Abstract
Protein carbonylation has been associated with various pathophysiological processes. A representative reactive carbonyl species (RCS), 4-hydroxy-2-nonenal (HNE), has been implicated specifically as a causative factor for the initiation and/or progression of various diseases. To date, however, little is known about the proteins and their modification sites susceptible to “carbonyl stress” by this RCS, especially in the liver. Using chemoprecipitation based on a solid phase hydrazine chemistry coupled with LC-MS/MS bottom-up approach and database searching, we identified several protein-HNE adducts in isolated rat liver mitochondria upon HNE exposure. The identification of selected major protein targets, such as the ATP synthase β-subunit, was further confirmed by immunoblotting and a gel-based approach in combination with LC–MS/MS. A network was also created based on the identified protein targets that showed that the main protein interactions were associated with cell death, tumor morphology and drug metabolism, implicating the toxic nature of HNE in the liver mitoproteome. The functional consequence of carbonylation was illustrated by its detrimental impact on the activity of ATP synthase, a representative major mitochondrial protein target for HNE modifications.
1. Introduction
Mitochondria are the major energy sources in mammalian cells by generating ATP via oxidative phosphorylation. The mitochondrial respiratory chain is also a key intracellular source of reactive oxygen species (ROS); therefore, mitochondria are important subcellular organelles to understand the impact of oxidative stress in the onset and/or progression of numerous diseases and aging. Oxidative stress occurs when the body’s well-designed proteolytic or other repair systems are overwhelmed by excess ROS [1]. Mitochondrial membranes, or in general membrane lipids rich in polyunsaturated fatty acids, are especially sensitive to lipid peroxidation in the bis-allylic positions of unsaturated fatty acids [2], which eventually leads to the formation of a great diversity of reactive carbonyl species (RCS) [3]. RCS as electrophiles, then, react with nucleophilic sites of cellular constituents, including proteins, not only locally but also farther from the site of their formation [4], propagating thereby oxidative cellular damage via “carbonyl stress” [5,6]. Cumulative damage may lead mitochondria to a state of dysfunction that, in time, may generate intracellular signals for lysosomal digestion and apoptosis [7–9].
In the proteome, carbonyl stress results in the formation of adducts and crosslinking having a wide range of downstream functional consequences and cellular dysfunctions [10–12]. Among RCS, 4-hydroxy-2-nonenal (HNE), formed during peroxidation of ω-6 fatty acids, has drawn particular attention as a specific biomarker indicated, among others, in atherosclerosis, cancer, diabetes, neurodegenerative disorders and liver diseases [13–18]. Due to its chemical nature, HNE easily undergoes Michael addition on the side chain functional group of Cys, His, and Lys, respectively [19]. It can also produce Schiff bases with the ε-amino group of Lys, but the corresponding kinetics are inherently slow and Schiff-base formation is also reversible [20,21] making Michael-adducts of HNE the predominant [21–23] and most reliable products for exploring protein carbonylation through proteomics [21,23,24].
Proteomic approaches have been successfully employed to identify HNE-derived protein carbonyls [6]. The HNE-directed posttranslational modification is neither enzymatic process nor genetically coded, hence lacks the consensus motif and, therefore, precludes the possibility of predicting the exact modification sites. Specific mass spectrometry techniques have become particularly powerful tools in this regard due to their ability to site-specifically locate amino acids targeted by RCS both in vitro and in vivo [21–24].
Liver is a major organ for a variety of pathophysiological processes. Levels of ROS production and, therefore, carbonylation have been found to increase with age, diabetes, inflammation, alcoholism, metabolic diseases, drug-induced toxicity, and various other conditions [25–32]. HNE has specifically been implicated as a contributor and/or marker of hepatic pathologies. However, little is known about the proteins and, more importantly, their exact sites susceptible to oxidative damages by this RCS in this organ. Although detection of HNE-modified proteins has been possible through exposure to the endogenously formed lipid peroxidation end-product [33], studies with isolated organelles, such as mitochondria, may allow for increased protein coverage by mass spectrometry-based proteomics [34], as well as permit straightforward and controlled follow-up experiments focusing on the verification of findings and/or possible consequences on mitochondrial functions. Enrichment of HNE-modified peptides also appears to be crucial for carbonyl-directed identifications by liquid chromatography–tandem mass spectrometry (LC–MS/MS) [35]. Accordingly, we exposed isolated liver mitochondrial proteins to HNE in the present study and, then, applied a “bottom-up” proteomics strategy utilizing enrichment of the tryptic peptide carbonyls via chemoprecipitation [35,36] followed by LC-MS/MS and database searching for site-specific identifications of protein carbonyls. A network implicating the main molecular functional interactions and biological connections among the identified major protein targets of HNE-induced carbonyl stress was also revealed. The functional consequence of HNE-modification was illustrated by its impact on the activity of ATP synthase, a representative protein target in this network.
2. Materials and methods
2.1.Chemicals
4-Hydroxy-2-nonenal (HNE) was obtained from Cayman Chemical Company (Ann Arbor, MI). Water, acetonitrile and methanol were of high-performance liquid chromatography (HPLC) grade and purchased from Honeywell Burdick and Jackson (Morristown, NJ, USA). Sequencing grade trypsin was obtained from Applied Biosystems (Foster City, CA). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO).
2.2. Preparation of rat liver mitochondria
All experiments were performed in compliance with the guidelines for the welfare of experimental animals issued by the National Institutes of Health and in accordance with the guidelines of Institutional Animal Care and Use Committee at the University of North Texas Health Science Center. Liver tissues were isolated from 12-week-old male Sprague Dawley rats (Harlan Laboratory, Indianapolis, IN) maintained on standard rodent chow and housed under specific pathogen-free conditions at a temperature of 25 ± 1.5 °C with 12:12-h light-dark cycles. Animals were euthanized by intraperitoneal injection of sodium pentobarbital (60 mg/kg body weight) and the liver was removed. Mitochondria were isolated using a Mitochondria Isolation Kit for Tissue (Piercenet, Rockford, IL) from 3.0 g of pooled tissue according to the manufacturer’s instruction. Mitochondrial pellets were resuspended in 250 µl of 10 mM PBS containing a protease inhibitor cocktail (Complete Mini, Roche, Indianapolis, IN) and, then, the mitochondria were ruptured with 3 freeze/thaw cycles followed by 10 bursts of sonication with chilling on ice after each burst. The resultant solution was clarified by centrifugation (11000g, 30 min, 4 °C) and the protein concentration was determined with the Micro BCA Protein Assay Kit (Piercenet, Rockford, IL) using BSA as a reference. Mitochondria were stored at −80 °C until the appropriate experiments were performed or used immediately for the measurement of ATP synthase activity upon exposure to various concentrations of HNE.
2.3. HNE-exposure of liver mitochondrial proteins
Approximately 1 mg of mitochondrial liver proteins in 0.5 mL of 10 mM PBS were incubated with 156 µg (2 mM) and 39 µg (0.5 mM) of HNE, respectively, for 2 h at room temperature [37,38] followed by precipitation through the addition of four volumes of − 20 °C acetone according to a standard protocol (http://www.piercenet.com/files/TR0049-Acetone-precipitation.pdf). The mixture was kept overnight at −20 °C, then centrifuged at 13000g for 10 min at 4 °C. The pellets were resuspended in 50 µL of 8M urea and 200 µL of 50 mM ammonium bicarbonate then treated with 5 µL of 250 mM DTT at 50 °C for 1 h. Carbamidomethylation was done with 10 µL of 250 mM iodoacetamide for 30 min at room temperature in the dark. Excess iodoacetamide was quenched with 10 µL of 250 mM DTT for 15 min. The sample was then diluted with 250 µL of 50 mM ammonium bicarbonate and digested with 8 µL of 0.5 µg/µL trypsin overnight (18 h) at 37 °C. The enzymatic reaction was terminated by acidifying the sample to pH <2.0, and the digest was loaded onto a Supelco C18 solid-phase extraction cartridge (Bellefonte, PA). The cartridge was first washed with 2 × 1 mL water containing 0.1% (v/v) acetic acid and, then samples were eluted with 2 × 300 µL of 80% v/v/ acetonitrile containing 0.1% v/v acetic acid. The desalted mitochondrial tryptic digests were dried with a SpeedVac (Thermo Scientific Savant, San Jose, CA) and stored at −80 °C until analyses.
2.4. Solid-phase enrichment of HNE-modified peptides
HNE-treated samples obtained from procedure above were dissolved in 150 µL of reaction buffer (pH 3.6) containing 10% (v/v) aqueous acetonitrile and 0.2% (v/v) acetic acid followed by the addition of 5 mg of hydrazide-coated glass beads (SPH) [35,36] to capture peptide carbonyls. The resulting mixture was rotated end-over-end overnight at room temperature followed by centrifugation (13400 rpm) to settle the beads. The supernatant was collected, dried by lyophilization, and saved for LC–MS/MS analysis. The beads were then thoroughly washed with 4 × 400 µL of reaction buffer followed by 1 M NaCl, distilled water, 80% (v/v) acetonitrile, and a second round of distilled water to remove any unmodified tryptic peptides. Then, the peptide carbonyls (captured as hydrazones) were released from the beads with 200 µL of formic acid (10%, v/v) at 60 °C for 30 min. This step was repeated one more time. The combined acidic solutions containing peptide carbonyls were lyophilized, and then reconstituted in 20 µL of 5% (v/v) acetonitrile in water containing 0.1% (v/v) acetic acid. Five µL aliquots were used for subsequent LC–MS/MS analyses.
2.5. LC–MS/MS
LC–MS/MS analysis of the peptide samples was performed using a hybrid linear ion trap–Fourier transform ion cyclotron resonance (7-T) mass spectrometer (LTQ-FT, Thermo Finnigan, San Jose, CA) equipped with a nano-electrospray ionization (ESI) source and operated with the Xcalibur (version 2.2) and Tune Plus (version 2.2) data acquisition software. Online reversed phase-high performance liquid chromatography (RP-HPLC) was performed with an Eksigent nano-LC-2D (Eksigent, Dublin, CA) system. Five µL of the sample was automatically loaded onto the IntegraFrit™ sample trap (2.5 cm × 75 µm) (New Objective, Woburn, MA) at a flow rate of 1.5 µL/min in a loading solvent containing 0.1% (v/v) acetic acid and 5% (v/v) acetonitrile in 94.9% (v/v) water for concentration and desalting prior to injection onto a 75 µm i.d. × 15 cm reverse-phase PepMap C18 analytical column (LC Packings, Sunnyvale, CA). Following peptide desalting and injection onto the analytical column, peptides were separated using the following gradient conditions: (1) 5 min in 95.2% solvent A [0.1% (v/v) acetic acid and 99.9% (v/v) water] for equilibration; (2) linear gradient to 40% solvent B [0.1% (v/v) acetic acid and 99.9% (v/v) acetonitrile] over 90 min and holding at 40% solvent B for isocratic elution for 5 min; (3) increasing the gradient to 90% solvent B and maintaining for 5 min; and finally (4) 95.2% solvent A in the next 20 min. The flow rate through the column was 250 nL/min. Peptides eluted through a Picotip emitter (internal diameter 10 ± 1 µm; New Objective) were directly supplied into the nanoelectrospray source of the mass spectrometer. Spray voltage and capillary temperature during the gradient run were maintained at 2.0 kV and 250 °C. The conventional data-dependent mode of acquisition was utilized in which an accurate m/z survey scan was performed in the FTICR cell followed by parallel MS/MS linear ion trap analysis of the top five most intense precursor ions. FTICR full-scan mass spectra were acquired at 50000 mass resolving power (m/z 400) from m/z 350 to 1500 using the automatic gain control mode of ion trapping. Peptide fragmentation was induced by collision-induced dissociation (CID) in the linear ion trap using a 3.0-Th isolation width and 35% normalized collision energy with helium as the target gas. The precursor ion that had been selected for CID was dynamically excluded from further MS/MS analysis for 60 s.
2.6. Sequence database search, peptide identification and pathway analysis
MS/MS data generated by data-dependent acquisition via the LTQ-FT were extracted by BioWorks version 3.3 and searched against a composite IPI rat (version 3.28) protein sequence database using the Mascot (version 2.2.2; Matrix Science, Boston, MA) search algorithm. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 25 ppm. Carbamidomethylation of Cys, oxidation of Met, HNE-Michael adducts formation on Cys, His and Lys were specified as variable modifications among the Mascot options. Trypsin was selected as the digesting enzyme allowing for the possibility of one missed cleavage site.
The software program Scaffold (version 2_00_06, Proteome Software Inc., Portland, OR) was then employed to compile and validate tandem MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm. Protein identifications including single peptide-based identifications, where protein probabilities were assigned by the Protein Prophet algorithm, were accepted at greater than 80.0% probability. Manual validation of tandem MS data of each HNE-modified peptide was performed to discard false positives and accept false negatives, if any. The MS-Product module of ProteinProspector (http://prospector.ucsf.edu) was used for validation of HNE-modified peptides by manually comparing the masses of b- and y-ions obtained during CID-MS/MS of peptides with the fragment ion masses generated from MS-Product. MS/MS spectra were also searched using SEQUEST (Thermo Finnigan, San Jose, CA) against a non-redundant rat proteome sequence database from the European Bioinformatics Institute (http://www.ebi.ac.uk/IPIrat.html). The differential search parameters were specified for detecting oxidation of Met (+16 Da), carbamidomethylation of Cys (+57.02 Da) and the addition of HNE (+156.12 Da) to Cys, His, and Lys residues. Additionally, Ingenuity Pathway Analysis 8.8 (Ingenuity Systems, Redwood City, CA) was utilized to derive potential protein interaction networks among the identified carbonylated mitochondrial proteins in the rat liver.
2.7. Gel electrophoresis and Western blots
Isolated liver mitochondria, approx. 100 µg of total protein in 100 µL of PBS, were treated with 0.2 µmol HNE and pelleted by centrifugation (13000 rpm, 5 min). The pellet was solubilized with 1% (w/v) lauryl maltoside and protein content was determined by Micro BCA Protein Assay Kit (Piercenet, Rockford, IL). Protein samples (10 µg per well) were separated by sodium dodecylsulphate−polyacrylamide gel electrophoresis (SDS-PAGE) using 12% polyacrylamide gels. Precision plus protein marker (Bio-Rad, Hercules, CA) was also run along with the samples. One gel was processed for Western blot analysis, while the other one was visualized with Coomassie Blue G-250 (Bio-Rad) per standard protocol. For the immunoblots, proteins were transferred to a nitrocellulose membrane (GE Healthcare, Piscataway, NJ) in a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad) using transfer buffer (25 mM Tris/192 mM glycine/15% v/v ethanol) with 300 mA for 2 h in a cold room. Afterward, the membrane was blocked with 5 ml 5% (w/v) nonfat milk powder in Tris-buffered saline (20 mM Tris/500 mM NaCl, pH 7.5) containing 0.1% Tween-20 (TTBS) for 30 min then incubated with mouse anti-HNE antibody (R&D Systems, Minneapolis, MN) diluted to 1:500 in TTBS containing 5% milk powder. The primary antibody was removed, and the blots were washed 3 times for 10 min with TTBS. The blots were then incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse secondary antibody (Invitrogen, Carlsbad, CA), at a dilution of 1:3000. After washing, the bands were visualized with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Rockford, IL). The same membrane was then stripped with Restore™ Western Blot stripping buffer (Pierce) according to manufacturer’s instruction and incubated with rabbit anti-ATP-synthase antibody (Aviva Systems Biology, San Diego, CA) diluted to 1:400. After washing, the blot was incubated at room temperature for 1 h with a 1:3000 dilution of peroxidase-conjugated goat anti-rabbit antibody (Invitrogen). After additional wash with TTBS, protein bands were visualized with SuperSignal West Pico chemiluminescent substrate and developed by LabWorks Image Acquisition and Analysis software v. 4.6 (Ultraviolet Products, Cambridge, UK).
2.8. In-gel digestion
Two gel bands of interest were excised with a clean scalpel and sliced into small pieces and, then, placed into siliconzed tubes followed by washing three times with 25 mM aqueous NH4HCO3 containing 50% acetonitrile. Gel pieces were then dried and proteins were reduced with 10 mM DTT (56 °C, 1 h) and, then, alkylated with 25 µL of 50 mM iodoacetamide (30 min, room temperature). The supernatant was removed and discarded. Gel pieces were washed with 25 mM aqueous NH4HCO3 and dried before trypsin digestion (10 ng/µL, approx. 3-times the volume of the gel, 37 °C, overnight). Tryptic peptides were extracted from the gels with water/acetonitrile (1:1, v/v) containing 5% formic acid and dried by Speedvac before C18 ZipTip (Millipore) cleanup and LC–MS/MS analysis.
2.9. ATP synthase activity
Enzyme activity in freshly prepared liver mitochondria upon HNE-exposure was monitored with an ATP Synthase Enzyme Activity Microplate Assay Kit (Mitosciences, OR, USA) according to the manufacturer’s instructions. The principle of the method is based on the hydrolysis of ATP to ADP by ATP-synthase that is coupled to the oxidation of NADH to NAD+ leading to a decrease in ultraviolet (UV) absorbance at 340 nm. Samples (approx. 200 µg protein) were treated with various concentrations of HNE (20–5000 µM) for 5 min at room temperature then loaded onto plates coated with anti-ATP synthase antibodies for β-subunit and F1-portion. The activity was expressed as the change in absorbance at 340 nm/min/amount of sample loaded into the well. Blank samples contained only reaction buffer, while control samples consisted of untreated liver mitochondria. Control activity was considered 100%, and activities of ATP synthase with varying concentrations of HNE treatment were normalized accordingly to yield percentage inhibition by the given concentration of the lipid peroxidation end-product. Data are represented as means ± SEM (n=4).
IC50 (the HNE concentration that results in 50% inhibition of the enzyme’s activity) was calculated using the Scientist software V2.01 (St. Louis, MI, USA) by fitting the results of the dose–response experiments to the equation
| (1) |
where Ci and h were the HNE concentration (µM) used and the Hill coefficient, respectively.
3. Results and Discussion
3.1. Identification of HNE-carbonylated peptides from rat liver mitochondria by LC–MS/MS
In this experiment, we treated proteins extracted from ruptured mitochondria with HNE to allow access of the RCS to as many proteins as possible for discovery-driven identification of potential targets through a proteomics approach striving for the unequivocal identification and localization of carbonylation sites. The relatively low abundance of HNE-protein adducts [11,20] required enrichment prior to mass spectrometric analyses to reduce sample complexity. SPH chemistry of peptide carbonyls [35,36] permitted the removal of non-modified species and, thereby, facilitated the identification of HNE-modified peptides in the proteome. We have previously shown that this enrichment technique significantly improved the quality of MS/MS spectra by increasing the number of ion counts and reducing interference during precursor-ion isolation and fragmentation in brain derived mitochondria [23,36]. We, again, utilized this enrichment method in this study to “fish-out” peptide carbonyls from the tryptic digest of HNE-treated mitochondria protein fraction of the rat liver for modification-directed identification.
Previous reports suggested that as high as 5 mM of HNE accumulates in the cellular lipid membranes under oxidative stress [39]. Specifically, it has been shown that the concentration of HNE in the lipid bilayer of isolated peroxidizing microsomes is about 4.5 mM [19]. Therefore, treatment of mitochondrial proteins with 2 mM of HNE may mimic the extreme situations of oxidative stress locally near the peroxidizing membranes. When 0.5 mM HNE was applied to mitochondria, proteins such as carbamoyl-phosphate synthase, catalase, glutamate dehydrogenase and ATP synthase were identified as modified proteins, indicating that these proteins are susceptible to HNE modifications even at a lower concentration of the lipid peroxidation product. Albeit fewer modified peptides were obtained due to their reduced concentration and given the assay sensitivity, they were consistent with the ones detected when higher HNE concentration (2 mM) was used.
Among the 26 identified proteins susceptible to HNE attack, the majority indeed were of mitochondrial origin (Table 1), albeit some non-mitochondrial proteins were also found (Table S1, Supporting Information) possibly due to co-isolation and/or association of these proteins with the mitochondria. In addition, it has to be acknowledged that coverage of protein modifications by HNE within a complex proteome is clearly incomplete [40] and, also, method-dependent with today’s technology. There may be even a bias to reveal certain targets for modification with preference over others. For example, although the highest reactivity has been reported for Cys residues [41], we and others have found that modified His residues were the most abundant in proteomic studies done without enrichment [21,23], or using SPH chemistry [42] and affinity columns made by immobilizing an antibody recognizing HNE-Michael adducts [33] to enrich these posttranslationally modified peptides for subsequent LC–ESI-MS/MS analysis. Loss of mild alkalinity in ruptured mitochondria might be another reason that could lead to a decrease of HNE’s reactivity towards Cys residues [43]. Nevertheless, our studies have revealed a meaningful subset of proteins susceptible for carbonylation by HNE in the liver mitochondria.
Table 1.
A list of HNE-modified tryptic peptides of liver mitochondrial proteins identified after SPH enrichment and data-dependent MS/MS acquisition.
| Protein namea | IPI accession number |
HNE-modified peptides | Modification site(s) |
|---|---|---|---|
| 3-ketoacyl-CoA thiolase (Acaa2) | IPI00201413 |
HNFTPLAR LDLDPSKTNVSGGAIALGHPLGGSGSR TNVSGGAIALGHPLGGSGSR |
271, 352 |
| Acyl-Coenzyme A dehydrogenase (Acads) | IPI00231359 | LHTVYQSVELPETHQMLR | 26 |
| Acetyl- CoAacetyltransferase (Acat1) | IPI00324302 | IHMGNCAENTAK | 189 |
| Methylmalonatesemialdehyde dehydrogenase [acylating] (Aldh6a1) | IPI00205018 | NHGVVMPDANKENTLNQLVGAAFGAAGQR | 289 |
| ATP synthase subunit beta (Atp5b) | IPI00551812 | AHGGYSVFAGVGER LVLEVAQHLGESTVR IMDPNIVGSEHYDVAR |
102, 227, 417 |
| Catalase (Cat) | IPI00231742 | NAIHTYVQAGSHIAAK NAIHTYVQAGSHIAAK GAGAFGYFEVTHDITR NFTDVHPDYGAR LFAYPDTHR AVKNFTDVHPDYGAR VWPHKDYPLIPVGK LCENIANHLKDAQLFIQR |
89,260, 305,362, 466, 486, 510,518 |
| Carbamoyl-phosphate synthase (Cps1) | IPI00210644 | AQTAHIVLEDGTK VSQEHPVVLTK VISHAISEHVEDAGVHSGDATLMLPTQTISQGAIEK MCHPSVDGFTPR FVHDNYVIR |
47, 817, 1162, 1202, 1447 |
| Glutamate dehydrogenase 1 (Glud1) | IPI00324633 |
HGGTIPVVPTAEFQDR ISGASEKDIVHSGLAYTMER DIVHSGLAYTMER |
481, 507 |
| Glutathione S-transferase kappa 1 (Gstk1) | IPI00327079 | AGMATAQAQHLLNK | 154 |
| Hydroxyacylglutathione hydrolase (Hagh) | IPI00214152 | TVQQHAGETDPVTTMR HVEPGNTAVQEK |
234, 284 |
| Pyruvate carboxylase (Pc) | IPI00210435 | VVEIAPATHLDPQLR | 282 |
| Stress protein-70 (Hspa9) | IPI00363265 | ASNGDAWVEAHGK | 157 |
| Sulfite oxidase (Suox) | IPI00193919 | VSVESEESYSHWQR | 395 |
| Transketolase (Tkt) | IPI00231139 | HQPTAIIAK | 233 |
| Thiosulfate sulfurtransferase (Rhodanese) (Tst) | IPI00366293 | YLGTQPEPDAVGLDSGHIR | 204 |
Gene symbols are given in parentheses.
One of the major targets of carbonylation was ATP synthase. Specifically, we localized three modification sites on the β-subunit of this enzyme (Atp5b, Table 1) that belongs to the catalytic core of the F1 complex [44]. A representative MS/MS spectrum for the [M+2H]2+ ion of one of the corresponding peptide-HNE adducts, LVLEVAQHLGESTVR (m/z 904.0259, where the residue in bold typeface represents modification by HNE as a Michael adduct), is shown in Fig. 1(a). The high sensitivity of this enzyme to carbonyl stress in various tissues has been reported [20,23,25] and, for example, carbonylated Atp5b has been found in the alcoholic liver [13]. The consequence of carbonylation on this subunit may be the disruption of the entire ATP synthase complex, possibly contributing to an impaired ATP production [45]. Insufficient ATP production can also affect glutamate dehydrogenase 1 (Glud1), an enzyme located in mitochondrial matrix and catalyses the reversible oxidative deamination of glutamate to α-ketoglutarate and free ammonia [45–47]. In agreement with these implications, here we identified two HNE modification sites (His-481 and His-507, Table 1) for Glud1 in our experimental model. In vivo, a significant decrease in Glud1 level and its increased carbonylation was detected in an animal model of chronic ethanol exposure [16].
Fig. 1.
(a) MS/MS spectrum for [M+2H]2+ ion of LVLEVAQHLGESTVR from the ATP synthase beta-subunit (m/z 904.0259); (b) MS/MS spectrum for [M+2H]2+ ion of AQTAHIVLEDGTK of carbamoyl-phosphate synthase (m/z 769.9283). HNE-modified residues are indicated by bold typeface).
We have also shown that carbamoyl-phosphate synthase (Cps1) was heavily modified by HNE (Table 1, Fig. 2); specifically, five Michael adducts of the enzyme primarily localized in liver mitochondria and pivotal in the production of urea and nitrogen metabolism [48] were detected. A representative MS/MS spectrum using the [M+2H]2+ ion of the tryptic peptide AQTAHIVLEDGTK (m/z 769.9283) is shown in Fig. 1(b). Increased levels of carbonylation and circulatory release of Cps1 have been shown to be directly associated with mitochondrial damage and/or impaired mitochondrial function in the liver during sepsis and upon alcohol-induced liver damage [16,48]. Several other enzymes identified here as targets of HNE-modification in the liver (Table 1), such as Acyl-CoA dehydrogenase, 3-ketoacyl-CoA, catalase, thiolase, glutamate dehydrogenase, etc., have also been specifically implicated in animal models of aging, alcoholism and metabolic diseases [13, 31,49–54].
Fig. 2.
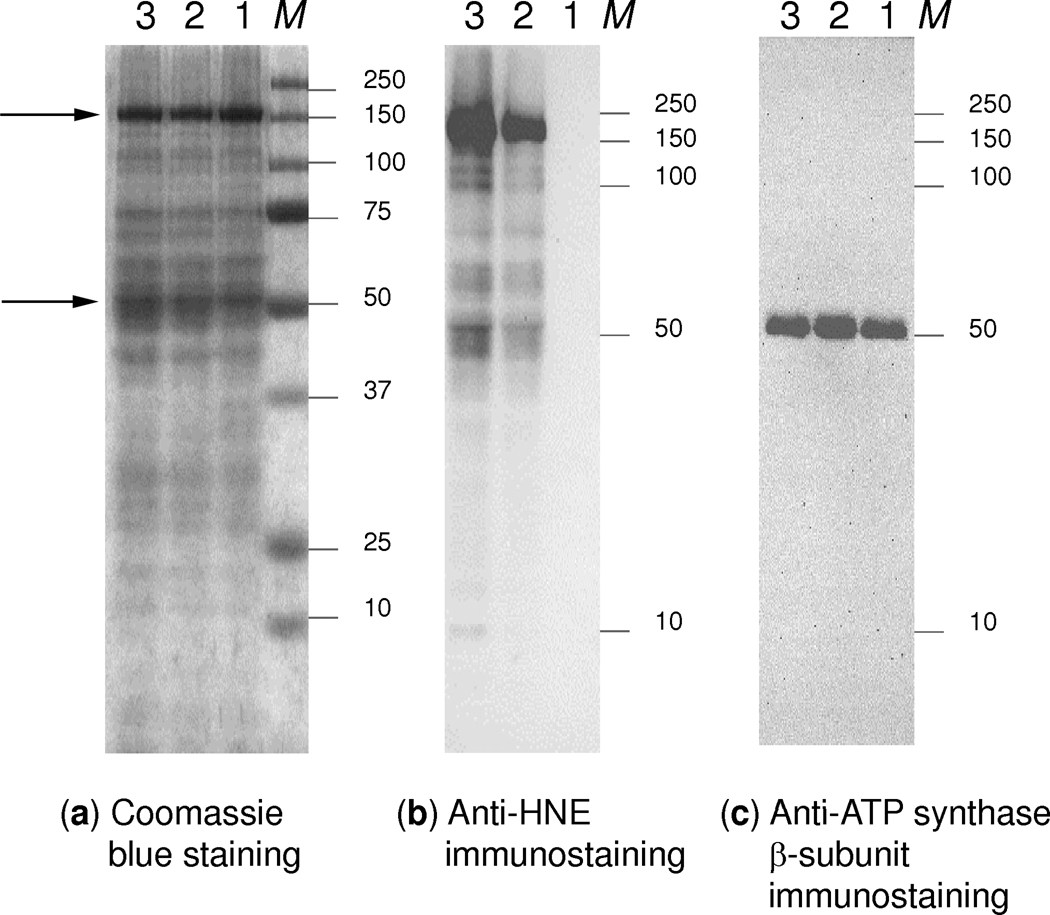
SDS-PAGE and immunoblots of HNE treated mitochondrial proteins from rat liver: (a) Coomassie-stained SDS-PAGE, (b) anti-HNE and (c) anti-ATP synthase β-subunit immunodetection from the SDS-PAGE. Lane 1, control sample without HNE treatment; Lane 2, liver mitochondria treated with 2 mM HNE for 10 min; Lane 3, sample treated with 2 mM HNE for 30 min; M, protein standards. Arrows indicate the bands cut from lanes 2 and 3 for in-gel digestion and protein identification.
Concurrently to the SPH-based enrichment coupled with LC–MS/MS identifications of HNE-carbonylated peptides, liver mitochondrial proteins were also separated, after treatment with the RCS, by SDS-PAGE and immunoblotted with anti-HNE antibody to furnish an independent verification of our findings (Fig. 2). Proteins around 150±25 kDa and 50±5 kDa were evidently the major targets for HNE modifications under the experimental conditions used. These bands were excised and, after in-gel digestion, proteins were identified by data-depended LC-MS/MS analyses followed by database search. In agreement with the SPH-based covalent enrichment technique (Table 1), carbamoyl-phosphate synthase (Cps1, 165 kDa) was identified from the band at 150±25 kDa (Fig. S1, Supporting Information) while Atp5b (52 kDa) was present as the predominant protein, in the band around 50±5 kDa (Fig. S2, Supporting Information). However, their HNE-adducts could not be identified from the SDS-PAGE bands because of the overwhelming presence of unmodified peptides demonstrating, again, the necessity of enrichment for the unequivocal identification/localization of peptide carbonyls prior to mass spectrometric analyses.
3.2. Pathway analysis of HNE-modified proteins from liver mitochondria
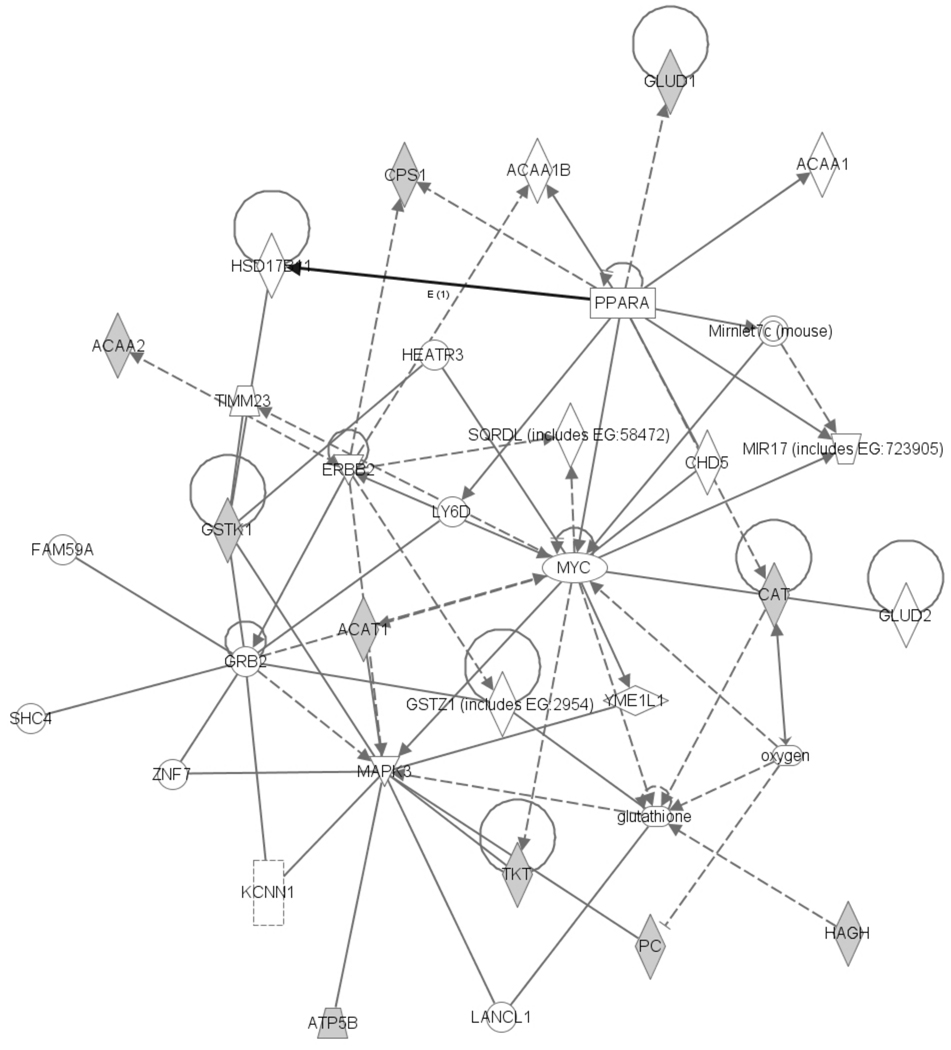
The identified 15 mitochondrial targets of HNE were submitted to Ingenuity Pathways Analysis to elucidate the main molecular functional interactions and biological connections, as well as to represent them by a network. The network with the highest score is shown in Fig. 3. It contains 33 peptides/proteins, a microRNA and oxygen, and is associated with cell death, tumor morphology and drug metabolism implicating the detrimental effect of HNE on the cellular proteome. HNE has been implicated to be a modulator in liver cancer development [55]; however, the mechanisms by which HNE insults may lead to tumor formation are yet to be elucidated. The top canonical pathways in the network based on HNE-modified liver mitochondrial proteins are Val, Leu and Ile degradation, propanoate metabolism, pyruvate metabolism, fatty acid metabolism, the urea cycle and metabolism of amino groups. Overall, these results support the cytotoxic nature of HNE modifications in biological contexts.
Fig. 3.
The highest scoring network constructed through Ingenuity Pathways Analysis based on protein targets for carbonylation by HNE in liver mitochondria (grey objects, input) shows 33 proteins, a microRNA (MIR17) and a signaling molecule, oxygen. Protein-protein interactions from the network diagram are represented by single lines and proteins/compounds that regulate another protein are indicated by arrows. Solid or dashed lines indicate direct or indirect interactions, respectively. The various shapes represent different protein functions: enzyme (diamond), ligand-dependent nuclear receptor (rectangle lying on its side), transcription regulator (oval), kinase (inverted triangle), ion channel (dashed rectangle), transporter (trapezoid), protein complex (concentric circle) and other (circle).
3.3. Effects of HNE exposure on ATP synthase activity
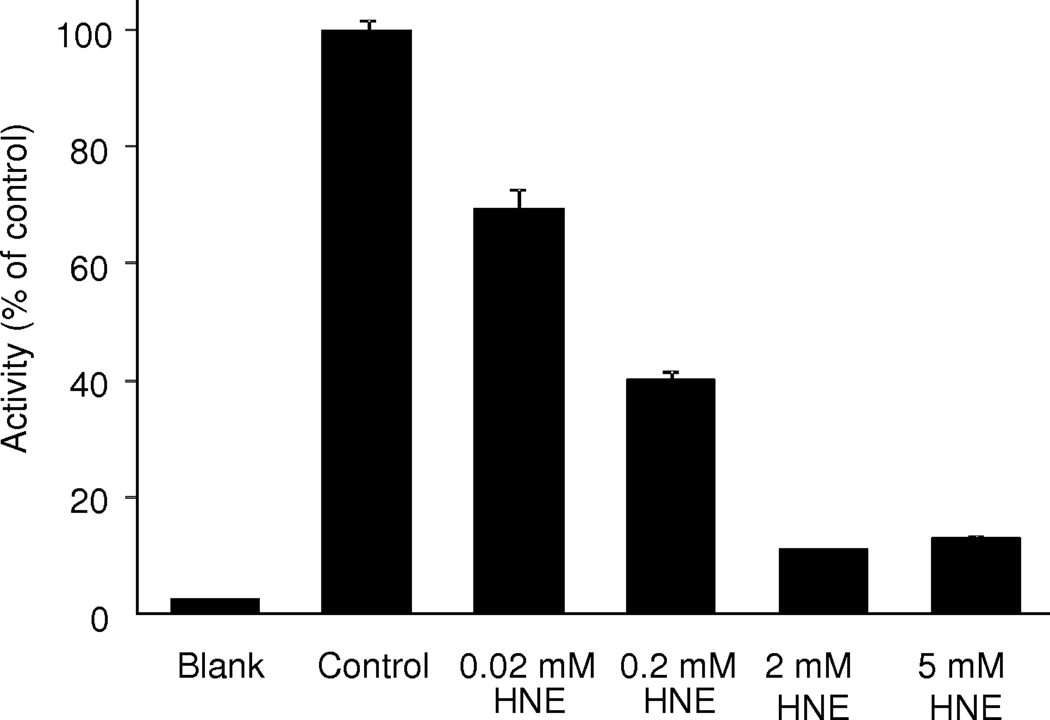
Since covalent protein modifications generally alter function [1,7], we selected a representative protein in the network revealed by the pathway analysis (Fig. 3) for further studies on the effects of HNE exposure to mitochondrial proteins of the rat liver. Specifically, ATP synthase that powers the synthesis of the energy currency of the cells was chosen. The high sensitivity of ATP synthase to oxidative stress has been implicated [20,23,24] and, for instance, the α-subunit of ATP synthase was found to undergo HNE adduction in the brain of patients with mild cognitive impairment that was also accompanied by a significant decrease of ATP synthase activity compared to the age-matched controls [56]. In the present study, the β-subunit of ATP synthase was revealed as a major HNE-modified protein in rat liver mitochondria (Table 1); however, the direct effect of HNE exposure on corresponding enzyme activity has not been reported. As shown in Fig. 4, a significant and dose-dependent inactivation of the enzyme with an apparent IC50 of 89±17 µM was measured, when freshly isolated liver mitochondria were treated with varying concentrations of HNE (0.02–5 mM) compared to control. The Hill coefficient (h=0.56±0.06) also indicated, among others, interactions of the lipid peroxidation end-product with several binding sites on the enzyme complex [57]. Indeed, multiple modification sites were observed through our enrichment-driven proteomics approach (Table 1). The clear benefit of our paradigm has been the rapid and straightforward translation of target identifications to focused interrogation of functional consequences. Specifically, the study has aptly exemplified that HNE modification(s), indeed, accompany functional consequence by inactivating ATP synthase that has also been speculated to result in electron leakage and eventually in enhanced ROS production [25]. Investigations of the impact of HNE modifications on the function of additional proteins implicated by our analyses (Table 1 and Fig. 3) are in progress and will be reported separately.
Fig. 4.
ATP synthase activity of rat liver mitochondria after treatment with increasing concentrations of HNE.
In summary, we have shown in the present study that selective capture and, then, release of HNE-modified tryptic peptides followed by LC–MS/MS analysis offer the benefit of unambiguous identification of protein targets as well as sites of modification. Here, we identified protein–HNE Michael adducts in rat liver mitochondria heavily implicated in pathological processes not only involving the liver, but also other organs, including the brain [24,56]. Our study also reinforces the concept that HNE-induced carbonylation is selective and not a stochastic phenomenon. Bioinformatics interrogating mitochondrial protein networks revealed cell death, tumor morphology and drug metabolism that may be influenced by HNE-modifications. As an example to the potential functional consequences of HNE-induced carbonyl stress, we have also shown in isolated rat liver mitochondria a dose-dependent loss of enzyme activity for ATP synthase, one of the major targets for posttranslational modification by HNE and associated with the highest scoring network from our pathway analysis.
Supplementary Material
Acknowledgements
This study was supported by the National Institutes of Health, grant number AG025384. The authors wish to thank to Shastazia White for her excellent technical assistance. Laszlo Prokai is the Robert A Welch Chair in Biochemistry at UNTHSC (endowment number BK-0031).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Shacter E. Quantification and significance of protein oxidation in biological sample. Drug Metabol Rev. 2000;32:307–326. doi: 10.1081/dmr-100102336. [DOI] [PubMed] [Google Scholar]
- 2.Porter NA, Caldwell SE, Mills KA. Mechanisms of free radical oxidation of unsaturated lipids. Lipids. 1995;30:277–290. doi: 10.1007/BF02536034. [DOI] [PubMed] [Google Scholar]
- 3.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxyalkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Pamplona R. Membrane phospholipids, lipoxidative damage and molecular integrity: a causal role in aging and longevity. Biochim Biophys Acta. 2008;1777:1249–1262. doi: 10.1016/j.bbabio.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carini M, Aldini G, Facino RM. Mass spectrometry for detection of 4-hydroxytrans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spec Reviews. 2004;23:281–305. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 7.Navarro A, Boveris A. Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am J Physiol Regul Integr Comp Physiol. 2004;287:R1244–R1249. doi: 10.1152/ajpregu.00226.2004. [DOI] [PubMed] [Google Scholar]
- 8.Marques C, Pereira P, Taylor A, Liang JN, Reddy VN, Szweda LI, et al. Ubiquitin-dependent lysosomal degradation of the HNE-modified proteins in lens epithelial cells. FASEB J. 2004;18:1424–1426. doi: 10.1096/fj.04-1743fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cecarini V, Gee J, Fioretti E, Amici M, Angeletti M, Eleuteri A, et al. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim Biophys Acta. 2007;1773:93–104. doi: 10.1016/j.bbamcr.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 10.Negre-Salvayre A, Coatrieux C, Ingueneau C, Salvayre R. Advanced lipid peroxidation end products in oxidative damage to proteins. Potential role in diseases and therapeutic prospects for the inhibitors. Br J Pharmacol. 2008;153:6–20. doi: 10.1038/sj.bjp.0707395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier CS, Chavez J, Wang J, Wu J. Protein adducts of aldehydic lipid peroxidation products: identification and characterization of protein adducts using an aldehyde/keto-reactive probe in combination with mass spectrometry. In: Cadenas E, Packer L, editors. Methods in Enzymology, vol 473: Thiol redox transitions in cell signaling, Part A: Chemistry and biochemistry of low molecular weight and protein thiols. San Diego: Academic Press/Elsevier Inc; 2010. pp. 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu GZ, Liu Y, Sayre LM. Independent synthesis, solution behavior, and studies on the mechanism of formation of a primary amine-derived fluorophore representing cross-linking of proteins by (E)-4-hydroxy-2-nonenal. J Org Chem. 1999;64:5732–5745. [Google Scholar]
- 13.Poli G, Biasia F, Leonarduzzi G. 4-Hydroxynonenal-protein adducts: A reliable biomarker of lipid oxidation in liver diseases. Mol Aspects Med. 2008;29:67–71. doi: 10.1016/j.mam.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Schneider C, Porter NA, Brash AR. Routes to 4-hydroxynonenal: fundamental issues in the mechanisms of lipid peroxidation. J Biol Chem. 2008;283:15539–15543. doi: 10.1074/jbc.R800001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Newton BW, Russell WK, Russell DH, Ramaiah SK, Jayaraman A. Liver proteome analysis in a rodent model of alcoholic steatosis. J Proteome Res. 2009;8:1663–1671. doi: 10.1021/pr800905w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone DL, Doorn JA, Kiebler Z, Ickes BR, Petersen DR. Modification of heat shock protein 90 by 4-hydroxynonenal in a rat model of chronic alcoholic liver disease. J Pharmacol Exp Ther. 2005;315:8–15. doi: 10.1124/jpet.105.088088. [DOI] [PubMed] [Google Scholar]
- 18.Dalle-Donne I, Aldini G, Carini M, Colombo R, Rossi R, Milzani A. Protein carbonylation, cellular dysfunction, and disease progression. J Cell Mol Med. 2006;10:389–406. doi: 10.1111/j.1582-4934.2006.tb00407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Rad Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 20.Bruenner BA, Jones AD, German JB. Direct characterization of protein adducts of the lipid peroxidation product 4-hydroxy-2-nonenal using electrospray mass spectrometry. Chem Res Toxicol. 1995;8:552–559. doi: 10.1021/tx00046a009. [DOI] [PubMed] [Google Scholar]
- 21.Rauniyar N, Prokai L. Detection and identification of 4-hydroxy-2-nonenal Schiff-base adducts along with products of Michael addition using data-dependent neutral loss-driven MS3 acquisition: method evaluation through an in vitro study on cytochrome c oxidase modifications. Proteomics. 2009;9:5188–5193. doi: 10.1002/pmic.200900116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauniyar N, Stevens SM, Prokai L. Fourier transform ion cyclotron resonance mass spectrometry of covalent adducts of protein and 4-hydroxy-2-nonenal, a reactive end-product of lipid peroxidation. Anal Bioanal Chem. 2007;389:1421–1428. doi: 10.1007/s00216-007-1534-2. [DOI] [PubMed] [Google Scholar]
- 23.Stevens SM, Rauniyar N, Prokai L. Rapid characterization of covalent modifications to rat brain mitochondrial proteins after ex vivo exposure to 4-hydroxy-2-nonenal by liquid chromatography–tandem mass spectrometry using data dependent and neutral loss-driven MS3 acquisition. J Mass Spectrom. 2007;42:1599–1605. doi: 10.1002/jms.1349. [DOI] [PubMed] [Google Scholar]
- 24.Prokai L, Yan LJ, Vera-Serrano JL, Stevens SM, Forster MJ. Mass spectrometry-based survey of age-associated protein carbonylation in rat brain mitochondria. J Mass Spectrom. 2007;42:1583–1589. doi: 10.1002/jms.1345. [DOI] [PubMed] [Google Scholar]
- 25.Uto H, Kanmura S, Takami Y, Tsubouchi H. Clinical proteomics for liver disease: a promising approach for discovery of novel biomarkers. Proteome Sci. 2010;8:70. doi: 10.1186/1477-5956-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young TA, Cunningham CC, Bailey SM. Reactive oxygen species production by the mitochondrial respiratory chain in isolated rat hepatocytes and liver mitochondria: studies using myxothiazol. Arch Biochem Biophys. 2002;405:65–72. doi: 10.1016/s0003-9861(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 27.Venkatraman A, Landar A, Davis AJ, Ulasova E, Page G, Murphy MP, Darley-Usmar V, Bailey SM. Oxidative modification of hepatic mitochondria protein thiols: effect of chronic alcohol consumption. Am J Physiol Gastrointest Liver Physiol. 2004;286:G521–G527. doi: 10.1152/ajpgi.00399.2003. [DOI] [PubMed] [Google Scholar]
- 28.Sharma R, Nakamura A, Takahashi R, Nakamoto H, Goto S. Carbonyl modification in rat liver histones: decrease with age and increase by dietary restriction. Free Rad Biol Med. 2006;40:1179–1184. doi: 10.1016/j.freeradbiomed.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Arnaiz S, Llesuy S, Cutrin J, Boveris A. Oxidative stress by acute acetaminophen administration in mouse liver. Free Rad Biol Med. 1995;19:303–310. doi: 10.1016/0891-5849(95)00023-q. [DOI] [PubMed] [Google Scholar]
- 30.Niemela O. Distribution of ethanol-induced protein adducts in vivo: relationship to tissue injury. Free Rad Biol Med. 2001;31:1533–1538. doi: 10.1016/s0891-5849(01)00744-4. [DOI] [PubMed] [Google Scholar]
- 31.Traverso N, Menini S, Odetti P, Pronzato MA, Cottalasso D, Marinari UM. Lipoperoxidation in hepatic subcellular compartments of diabetic rats. Free Rad Biol Med. 1999;26:538–547. doi: 10.1016/s0891-5849(98)00238-x. [DOI] [PubMed] [Google Scholar]
- 32.Chen JJ, Schenker S, Henderson GI. 4-hydroxynonenal levels are enhanced in fetal liver mitochondria by in utero ethanol exposure. Hepatology. 1997;25:142–147. doi: 10.1002/hep.510250126. [DOI] [PubMed] [Google Scholar]
- 33.Mendez D, Hernaez ML, Diez A, Puyet A, Bautista JM. Combined proteomic approaches for the identification of specific amino acid residues modified by 4-hydroxy-2-nonenal under physiological conditions. J Proteome Res. 2010;9:5770–5781. doi: 10.1021/pr100555v. [DOI] [PubMed] [Google Scholar]
- 34.Brunet S, Thibault P, Gagnon E, Kearney P, Bergeron JJM, Desjardins M. Organelle proteomics: looking at less to see more. Trends Cell Biol. 2003;13:629–638. doi: 10.1016/j.tcb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal Chem. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauniyar N, Prokai-Tatrai K, Prokai L. Identification of carbonylation sites in apomyoglobin after exposure to 4-hydroxy-2-nonenal by solid-phase enrichment and liquid chromatography-electrospray ionization tandem mass spectrometry. J Mass Spectrom. 2009;45:398–410. doi: 10.1002/jms.1725. [DOI] [PubMed] [Google Scholar]
- 37.Uchida K, Stadtman ER. Modification of histidine residues in proteins by reaction with 4-hydroxynonenal. Proc Natl Acad Sci USA. 1992;89:4544–4548. doi: 10.1073/pnas.89.10.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchida K, Stadtman ER. Selective cleavage of thioether linkage in proteins modified with 4-hydroxynonenal. Proc Natl Acad Sci USA. 1992;89:5611–5615. doi: 10.1073/pnas.89.12.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 40.Roe MR, McGowan TF, Thomas F, Thompson LV, Griffin TJ. Targeted O-18-labeling for improved proteomic analysis of carbonylated peptides by mass spectrometry. J Am Soc Mass Spectrom. 2010;21:1190–1203. doi: 10.1016/j.jasms.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doorn JA, Petersen DR. Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem Biol Interact. 2003;143–144:93–100. doi: 10.1016/s0009-2797(02)00178-3. [DOI] [PubMed] [Google Scholar]
- 42.Roe MR, Xie H, Bandhakavi S, Griffin TJ. Proteomic mapping of 4-hydroxynonenal protein modification sites by solid-phase hydrazide chemistry and mass spectrometry. Anal Chem. 2007;79:3747–3756. doi: 10.1021/ac0617971. [DOI] [PubMed] [Google Scholar]
- 43.Lopachin RM, Geohagen BC, Gavin T. Synaptosomal toxicity and nucleophilic targets of 4-hydroxy-2-nonenal. Toxicol Sci. 2009;107:171–181. doi: 10.1093/toxsci/kfn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohta S, Kagawa Y. Human F1-ATPase: molecular cloning of cDNA for the beta subunit. J Biochem. 1968;99:135–141. doi: 10.1093/oxfordjournals.jbchem.a135452. [DOI] [PubMed] [Google Scholar]
- 45.Del Rio P, Montiel T, Chagoya V, Massieu L. Exacerbation of excitotoxic neuronal death induced during mitochondrial inhibition in vivo: relation to energy imbalance or ATP depletion? Neuroscience. 2007;146:1561–1570. doi: 10.1016/j.neuroscience.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 46.Smith DD, Campbell JW. Distribution of glutamine synthetase and carbamoylphosphate synthetase I in vertebrate liver. Proc Natl Acad Sci U S A. 1988;85:160–164. doi: 10.1073/pnas.85.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang J, Hsu BYL, Macmullen CM, Poncz M, Smith TJ, Stanley CA. Expression, purification and characterization of human glutamate dehydrogenase (GDH) allosteric regulatory mutations. Biochem J. 2002;363:81–87. doi: 10.1042/0264-6021:3630081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crouser ED, Julian MW, Huff JE, Struck J, Cook CH. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Critical Care Med. 2006;34:2439–2446. doi: 10.1097/01.CCM.0000230240.02216.21. [DOI] [PubMed] [Google Scholar]
- 49.Radi R, Turrens JF, Chang LY, Bush KM, Crapo JD, Freeman BA. Detection of catalase in rat heart mitochondria. J Biol Chem. 1991;266:22028–22034. [PubMed] [Google Scholar]
- 50.Jiang XS, Dai J, Sheng QH, Zhang L, Xia QC, Wu JR, et al. A comparative proteomic strategy for subcellular proteome research: ICAT approach coupled with bioinformatics prediction to ascertain rat liver mitochondrial proteins and indication of mitochondrial localization for catalase. Mol Cell Proteomics. 2005;4:12–34. doi: 10.1074/mcp.M400079-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Semsei I, Rao G, Richardson A. Changes in the expression of superoxide dismutase and catalase as a function of age and dietary restriction. Biochem Biophys Res Commun. 1989;164:620–625. doi: 10.1016/0006-291x(89)91505-2. [DOI] [PubMed] [Google Scholar]
- 52.D'Souza A, Kurien B, Rodgers R, Shenoi J, Kurono S, Matsumoto H, et al. Detection of catalase as a major protein target of the lipid peroxidation product 4-HNE and the lack of its genetic association as a risk factor in SLE. BMC Med Genetics. 2008;9:62. doi: 10.1186/1471-2350-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuler AM, Wood PA. Mouse models for disorders of mitochondrial fatty acid beta-oxidation. ILAR J. 2002;43:57–65. doi: 10.1093/ilar.43.2.57. [DOI] [PubMed] [Google Scholar]
- 54.Mi J, Garcia-Arcos I, Alvarez R, Cristobal S. Age-related subproteomic analysis of mouse liver and kidney peroxisomes. Proteome Sci. 2007;5:19–34. doi: 10.1186/1477-5956-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marquez A, Villa-Treviño S, Guéraud F. The LEC rat: a useful model for studying liver carcinogenesis related to oxidative stress and inflammation. Redox Rep. 2007;12:35–39. doi: 10.1179/135100007X162220. [DOI] [PubMed] [Google Scholar]
- 56.Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, et al. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiol Disease. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Mailman RB. Toxicant–receptor interactions: fundamental principles. In: Smart RC, Hodgson E, editors. Molecular and Biochemical Toxicology. 4th Edition. Chichester: John Wiley & Sons, Inc; 2008. pp. 359–390. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.