and performing transcriptional profiling of these tissues for multiple transcription factors that have been shown to be intimately involved in tooth development (Thesleff, 2003). These studies revealed that five critical odontogenic-related genes - Egr1, Lhx6, Lhx8, Msx1, and Pax9 - are upregulated in E13 DM in vivo when mesenchymal condensation reaches maximal levels (Fig. S1d,e). Among these markers, Pax9 has been shown to be critical for formation of tooth as well as other organs (Peters et al., 1998), and it is specifically enriched in the region of the condensed mesenchyme in the tooth germ at E13 (not shown). Based on its preferential expression in condensed mesenchyme in vivo, Pax9 expression was used as an initial marker for odontogenic induction in our in vitro condensation model. These studies revealed that DE isolated at E11 (when it is most effective at inducing mesenchymal condensation in vivo) also induced the highest levels of Pax9 protein and mRNA expression in condensed mesenchymal cells after 2 days of culture (Fig. 1g and Fig. S1f–h). Thus, this in vitro model recapitulates the full developmental cascade from mesenchymal condensation through the cell fate switching that triggers odontogenic induction in vivo.
Mesenchymal condensation is induced by Fgf8 and Sema3f
We carried out transcriptional profiling of various morphogen classes and subtypes (e.g., Fgfs, Bmps, Shh, Wnts, CXCLs) expressed in DE isolated at different stages of development to identify cues specifically produced by E11 DE that might be responsible for the opposing motility to drive mesenchymal condensation and Pax9 induction in vitro. Five morphogens (Bmp4, CXCL12, Fgf8, Shh, Wnt5a) were expressed at higher levels in E10 and E11 DE relative to E13 DE (Fig. S2a). When in vitro migration assays were carried out with these five factors, only Fgf8, which is known to both play a critical role during formation of many embryonic organs, including tooth and induce Pax9 (Neubuser et al., 1997), was able to attract mesenchymal cells (Fig. S2b). We also confirmed that Fgf8 was expressed at higher levels in E10 and E11 DE, whereas its mRNA levels were suppressed in E13 DE when embryonic tooth germ was analyzed using qRT-PCR and in situ hybridization (ISH) (Fig. S2c,d). In contrast, immunohistochemical analysis revealed that Fgf8 protein was present in tissues at E13, and it appears to be stored within the BM where it co-localized with its binding partner, heparan sulfate proteoglycan (HSP) (Fig. S2e). As Fgfs can be stored in BM and released later times (Folkman et al., 1988), these findings suggested that the BM could serve as an Fgf8 storage site for sustained release of this morphogen even after the DE ceases its production at later stages of development. Indeed, our finding that E13 DE cannot induce mesenchymal condensation in our in vitro model after the BM is removed with Dispase enzyme supports this concept that Fgf8 stored within the BM is functionally critical for the mesenchymal condensation (Fig. S2f).
We also performed transcriptional profiling to identify repulsive molecules that are more highly expressed in E11 and E13 DE compared to E10, which could be responsible for the cell clearing activity we observed at these times. Two semaphorin molecules that have been shown to act as repulsive molecules and guide pattern formation during nerve and vascular development (Carmeliet and Tessier-Lavigne, 2005) —semaphorin-3f (Sema3f) and semaphorin-6a (Sema6a) —were preferentially upregulated in E11 and E13 DE (Fig. S2g). Given that a continuous BM separates the DE and DM in vitro and in vivo during tooth development, we focused on Sema3f because it acts in a soluble form, whereas Sema6a is a transmembrane protein that is critical for direct cell contact-mediated repulsion (Toyofuku et al., 2008). Sema3f was upregulated in E11 and E13 DE and its counter-receptor, neuropilin 2 (Nrp2) was also expressed at high levels in DM at these same times (Fig. S2h,i). Importantly, non-dental epithelium isolated from the back region of an E11 embryo (dorsal paravertebral ectoderm), which has significantly less expression of Fgf8 and similar expression of Sema3f compared with E11DE, can repulse DM cells, but it does not attract DM cells and thus, it fails to induce effect mesenchymal condensation in our model system (Fig. S3a). Thus, both Fgf8 and Sema3f seem to be required for this DM cell compaction response in tooth.
We then used microfluorimetry to quantitate the distribution of Fgf8 and Sema3f at different stages of tooth development. These studies confirmed that Fgf8 is distributed in a shallow gradient with highest intensity being observed in the epithelial BM and progressively lower staining levels as one moves out over longer distances from the BM and through the mesenchyme (Fig. 2a). These results raise the possibility that this Fgf8 gradient could drive mesenchymal cell migration towards the epithelial-mesenchymal interface. Indeed, cultured mesenchymal cells were able to migrate up a similar shallow gradient of Fgf8 generated in vitro in a Transwell migration assay (Fig. 2b).
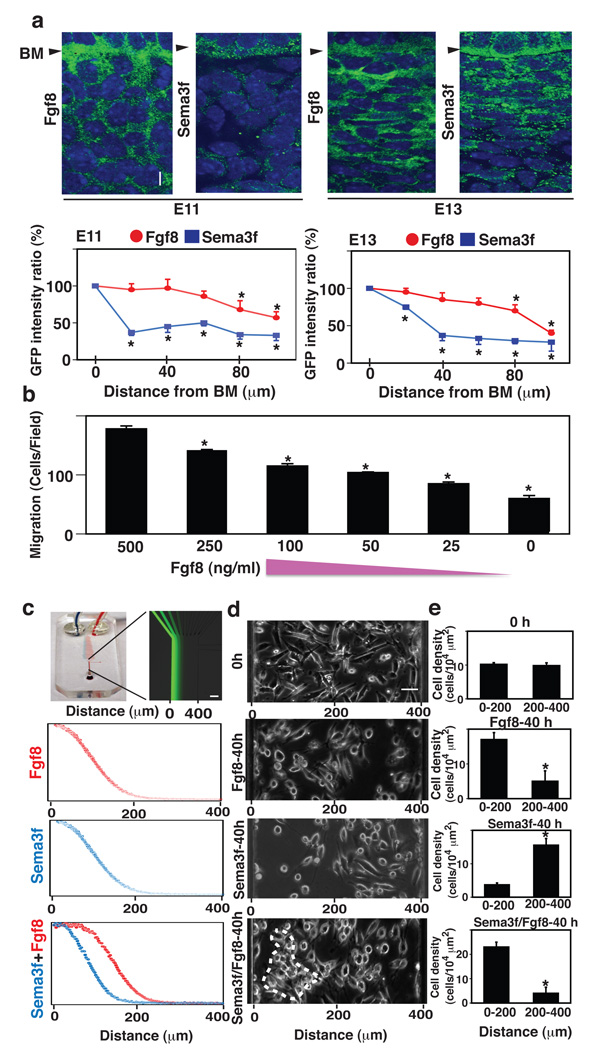
Figure 2. Fgf8 and Sema3f act as opposing short- and long-range morphogens that respectively stimulate and repulse mesenchymal cell migration.
a, Fluorescence micrographs (top) showing the localization of Fgf8 (left) and Sema3f (right) proteins in mesenchyme at E11 (left) and E13 (right) (see also Fig.S2). Graphs (bottom) show the ratios of GFP intensities for Fgf8 (red circle) and Sema3f (blue square) measured in the mesenchyme at the indicated distances from BM, and in the DE adjacent to the BM (distance 0). b, The effects of different concentrations of Fgf8 (500, 250, 100, 50, 25 and 0 ng/ml) on mesenchymal cell migration for 16 h quantified using a Transwell migration assay. c, A microfluidic device fabricated in PDMS (left) containing microfluidic mixers that was used to create gradients of soluble molecules within a single flow channel, as visualized here by infusing FITC-dextran (Mr 70,000) (right). Graphs show the gradients of Fgf8 (top), Sema3f (middle) or Fgf8/Sema3f (bottom) that were applied to the cells in the device. Overlapping gradients of Fgf8 and Sema3f (bottom) were generated by adding an additional inlet channel to the left side of the last branching point of the original microfluidic gradient generator. d, The mesenchymal cells were plated in the microfluidic device and exposed to the gradients of Fgf8 and/or Sema3f with high concentrations at the left and low at the right, that was generated in the device by laminar flow. The upper phase contrast micrograph shows the localization of cells at time zero and 40 h after exposure to the gradients of Fgf8 (top), Sema3f (middle) and Fgf8/Sema3f (bottom); dashed circle indicates region of cell compaction. e, Graph showing the cell densities in the corresponding areas from e at 0 (top) or 40 h (bottom) in each treatment. Scale bars = 20 µm for a; 100 µm for c; 50 µm for d; *, p < 0.01.
In contrast, similar microfluorimetric analysis revealed the presence of a much steeper gradient of Sema3f protein in vivo (Fig. 2a), with Sema3f being highly concentrated near the BM. This high local concentration of Sema3f would be expected to act locally to repulse migratory mesenchymal cells attracted by Fgf8 so that the cells pile up at the epithelial boundary if Sema3f influences mesenchymal cell migration in a similar inhibitory manner as it does endothelial cells and nerve cells (Carmeliet and Tessier-Lavigne, 2005). To test this directly, we exposed cultured mesenchymal cells to gradients of Sema3f and/or Fgf8 generated and sustained for 40 h in a microfluidic system (Fig. 2c). As predicted, the cells migrated towards the region of the higher Fgf8 concentration or lower Sema3f concentration (Fig. 2 c–e). Moreover, when we exposed cells to overlapping gradients of Sema3f (shorter-range) and Fgf8 (longer-range) simultaneously by adding an additional inlet channel to the left side of the last branching point of the microfluidic gradient generator (Dertinger et al., 2001) (Fig. 2 c), cell compaction was observed between the two gradients (Fig. 2 d, e). These results confirm that a combination of Fgf8 and Sema3f is sufficient to drive mesenchymal cell compaction whereas either molecule alone cannot drive the entire condensation response.
To further test this hypothesis, we transfected Fgf8, Sema3f or both into HEK 293 cells (Bielenberg et al., 2004) that were labeled with tracking dye, and then we overlaid these cell pellets on confluent monolayers of cultured GFP-labeled E10 mesenchymal cells. Over-expression of Fgf8 induced a low level of mesenchymal cell aggregation around individual HEK 293 producer cells, whereas transfection of Sema3f caused the mesenchymal cells to disperse without producing mesenchymal condensation (Fig. 3a). Efficient mesenchymal condensation of a similar size and density to that induced by an E11 DE overlay, only was observed when both of the antagonistic morphogens (Fgf8 and Sema3f) were over-expressed simultaneously in the same cells (Fig. 3a and Fig. S3b). Thus, these data are consistent with a model in which simultaneous exposure of the mesenchymal cells to a gradient of Fgf8 that attracts the migratory cells from long distances, and a steep gradient of Sema3f that repulses these same cells, collectively induce the what we call the ‘mesenchymal condensation’ response
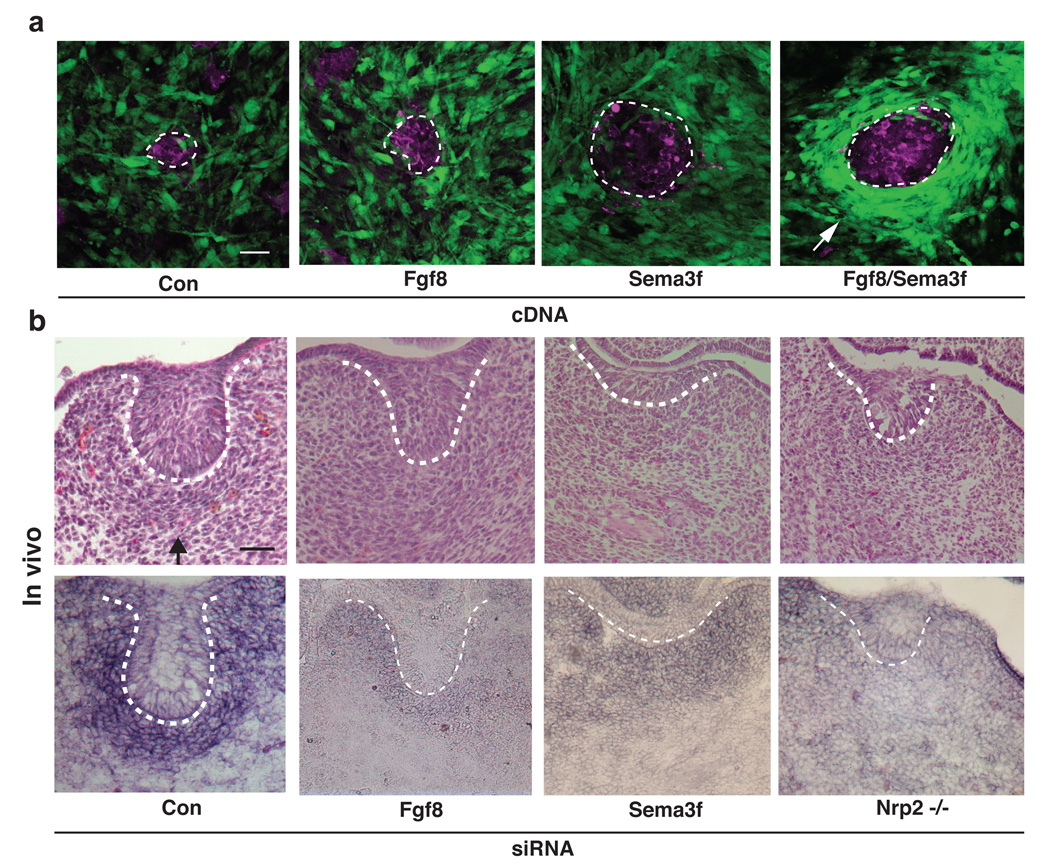
Figure 3. Early dental epithelium uses Fgf8 and Sema3f to induce mesenchymal condensation and Pax9 expression.
a, Fluorescence (top) micrographs showing cultures containing labeled HEK 293 cell pellets that were being transfected with control (Con), Fgf8, Sema3f or both Fgf8 and Sema3f cDNAs, and overlaid on top of a GFP-labeled mesenchymal cell monolayer for 2 Ds (magenta, labeled HEK293 cell pellets; dashed line indicates pellet-mesenchymal cell interface). Note that only cells expressing both Fgf8 and Sema3f caused mesenchymal condensation around the pellet (white arrow) (see also Fig. S3b). b, Light micrographs of H/E-stained tissue sections (top) and Pax9 ISH images (bottom) showing tooth germs in E13 embryos treated with control (Con), Fgf8 or Sema3f siRNA in utero, or similar stage tooth germs from Nrp2 knockout (−/−) mice (see also Fig.S3c–e). Dashed lines indicate the epithelial-mesenchymal interface and tip of black arrows abut on the lower edge of the condensed mesenchyme. In all figures: scale bars=50 µm.
To determine the physiological relevance of these findings in vivo, we then knocked down Fgf8 or Sema3f in living embryos in utero by delivering siRNAs transplacentally (Gratsch et al., 2003; O'Shea K et al., 2006) (Fig. S3c). Suppressing expression of either Fgf8 or Sema3f inhibited mesenchymal condensation and Pax9 induction at E13 in the first pharyngeal arch, and knockdown of Sema3f also prevented DE budding at E13 in vivo (Fig. 3 b and Fig. S3d). Nrp 2 knockout (Nrp 2 −/−) embryos that lack this Sema3f receptor also failed to induce mesenchymal condensation, Pax9 expression or DE budding at E13 when compared to wild type embryos (Fig. 3b and Fig. S3e). Importantly, mesenchymal condensation and Pax9 induction were also inhibited when DE isolated at E11 from Sema3f-knockdown embryos was overlaid on DM cell monolayers in vitro, and DE from Sema3f knockdown embryos exhibited significantly reduced repulsion of underlying DM cells (data not shown). These results are consistent with past proposals that short-range repulsion by semaphorins not only defines the epithelial boundary, but also dictate the patterns and size of epithelial buds during development of other ectomesenchymal organs (Chung et al., 2007; Korostylev et al., 2008; Horowitz and Simons, 2008). Taken together, these results confirm that mesenchymal condensation is critical for Pax9 induction, and that it is driven by the opposing actions of Fgf8 and Sema3f that act respectively as attractive and repulsive cues for mesenchymal cell migration in vitro and in vivo.
Cell fate switching is induced by changes in cell shape and RhoA activity
Computerized morphometric analysis of histological sections of whole embryonic tooth germs and overlay cultures revealed that cells at E12–E13 within the condensed mesenchyme are significantly smaller (~ 25% reduction in projected cell area and volume) relative to cells in non-condensed regions of the mesenchyme, both in vivo and in vitro (Fig. S4a,b). Changes in cell shape modulate cell sensitivity to chemical cues and thereby, switch cells between different fates in vitro (Chen et al., 1997; Dike et al., 1999; Singhvi et al., 1994); however, alterations in cell-cell contact formation also can influence cell differentiation (Okamura et al., 2003). To explore whether changes in cell shape or cell-cell adhesion contribute to cell fate switching in embryonic mesenchyme, we controlled these parameters independently using microengineered adhesive islands (Chen et al., 1997; Dike et al., 1999; Singhvi et al., 1994) coated with the ECM protein, fibronectin (FN), which is present throughout E13 tooth mesenchyme in vivo (not shown). When mesenchymal cells were cultured for 16 h on large circular FN islands (500 µm diameter) at a high plating density (2.4 × 105 cells/cm2) that produced cell compaction and resulted in reductions in cell size similar to those observed in regions of mesenchymal condensation induced by E11 DE in vitro (Fig. 4a and Fig. S4b), mesenchymal Pax9 mRNA levels increased by 3- to 4-fold (Fig. 4b). In contrast, Pax9 was not induced in cells cultured on the same FN islands at a lower plating density (2 × 104 cells/cm2) that failed to constrain cell shape or promote high levels of cell-cell adhesion (Fig. 4a,b). Interestingly, soluble Fgf8 and Sema3f had no effect on Pax9 expression when cell compaction was suppressed by culturing cells at low plating density, and there was no additive effect on Pax9 induction when these morphogens were added to cells cultured at high densities that physically promoted high levels of cell rounding because these artificially condensed cell clumps induced Pax9 expression even in the absence of these soluble morphogens (Fig. 4b). Importantly, other critical odontogenic markers, including Lhx8, Msx1 and BMP4, were similarly upregulated in a cell density-dependent manner (Fig. 4c).
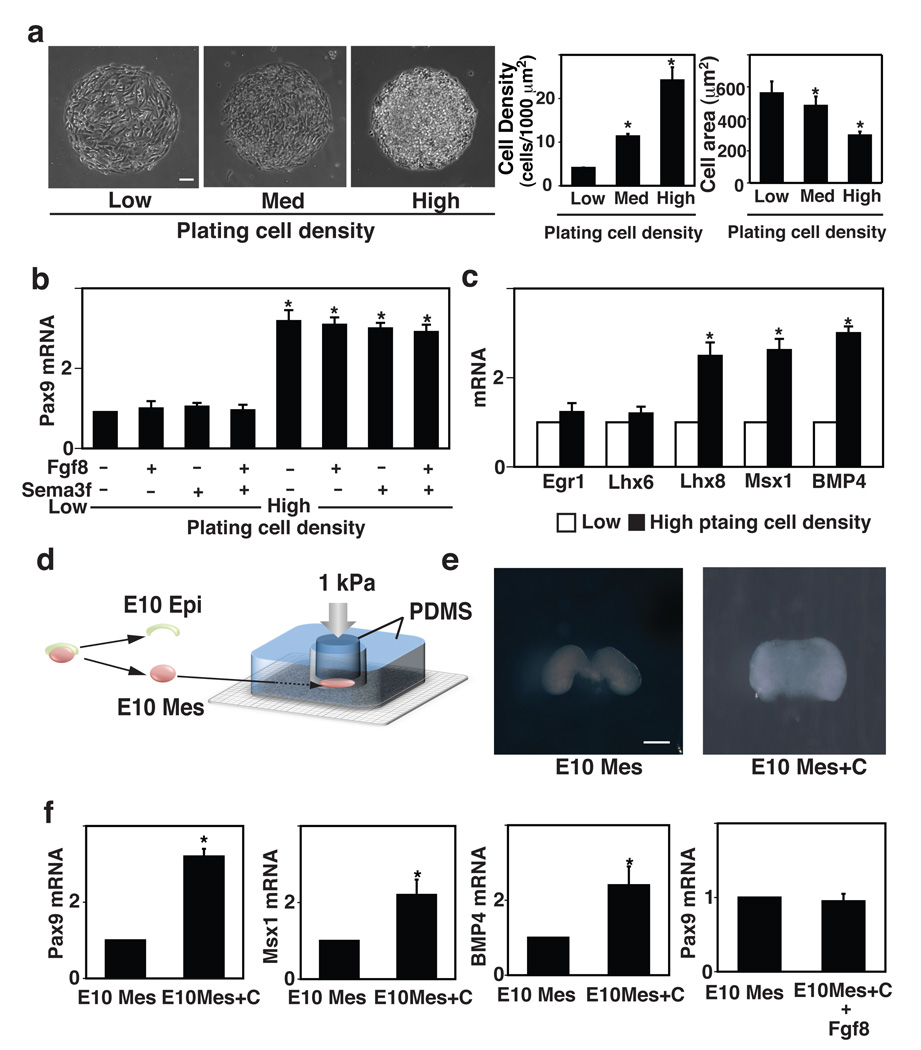
Figure 4. Mechanical control of odontogenic transcription factors during tooth development.
a, Phase contrast micrographs (left) showing mesenchymal cells cultured for 16 h on microfabricated circular FN islands (500 µm diameter) in vitro at low, medium or high plating cell density (0.2, 1.2 or 2.4 × 105 cells/cm2, respectively), and graphs at right showing corresponding cell densities and projected cell areas measured under each condition. b, Graph showing Pax9 induction in mesenchymal cells cultured for 16 h on the circular FN islands (500 µm diameter) at low or high plating density with or without Fgf8 (150 ng/ml) and/or Sema3f (150 ng/ml). c, Graph showing induction of additional odontogenic factors (Egr1, Lhx6, Lhx8, Msx1 and BMP4) in mesenchymal cells cultured under the same conditions as in b. d, Freshly isolated E10 mesenchyme from first pharyngeal arch was physically compressed (1 kPa) for 16 h using a mechanical compressor composed of two pieces of PDMS polymer that are overlaid with a metal weight (see also Fig.S4f). e, Macroscopic images of the mesenchyme that was cultured ex vivo for 16 h in the absence (E10 Mes) or presence of compression (E10 Mes+C). f, Graph showing expression of Pax9, Msx1 and BMP4 mRNAs in control (E10 Mes) versus compressed mesenchyme (E10 Mes+C) and expression of Pax9 in control versus mesenchyme treated with soluble Fgf8 (150 ng/ml) for 16 h. In all figures: scale bars = 50 µm for a; 500 µm for e; *, p < 0.01.
ECM components, such as fibronectin, are deposited locally during mesenchymal condensation and cell-ECM interactions appear to mediate condensation during chondrogenesis (Frenz et al., 1989; Knudson and Toole, 1985; Kulyk et al., 1989). Therefore, we analyzed changes in ECM production using our gene microarray data, and found that collagen VI is specifically upregulated and deposited around individual cells within the condensed mesenchyme at E13 in vivo and in vitro (Fig. S4c,d). These results suggest that ECM remodeling might serve to physically stabilize inductive cell shapes during tooth development. However, variations in ECM density also can influence the shape and growth of cultured cells (Ingber, 1990), and thus changes cell-ECM binding interactions could potentially influence Pax9 induction in our system. To explore this possibility, we plated mesenchymal cells on dishes coated with poly-L-lysine (PLL) or collagen VI (low: 30 µg/cm2, medium: 500 µg/cm2, high: 1800 µg/cm2). These variations in ECM density produced minimal changes in cell area or Pax9 induction after 16 h of culture (Fig. S4e). Thus, cell compaction and changes in cell size appear to be more critical for control of Pax9 induction in mesenchymal cells than variations in cell-ECM binding interactions.
To directly explore whether physical compaction of cells per se is sufficient to induce differentiation of the mesenchyme, we mechanically compressed freshly isolated whole E10 mandibular mesenchyme (E10 Mes) between two blocks of poly-dimethylsiloxane (PDMS) polymer for 16 h using constant pressure (1 kPa) (Fig. 4d). This level of pure mechanical compression produced cell densities and sizes similar to those observed during mesenchymal condensation at E13 in vivo (Fig. 4e and Fig. S4f). These studies revealed that Pax9 and other critical odontogenic markers, including Msx1 and BMP4, were up-regulated in the compressed DM tissue compared to unloaded controls (Fig. 4f), whereas Pax9 was not induced in freshly isolated E10 mesenchyme when it was treated with soluble Fgf8 for a similar time (Fig. 4f, right). Interestingly, mechanical compression of the DM ex vivo also upregulated expression of collagen VI and increased its deposition around individual cells (Fig. S4g), as we previously observed in vitro and in vivo.
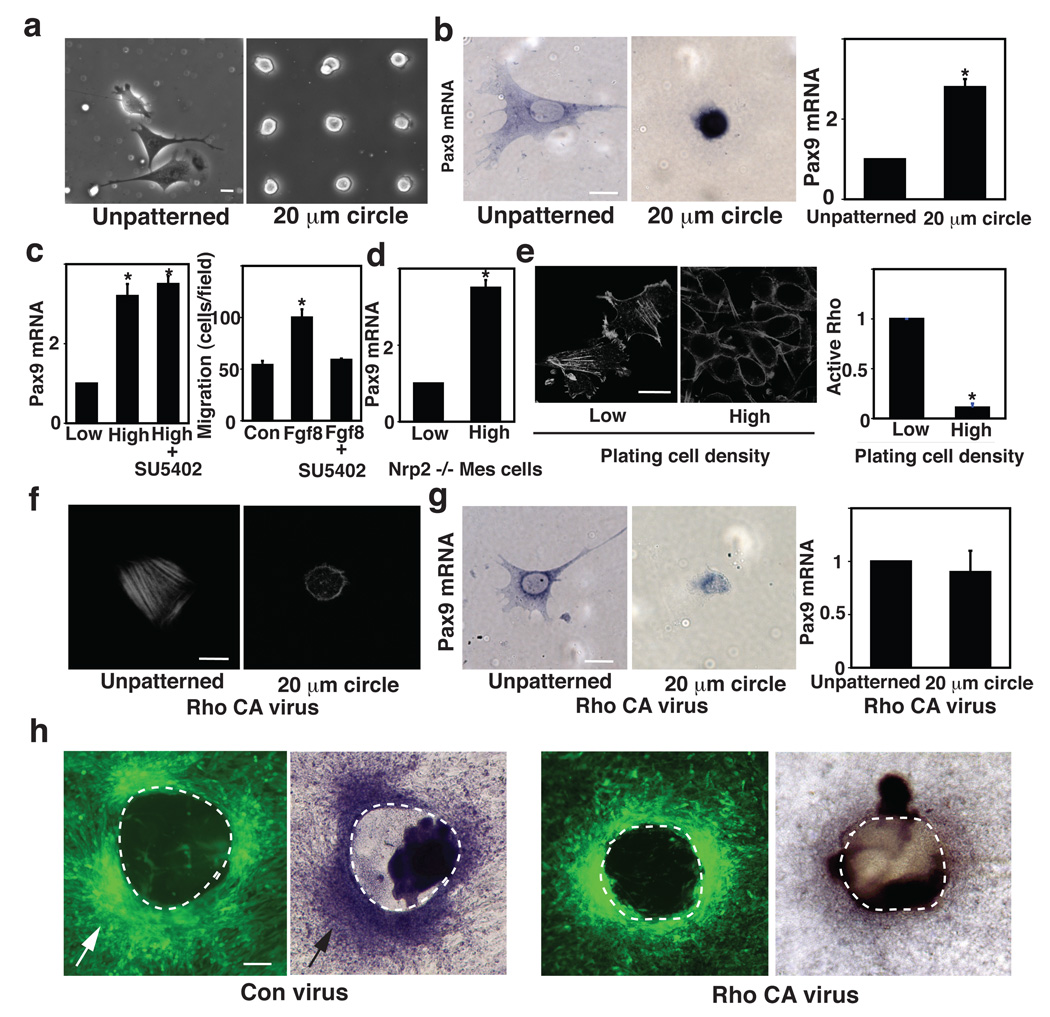
These results were also highly consistent with those obtained using micropatterned ECM islands that induce a similar high degree of compaction of cultured mesenchymal cells, and stimulated Pax9 expression (Fig. 4a–c). But it was still not possible to separate effects induced by mechanical compression-dependent changes in cell shape (i.e., cell rounding) from those induced by increased cell-cell contact formation that might result from greater surface contact area between neighboring cells in this assay. To unequivocally identify the critical inducer of cell fate switching that mediates compaction and mechanical compression-dependent induction of odontogenic cell fate switching, we cultured single mesenchymal cells for 16 h on small, circular FN islands (20 µm diameter) created with microcontact printing that are separated by non-adhesive barrier regions, which physically prevent formation of cell-cell contacts (Chen et al., 1997; Dike et al., 1999). These islands (Fig. 5a) also restrict the spreading of individual adherent cells, and hold them in a round shape and size (~25% smaller in volume than spread cells, as determined by confocal microscopic analysis) similar to that exhibited by individual mesenchymal cells within condensed mesenchyme induced to form by contact with E11 DE in vitro (Fig. S4b). Importantly, single rounded cells cultured on these islands free of any cell-cell contacts increased Pax9 expression even in the absence of soluble Fgf8 and Sema3f, when analyzed by ISH and qRT-PCR (Fig. 5b). Thus, these data indicate that the soluble epithelial morphogens, Fgf8 and Sema3f, induce Pax9-dependent tooth differentiation through their physical effects on cell shape that result from mesenchymal cell compaction during the migration-driven condensation response, rather than through a mechanism that involves direct chemical signaling or cell-cell adhesion-dependent signaling mechanisms.
Figure 5. Odontogenic induction is controlled by cell shape distortion and mediated by RhoA.
a, Phase contrast micrographs showing spread cells cultured for 16 h on an unpatterned FN substrate (left) versus round mesenchymal cells cultured on small (20 µm diameter) circular FN islands (right). b, ISH views (left) and qRT-PCR (right) analysis showing Pax9 induction in mesenchymal cells cultured under the same conditions as in a. c, Graphs showing Pax9 induction in cells cultured for 16 h at low or high density (0.2 or 2.4 × 105 cells/cm2, respectively) on the circular FN island (500 µm diameter) in the absence or presence of SU5402 (left), and migration of the mesenchymal cells towards Fgf8 (150 ng/ml) measured using a Transwell migration assay with or without SU5402 (25 µM) (right). d, Graph showing Pax9 induction in Nrp2 −/− mesenchymal cells isolated at E10 from Nrp2 −/− embryos is preserved when the cells were cultured under the same conditions as in c. e, Fluorescence immunomicrographs showing the actin cytoskeleton in mesenchymal cells cultured under the same conditions as in c. Graph shows quantification of RhoA activity (ratio of active RhoA to total RhoA protein) in these mesenchymal cells (see also Fig.S5b). Cells were treated with virus encoding constitutively active RhoA (Rho CA virus) and analyzed as in b. Note that even though stress fibers do not form in the compact round cells (f), the Pax9 induction response was inhibited by constitutively active RhoA (g). h, Fluorescence micrographs (left) and ISH (right) views showing mesenchymal cell condensation (white arrows) and Pax9 induction (black arrow), respectively, in cultured GFP-labeled mesenchymal cells transduced with control (Con) virus or Rho CA virus and overlaid with E11 DE for 2 days. Note that increased RhoA activity suppressed Pax9 induction (see also Fig.S5d). In all figures: dashed lines indicate the epithelial-mesenchymal interface; scale bars = 20 µm for a, b, e, f and g; 50 µm for h; *, p < 0.01.
This mechanical mechanism of developmental control is supported by the finding that the Fgf receptor inhibitor, SU5402, had little effect on cell compaction-dependent induction of Pax9, although it inhibited mesenchymal cell migration towards Fgf8 (Fig. 5c). Cell rounding-dependent induction of Pax9 was also observed in Nrp2 −/− mesenchymal cells constrained from spreading on micropatterned adhesive islands (Fig. 5d), further demonstrating the physical nature of this signaling response as it functions efficiently even in cells that can not respond to Sema3f. These responses, however, do not perfectly mimic those observed in vivo; for example, mesenchymal Pax9 increases 3-to 4-fold relative to E10 when the cells are physically distorted in vitro or ex vivo, whereas levels rise about 8-fold at E13 in vivo. Thus, it is possible that other morphogens or ECM molecules which may be present in vivo could synergize with physical cues during normal tooth development, or we might not fully reproduce the physical, ECM or soluble factor environment necessary to perfectly mimic this complex developmental process in vitro.
Cell shape distortion often influences cell behavior and developmental control by altering mechanical signaling pathways associated with the cytoskeleton (Chen et al., 1997; McBeath et al., 2004). We therefore tested the ability of various modulators of mechanotransduction that can be involved in control of organ and tissue development (Mammoto and Ingber, 2010) to determine whether any interfere with cell compaction-dependent Pax9 induction in mesenchymal cells cultured at high density on the large circular FN islands. We tested the following compounds: Myosin light chain kinase inhibitors (Blebbistatin, ML7), Rho/ROCK inhibitors (C3, Y27632), Rho activator (cytotoxic necrotizing factor 1: CNF1), N-cadherin neutralizing antibody, Rac1 inhibitor (NSC23766), Rac and Cdc42 activator (CN02), PI3 kinase inhibitor (LY294002), MEK 1/2 inhibitor (U0126), G-protein coupled receptor inhibitor (pertussis toxin), Src inhibitor (PP1), PKC inhibitor (Ro31-8220), NO inhibitor (L-name) and p38MAPK inhibitor (SB203580); however, only RhoA activation inhibited this response (Fig. S5a). Consistent with this result, condensed mesenchymal cells exhibited decreased RhoA activity and loss of actin stress fibers (which require RhoA-dependent increases in cytoskeletal tension generation for their formation) compared with spread cells cultured at a low density (Fig. 5e and Fig. S5b). To directly test the possibility that RhoA mediates this mechanochemical control induction mechanism, we used mesenchymal cells that were transduced with a retroviral vector encoding constitutively active RhoA; this increased stress fiber formation in spread cells on unpatterned substrates but not in round cells on the small circular FN islands (Fig. 5f). Nevertheless, over-expression of RhoA in round cells completely suppressed Pax9, Msx1 and BMP 4 expression to low levels similar those seen in spread cells on unpatterned substrates (Fig. 5g versus 5b and Fig. S5c).
Pax9 induction was also impaired in the mesenchymal cells expressing constitutively active RhoA when they were overlaid with E11DE, even though mesenchymal condensation was still observed (Fig. 5h and Fig. S5d). Furthermore, when we compressed cell pellets made from DM cells expressing constitutively active RhoA ex vivo, compression-dependent odontogenic induction was impaired compared with pellets made from control cells (not shown). Thus, suppression of RhoA activity induced by cell rounding appears to mediate the process by which mesenchymal cells sense mechanical changes in cell shape and cytoskeletal organization produced by cell compaction during the mesenchymal condensation response, and it helps to transduce these physical signals into changes in Pax9, Msx1 and BMP4 expression that alter cell fate and promote tooth differentiation. This finding is consistent with s past study which showed that changes in cell shape and RhoA-dependent alterations of cytoskeletal prestress (tension generation) play a central role in control of cell fate switching in adult mesenchymal stem cells (McBeath et al., 2004).
Discussion
In the embryo, odontogenic potential is believed to reside primarily in the early DE, and then it is subsequently transferred to the underlying DM that directs later stages of tooth development (Mina and Kollar, 1987; Ohazama et al., 2004). Several morphogens, such as Fgfs, BMPs and TGF-β, have been shown to be required for mesenchymal condensation during kidney and limb formation (Hall and Miyake, 1995; Hentschel et al., 2004; Urban et al., 2006), and cell-generated contractile forces and differential cell adhesiveness have been proposed to mediate mesenchymal compaction in combination with chemical cues in some organ systems (Forgacs, 2005; Oster et al., 1983). Our results suggest that mesenchymal condensation during tooth development is indeed controlled by a mechanism that is both chemical and mechanical, but it is not mediated by any of these previously proposed mechanisms.
Past studies have revealed that combinations of spatiotemporal attractive and repulsive cues mediate pattern formation of various organs (Yang et al., 2002; Tran et al., 2007; Klein, 2004). For example, mesenchymal cells undergo physical compaction, followed by the subsequent morphological changes leading to rounded somites during somitogenesis. One model proposed to explain this mechanism has suggested that long-range attraction forces might cause the cells to squeeze themselves together within a compacted aggregate (Grima and Schnell, 2007), whereas another model predicts that short-range repulsion forces can help the cells define a certain radius and degree of compressibility (Newman and Grima, 2004). Consistent with these general models, the present experimental results indicate that early (E11) DE induces underlying mesenchymal cells to form a condensed mass at the epithelial-mesenchymal interface by producing Fgf8 and Sema3f that simultaneously attract and repulse the mesenchymal cells, respectively. The opposing movements of these cells results in formation of a dense cell mass composed of tightly packed mesenchymal cells, and Pax9, Msx1 and BMP4 are mechanically induced through physical compression of the cells (Fig. 6). Given that induction of Msx1 and BMP4 are critical for shifting the odontogenic inductive capability from the early DE to the DM at later stages of development (Bei et al., 2000; Mina and Kollar, 1987), these findings also suggest that the mechanical compaction of cells that occurs during mesenchymal condensation may serve as an early initiator of this transition process at earlier times (i.e., before E13).
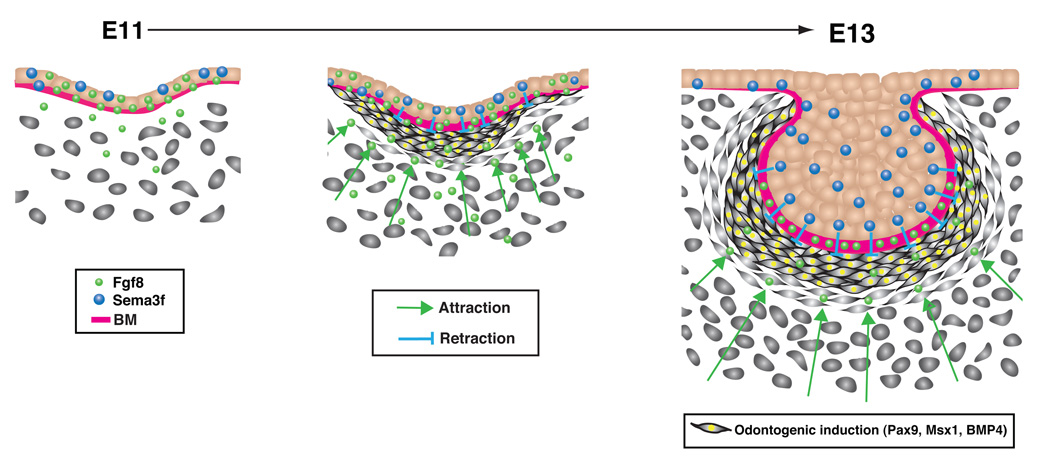
Figure 6. Mechanochemical control of tooth organ development.
Fgf8 is produced by the dental epithelium (DE) and deposited in the basement membrane (BM) by E11 of development (left). The stored Fgf8, which is released over time (left to right), acts as a long-range morphogen to promote mesenchymal cell migration and to attract increased numbers of these cells toward the epithelial boundary. At the same time, the DE also produces local high concentrations of the repulsive morphogen Sema3f, which acts locally to repulse the migrating cells, causing them to crowd at the epithelial-mesenchymal interface (middle) and to form a well developed 'condensed mesenchyme' by E13 (right). The resulting compaction and physical compression of the mesenchymal cells (middle to right) is sufficient to induce expression of critical odontogenic genes, including Pax9, Msx1 and BMP4, which drive subsequent tooth organ formation. This mechanotransduction response is mediated by the cytoskeletal signaling molecule RhoA, which is suppressed in compressed cells within the condensed mesenchyme (not shown).
Pattern formation during formation of elaborate neuronal and vascular networks (Carmeliet and Tessier-Lavigne, 2005) is tightly controlled by a balance between attractive and repulsive cues from the extracellular environment. However, an antagonistic balance between motility factors of this type has not previously been shown to mediate cell fate switching during epithelial-mesenchymal interactions that mediate early organ formation. Importantly, the compaction of mesenchymal cells produced by these opposing cell migratory movements appears to be the crucial developmental trigger as mechanical compression of the mesenchyme is sufficient to induce instructive signals for odontogenesis, and stimulation with Fgf8 and Sema3f morphogens alone fails to induce this odontogenic response in the absence of cell compaction. In addition, we found that RhoA, which is important for many shape-dependent control mechanisms in vitro, is critical for induction of odontogenic differentiation in this organ rudiment model.
Although the concept that body patterns are shaped by soluble morphogen gradients is well documented and accepted, it is unclear how these gradients spatially limit expression of target differentiation genes and produce distinct cell fates within precise tissue boundaries during organogenesis. The present findings suggest that these boundaries are defined by the action of different soluble chemical factors that interact to alter mechanical forces and induce physical changes in cell shape at distinct spatial locations during mesenchymal condensation in the developing tooth. Once the DM cells physically condensed or rounded, Fgf8 and Sema3f had no influence on subsequent cell fate switching, and thus, these morphogens appeared to manifest their pattern-generating activities entirely through this mechano-chemical transduction mechanism.
Interestingly, Fgfs or semaphorins are also involved in mesenchymal condensation during chondrogenesis (Newman and Bhat, 2007), nephorogenesis (Gammill et al., 2007) and neurogenesis (Urban et al., 2006), and mechanical compression also contributes formation of both cartilage and bone (Angele et al., 2004; Roelofsen et al., 1995). The transcription factors and chemical cues examined in the present study (e.g., Pax9, Msx1 and BMP4) are also widely expressed and critical for morphogenesis in many other organs during embryogenesis. Thus, this type of orchestrated interplay between chemical and physical cues could be utilized for control of other organogenetic processes that involve a similar mesenchymal condensation step. Finally, given that early DE can induce differentiated tooth formation when recombined with non-dental adult bone marrow-derived stem cells (Ohazama et al., 2004), these new insights into the molecular biophysical mechanisms that underlie tooth induction also might facilitate future development of biomimetic materials for whole organ engineering that mimic the inductive properties of embryonic tissues.
Methods
Animal experiments
All animal studies were reviewed and approved by the Animal Care and Use Committee of Children's Hospital Boston. Histological assays were performed as described previously (Mammoto et al., 2009) and morphometric analysis was performed using Image J software (http://rsbweb.nih.gov/ij/) (National Institute of Health, USA). Mouse WG-6v11 Illumina microbead chips were used with total RNA extracted from the isolated epithelium and mesenchyme for transcriptional profiling. For cell growth analysis, timed-pregnant mice were injected intraperitoneally with 5-bromo-2’-deoxyuridine (BrdU) solution (Invitrogen) at 100 mg/kg body weight and sacrificed 2 h after the injection. Embryos were then harvested and processed for cryosections (7 µm), and proliferating cells were detected using a BrdU staining kit (Invitrogen). For transplacental gene delivery to living embryos in utero (Gratsch et al., 2003; O'Shea K et al., 2006), a complex of siRNA (10 µg) and ExGen transfection reagent (Fermentas) was prepared according to manufacturer’s instruction and the mixture was injected intravenously through the retro-orbital vein into timed-pregnant dams (E9) under isoflurane anesthesia. siRNA sequences are shown in Supplemental Methods Table 1. RNA was purified from the first pharyngeal arch of E11 embryos and gene knockdown was confirmed using qRT-PCR. Nrp2 −/− mice were a gift from S. Takashima (Takashima et al., 2002).
Cell culture
Using sterile technique, the first pharyngeal arch was dissected from E10 embryos and treated with dispase II (2.4 U/ml; Roche) and DNase I (Qiagen) at 37 C for 23 min. After the epithelium and mesenchyme were separated using fine forceps, the presumptive dental mesenchyme (DM) was dissected out and physically triturated several times using fire-polished Pasteur pipette before being cultured on fibronectin (FN) (Becton Dickinson)-coated glass bottom dishes (MatTek Corporation) in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FCS. We confirmed the purity of the isolated DE for cell culture and DM overlay studies using GFP-labeled dental epithelial cells isolated from keratin (K)-14/GFP transgenic mice from Jackson Laboratory. The DM cells were passaged by using FN-coated microcarrier beads for first several passages (Thermo Scientific). DM functional capacity, as measured by their ability to undergo cell compaction-dependent odontogenic (Pax9/Msx1/BMP4) induction, is preserved in cultured DM cell for at least 7 cell passages, and all studies utilized cells at passage 4.
In Vitro Mesenchymal Induction Assay
Mesenchymal cells were GFP-labeled with retroviral transduction (Mammoto et al., 2009) and plated on FN-coated glass bottom dishes at the density of 1.5 × 105 cells/cm2. After culturing the mesenchymal cells for 16 h at a point when they reached ~90% confluence, freshly isolated dental epithelium (DE) was overlaid on top of the monolayer. In some experiments, isolated DE was labeled with CellTracker Red CMTPX (Invitrogen) according to the manufacturer’s instructions before the overlay. Preparations were maintained using a LiveCell™ Stage Top Incubation System (Pathology Devices, Inc.) and images were acquired every 15 min using IP lab software.
Cellular and Molecular Biological Methods
Quantitative RT–PCR (qRT-PCR) was performed with the Quantitect SYBR Green RT–PCR kit (Qiagen) using ABI7300 real-time PCR system (Applied Biosystems); the primers used are shown in Supplemental Methods Table 2. Methods for construction of constitutively active RhoA and generation of viral vectors were previously described (Mammoto et al., 2007). HEK 293 cells were transiently transfected with Fgf8 and/or Sema3F cDNA using Superfect (Qiagen) and labeled with CellTracker Red CMTPX according to the manufacturer’s instructions. A small pellet was made from 1×103 HEK293 cells in polydimethylsiloxane (PDMS) molds for overlay experiments. RhoA activity assay was performed using Rho activation assay kit (Cytoskeleton). For cell migration assay, Transwell membranes (Coster) were coated with 0.5% gelatin, and cells were seeded (105 cells per 100 µl) with DMEM supplemented with 0.2% FCS. Cells were stained with Giemsa solution 16 h later, and counted in ten random fields (at 200× magnification).
Fabrication of Microfluidic Devices
For analysis of the effects of Fgf8 and/or Sema3F gradients on cell packing, a microfluidic device was fabricated from PDMS (Wang et al., 2004) and bonded to a glass cover slip on which FN was coated and cells were plated by introduction through the microfluidic channel. After 16 h incubation, cells were exposed to gradients of Fgf8 and/or Sema3f for 40 h by infusing Fgf8 and/or Sema3f (1 µg/ml in DMEM with 0.5 % FCS) and control medium into two inlets of a gradient generator chamber at a flow rate of 1 µl/min. The overlapping gradient of Fgf8 and Sema3f was generated by adding an additional inlet channel to the left side of the last branching point of the original microfluidic gradient generator (Fig. 2c top) (Dertinger et al., 2001). These devices use laminar flow streams to maintain a constant gradient over time.
Fabrication of Micropatterned Substrates
Our microcontact printing method for producing glass substrates containing micrometer-sized ECM islands has been described previously (Chen et al., 1997; Dike et al., 1999; Singhvi et al., 1994). In brief, PDMS stamps were cast, cured, and removed from master templates, which were created using photolithographic methods. Stamps were coated with FN (50 µg/ml) for 1 hr, dried with compressed nitrogen. Flat and thin PDMS substrate were prepared on the cover glass and UV oxidized for 5 min (Ellsworth Adhesives), stamped with FN, blocked with Pluronic F108 (Sigma) for 2 hr, and rinsed three times with PBS before plating the cells. Cells on the micropatterned substrates were lysed and harvested using a cell scraper, and odontogenic induction was analyzed by qRT-PCR.
Mechanical Compression of Mesenchymal tissues
We fabricated a mechanical compressor (base and piston) using flexible, gas-permeable PDMS elastomer. One PDMS membrane was sealed to the bottom of the base and the entire compressor rested on a fine wire mesh for support (Fig. 4d). Freshly dissected DM from E11 embryos was placed in the chamber of the device, and a metal weight (30g) was then placed on the PDMS piston to generate a constant pressure (1 kPa) on the tissue for 16–48 h of culture.
Statistical analysis
Statistical significance was evaluated using an unpaired Student’s t-test after ANOVA analysis (all data are presented as mean ± S.E.M).
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Gee, J. Sero, D. Bielenberg, A. Shimizu, E. Jiang, A. Jiang, YA Minamishima, K. Otsu, K Johnson, X. Liu, D. Stanton, B. Hashmi, and X. Wang for the technical assistance and R. Maas, D. O’Connell, B. Gerami-Naini, I. Agoulnik, D. Nusinow, O. Campas, J. Costello and H. Li for helpful discussions and comments on the manuscript. This work was supported by grants from NIH (RL1 DE019023 to D.E.I), AHA (to A.M.), Hearst Foundation (to A.M), Children’s Hospital House Officer Development Award (to A.M.) and by the Wyss Institute for Biologically Inspired Engineering at Harvard University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions: T.M. conceived of the experiments, and performed experiments, designed research and analyzed data with assistance from A.M., Y.T., A.G., T.T., R.D., R.M., M.D.B., C.W.Y., D.H. and D.E.I. T.M. wrote the manuscript with D.E.I.
REFERENCES
- Angele P, Schumann D, Angele M, Kinner B, Englert C, Hente R, Fuchtmeier B, Nerlich M, Neumann C, Kujat R. Cyclic, mechanical compression enhances chondrogenesis of mesenchymal progenitor cells in tissue engineering scaffolds. Biorheology. 2004;41:335–346. [PubMed] [Google Scholar]
- Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–4718. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- Bielenberg DR, Hida Y, Shimizu A, Kaipainen A, Kreuter M, Kim CC, Klagsbrun M. Semaphorin 3F, a chemorepulsant for endothelial cells, induces a poorly vascularized, encapsulated, nonmetastatic tumor phenotype. J Clin Invest. 2004;114:1260–1271. doi: 10.1172/JCI21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- Carter DR, Wong M. Mechanical stresses and endochondral ossification in the chondroepiphysis. J Orthop Res. 1988;6:148–154. doi: 10.1002/jor.1100060120. [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- Christley S, Alber MS, Newman SA. Patterns of mesenchymal condensation in a multiscale, discrete stochastic model. PLoS Comput Biol. 2007;3:e76. doi: 10.1371/journal.pcbi.0030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, Yang TL, Huang HR, Hsu SM, Cheng HJ, Huang PH. Semaphorin signaling facilitates cleft formation in the developing salivary gland. Development. 2007;134:2935–2945. doi: 10.1242/dev.005066. [DOI] [PubMed] [Google Scholar]
- Dertinger SKW, Chiu DT, Jeon NL, Whitesides GM. Generation of Gradients Having Complex Shapes Using Microfluidic Networks. Anal Chem. 2001;73:1240–1246. [Google Scholar]
- Dike LE, Chen CS, Mrksich M, Tien J, Whitesides GM, Ingber DE. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. In Vitro Cell Dev Biol Anim. 1999;35:441–448. doi: 10.1007/s11626-999-0050-4. [DOI] [PubMed] [Google Scholar]
- Downie SA, Newman SA. Different roles for fibronectin in the generation of fore and hind limb precartilage condensations. Dev Biol. 1995;172:519–530. doi: 10.1006/dbio.1995.8068. [DOI] [PubMed] [Google Scholar]
- Farge E. Mechanical induction of Twist in the Drosophila foregut/stomodeal primordium. Curr Biol. 2003;13:1365–1377. doi: 10.1016/s0960-9822(03)00576-1. [DOI] [PubMed] [Google Scholar]
- Folkman J, Klagsbrun M, Sasse J, Wadzinski M, Ingber D, Vlodavsky I. A heparin-binding angiogenic protein--basic fibroblast growth factor--is stored within basement membrane. Am J Pathol. 1988;130:393–400. [PMC free article] [PubMed] [Google Scholar]
- Forgacs G, Newman SA. Biological Physics of the Developing Embryo. Cambridge University Press; 2005. [Google Scholar]
- Frenz DA, Jaikaria NS, Newman SA. The mechanism of precartilage mesenchymal condensation: a major role for interaction of the cell surface with the amino-terminal heparin-binding domain of fibronectin. Dev Biol. 1989;136:97–103. doi: 10.1016/0012-1606(89)90133-4. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Gonzalez C, Bronner-Fraser M. Neuropilin 2/semaphorin 3F signaling is essential for cranial neural crest migration and trigeminal ganglion condensation. Dev Neurobiol. 2007;67:47–56. doi: 10.1002/dneu.20326. [DOI] [PubMed] [Google Scholar]
- Gratsch TE, De Boer LS, O'Shea KS. RNA inhibition of BMP-4 gene expression in postimplantation mouse embryos. Genesis. 2003;37:12–17. doi: 10.1002/gene.10221. [DOI] [PubMed] [Google Scholar]
- Grima R, Schnell S. Can tissue surface tension drive somite formation? Dev Biol. 2007;307:248–257. doi: 10.1016/j.ydbio.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK, Miyake T. The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis. Anat Embryol (Berl) 1992;186:107–124. doi: 10.1007/BF00174948. [DOI] [PubMed] [Google Scholar]
- Hall BK, Miyake T. Divide, accumulate, differentiate: cell condensation in skeletal development revisited. Int J Dev Biol. 1995;39:881–893. [PubMed] [Google Scholar]
- Hall BK, Miyake T. All for one and one for all: condensations and the initiation of skeletal development. Bioessays. 2000;22:138–147. doi: 10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hentschel HG, Glimm T, Glazier JA, Newman SA. Dynamical mechanisms for skeletal pattern formation in the vertebrate limb. Proc Biol Sci. 2004;271:1713–1722. doi: 10.1098/rspb.2004.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Simons M. Branching morphogenesis. Circ Res. 2008;103:784–795. doi: 10.1161/CIRCRESAHA.108.181818. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Fibronectin controls capillary endothelial cell growth by modulating cell shape. Proc Natl Acad Sci U S A. 1990;87:3579–3583. doi: 10.1073/pnas.87.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr Opin Cell Biol. 2004;16:580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Knudson CB, Toole BP. Changes in the pericellular matrix during differentiation of limb bud mesoderm. Dev Biol. 1985;112:308–318. doi: 10.1016/0012-1606(85)90401-4. [DOI] [PubMed] [Google Scholar]
- Korostylev A, Worzfeld T, Deng S, Friedel RH, Swiercz JM, Vodrazka P, Maier V, Hirschberg A, Ohoka Y, Inagaki S, et al. A functional role for semaphorin 4D/plexin B1 interactions in epithelial branching morphogenesis during organogenesis. Development. 2008;135:3333–3343. doi: 10.1242/dev.019760. [DOI] [PubMed] [Google Scholar]
- Kulyk WM, Upholt WB, Kosher RA. Fibronectin gene expression during limb cartilage differentiation. Development. 1989;106:449–455. doi: 10.1242/dev.106.3.449. [DOI] [PubMed] [Google Scholar]
- Mammoto A, Connor KM, Mammoto T, Yung CW, Huh D, Aderman CM, Mostoslavsky G, Smith LE, Ingber DE. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407–1420. doi: 10.1242/dev.024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammoto T, Parikh SM, Mammoto A, Gallagher D, Chan B, Mostoslavsky G, Ingber DE, Sukhatme VP. Angiopoietin-1 requires p190RhoGAP to protect against vascular leakage in vivo. J Biol Chem. 2007;282:23910–23918. doi: 10.1074/jbc.M702169200. [DOI] [PubMed] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–127. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- Neubuser A, Peters H, Balling R, Martin GR. Antagonistic interactions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Newman SA, Bhat R. Activator-inhibitor dynamics of vertebrate limb pattern formation. Birth Defects Res C Embryo Today. 2007;81:305–319. doi: 10.1002/bdrc.20112. [DOI] [PubMed] [Google Scholar]
- Newman SA, Christley S, Glimm T, Hentschel HG, Kazmierczak B, Zhang YT, Zhu J, Alber M. Multiscale models for vertebrate limb development. Curr Top Dev Biol. 2008;81:311–340. doi: 10.1016/S0070-2153(07)81011-8. [DOI] [PubMed] [Google Scholar]
- Newman SATJ. Extracellular Matrices. Vol 2. Peading, Mass: Harwood Academic; 1996. Morphogenesis of connective tissues. [Google Scholar]
- Newman TJ, Grima R. Many-body theory of chemotactic cell-cell interactions. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;70 doi: 10.1103/PhysRevE.70.051916. 051916. [DOI] [PubMed] [Google Scholar]
- O'Shea KS, De Boer LS, Slawny NA, Gratsch TE. Transplacental RNAi: deciphering gene function in the postimplantation-staged embryo. J Biomed Biotechnol. 2006;2006:1–12. doi: 10.1155/JBB/2006/18657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlender SA, Tuan RS. Expression and functional involvement of N-cadherin in embryonic limb chondrogenesis. Development. 1994;120:177–187. doi: 10.1242/dev.120.1.177. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Modino SA, Miletich I, Sharpe PT. Stem-cell-based tissue engineering of murine teeth. J Dent Res. 2004;83:518–522. doi: 10.1177/154405910408300702. [DOI] [PubMed] [Google Scholar]
- Okamura D, Kimura T, Nakano T, Matsui Y. Cadherin-mediated cell interaction regulates germ cell determination in mice. Development. 2003;130:6423–6430. doi: 10.1242/dev.00870. [DOI] [PubMed] [Google Scholar]
- Oster GF, Murray JD, Harris AK. Mechanical aspects of mesenchymal morphogenesis. J Embryol Exp Morphol. 1983;78:83–125. [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofsen J, Klein-Nulend J, Burger EH. Mechanical stimulation by intermittent hydrostatic compression promotes bone-specific gene expression in vitro. J Biomech. 1995;28:1493–1503. doi: 10.1016/0021-9290(95)00097-6. [DOI] [PubMed] [Google Scholar]
- Shigemura N, Kiyoshima T, Kobayashi I, Matsuo K, Yamaza H, Akamine A, Sakai H. The distribution of BrdU- and TUNEL-positive cells during odontogenesis in mouse lower first molars. Histochem J. 1999;31:367–377. doi: 10.1023/a:1003796023992. [DOI] [PubMed] [Google Scholar]
- Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–698. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- Smith MM, Hall BK. Development and evolutionary origins of vertebrate skeletogenic and odontogenic tissues. Biol Rev Camb Philos Soc. 1990;65:277–373. doi: 10.1111/j.1469-185x.1990.tb01427.x. [DOI] [PubMed] [Google Scholar]
- Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji J, Hirota S, Kitamura Y, et al. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc Natl Acad Sci U S A. 2002;99:3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I. Developmental biology and building a tooth. Quintessence Int. 2003;34:613–620. [PubMed] [Google Scholar]
- Toyofuku T, Yoshida J, Sugimoto T, Yamamoto M, Makino N, Takamatsu H, Takegahara N, Suto F, Hori M, Fujisawa H, et al. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev Biol. 2008;321:251–262. doi: 10.1016/j.ydbio.2008.06.028. [DOI] [PubMed] [Google Scholar]
- Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu Rev Cell Dev Biol. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- Urban AE, Zhou X, Ungos JM, Raible DW, Altmann CR, Vize PD. FGF is essential for both condensation and mesenchymal-epithelial transition stages of pronephric kidney tubule development. Dev Biol. 2006;297:103–117. doi: 10.1016/j.ydbio.2006.04.469. [DOI] [PubMed] [Google Scholar]
- Wang SJ, Saadi W, Lin F, Minh-Canh Nguyen C, Li Jeon N. Differential effects of EGF gradient profiles on MDA-MB-231 breast cancer cell chemotaxis. Exp Cell Res. 2004;300:180–189. doi: 10.1016/j.yexcr.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.