Introduction
Reactive nitrogen species (RNS) are formed in a wide array of diseases, including Alzheimer’s disease, atherosclerosis, and stroke [1]. In these settings, there is robust production of both nitric oxide (NO) and superoxide (O2−), which combine in a near diffusion-rate limited reaction to form peroxynitrite (ONOO−) [2]. In turn, ONOO− has the ability to modify a variety of amino acids on proteins, including oxidation of sulfur-containing amino acids (Cys and Met) and nitration of aromatic amino acids (Tyr, Trp, Phe and His), often resulting in modulation of the modified protein’s function [3].
To date, the vast majority of research on RNS-derived protein modifications has focused on the nitration of Tyr residues to form 3-nitrotyrosine (3-NT). However, there is growing evidence that another nitrated amino acid, nitrotryptophan (NO2-Trp), may also play a significant role in the cellular regulation during nitrosative stress (reviewed previously, see [4]). The goal of this review is to describe our current knowledge regarding the frequency and consequences of Trp nitration in order to enhance our understanding of the full impact of RNS in health and disease.
Modification of Free Tryptophan by Reactive Nitrogen Species
In order to understand how NO2-Trp is formed, it is useful to compare it with what is already known about the formation of 3-NT. For example, in the case of in vitro 3-NT formation by ONOO−, it has been shown that ONOO− does not directly react with Tyr residues [2]. Instead, ONOO− is rapidly protonated at neutral pH to form its conjugate acid, peroxynitrous acid (ONOOH), which undergoes rapid decomposition to form hydroxyl radical (OH·) and nitrogen dioxide (·NO2). Either of these radicals can abstract a hydrogen atom from the 3-carbon of Tyr’s benzene ring to form a tyrosyl radical, which then reacts with either ·NO2 (to form 3-nitrotyrosine), OH· (to form 3-hydroxytyrosine), or with a second tyrosyl radical (to form 3,3-dityrosine) [2].
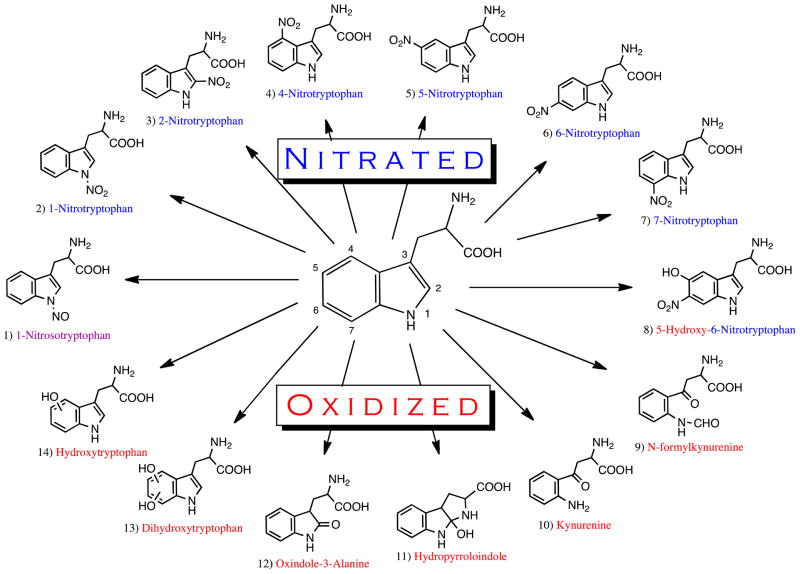
Although far less is known about the reaction mechanism between ONOO− and Trp, it is likely that a similar sequence of events occurs as that which has been reported for 3-NT formation. Indeed, tryptophanyl radicals have been observed in response to ONOO− treatment [5], hinting that Trp modification by ONOO− also proceeds through a radical intermediate. However, unlike Tyr, which is only susceptible to modification at a single carbon on its benzene ring, the indole side-chain of Trp has numerous reactive sites including the 2-, 4-, 5-, 6-, and 7- carbons, as well as the 1- nitrogen [4]. Thus, 1-, 2-, 4-, 5-, 6- and 7-nitrotryptophan are all possible products of ONOO− reactivity with Trp, in addition to the formation of diverse oxidation products that also proceed through a tryptophanyl radical at one of these sites (Figure 1). In addition, the 1-nitrogen site has also been shown to undergo nitrosation (addition of NO) in response to ONOO− treatment [6, 7]. This occurs when ONOO− reacts with OH− to form a peroxynitrite radical (ONOO·), followed by decomposition to NO, which then reacts with Trp [8]. Finally, unlike Tyr residues, Trp can also react directly with ONOO−, which likely results in additional oxidation products.
Figure 1. Potential tryptophan modifications by nitrating agents.
A diagram of all tryptophan modifications that have been identified to date.
As evidenced by the myriad of possible nitration and oxidation products listed in Figure 1, Trp is clearly susceptible to a far greater array of RNS-mediated modifications than is tyrosine. While this may add complications to the investigation of Trp nitration/oxidation, it also presents the possibility that modification of Trp may lead to a more diverse set of biological consequences than is possible for Tyr modification. For this reason, it is important to recognize conditions that preferentially lead to each potential Trp modification. To do this, we will first consider the reaction of free Trp with different nitrating species (Table 1) followed by the modifications of protein-bound Trp in the next section (Table 2).
Table 1.
Publications describing modifications of non-proteinaceous tryptophan by nitrating agents
| Starting Molecule | Nitrating Agent | Modifications | Detection Method | Reference |
|---|---|---|---|---|
| 5mM L-Trp | 0.8mM ONOO− | 6-NO2-Trp, 5-NO2-Trp, 4-NO2-Trp, 7-NO2-Trp | HPLC/UV-Vis | [10] |
| 10mM L-Trp | 0.5–5mM ONOO− | 6-NO2-Trp | HPLC/UV-Vis | [11] |
| 10mM ONOO− | 6-NO2-Trp, 2 x-NO2-Trps, x-OH-Trp, N-formylkinurenine or dihydrotryptophan | |||
| Boc-Trp | 1mM ONOO− | Oxindole, hydropyrroloindole, N-formylkinurenine | HPLC/UV-Vis | [12] |
| 6mM N-Ac-L-Trp | 33.4mM ONOO− | 6-NO2-Trp, 4-NO2-Trp, 7-NO2-Trp, 1-NO-Trp, 1-NO2-Trp | HPLC/UV-Vis | [6] |
| Gaseous ·NO2 | ||||
| LPO/H2O2/NO2− | ||||
| HRP/H2O2/NO2− | ||||
| NO2+ | 6-NO2-Trp, 4-NO2-Trp, 7-NO2-Trp, 5-NO2-Trp | |||
| 0.5mM N-Ac-L-Trp | 0.5mM ONOO− | 6-NO2-Trp, 1-NO-Trp, 1-NO2-Trp, N-formylkinurenine | HPLC/UV-Vis | [7] |
| MPO/H2O2/NO2− | 6-NO2-Trp, 1-NO2-Trp, N-formylkinurenine | |||
| 0.5mM SIN-1 | 1-NO-Trp, N-formylkinurenine | |||
| 0.5mM Angeli’s Salt | ||||
| 0.5mM spermine NONOate | 1-NO-Trp | |||
| 250uM L-Trp | metMb/H2O2/NO2− | 6-NO2-Trp, 4-NO2-Trp, 5-NO2-Trp | HPLC/UV-Vis | [15] |
| 2mM N-Ac-L-Trp | Gaseous ·NO2 | 1-NO-Trp | UV-Vis and NMR | [13] |
| NaNO2/CH3COOH | ||||
| 0.5mM MAMA NONOate | ||||
| N2O3 | ||||
| L-Gly-L-Trp | NaNO2/HCl | 1-NO-Trp | UV-Vis and MS | [16] |
Table 2.
Publications describing modifications of proteinaceous-tryptophan by nitrating agents
| Protein | Source | Nitrating Agent | Tryptophan Modifications | Modification Site(s) | Detection Method | Reference |
|---|---|---|---|---|---|---|
| BSA | Pure protein | ONOO− | 6-NO2-Trp | Not identified | HPLC-ECD | [7] |
| BSA | Pure protein | ONOO- | 6-NO2-Trp,4-NO2-Trp, 2-NO2-Trp | Not identified | MRM with LC-MS/MS | [21] |
| Apo, met, and oxyMyoglobin | Pure protein | ONOO- | 6-NO2-Trp, 4-NO2-Trp, 5-NO2-Trp | Not identified | HPLC/UV-Vis | [15] |
| apoHemoglobin | ||||||
| Met and oxyHemoglobin | 6-NO2-Trp, 4-NO2-Trp | |||||
| FGF-1 | Pure protein | ONOO- | x-NO2-Trp | NW121FVGLK | ESI Mass Spec | [23] |
| Mitochondrial creatine kinase | Pure protein | ONOO- | x-NO2-Trp | GW264EFMW268NER | MALDI-TOF | [24] |
| human frataxin | Pure protein | ONOO- | x-NO2-Trp | QIW155LSSPSSGPK | LC-MS/MS | [25] |
| human Cu,Zn-SOD | Pure protein | ONOO−/CO2 | 6-NO2-Trp, 5-NO2-Trp, kinurenine, oxindole-3-alanine, dihydroxytryptophan | VW32GSIK | HPLC/UV-Vis | [20] |
| MPO/H2O2/NO2 | ||||||
| Hen egg-white lysozyme | Pure protein | ONOO−/CO2 | 6-NO2-Trp | W62W63CNDGR | HPLC/UV-Vis | [26] |
| GTDVQAW123IR | ||||||
| M2 pyruvate kinase | PC12 cells | ONOO- | 6-NO2-Trp | Not identified | 2D-gel electrophoresis/MS | [61] |
| Mitochondrial aconitase 2 | ||||||
| Eukatyotic translation elongation factor 2 | ||||||
| Pyruvate carboxylase | ||||||
| Vinculin | ||||||
| Sarcosine dehydrogenase | ||||||
| Heat shock protein 90α | ||||||
| L-lactate dehydrogenase A | NGF and untreated-PC12 cell lysates | ONOO- | 6-NO2-Trp | SLNPQLGTDADKEQW227K | 2D-gel electrophoresis/MS | [58] |
| SADTLW324GIQK | ||||||
| NGF-treated PC12 cells | LLIVSNPVDILTYVAW148 K | |||||
| GYTSW250AIGLSVADLAESIMK | ||||||
| Malate dehydrogenase 1 | NGF and untreated-PC12 cell lysates | EVGVYEALKDDSW218LK | ||||
| Transaldolase | NGF-treated PC12 cell lysates | LSSTW147EGIQAGK | ||||
| Lactoylglutathione lyase | NGF-treated PC12 cell lysates | GLAFVQDPDGYW171IEILNPNK | ||||
| M2 pyruvate kinase | Untreated PC12 cell lysates | DAVLDAW482AEDVDLR | ||||
| Heat shock protein 90α | Untreated PC12 cell lysated | HNDDEQYAW162ESSAGGFTVR | ||||
| SLTNDW321EEHLAVK |
Since the first report that Trp fluorescence decreases in response to gaseous ·NO2 [9], numerous groups have attempted to elucidate the precise products resulting from the reaction of free Trp with various nitrating species, often arriving at contradictory results. The earliest of these studies focused on the reaction of L-Trp with ONOO− and used high performance liquid chromatography (HPLC) coupled to ultraviolet-visible light (UV-Vis) spectrophotometry to identify the reaction products. In 1996, Padmaja et al. [10] found that treatment of 5mM L-Trp with 0.8mM ONOO− resulted in the formation of 5-nitrotryptophan (5-NO2-Trp; Figure 1, compound 5) and 6-nitrotryptophan (6-NO2-Trp; Figure 1, compound 6), with 5-NO2-Trp being the major product observed at lower pH and 6-NO2-Trp the major product at neutral pH. Also in 1996, Alvarez et al. [11] reported that the products of this same reaction were highly concentration-dependent. When ONOO− concentrations were lower than L-Trp (0.5–5mM ONOO− to 10mM L-Trp), they found that 6-NO2-Trp was the only product formed. However, when ONOO− and L-Trp concentrations were equal (10mM each), numerous additional products were formed, including two unidentified NO2-Trp isomers, hydroxytryptophan (Figure 1, compound 14), and either N-formylkynurenine (a Trp indole ring-open product; Figure 1, compound 9) or dihydroxytryptophan (Figure 1, compound 13). On the other hand, Kato et al. [12] reported that equimolar concentrations of ONOO− and tert-butoxycarbonyl-L-tryptophan (Boc-Trp) only resulted in oxindole-3-alanine (Figure 1, compound 12), hydropyrroloindole (Figure 1, compound 11), and N-formylkynurenine, with no nitration products observed. However, the latter authors indicated that they did not attempt to identify any additional products that eluted at later retention times than Boc-Trp during separation on their C18 column by HPLC, which would be expected for any NO2-Trp isomers [6].
In addition to these earlier studies, two investigations have since reported on the reaction between ONOO− and N-acetyl-L-tryptophan (N-Ac-Trp). In 2004, Sala et al. [6] found that treatment with excess ONOO− resulted in the production of 6-NO2-Trp, 4-nitrotryptophan (4-NO2-Trp; Figure 1, compound 4), and 7-nitrotryptophan (7-NO2-Trp; Figure 1, compound 7), plus two species with modifications on the 1-nitrogen: 1-nitrotryptophan (1-NO2-Trp; Figure 1, compound 2) and 1-nitrosotryptophan (1-NO-Trp; Figure 1, compound 1). Also in 2004, Suzuki et al. [7] observed four primary products when they reacted equimolar concentrations of ONOO− and N-Ac-Trp: 6-NO2-Trp, 1-NO-Trp, 1-NO2-Trp, and N-formylkynurenine,.
It is interesting that 1-NO-Trp and 1-NO2-Trp were found as major products in the Sala et al. and Suzuki et al. studies, but were not identified at all in the earlier studies. This may be due to the relatively low stability and chromatographic characteristics of 1-NO-Trp and 1-NO2-Trp. Notably, while 6-NO2-Trp is stable for up to 6 days in pH 7.4 buffer at 37°C, 1-NO-Trp and 1-NO2-Trp have half-lives of 1.5 hours and 18 hours under these conditions, respectively [7]. Additionally, 1-NO-Trp and 1-NO2-Trp consistently eluted after much longer retention times during separation on C18 columns by HPLC [6, 7]. Therefore, it is possible that the earlier studies either analyzed their samples too late or neglected to analyze compounds with longer chromatographic retention times. Also, in the case of 1-NO-Trp formation, it has been shown that N-Ac-Trp is more likely to be nitrosated than L-Trp, due to interference from the unblocked N-terminal amino group on L-Trp [13]. This is an important distinction, however, since N-Ac-Trp is more structurally related to the protein-bound Trp residues modified in vivo, which also do not possess a free N-terminal amino group.
In addition, there are other disparities between the in vitro experiments described above and what may actually occur with proteinaceous Trp in vivo. For example, in all of the reactions described above, the reported oxidation and nitration of Trp is caused either by direct reaction with ONOO−, or by reaction with the breakdown products of ONOO− at neutral pH (i.e. OH· and ·NO2). However, neither of these reactions is likely to occur in a cellular system, given the high levels of CO2 in vivo [2]. It has been reported that ONOO− reacts readily with CO2 to form nitrosoperoxycarbonate (ONOOCO2−), which then rapidly decomposes to ·NO2 and carbonate radical (CO3·−) [2]. Given the fact that the cellular concentration of CO2 is nearly 1mM, this reaction of ONOO− with CO2 will readily occur in vivo and at a faster rate than either the direct reaction of ONOO− with amino acids or the homologous decomposition of ONOO− to OH· and ·NO2 [2]. Since CO3·−, much like OH·, has the ability to abstract hydrogen atoms from Tyr aromatic rings (and thus, probably from Trp as well), the primary mechanism of both Tyr and Trp nitration in vivo is expected to be CO3·−-induced formation of a tyrosinyl or tryptophanyl radical, followed by the direct addition of ·NO2 to form the nitrated compound [2].
Given this information, it is important to investigate the reaction of free Trp with ONOO− in the presence of CO2, especially since it has been shown that reaction with CO2 enhances the aromatic nitration activity of ONOO−, while minimizing its oxidation activity [14]. In agreement with this, Suzuki et al. [7] found that the nitration profile of N-Ac-Trp changed dramatically when the ONOO− reaction was performed in the presence of 10mM sodium bicarbonate (which is in equilibrium with CO2). In the absence of bicarbonate, the ratio of ONOO−-induced products was 10% 1-NO-Trp, 3.1% 6-NO2-Trp, 2.6% N-formylkynurenine, and 1.7% 1-NO2-Trp. However, in the presence of bicarbonate, 6-NO2-Trp was 3-times more abundant than both 1-NO-Trp and N-formylkynurenine, and 1-NO2-Trp was 2-times more abundant than the two non-nitrated products. Thus, CO2 pushed the ONOO− reaction towards nitration of Trp.
In addition to ONOO−-derived ·NO2, certain hemoproteins (including numerous peroxidases) have been shown to produce ·NO2 in the presence of nitrite (NO2−) and hydrogen peroxide (H2O2), and this can lead to nitration of tyrosine residues [2]. Thus, it is important to determine whether this nitration mechanism can also result in Trp nitration and, if so, whether the products differ from those found with ONOO−-mediated nitration of Trp. To this end, Sala et al. [6] found that lactoperoxidase (LPO) and horseradish peroxidase (HPO), in the presence of NO2− and H2O2, both produced the exact same ratios of nitration/oxidation products as ONOO−-treatment. Suzuki et al. [7] also showed that myeloperoxidase (MPO), NO2− and H2O2 produced the same products as ONOO−-treatment, with the exception of 1-NO-Trp, which they did not observe. Furthermore, Herold [15] showed that metmyoglobin (metMb), NO2− and H2O2 produced 6-NO2-Trp as a major product and 4-NO2-Trp and 5-NO2-Trp as minor products.
Finally, Sala et al. [6] demonstrated that ·NO2 gas and NO2+ produce the same Trp modifications as with ONOO−-treatment, although 1-NO-Trp was not detected. Interestingly, when Suzuki et al. [7] used the ONOO−-generator 3-morpholinosydnonimime (SIN-1), which produces NO and O2− simultaneously, as well as Angeli’s salt, an HNO donor, they only discovered the formation of 1-NO-Trp and N-formylkynurenine, but no nitrated Trp species.
Although these disparate results may seem perplexing and may even portray Trp nitration as a random process, it is likely that the variability in the reported reaction products by each group arises mainly from variations in experimental parameters, i.e. conditions used for the reactions and the chromatographic detection of compounds. One similarity that is seen, however, is that Trp treatment with equimolar or excess concentrations of ONOO− engenders the widest range of products, including both nitrated and oxidated species. However, given the instability of ONOO−, along with the abundance of amino acids in cells, it is inconceivable that the concentration of ONOO− will ever exceed, or even approach, the level of Trp residues in cellular proteins. Therefore, the most biologically relevant results might be represented in the work from Padmaja et al. [10] and Alvarez et al. [11], who each found that 6-NO2-Trp was either the only product or the major product when Trp was present in excess of ONOO−. Of course, it is possible that 1-NO2-Trp may also be a major product, but was not observed in their studies due to methodological limitations for detection. To this end, Sala et al. [6] determined that both 6-NO2-Trp and 1-NO2-Trp were the only significant products formed when low levels of NO2− were used in their peroxidase/NO2−/H2O2 system of nitration.
Notably, 1-NO-Trp, which was shown to be a significant nitrosation product in some experiments, seemed to decrease in abundance in the presence of physiological concentrations of CO2, which suggests that its role may be limited during periods of in vivo RNS-mediated nitration. Indeed, 1-NO-Trp may be a more relevant species in physiological settings where NO production is uncompromised by the production of excess reactive oxygen species, particularly since 1-NO-Trp has been shown to be the only major product in the reaction of Trp with various NO-donating species, including spermine NONOate [7], MAMA NONOate [13], acidified NO2− [13, 16], gaseous NO [13] and N2O3 [13].
Modification of Tryptophan Residues in Proteins
As was the case with free Trp, fluorescence-studies offered the first evidence that protein-bound Trp can be modified by nitrating agents. Notably, Trp exhibits the strongest fluorescence of any proteinaceous amino acid, with an excitation maximum of 280nm and an emission maximum between 305nm and 355nm, depending on its local environment [17]. Thus, numerous groups have exploited this fluorescence in order to investigate protein-Trp modifications after treatment with nitrating agents.
Kikugawa et al. [9] found that gaseous ·NO2 could decrease Trp-derived fluorescence of bovine serum albumin (BSA), γ-globulin, and α-crystallin. Ischiropoulos and Mehdi [18] also reported decreased fluorescence in ONOO−-treated BSA, with the fluorescence decreasing 12%, 29% and 45% in response to increasing ONOO−:protein concentrations of 1:1, 5:1 and 10:1, respectively. Additionally, Kato et al. [12] reported a decrease in Trp-derived fluorescence in ONOO−-treated collagen IV, as well as BSA. And Yamakura et al. [19] demonstrated that ONOO− treatment leads to a quenching of the fluorescence of human Cu, Zn-SOD, which contains one Trp residue (Trp32) and no Tyr residues. The latter group found that this decrease in Cu, Zn-SOD Trp fluorescence was associated with a 30% loss in enzymatic activity [19].
As with free Trp, it is important to elucidate the Trp modifications that actually occur in proteins, especially since a wide array of nitration and oxidation products are possible. To this end, Yamakura et al. [20] later demonstrated that the ONOO−-induced Trp modifications in human Cu,Zn-SOD included 5- and 6-NO2-Trp, as well as kynurenine (Figure 1, compound 10), oxindole-3-alanine and dihydroxytryptophan. In the case of BSA, Ishii et al. [21] showed that ONOO−-treatment resulted in 2-, 4-, and 6-NO2-Trp formation, with 6-NO2-Trp being the major product. Furthermore, Suzuki et al. [7] demonstrated that 6-NO2-Trp can also form in BSA following treatment with Sin-1, Angeli’s Salt, Spermine NONOate, and acidified NO2−. Additionally, 4-, 5-, and 6-NO2-Trp were identified in ONOO−-treated apoHemoglobin and met-, oxy-, and apo-Myoglobin, while only 4- and 6-NO2-Trp were identified in ONOO-treated met and oxyHemoglobin [22]. Table 2 provides a comprehensive summary of the identified Trp modifications in proteins.
With respect to site-identification, it was shown that Trp121 of acidic fibroblast growth factor (FGF-1) is a target of ONOO−-induced nitration, though the specific nitrated isomer was not identified [23]. Similarly, Trp264 and Trp268 of mitochondrial creatine kinase were identified as targets of ONOO-induced nitration [24], as was Trp155 of human frataxin [25]. Additionally, hen egg-white lysozyme was shown to be nitrated by ONOO−/CO2 on Trp62, Trp63 and Trp123, which was associated with a decrease in enzyme activity [26].
Although only 16 NO2-Trp sites have been specified on proteins to date, the current inventory (summarized in Table 2) still begs the question of whether similarities can be inferred that help explain the basis for specificity of protein-bound Trp nitration. Over the years, a vigorous debate has ensued over the determining factors of Tyr nitration, a topic we discussed elsewhere [27]. While most researchers agree with the assessment by Ischiropoulos and colleagues that a Tyr residue’s solvent-exposure and positioning on a peptide loop structure both confer a greater likelihood of nitration [28], there is some disagreement over the assertion that this likelihood also increases when an acidic amino acid (Glu or Asp) is present nearby or when nearby Cys or Met are absent [29]. Despite uncertainties regarding the actual determinants of Tyr nitration, it is helpful to consider these same assertions as a framework for looking at the determinants of Trp nitration.
As with protein Tyr nitration, it does appear that solvent-exposure is indeed a major factor for determining the susceptibility of protein Trp residues to nitration. For example, in hen egg-white lysozyme, only three of six Trp residues are apparently nitrated after ONOO−/CO2 treatment, and each of these are solvent-exposed [26]. Intriguingly, Trp111 is partially solvent-exposed but does not get nitrated, indicating that solvent-exposure may not be the sole determinant factor in Trp nitration [26]. The presence of adjacent acidic amino acid residues, however, does not appear to have a major contributing role, since only 11 out of 16 (68.75%) of the peptides listed in Table 2 contain a Glu or Asp within four residues of the nitrated Trp residue, which can be explained by chance and is approximately the same as the number of basic amino acid residues (Lys or Arg) found within four residues of the NO2-Trp (10/16 or 62.5%). These percentages are similar to what we previously measured for 3-NT residues, where 73% of identified peptides contained acidic residues within four amino acids, and 73% had basic residues within four amino acids [27]. Nevertheless, one cannot rule out Ischiropoulos and colleagues’ assertion that an adjacent Cys or Met residue may decrease the probability of Tyr or Trp nitration by outcompeting the reaction with nitrating agents. To this end, of the 16 NO2-Trp-containing peptides listed in Table 2, only one peptide has a Cys residue and another has a Met residue within four amino acids of NO2-Trp. This represents 6.25% for Cys and 6.25% for Met, which is slightly less than the 13% of 3-NT-containing peptides where the modified residue was found to be adjacent to Cys, and 13% adjacent to Met [27]. Of course, given the small number of Trp nitration sites identified to date, any statements about determinants of Trp nitration remain speculative. Also, as previously concluded for Tyr nitration [27], it is likely that a Trp residue’s 3D structural environment is an equally important feature in determining its susceptibility to nitration.
Nitrotryptophan Formation In Vivo
The in vitro studies discussed above can be important in establishing the patterns and susceptibilities of Trp nitration elicited by various nitrating species and agents. However, given the multitude of additional variables present in cellular systems, it is vital to investigate whether Trp nitration actually occurs in vivo, and if so, what are the patterns, frequency and consequences of this nitration. Published results are still sparse; however, a few reports have emerged in recent years that are beginning to elucidate the mechanism of in vivo formation of NO2-Trp.
In 2007, Ishii et al. [21] reported the accumulation of protein-bound 4-NO2-Trp and 6-NO2- Trp residues in the livers of mice treated with toxic levels of acetaminophen, with 6-NO2-Trp being the predominant modification. Indeed, acetaminophen toxicity had previously been associated with the formation of 3-NT residues in the liver [30]. Thus, this study offers the first validation that Trp nitration can occur concurrently with Tyr nitration during periods of RNS in vivo. Importantly, Rebrin et al. [31] also reported in 2007 the first identification of a Trp-nitrated protein in vivo, demonstrating a 5-hydroxy-6-nitrotryptophan (5-OH-6-NO2-Trp; Figure 1, compound 8) modification of Trp372 on the mitochondrial metabolic enzyme Succinyl-CoA:3-ketoacid coenzyme A transferase (SCOT) in numerous organs from 4-month old rats. Furthermore, they observed an age-dependent increase in this modification in hearts and brains of these rats, coinciding with a 30% increase in enzymatic activity at 24-months. The authors speculated that this age-dependent accumulation of 5-OH-6-NO2-Trp on SCOT may be a protective mechanism, allowing the heart to better utilize ketone metabolism for energy production at a time when other metabolic processes may be diminished [31].
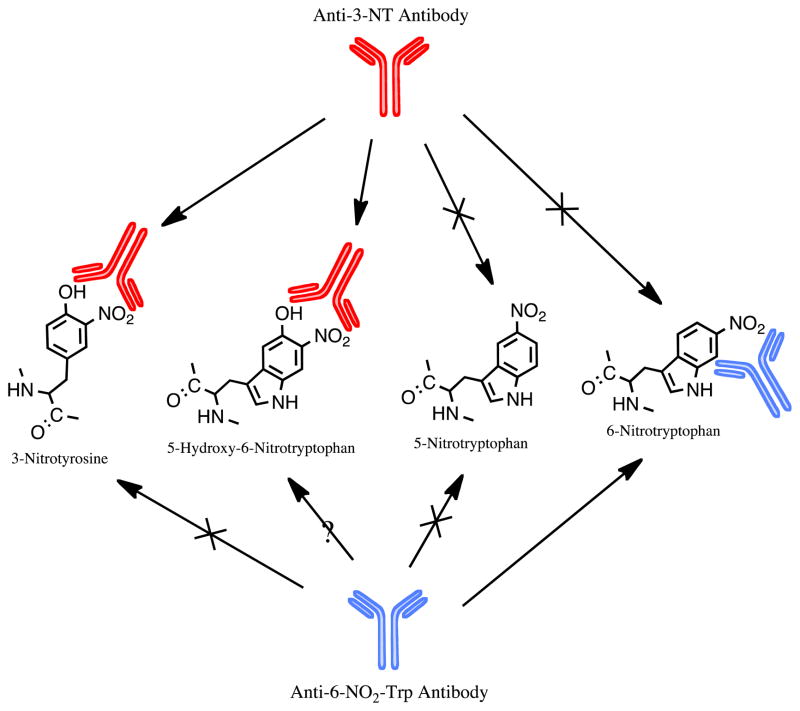
In addition to presenting the first identification of a nitrated Trp-containing protein in vivo, these results from Rebrin et al. [31] also illuminated a number of other interesting characteristics of NO2-Trp biology. For one, they discovered the formation of a 5-OH-6-NO2-Trp residue, thus increasing the diversity of possible Trp modifications that may occur in response to RNS in vivo. Interestingly, the authors demonstrated cross-reactivity for this 5-OH-6-NO2-Trp modification with at least one commercially available antibody directed against 3-NT (clone 1A6) [31]. Indeed, SCOT nitration had been reported previously [32], although it was recognized solely as Tyr nitration based on the reactivity of SCOT with this anti-3-NT antibody. While this cross-reactivity of 3-NT antibodies can be reconciled by the structural similarities between 3-NT and 5-OH-6-NO2-Trp (Figure 2), this finding calls into question the interpretation of other protein nitration sites in which protein 3-NT was identified solely on the basis of its reactivity with an anti-3-NT antibody.
Figure 2. Specificity of antibodies reported to react with nitrotryptophan.
Two antibodies have been shown to recognize NO2-Trp residues in proteins. An anti-3-NT antibody (clone 1A6) has been shown to have affinity for 5-OH-6-NO2-Trp residues, and an anti-6-NO2-Trp antibody has been shown to have affinity for 6-NO2-Trp.
A second intriguing observation from Rebrin et al. was the fact that the nitration and resulting activity changes in SCOT were highly dependent on the concentration of nitrating agent employed. While studying the in vitro effects of ONOO−-treatment on purified SCOT, the authors found that 25μM ONOO− resulted in formation of the expected 5-OH-6-NO2-Trp modification and a concomitant 24% increase in enzyme activity. However, using ONOO− concentrations higher than 50μM resulted in attenuated enzyme activity, coincident with additional post-translational modifications that include oxidation and/or nitration products of various Cys, Met, Tyr, and His residues [31]. It should be noted, however, that in a more recent study on SCOT nitration, Wang et al. observed Tyr nitration, but not Trp nitration, after treatment of mouse-derived recombinant SCOT protein with 20uM ONOO− [33]. It is unclear why these authors were unable to identify the 5-OH-6-NO2-Trp modification reported by Rebrin et al., but it may be due to the fact that the Wang et al. study relied on nitration of a purified, recombinant SCOT protein, whereas the Rebrin et al. study measured nitration of the endogenous protein present in a milieu of other soluble heart mitochondrial proteins. Thus, the specific cellular environment where nitration occurs seems to have a large impact on the nitration patterns of given proteins, a fact that further demonstrates that any in vitro data on protein nitration must be viewed with a measure of caution.
Although the two studies discussed above represent the only in vivo results on NO2-Trp formation reported to date, some additional published studies raise important questions about the biological consequences of NO2-Trp formation. For example, Southan et al. [34] reported that high levels of free 6-NO2-Trp can inhibit indoleamine 2,3-dioxygenase (IOD), which is the rate-limiting enzyme in Trp metabolism and has been shown to play an immunomodulatory role in diverse disease settings such as viral/bacterial host defense, fetal rejection and tumor responses [35, 36]. While the concentrations of 6-NO2-Trp necessary to inhibit IOD (1mM for 52% inhibition) is unlikely to be achieved in vivo, this finding highlights an important and as yet unanswered question: what is the fate of NO2-Trp amino-acids following cellular degradation of the nitrated proteins? Since 6-NO2-Trp inhibits the crucial enzyme in Trp metabolism, it is evident that this and perhaps other NO2-Trp derivatives may not undertake the same metabolic paths as unmodified Trp. Therefore, it will be important to determine the ultimate fate of these modified amino acids. Do they accumulate in cells after acute or chronic periods of nitrosative stress? And if so, do they damage the cells in which they accumulate?
Interestingly, one Trp derivative, 1-NO-Trp, has been shown to at least hold the potential to damage cells. 1-NO-Trp is technically a nitrosamine, which is a class of compounds known to be highly carcinogenic in animals [37]. And similar to other nitrosamines, the 1-NO-Trp form of N-Ac-Trp (N-Ac-NO-Trp) was found to be mutagenic in numerous bacterial strains, including one strain of E. coli and two strains of S. typhimurium [38, 39]. However, it is still unclear whether 1-NO-Trp is also carcinogenic in animals, and if so, whether the accumulating cellular concentration of free 1-NO-Trp could ever rise to levels necessary to promote oncogenesis. An additional intriguing concept is the possibility of a role for either free or protein-bound 1-NO-Trp as a NO donor in normal cellular functioning. Kirsch et al. [13] have shown that protein-bound Trp is nitrosated by N2O3 at a 63% faster rate than Cys, and that N-Ac-NO-Trp releases 5-times more free NO than S-nitrosoglutathione (GSNO) in the presence of ascorbate. Furthermore, Sonnenschein et al. [40] found that N-Ac-NO-Trp could efficiently transnitrosate NO onto glutathione. Additionally, Zhang et al. [16] found that both peptide-bound 1-NO-Trp and Trp-nitrosated BSA had the ability to induce vasorelaxation of rabbit aortic rings. Thus, 1-NO-Trp formation, either during nitrosative stress or during physiological NO production, may play a role in vasorelaxation or other NO-mediated events.
Nitrotryptophan in Bacteria
Given the small but growing evidence that NO2-Trp is indeed formed in biological systems, it is important to question its physiological role. Indeed, during the early phases of 3-NT research, 3-NT accumulation in proteins was simply viewed as an unfortunate byproduct of nitrosative stress, whose only valuable attribute was that it could be used as a tissue biomarker for monitoring exposure to RNS in cells [41]. However, numerous reports in recent years indicate that 3-NT may convey important functional changes to the proteins that get modified during these periods of nitrosative stress [42–44]. A few reports have even hinted that 3-NT may be a specific and reversible modification involved in signal transduction during normal physiology, akin to phosphorylation [45–47]. In terms of NO2-Trp formation, these same possibilities are only beginning to be considered. In this regard, the Ishii et al. study [21] clearly verifies that NO2-Trp can be used as a biomarker of nitrosative stress. Furthermore, the Rebrin et al. study [31] demonstrates that NO2-Trp formation can occur during normal physiological processes and may even be attributed to specific biological effects. However, there are still far too few reports of in vivo NO2-Trp formation to be able to draw firm conclusions about a possible biological role.
Unlike the currently ambiguous role of NO2-Trp in mammals, however, emerging evidence suggests that NO2-Trp may indeed have conserved roles in the normal physiological functioning of prokaryotic systems. In fact, some of the most exciting recent findings in the field of NO2-Trp research have emerged from the study of bacteria. In 2004, a unique tryptophanyl-tRNA synthetase (TrpRS II) from the radioresistant bacterium Deinoccocus radiodurans (D rad) was shown to form a functional complex with D rad nitric oxide synthase (deiNOS), resulting in the regioselective nitration of Trp to form 4-NO2-Trp [48, 49]. The selective production of this 4-NO2-Trp isomer is intriguing, given the aforementioned results showing that 6-NO2-Trp is the predominant species generated from the in vitro reaction of Trp with various nitrating agents [11, 20]. The physiological role of 4-NO2-Trp in D rad has not yet been determined, but the authors speculate that it is unlikely that 4-NO2-Trp is incorporated into proteins during canonical D rad ribosomal synthesis [48]. Instead, they have suggested that TrpRS II and 4-NO2-Trp may be involved in an as yet uncharacterized D rad metabolic biosynthesis pathway [48, 50].
Notwithstanding the undetermined role for 4-NO2-Trp in D rad, 4-NO2-Trp has been shown to play a role in non-ribosomal biosynthesis pathways in other bacterial species. Johnson et al. [51] have demonstrated that in various plant-pathogenic species of the bacterium Streptomyces, 4-NO2-Trp is incorporated into the phytotoxic cyclic dipeptide, thaxtomin A, which is the pathogenicity determinant for infection of potato tubers by these bacteria [52]. Although it had previously been reported that Trp residues were nitrated by Streptomyces NOS (stNOS) during biosynthesis of thaxtomin A [53, 54], Johnson et al. were the first to explicitly demonstrate that the site-specific nitration of Trp occurs prior to peptide assembly. This study represents the first example of a nitrated Trp, whose regioselective nitration requires NOS, being directly utilized in a biosynthesis pathway to produce a bioactive molecule [51].
It is still unclear if stNOS associates with Trp in a multimeric functional complex, or if diffusible free RNS are responsible for the site-specific nitration of Trp during thaxtomin A synthesis. However, it seems unlikely that the less common 4-NO2-Trp isomer would be selectively produced without some level of enzymatic control [53, 54]. It is enticing to speculate that stNOS in complex with a TrpRS II-like synthetase catalyzes Streptomyces 4-NO2-Trp synthesis, as has been reported in D rad, but this has yet to be demonstrated.
Methods of Detecting Nitrotryptophan
A May 2011 Pubmed literature survey revealed 4240 manuscripts published with the term “nitrotyrosine” in either the title or the abstract, but only 23 with “nitrotryptophan.” This begs the question of whether 3-NT is a more abundant and more biologically relevant modification than NO2-Trp, as the publication imbalance would suggest, or if there may be other explanations for why protein 3-NT research has markedly eclipsed NO2-Trp research. One possible explanation is differences in the ability of researchers to identify and quantify nitrated Tyr residues, as compared to nitrated Trp.
In the case of Tyr, the earliest reports of 3-NT detection in proteins used HPLC/UV-Vis and amino acid analysis [55, 56]. Although these studies began in the early 1970’s, as compared to the 1990’s for NO2-Trp research, the earlier start date alone cannot account for the vast disparity in reports focusing on 3-NT vs. NO2-Trp. Indeed, in the first 24 years of research involving 3-NT, only about 100/4240 current 3-NT papers had been published. Then, in 1994, Beckman et al. [57] reported their effective use of monoclonal and polyclonal antibodies for the detection of 3-NT-containing proteins, and this was followed by a virtual explosion in the number of manuscripts published on 3-NT in the ensuing years. This influx of publications on 3-NT was likely due to the relative ease and confidence with which these new antibodies allowed researchers to identify 3-NT-containing proteins. Since that time, numerous additional methods have been developed for identifying 3-NT, including HPLC-based techniques for quantifying total 3-NT in tissues and lysates, and various mass spectrometry (MS) techniques for identifying 3-NT sites in proteins [58].
As is evident from the timeline above, the development of methods to analyze specific modifications plays a critical role in setting the pace for future discoveries. Unfortunately, the methodology for NO2-Trp detection has lagged far behind that of 3-NT detection. The good news, however, is that numerous methods have been developed recently that are anticipated to accelerate protein NO2-Trp research in the years to come.
As it was with 3-NT, the earliest approach used to detect free NO2-Trp relied on HPLC combined with UV-Vis spectrophotometry (Table 1) [6, 7, 10–12]. However, since there are numerous isomers of NO2-Trp, confident identification is still not as straightforward as it is for 3-NT. Indeed, identification of each NO2-Trp isomer requires knowledge of their individual column retention time and optical absorption characteristics. However, chemical standards are not available commercially and thus require custom-synthesis.
Even with synthetic NO2-Trp standards, the selectivity and sensitivity of HPLC/UV-Vis are generally too low to reliably quantify total levels of the various nitrated Trp isomers in biological lysates or extracts. For this reason, researchers have sought more sensitive systems for NO2-Trp detection. For example, Suzuki et al. [7] successfully used HPLC coupled with electrochemical detection (HPLC-ECD) to quantify 6-NO2-Trp in BSA treated with various nitrating agents, and Kawasaki et al. [59] used HPLC-ECD to quantify 6-NO2-Trp in peroxynitrite-treated rat pheochromocytoma (PC12) cell lysates (Table 2). However, HPLC-ECD may still not be sensitive enough to measure the relatively low levels of NO2-Trp that are expected to occur in settings of endogenous protein nitration. For this reason, Ishii et al. [21] recently developed a method for identifying various NO2-Trp isomers using multiple reaction monitoring (MRM) on a liquid chromatography-tandem mass spectrometry (LC-MS/MS) system. Using this approach, they were able to confidently differentiate the parent ions of 2-, 4-, and 6-NO2-Trp, which all have the same mass-to-charge ratio (m/z), based on the distinct m/z’s of their respective daughter ions, formed by collision-induced dissociation. More importantly, Ishii et al. were able to reproducibly quantify endogenous, nanomolar quantities of 4- and 6-NO2-Trp from mouse livers damaged by acetaminophen-toxicity [21].
The above methods can also be used to identify individual NO2-Trp-containing proteins, following complete proteolysis to free amino acids [22, 26]. A limitation, however, is the failure of this approach to identify the specific Trp residues that are nitrated. To overcome this problem, researchers have begun to apply proteomic MS methods to identify NO2-Trp residues in peptides and proteins [23–25, 31]. By trypsinizing the protein prior to MS analysis, specific NO2-Trp-containing peptides can be tentatively identified based on the addition of 45 Da (a nitro- group) [23–25] or 61 Da (a nitro- plus a hydroxyl- group) and confirmed by manual inspection of the MS/MS data [31]. One note of caution, however, is that utilizing SDS-page separation of proteins prior to analysis by Mass Spec has been shown to cause artifactual oxidation of Trp residues, including the formation of kynurenine, hydroxytryptophan, and N-formylkynurenine [60].
Although this has not yet been implemented, a useful approach for validation of the NO2-Trp-peptides identified by MS may be to monitor the formation of an immonium ion that results from free NO2-Trp, produced in the collision cell during an MS/MS experiment. The frequency that a NO2-Trp immonium ion can be detected for diverse NO2-Trp-containing peptides and the optimal collision energy for formation remain to be determined. Notably, a similar strategy has been used to validate 3-NT-containing peptides [61, 62], and it will be important to assess whether the same technique could similarly be used for confident sequencing of NO2-Trp-containing peptides.
Although MS analysis offers a powerful proteomic tool, given the low abundance of endogenous protein nitration in most settings, Mass Spec alone can be insufficient when it comes to identifying NO2-Trp-containing proteins/peptides in complex biological mixtures. For this reason, specific enrichment of the NO2-Trp-containing proteins is usually necessary prior to identification by MS. Because of this, one of the most important advances in the NO2-Trp field may be the recent development of an anti-nitrotryptophan antibody. In 2007, Ikeda et al. [63] reported the first ever production of a polyclonal antibody to 6-NO2-Trp-containing proteins. Since in vitro nitrated proteins regularly show multiple modifications on Trp, they opted to use as their antigen a custom-made 6-NO2-Trp peptide conjugated to keyhole limpet hemocyanin (KLH), for generation of a polyclonal rabbit antibody [63]. This antibody proved to exhibit high affinity and specificity for 6-NO2-Trp. Furthermore, it did not recognize 5-NO2-Trp or 3-NT (Figure 2), and immunoreactivity was lost when the antibody was either saturated with free 6-NO2-Trp or when nitrated moieties of Trp-nitrated proteins were reduced by pretreatment with sodium dithionite [63].
To prove the utility of this new antibody, the authors used two-dimensional (2D) gel electrophoresis coupled with mass spectrometry to identify five previously undiscovered NO2-Trp-containing proteins from ONOO−-treated PC12 cells [63]. They were also able to identify specific sites of Trp nitration in six proteins from ONOO−-treated PC12 lysates, also by using 2D-gel electrophoresis coupled with MS (Table 2) [59]. These experiments demonstrate the potential utility of this antibody to help identify new NO2-Trp-containing proteins. Unfortunately, this 6-NO2-Trp antibody is not yet available commercially.
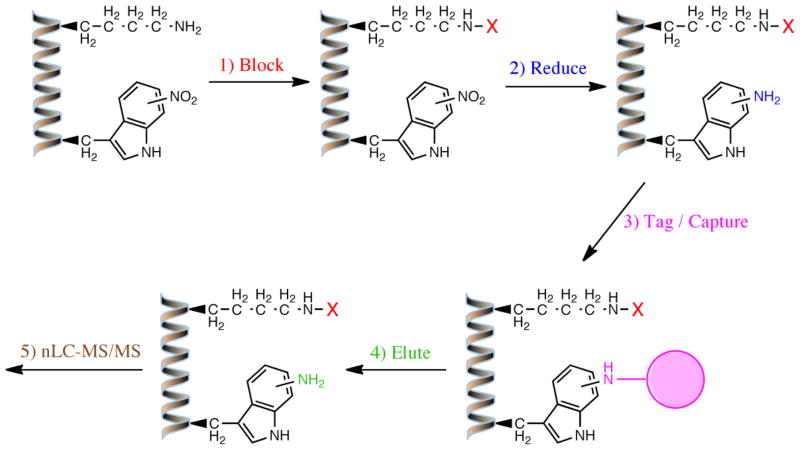
Although this new antibody represents a major step forward, it is not the only methodological advancement that is needed to advance protein NO2-Trp research. Antibody-based proteomic methods such as 2D-gel electrophoresis coupled to MS have been shown to suffer from multiple shortcomings when it comes to unbiased identification of modified proteins in complex biological mixtures, especially with regards to identifying the specific sites of modification [64]. For this reason, our laboratory has sought to develop a new proteomic approach that relies on a solid-phase, chemical-capture strategy for isolation and identification of both 3-NT and NO2-Trp-containing proteins in complex biological samples, as well as specifying cognate sites of nitration [58]. In brief, this method involves blocking all primary amines, reducing 3-NT and NO2-Trp residues to their amino- counterparts, selectively attaching agarose beads to the resulting aromatic amines, and finally, eluting and analyzing the amino acid sequence of captured peptides by nanoLC-MS/MS and database searching (Figure 3). Although this method is still in the optimization phase, we have validated its ability to identify NO2-Trp sites using ONOO−-treated brain homogenates (unpublished results), and we are currently testing it on various endogenously nitrated tissues. Once established, this and related proteomic methods are anticipated to greatly facilitate the identification of NO2-Trp-containing proteins and modification sites, accelerating research on the biological significance of Trp nitration in physiology and pathophysiology.
Figure 3. Solid-phase, chemical-capture strategy for proteome-wide site identification of nitrotryptophan residues in proteins and peptides.
The overall strategy is: (1) block free amines (i.e., N-terminal and lysine); (2) reduce the NO2-group in NO2-Trp to create a new amine; (3) tag/capture with amine-selective agarose beads. The captured peptides are then (4) eluted and (5) analyzed by nano-LC-MS/MS. “X” = chemical moiety used for amine blocking.
Final Analysis
Even with the delay in development of NO2-Trp detection methodologies, there is reason to believe that NO2-Trp modifications do not occur as frequently as 3-NT modifications. For one, Trp is the least prevalent of all the amino acids, representing only ~1% of total amino acid residues in proteins, compared to Tyr, which is ~3–4% prevalent [65]. Secondly, Trp, which has a hydrophobicity value of −2.13 kcal/mol, is more hydrophobic than tyrosine (hydrophobicity value of −1.47kcal/mol). Because of this, the fraction of Trp residues that are found within a protein’s interior is 0.27, compared to 0.15 for Tyr. In other words, Trp is less likely to be solvent-exposed and vulnerable to nitrating agents than Tyr [65]. Lastly, Trp has a slightly higher redox potential (1.015V) than Tyr (0.93V) at pH 7.0 [66], which may explain the slightly slower reaction rate constant of free Trp with ONOO− (1.3 × 102 M−1 s−1) [10], as compared to that for free Tyr with ONOO− (3.63 × 102 M−1 s−1) [67].
Some recent studies confirm that Trp nitration occurs less frequently than Tyr nitration. By comparing 6-NO2-Trp levels to 3-NT levels in nitrated BSA, Suzuki et al. [7] demonstrated that 3-NT was more prevalent than 6-NO2-Trp in the majority of nitrating conditions employed. Similarly, using PC12 cell lysates nitrated with ONOO−, Kawasaki et al. [59] reported a six-fold increase in 3-NT formation relative to 6-NO2-Trp. Plus, in our own preliminary investigations using the affinity-tag method for discovery of nitration sites from ONOO−-treated rat brain homogenates, we observed that, out of 69 total nitrated peptides identified, ~9% contained NO2-Trp, as compared to ~91% with 3-NT (unpublished results). However, the strongest reported difference between 3-NT and NO2-Trp levels comes from the study of acetaminophen-induced liver toxicity by Ishii et al. [21], which stated that measured levels of 6-NO2-Trp and 4-NO2-Trp (nmol range) were 1000-fold less than the 3-NT levels (μmol range) that they had measured in a similar, previously reported acetaminophen-toxicity study [30].
Despite the apparently lower incidence of protein NO2-Trp relative to 3-NT, there is still reason to believe that NO2-Trp formation may play a significant role in physiological and pathophysiological processes in mammalian systems. After all, the biological impact of a modification is not dependent on the shear number of amino acids that are modified, but on whether the specific amino acids that become modified are functionally significant. Thus, even if there are more Tyr residues than Trp residues nitrated in vivo, the real question is how vital to protein function are the Trp residues that become nitrated. To this end, Yamakura and Ikeda [4] have proposed that nitration of solvent-exposed Trp residues may be particularly disruptive since they often serve to facilitate specific interactions with other molecules. Indeed, there is plenty of evidence to support this theory. Since Trp’s indole side-chain has the largest surface area of any amino acid, Trp is often utilized by proteins to help form stable bridges with neighboring molecules, both through hydrogen bonds and hydrophobic interactions with atoms on the indole ring [68, 69]. For example, Trp residues are often present in protein domains used for the binding of various small molecules (Sushi/CCP/SCR domains), carbohydrates (C-type lectin-like domains), nucleotides (BSD domains), and other proteins (WWE domains, WD repeats) [70]. Thus, even though Trp residues may not be nitrated as frequently as Tyr residues, the impact of Trp nitration may be equivalent due to the potential extent of functional disruptions in complex formation and protein signaling.
Lastly, there is the question of the metabolic fate of NO2-Trp-modified proteins. This issue is brought to the forefront by the findings of Rebrin et al. [31], who showed that mitochondrial SCOT, with its 5-OH-6-NO2-Trp modification, is the only identifiable nitrated protein in mitochondria from a variety of rat organs, as detected using an anti-3-NT antibody. This finding is somewhat curious, since mitochondria are known to be a robust source of ONOO− production [2], given the fact that O2− is a natural byproduct of the mitochondrial respiratory chain [2], and NO has been shown to be produced by a putative mitochondrial NOS isoform (mtNOS) [71]. So how do we reconcile the fact that the only nitrated protein that Rebrin et al. observed in these rat mitochondria was actually nitrated on a Trp residue and not a Tyr residue?
One possible explanation may have to do with the protein disposal and repair mechanisms that cells utilize to rehabilitate themselves after periods of nitrosative stress. In the case of 3-NT, for example, it has been reported that Tyr nitration can accelerate a protein’s degradation by the proteasome [72, 73]. Additionally, it has been reported that numerous cells and tissues possess a “denitrase” activity that is capable of either reducing or reversing a 3-NT modification in proteins [45, 74, 75], and that this activity is robustly present in mitochondria [47]. Although the details of these mechanisms have yet to be elucidated for 3-NT (and have not been addressed at all for NO2-Trp), it is certainly possible that the proteins involved in the repair and disposal of 3-NT-containing proteins may not recognize NO2-Trp-containing proteins. This may explain why Trp-nitrated SCOT was found to be abundant in rat mitochondria, while no 3-NT-containing proteins were detected [31]. Indeed, this would have wide-ranging implications for the influence that NO2-Trp modifications may play in the long term, especially after periods of prolonged nitrosative stress.
Clearly much has yet to be learned about the frequency and consequences of NO2-Trp modifications in proteins. At present, the extent to which this modification may impact on normal cellular function or any given disease state is unknown. Nevertheless, there is certainly enough evidence to endorse the need for further research into the biological significance of protein Trp nitration, including in vivo mechanisms and sites of modification. With key advances in immunological and proteomic technologies for enrichment and site specification, along with MRM-based quantification, the pace of research on protein Trp nitration is expected to accelerate markedly during the next few years, providing answers to fundamental questions highlighted in this review.
Acknowledgments
We gratefully acknowledge Dr. Ruba S. Deeb for her helpful discussions and comments in preparing this manuscript. Support was provided by NIH research grants HL087062 and HL046403 (SSG) and predoctoral training grants AG032195-01A1 (TN) and GM073564-04S1 (AH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greenacre CB, Young DW, Behrend EN, Wilson GH. Validation of a novel high-sensitivity radioimmunoassay procedure for measurement of total thyroxine concentration in psittacine birds and snakes. Am J Vet Res. 2001;62:1750–1754. doi: 10.2460/ajvr.2001.62.1750. [DOI] [PubMed] [Google Scholar]
- 2.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- 4.Yamakura F, Ikeda K. Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species. Nitric Oxide. 2006;14:152–161. doi: 10.1016/j.niox.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Pietraforte D, Minetti M. One-electron oxidation pathway of peroxynitrite decomposition in human blood plasma: evidence for the formation of protein tryptophan-centred radicals. Biochem J. 1997;321(Pt 3):743–750. doi: 10.1042/bj3210743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sala A, Nicolis S, Roncone R, Casella L, Monzani E. Peroxidase catalyzed nitration of tryptophan derivatives. Mechanism, products and comparison with chemical nitrating agents. Eur J Biochem. 2004;271:2841–2852. doi: 10.1111/j.1432-1033.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki T, Mower HF, Friesen MD, Gilibert I, et al. Nitration and nitrosation of N-acetyl-L-tryptophan and tryptophan residues in proteins by various reactive nitrogen species. Free Radic Biol Med. 2004;37:671–681. doi: 10.1016/j.freeradbiomed.2004.05.030. [DOI] [PubMed] [Google Scholar]
- 8.Uppu RM, Squadrito GL, Bolzan RM, Pryor WA. Nitration and Nitrosation by Peroxynitrite: Äâ Role of CO2 and Evidence for Common Intermediates. Journal of the American Chemical Society. 2000;122:6911–6916. [Google Scholar]
- 9.Kikugawa K, Kato T, Okamoto Y. Damage of amino acids and proteins induced by nitrogen dioxide, a free radical toxin, in air. Free Radic Biol Med. 1994;16:373–382. doi: 10.1016/0891-5849(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 10.Padmaja S, Ramazenian MS, Bounds PL, Koppenol WH. Reaction of peroxynitrite with L-tryptophan. Redox Report. 1996;2:173–177. doi: 10.1080/13510002.1996.11747045. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez B, Rubbo H, Kirk M, Barnes S, et al. Peroxynitrite-dependent tryptophan nitration. Chem Res Toxicol. 1996;9:390–396. doi: 10.1021/tx950133b. [DOI] [PubMed] [Google Scholar]
- 12.Kato Y, Kawakishi S, Aoki T, Itakura K, Osawa T. Oxidative modification of tryptophan residues exposed to peroxynitrite. Biochem Biophys Res Commun. 1997;234:82–84. doi: 10.1006/bbrc.1997.6587. [DOI] [PubMed] [Google Scholar]
- 13.Kirsch M, Fuchs A, de Groot H. Regiospecific nitrosation of N-terminal-blocked tryptophan derivatives by N2O3 at physiological pH. J Biol Chem. 2003;278:11931–11936. doi: 10.1074/jbc.M300237200. [DOI] [PubMed] [Google Scholar]
- 14.Lemercier JN, Padmaja S, Cueto R, Squadrito GL, et al. Carbon dioxide modulation of hydroxylation and nitration of phenol by peroxynitrite. Arch Biochem Biophys. 1997;345:160–170. doi: 10.1006/abbi.1997.0240. [DOI] [PubMed] [Google Scholar]
- 15.Herold S. Nitrotyrosine, dityrosine, and nitrotryptophan formation from metmyoglobin, hydrogen peroxide, and nitrite. Free Radic Biol Med. 2004;36:565–579. doi: 10.1016/j.freeradbiomed.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YY, Xu AM, Nomen M, Walsh M, et al. Nitrosation of tryptophan residue(s) in serum albumin and model dipeptides. Biochemical characterization and bioactivity. J Biol Chem. 1996;271:14271–14279. [PubMed] [Google Scholar]
- 17.Vivian JT, Callis PR. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys J. 2001;80:2093–2109. doi: 10.1016/S0006-3495(01)76183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ischiropoulos H, al-Mehdi AB. Peroxynitrite-mediated oxidative protein modifications. FEBS Lett. 1995;364:279–282. doi: 10.1016/0014-5793(95)00307-u. [DOI] [PubMed] [Google Scholar]
- 19.Yamakura F, Matsumoto T, Fujimura T, Taka H, et al. Modification of a single tryptophan residue in human Cu,Zn-superoxide dismutase by peroxynitrite in the presence of bicarbonate. Biochim Biophys Acta. 2001;1548:38–46. doi: 10.1016/s0167-4838(01)00212-6. [DOI] [PubMed] [Google Scholar]
- 20.Yamakura F, Matsumoto T, Ikeda K, Taka H, et al. Nitrated and oxidized products of a single tryptophan residue in human Cu,Zn-superoxide dismutase treated with either peroxynitrite-carbon dioxide or myeloperoxidase-hydrogen peroxide-nitrite. J Biochem. 2005;138:57–69. doi: 10.1093/jb/mvi095. [DOI] [PubMed] [Google Scholar]
- 21.Ishii Y, Ogara A, Katsumata T, Umemura T, et al. Quantification of nitrated tryptophan in proteins and tissues by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J Pharm Biomed Anal. 2007;44:150–159. doi: 10.1016/j.jpba.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Herold S, Shivashankar K, Mehl M. Myoglobin scavenges peroxynitrite without being significantly nitrated. Biochemistry. 2002;41:13460–13472. doi: 10.1021/bi026046h. [DOI] [PubMed] [Google Scholar]
- 23.Bagnasco P, MacMillan-Crow LA, Greendorfer JS, Young CJ, et al. Peroxynitrite modulates acidic fibroblast growth factor (FGF-1) activity. Arch Biochem Biophys. 2003;419:178–189. doi: 10.1016/j.abb.2003.08.025. [DOI] [PubMed] [Google Scholar]
- 24.Wendt S, Schlattner U, Wallimann T. Differential effects of peroxynitrite on human mitochondrial creatine kinase isoenzymes. Inactivation, octamer destabilization, and identification of involved residues. J Biol Chem. 2003;278:1125–1130. doi: 10.1074/jbc.M208572200. [DOI] [PubMed] [Google Scholar]
- 25.Correia AR, Ow SY, Wright PC, Gomes CM. The conserved Trp155 in human frataxin as a hotspot for oxidative stress related chemical modifications. Biochem Biophys Res Commun. 2009;390:1007–1011. doi: 10.1016/j.bbrc.2009.10.095. [DOI] [PubMed] [Google Scholar]
- 26.Yamakura F, Ikeda K, Matsumoto T, Taka H, Kaga N. Formation of 6-nitrotryptophan in purified proteins by reactive nitrogen species: A possible new biomarker. International Congress Series. 2007;1304:22–32. [Google Scholar]
- 27.Deeb R, Nuriel T, Gross SS. In: Nitric Oxide: Biology and Pathology. 2. Ignarro LJ, editor. Academic Press; 2010. pp. 327–389. [Google Scholar]
- 28.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 29.Sacksteder CA, Qian WJ, Knyushko TV, Wang H, et al. Endogenously nitrated proteins in mouse brain: links to neurodegenerative disease. Biochemistry. 2006;45:8009–8022. doi: 10.1021/bi060474w. [DOI] [PubMed] [Google Scholar]
- 30.Ishii Y, Iijima M, Umemura T, Nishikawa A, et al. Determination of nitrotyrosine and tyrosine by high-performance liquid chromatography with tandem mass spectrometry and immunohistochemical analysis in livers of mice administered acetaminophen. J Pharm Biomed Anal. 2006;41:1325–1331. doi: 10.1016/j.jpba.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 31.Rebrin I, Bregere C, Kamzalov S, Gallaher TK, Sohal RS. Nitration of tryptophan 372 in succinyl-CoA:3-ketoacid CoA transferase during aging in rat heart mitochondria. Biochemistry. 2007;46:10130–10144. doi: 10.1021/bi7001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcondes S, Turko IV, Murad F. Nitration of succinyl-CoA:3-oxoacid CoA-transferase in rats after endotoxin administration. Proc Natl Acad Sci U S A. 2001;98:7146–7151. doi: 10.1073/pnas.141222598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Peng F, Tong W, Sun H, et al. The nitrated proteome in heart mitochondria of the db/db mouse model: characterization of nitrated tyrosine residues in SCOT. J Proteome Res. 2010;9:4254–4263. doi: 10.1021/pr100349g. [DOI] [PubMed] [Google Scholar]
- 34.Southan MD, Truscott RJW, Jamie JF, Pelosi L, et al. Structural requirements of the competitive binding site of recombinant human indoleamin 2,3-dioxygenase. Medicinal Chemistry Research. 1996;6:343–352. [Google Scholar]
- 35.Grohmann U, Fallarino F, Puccetti P. Tolerance, DCs and tryptophan: much ado about IDO. Trends Immunol. 2003;24:242–248. doi: 10.1016/s1471-4906(03)00072-3. [DOI] [PubMed] [Google Scholar]
- 36.Lob S, Konigsrainer A, Rammensee HG, Opelz G, Terness P. Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees? Nat Rev Cancer. 2009;9:445–452. doi: 10.1038/nrc2639. [DOI] [PubMed] [Google Scholar]
- 37.Brown JL. N-Nitrosamines. Occup Med. 1999;14:839–848. [PubMed] [Google Scholar]
- 38.Venitt S, Crofton-Sleigh C, Ooi SL, Bonnett R. Mutagenicity of nitrosated alpha-amino acid derivatives N-acetyl-N′-nitrosotryptophan and its methyl ester in bacteria. Carcinogenesis. 1980;1:523–532. doi: 10.1093/carcin/1.6.523. [DOI] [PubMed] [Google Scholar]
- 39.Gatehouse D, Wedd D. The bacterial mutagenicity of three naturally occurring indoles after reaction with nitrous acid. Mutat Res. 1983;124:35–51. doi: 10.1016/0165-1218(83)90183-0. [DOI] [PubMed] [Google Scholar]
- 40.Sonnenschein K, de Groot H, Kirsch M. Formation of S-nitrosothiols from regiospecific reaction of thiols with N-nitrosotryptophan derivatives. J Biol Chem. 2004;279:45433–45440. doi: 10.1074/jbc.M405987200. [DOI] [PubMed] [Google Scholar]
- 41.Halliwell B. What nitrates tyrosine? Is nitrotyrosine specific as a biomarker of peroxynitrite formation in vivo? FEBS Lett. 1997;411:157–160. doi: 10.1016/s0014-5793(97)00469-9. [DOI] [PubMed] [Google Scholar]
- 42.MacMillan-Crow LA, Crow JP, Kerby JD, Beckman JS, Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allografts. Proc Natl Acad Sci U S A. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abello N, Kerstjens HA, Postma DS, Bischoff R. Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J Proteome Res. 2009;8:3222–3238. doi: 10.1021/pr900039c. [DOI] [PubMed] [Google Scholar]
- 44.Deeb RS, Hao G, Gross SS, Laine M, et al. Heme catalyzes tyrosine 385 nitration and inactivation of prostaglandin H2 synthase-1 by peroxynitrite. J Lipid Res. 2006;47:898–911. doi: 10.1194/jlr.M500384-JLR200. [DOI] [PubMed] [Google Scholar]
- 45.Kamisaki Y, Wada K, Bian K, Balabanli B, et al. An activity in rat tissues that modifies nitrotyrosine-containing proteins. Proc Natl Acad Sci U S A. 1998;95:11584–11589. doi: 10.1073/pnas.95.20.11584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Go YM, Patel RP, Maland MC, Park H, et al. Evidence for peroxynitrite as a signaling molecule in flow-dependent activation of c-Jun NH(2)-terminal kinase. Am J Physiol. 1999;277:H1647–1653. doi: 10.1152/ajpheart.1999.277.4.H1647. [DOI] [PubMed] [Google Scholar]
- 47.Koeck T, Fu X, Hazen SL, Crabb JW, et al. Rapid and selective oxygen-regulated protein tyrosine denitration and nitration in mitochondria. J Biol Chem. 2004;279:27257–27262. doi: 10.1074/jbc.M401586200. [DOI] [PubMed] [Google Scholar]
- 48.Buddha MR, Keery KM, Crane BR. An unusual tryptophanyl tRNA synthetase interacts with nitric oxide synthase in Deinococcus radiodurans. Proc Natl Acad Sci U S A. 2004;101:15881–15886. doi: 10.1073/pnas.0405483101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buddha MR, Tao T, Parry RJ, Crane BR. Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase. J Biol Chem. 2004;279:49567–49570. doi: 10.1074/jbc.C400418200. [DOI] [PubMed] [Google Scholar]
- 50.Buddha MR, Crane BR. Structure and activity of an aminoacyl-tRNA synthetase that charges tRNA with nitro-tryptophan. Nat Struct Mol Biol. 2005;12:274–275. doi: 10.1038/nsmb907. [DOI] [PubMed] [Google Scholar]
- 51.Johnson EG, Krasnoff SB, Bignell DR, Chung WC, et al. 4-Nitrotryptophan is a substrate for the non-ribosomal peptide synthetase TxtB in the thaxtomin A biosynthetic pathway. Mol Microbiol. 2009;73:409–418. doi: 10.1111/j.1365-2958.2009.06780.x. [DOI] [PubMed] [Google Scholar]
- 52.Loria R, Kers J, Joshi M. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol. 2006;44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 53.Kers JA, Wach MJ, Krasnoff SB, Widom J, et al. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 54.Wach MJ, Kers JA, Krasnoff SB, Loria R, Gibson DM. Nitric oxide synthase inhibitors and nitric oxide donors modulate the biosynthesis of thaxtomin A, a nitrated phytotoxin produced by Streptomyces spp. Nitric Oxide. 2005;12:46–53. doi: 10.1016/j.niox.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 55.Furth AJ, Hope DB. Studies on the chemical modification of the tyrosine residue in bovine neurophysin-II. Biochem J. 1970;116:545–553. doi: 10.1042/bj1160545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beeckmans S, Kanarek L. The modification with tetranitromethane of an essential tyrosine in the active site of pig fumarase. Biochim Biophys Acta. 1983;743:370–378. doi: 10.1016/0167-4838(83)90395-3. [DOI] [PubMed] [Google Scholar]
- 57.Beckmann JS, Ye YZ, Anderson PG, Chen J, et al. Extensive Nitration of Protein Tyrosines in Human Atherosclerosis Detected by Immunohistochemistry. Biological Chemistry Hoppe-Seyler. 1994;375:81–88. doi: 10.1515/bchm3.1994.375.2.81. [DOI] [PubMed] [Google Scholar]
- 58.Nuriel T, Deeb RS, Hajjar DP, Gross SS. Protein 3-nitrotyrosine in complex biological samples: quantification by high-pressure liquid chromatography/electrochemical detection and emergence of proteomic approaches for unbiased identification of modification sites. Methods Enzymol. 2008;441:1–17. doi: 10.1016/S0076-6879(08)01201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawasaki H, Ikeda K, Shigenaga A, Baba T, et al. Mass spectrometric identification of tryptophan nitration sites on proteins in peroxynitrite-treated lysates from PC12 cells. Free Radic Biol Med. 2010 doi: 10.1016/j.freeradbiomed.2010.10.688. [DOI] [PubMed] [Google Scholar]
- 60.Perdivara I, Deterding LJ, Przybylski M, Tomer KB. Mass spectrometric identification of oxidative modifications of tryptophan residues in proteins: chemical artifact or post-translational modification? J Am Soc Mass Spectrom. 2010;21:1114–1117. doi: 10.1016/j.jasms.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersson AS, Steen H, Kalume DE, Caidahl K, Roepstorff P. Investigation of tyrosine nitration in proteins by mass spectrometry. J Mass Spectrom. 2001;36:616–625. doi: 10.1002/jms.161. [DOI] [PubMed] [Google Scholar]
- 62.Sarver A, Scheffler NK, Shetlar MD, Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 63.Ikeda K, Yukihiro Hiraoka B, Iwai H, Matsumoto T, et al. Detection of 6-nitrotryptophan in proteins by Western blot analysis and its application for peroxynitrite-treated PC12 cells. Nitric Oxide. 2007;16:18–28. doi: 10.1016/j.niox.2006.04.263. [DOI] [PubMed] [Google Scholar]
- 64.Kanski J, Schoneich C. Protein nitration in biological aging: proteomic and tandem mass spectrometric characterization of nitrated sites. Methods Enzymol. 2005;396:160–171. doi: 10.1016/S0076-6879(05)96016-3. [DOI] [PubMed] [Google Scholar]
- 65.Creighton TE. Proteins: structure and molecular properties. Freeman; New York: 1993. [Google Scholar]
- 66.Harriman A. Further comments on the redox potentials of tryptophan and tyrosine. J Phys Chem. 1987;91:6102–6104. [Google Scholar]
- 67.Nakagawa H, Takusagawa M, Arima H, Furukawa K, et al. Selective scavenging property of the indole moiety for the nitrating species of peroxynitrite. Chem Pharm Bull (Tokyo) 2004;52:146–149. doi: 10.1248/cpb.52.146. [DOI] [PubMed] [Google Scholar]
- 68.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 69.Samanta U, Pal D, Chakrabarti P. Environment of tryptophan side chains in proteins. Proteins. 2000;38:288–300. [PubMed] [Google Scholar]
- 70.Ikeda K, Iwai H, Matsumoto T, Mineki R, et al. Proteomic analysis of 6-nitrotryptophan-containing proteins in peroxynitrite-treated PC12 cells. International Congress Series. 2007;1304:33–40. [Google Scholar]
- 71.Kato K, Giulivi C. Critical overview of mitochondrial nitric-oxide synthase. Front Biosci. 2006;11:2725–2738. doi: 10.2741/2002. [DOI] [PubMed] [Google Scholar]
- 72.Grune T, Blasig IE, Sitte N, Roloff B, et al. Peroxynitrite increases the degradation of aconitase and other cellular proteins by proteasome. J Biol Chem. 1998;273:10857–10862. doi: 10.1074/jbc.273.18.10857. [DOI] [PubMed] [Google Scholar]
- 73.Souza JM, Choi I, Chen Q, Weisse M, et al. Proteolytic degradation of tyrosine nitrated proteins. Arch Biochem Biophys. 2000;380:360–366. doi: 10.1006/abbi.2000.1940. [DOI] [PubMed] [Google Scholar]
- 74.Kuo WN, Kanadia RN, Shanbhag VP, Toro R. Denitration of peroxynitrite-treated proteins by ‘protein nitratases’ from rat brain and heart. Mol Cell Biochem. 1999;201:11–16. doi: 10.1023/a:1007024126947. [DOI] [PubMed] [Google Scholar]
- 75.Irie Y, Saeki M, Kamisaki Y, Martin E, Murad F. Histone H1.2 is a substrate for denitrase, an activity that reduces nitrotyrosine immunoreactivity in proteins. Proc Natl Acad Sci U S A. 2003;100:5634–5639. doi: 10.1073/pnas.1131756100. [DOI] [PMC free article] [PubMed] [Google Scholar]