Abstract
Abscisic acid (ABA) is a key hormone regulating plant growth, development and the response to biotic and abiotic stress. ABA binding to pyrabactin resistance (PYR)/PYR1-like (PYL)/Regulatory Component of Abscisic acid Receptor (RCAR) intracellular receptors promotes the formation of stable complexes with certain protein phosphatases type 2C (PP2Cs), leading to the activation of ABA signalling. The PYR/PYL/RCAR family contains 14 genes in Arabidopsis and is currently the largest plant hormone receptor family known; however, it is unclear what functional differentiation exists among receptors. Here, we identify two distinct classes of receptors, dimeric and monomeric, with different intrinsic affinities for ABA and whose differential properties are determined by the oligomeric state of their apo forms. Moreover, we find a residue in PYR1, H60, that is variable between family members and plays a key role in determining oligomeric state. In silico modelling of the ABA activation pathway reveals that monomeric receptors have a competitive advantage for binding to ABA and PP2Cs. This work illustrates how receptor oligomerization can modulate hormonal responses and more generally, the sensitivity of a ligand-dependent signalling system.
Keywords: hormone receptor, protein phosphatase 2C, PYR-PYL-RCAR, START domain, stress response
Introduction
The plant hormone abscisic acid (ABA) is an important regulator of plant growth and development and plays a key role in the control of the plant stress response (Verslues and Zhu, 2007; Cutler et al, 2010). Under adverse environmental conditions, particularly drought and salinity, the levels of ABA increase in the plant, triggering a series of adaptive responses required for plant survival (Buchanan et al, 2000; Nambara and Marion-Poll, 2005; Lee et al, 2006). Recently, a family of intracellular ABA receptors, named PYR/PYL (pyrabactin resistance/PYR1-like) (Park et al, 2009) or RCAR (Regulatory Component of Abscisic acid Receptor) (Ma et al, 2009) have been shown to play a crucial role in this response. The PYR/PYL/RCAR proteins are able to bind to ABA and regulate the activity of clade A serine/threonine protein phosphatases 2C (PP2Cs) including ABI1, ABI2 and HAB1 (Leung et al, 1994; Meyer et al, 1994; Leonhardt et al, 2004; Saez et al, 2004). This in turn regulates phosphorylation of some serine/threonine protein kinases in the sucrose non-fermenting1-related subfamily 2 (SnRK2; Mustilli et al, 2002; Yoshida et al, 2002). Under non-stress conditions, clade A PP2Cs dephosphorylate certain SnRK2, keeping them in an inactive state (Umezawa et al, 2009; Vlad et al, 2009). Under conditions of stress, ABA levels increase and induce the formation of a stable complex between PYR/PYL/RCARs and PP2Cs, which inactivates PP2C phosphatase activity and enables accumulation of active, phosphorylated SnRK2s (Cutler et al, 2010). Once activated, SnRK2s phosphorylate key mediators of stress adaptation such as the SLAC1 anion channel, involved in stomatal closure, and the ABF/AREB transcription factors, including ABF2, required for transcriptional activation of stress responsive genes (Fujii et al, 2009; Fujita et al, 2009; Geiger et al, 2009, 2011; Lee et al, 2009). Recently, it has been shown that the PYL/RCAR receptors and ABI1 regulate the activity of the CPK23 and CPK21 protein kinases, which also have SLAC1 and its homologue SLAH3 as substrates (Geiger et al, 2011).
Recent structural analyses of PYR/PYL/RCAR proteins alone and in complex with ABA and PP2Cs have provided significant insight into the molecular mechanism of ABA signalling (Melcher et al, 2009; Miyazono et al, 2009; Nishimura et al, 2009; Santiago et al, 2009a; Yin et al, 2009; Dupeux et al, 2011) (for review, see Cutler et al, 2010; Melcher et al, 2010b; Weiner et al, 2010). ABA binds inside a conserved hydrophobic cavity, which is surrounded by two flexible loops, the β3/β4 and β5/β6 loops (called the gating loops or the gate and latch) that control access to the ABA-binding pocket. In the ABA-free form, these loops adopt an open conformation leaving a free passage into the cavity. However, once ABA is bound, these loops close over the hormone stabilizing it inside the cavity. These ABA-induced conformational changes promote the formation of a stable complex between the ligand-bound receptor and the catalytic domain of clade A PP2Cs. In this complex, the β3/β4 gate loop inserts in the PP2C catalytic site and inhibits its activity, acting as a competitive inhibitor that blocks substrate access (Melcher et al, 2009; Miyazono et al, 2009; Yin et al, 2009; Dupeux et al, 2011). At the same time, a conserved tryptophan residue in the flap subdomain of the clade A PP2C inserts between the gating loops stabilizing them in the closed conformation and trapping the hormone inside. These interactions explain the increased stability of the ternary receptor–hormone–phosphatase complex as compared with that of the receptor–hormone complex alone.
Despite these molecular level details, major questions remain poorly understood. For example, PYR/PYL/RCAR proteins form multigene families in plants. Microarray data sets suggest that several different receptor proteins are often co-expressed in a single cell (Kilian et al, 2007), but it is still unclear whether they carry out fully redundant or specialized functions. Furthermore, molecular details of the receptor activation process are still poorly understood. For example, although the receptors studied crystallographically to date, including PYR1, PYL1 and PYL2, are dimeric, their receptor–ABA–phosphatase complexes show 1:1:1 stoichiometry. This implies that dissociation of the receptor dimer is necessary for receptor activation and signalling (Melcher et al, 2009; Miyazono et al, 2009; Nishimura et al, 2009; Santiago et al, 2009a; Yin et al, 2009). Moreover, the receptor–ABA–PP2Cs complexes are very stable, with apparent affinities for ABA in the low nanomolar range. However, ABA levels in vivo have been observed to vary over three orders of magnitude (from nanomolar to micromolar) (Harris et al, 1988; McCourt and Creelman, 2008). This raises the question as to whether the proposed mechanism for ABA perception, that is, a low-nanomolar sensing system, is enough to account for the perception of changes across the whole range of physiological ABA concentrations (McCourt and Creelman, 2008). Interestingly, when hormone binding is measured for the PYRL/PYL/RCAR proteins alone, the affinity for ABA is not only lower than that measured in the presence of PP2Cs, but the values of the Kd vary significantly among different receptor proteins. For example, PYL1 and PYL2 have Kds for ABA of 52 and 59 μM, respectively (Miyazono et al, 2009; Yin et al, 2009), while PYL5 and PYL8 have affinities of 1.0 and 0.9 μM, respectively (Santiago et al, 2009b; Szostkiewicz et al, 2010). These differences in affinity are unlikely to be explained by differences in the ABA-binding pocket, since critical amino acids involved in receptor–hormone contacts and in the gating loops are highly conserved among PYR/PYL/RCAR proteins. Here, we present a detailed analysis of several members of the PYRL/PYL/RCAR family and provide new insight into the receptor activation mechanisms. Moreover, the biochemical and structural characteristics of these proteins suggest the existence of two distinct classes of ABA receptors characterized by the oligomeric states of their apo forms. The differential properties of these two types of ABA receptors might lead to distinct responses when they are considered in the context of the plant.
Results
ABA induces dissociation of the PYR1 dimer
The PYR/PYL/RCAR proteins studied at a structural level to date (PYR1, PYL1 and PYL2) are homodimeric in the absence of ABA, but form 1:1 monomeric complexes with PP2Cs after ABA binding (Melcher et al, 2009; Miyazono et al, 2009; Nishimura et al, 2009; Yin et al, 2009; Santiago et al, 2009a). Interestingly, the homodimerization and PP2C-binding interfaces of these receptors are largely overlapping (Figure 1). This structural organization suggests that homodimerization and PP2C binding are in competition with one another. Thus, dimerization, together with the conformational changes in the gating loops, contributes to block receptor–PP2C interactions in the absence of ABA, presumably functioning to prevent basal activation of the pathway. Hence, dissociation of the PYR1 receptor dimer, likely induced by ABA binding, should precede the formation of the ternary complex. However, these conclusions are based largely on crystallographic observations of receptors at high concentration that shift equilibria towards dimer formation (Nishimura et al, 2009; Santiago et al, 2009a). To address this, we analysed the oligomeric state of several PYR/PYL/RCAR proteins by Size-Exclusion Chromatography (SEC) coupled to Multiple Angle Laser Light Scattering (MALLS). In this approach, SEC followed by SDS–PAGE analysis of the eluted fractions reveals the formation of complexes, while the MALLS analysis provides an independent determination of the molecular mass of the eluting species which, unlike SEC, is not affected by deviations from globularity (Wyatt, 1998). Additionally, with this technique it is possible to work at moderate protein concentrations. For all the MALLS experiments described below, proteins were injected at a concentration of 80 μM (see Materials and methods).
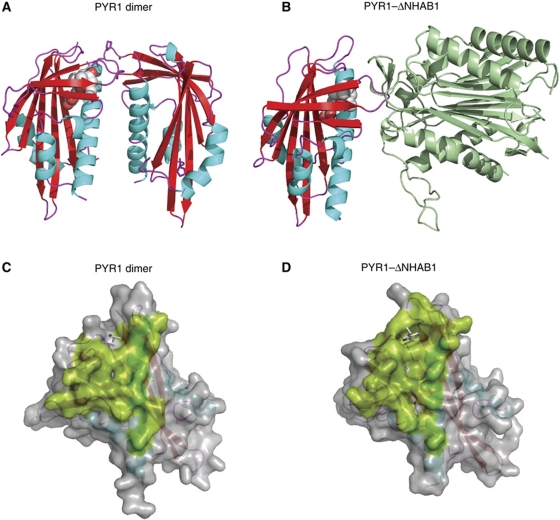
Figure 1.
The PYR/PYL/RCAR dimerization and phosphatase interaction regions. The structures of the PYR1 dimer (3K90) (A) and the PYR1–ABA–ΔNHAB1 ternary complex (3QN1) (B) are shown. The abscisic acid molecule is shown in CPK representation. The PYR1 molecular surfaces involved in dimerization (C) and ΔNHAB1 interaction (D) are shown in green colour. The side chains of the Leu 87, from the ABA-free subunit of the PYR1 dimer (C) and the side chain of Trp 385 from ΔNHAB1 (D) that occupy similar positions near the PYR1 gating loops are indicated as stick models. The large degree of overlap between the two surfaces indicates that the PP2C-interaction region is occluded in the PYR1 dimer and that receptor dissociation would be required for the formation of the ternary complex.
Using this approach, we studied the oligomeric state, PP2C interactions and effects of ABA for five members of the receptor family: PYR1, PYL1, PYL5, PYL6 and PYL8. The analyses of these proteins in the absence of ABA indicate that PYR1 and PYL1 are dimeric proteins (apparent Mr of 39 and 50 kDa; predicted 44 and 51 kDa, respectively; Figure 2; Supplementary Figure S1). Unexpectedly, PYL5, PYL6 and PYL8 are monomeric proteins (Figure 2; Supplementary Figure S1). When the same experiments are performed in the presence of ABA, PYR1 eluted at higher volumes (indicative of a decrease in molecular size) and the apparent molecular mass was 21 kDa, which coincides with that of a PYR1 monomer (predicted=22 kDa). On the contrary, analysis of the PYR1Y120A, a mutant defective in ABA binding (Dupeux et al, 2011), indicates that this mutant does not become monomeric in response to ABA (Figure 2). Together, these results indicate that ABA binding is both necessary and sufficient for dissociation of the PYR1 dimer. Similarly, PYL1 showed a reduction of its apparent Mr in the presence of ABA (Supplementary Figure S1), although it was less completely dissociated by ABA than PYR1. As expected, the apparent masses for monomeric receptors PYL5, PYL6 and PYL8 did not shift in the presence of ABA (Figure 2; Supplementary Figure S1). Thus, ABA receptors can exist in either monomeric or dimeric forms and ABA promotes dissociation of the dimeric receptors.
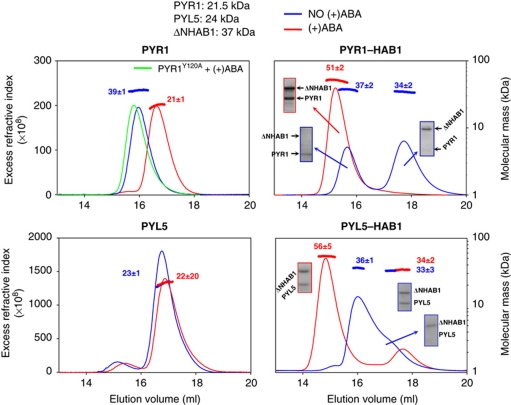
Figure 2.
ABA receptors exist in dimeric and monomeric forms. SEC-MALLS analysis of PYR1 (top) and PYL5 (bottom) alone (left) and in the presence of equimolar concentrations of ΔNHAB1 (right) as described in Materials and methods. The experiments were done in the absence (blue) and presence (red) of 1 mM (+)ABA. SDS–PAGE analysis of eluted fractions is shown (boxes). Both the SEC elution profiles (monitored through the excess refractive index, which is proportional to the protein concentration) and the molecular size calculated by MALLS (shown above the peaks for each species) indicate dissociation of the PYR1 dimer in the presence of ABA (expected Mw 42 and 21 kDa for dimers and monomers, respectively). The PYR1Y120A mutant, defective in ABA binding, remains as a dimer in the presence of ABA (green line). PYL5 is monomeric (expected Mw 24 kDa) and is not affected by the presence of (+)ABA. Both proteins form 1:1 complexes when combined with ΔNHAB1 in the presence of ABA (right panels); however, while PYR1 does not interact with ΔNHAB1 in the absence of ABA the interaction between PYL5 and ΔNHAB1 in the same conditions is revealed by a decrease in the height of the peak corresponding to monomeric ΔNHAB1 and the appearance of a peak containing both PYL5 and ΔNHAB1. See Supplementary Figure S1 for similar experiments with PYL1, PYL6 and PYL8.
We also examined interactions between receptors and the PP2C HAB1 in the absence of ABA. For this purpose, we used the catalytic core of HAB1 (ΔNHAB1, amino acids 179–511), rather than the full-length protein, which is difficult to produce in recombinant expression systems. These experiments revealed differences in basal PP2C interactions between monomeric and dimeric receptors. In agreement with crystallographic studies, all receptors tested form stable complexes with ΔNHAB1 in the presence of ABA at 1:1 stoichiometry, as indicated by the apparent Mr of the complexes (right panels in Figure 2; Supplementary Figure S1). Importantly, PYR1 and PYL1 do not interact with PP2C in the absence of ABA, while PYL5, PYL6 and PYL8 show partial interactions (Figure 2; Supplementary Figure S1). The MALLS analyses show intermediate molecular masses, indicative of a fast exchange between complexed and uncomplexed forms. These results indicate that monomeric receptors are capable of establishing weak but clearly detectable interactions with ΔNHAB1 in vitro in the absence of ABA, and suggest that a key function of dimerization in receptors like PYR1 and PYL1 is to prevent basal interactions with PP2Cs in the absence of ABA.
Monomeric and dimeric receptors have different intrinsic affinities for ABA
The apparent ABA dissociation constants for PYL1, PYL2, PYL5 and PYL8 have been previously determined (Miyazono et al, 2009; Santiago et al, 2009b; Yin et al, 2009; Szostkiewicz et al, 2010) and are shown in Table I. We carried out Isothermal Titration Calorimetry (ITC) experiments to measure the apparent ABA-binding affinity of PYL6 and PYR1. The Kd for PYL6 was 1.1 μM (±0.01) with a negative enthalpy, indicating an exothermic binding process (see Table I). Under the same conditions, PYR1 did not produce significant heat signals indicating a much lower affinity. When PYR1 was assayed at higher protein concentrations (200 μM), an endothermic binding curve was obtained (see below). Due to the low apparent binding affinity, the exact value of the apparent Kd could not be precisely determined. However, given the experimental conditions used, it can be estimated to be greater than 50 μM. NMR experiments in which binding was followed through ABA-induced resonance shifts on a 15N-labelled PYR1 sample were used to obtain an independent estimation of the dissociation constant. The value of the Kd obtained was 97±36 μM (Supplementary Figure S2). Thus, the dimeric receptor PYR1 has an apparent affinity for ABA that is almost two orders of magnitude lower than that of the monomeric receptors.
Table 1. Apparent ABA-binding affinities and oligomeric state of PYR/PYL/RCAR receptors.
| Kd (μM) | ΔH (kcal/mol) | Oligomeric state | Reference | |
|---|---|---|---|---|
| PYR1 | 97 (±36) | Endothermic | Dimer | This study |
| PYR1H60P | 3.0 (±0.26) | −2.5 (±0.035) | Monomer | This study |
| PYL1 | 52.0 | +1.4 | Dimer | Miyazono et al and this study |
| PYL2 | 59.1 (±2.5) | Endothermic | Dimer | Yin et al |
| PYL5 | 1.0 (±0.06) | −7.9 (±0.2) | Monomer | Santiago et al |
| PYL6 | 1.1 (±0.01) | −3.573 (±0.1) | Monomer | This study |
| PYL8 | 0.9 (±0.15) | −10.4 (±0.3) | Monomer | Szostkiewicz et al and this study |
The oligomeric state and apparent affinities for ABA of the PYR/PYL/RCAR receptors studied here and elsewhere are summarized in Table I. Comparison of these parameters indicates a strong correlation between the ABA affinities and the oligomeric state of the apo form of the receptor, with monomeric receptors showing Kds in the range of 1 μM and negative enthalpies, and dimeric receptors showing much lower apparent affinities and positive binding enthalpies for ABA. As mentioned above, the amino-acid residues involved in hormone binding are highly conserved in the PYR/PYL/RCAR family. Hence, the observed differences in apparent enthalpy and binding affinity are unlikely to be the result of differences in the ABA-binding pocket. The lower apparent affinity of dimeric receptors could be explained by the unfavourable contribution of dimer dissociation to the thermodynamics of the receptor activation process. Ligand binding is typically an exothermic process, as it normally results in the net creation of chemical bonds between ligand and protein, favouring the binding reaction. On the contrary, protein dissociation is often endothermic as it usually involves a net breakage of chemical bonds between protein monomers. We hypothesized that the unfavourable contribution of dimer dissociation to the thermodynamics of the receptor activation process could explain the correlation between the oligomeric state of the apo form of the receptor and their intrinsic hormone binding affinities. In order to test this, we generated a PYR1 variant that is impaired for dimerization but is not compromised in ABA or PP2C binding.
A histidine 60 to proline substitution destabilizes the PYR1 dimer and increases its apparent ABA affinity
As shown above, monomeric receptors have a certain capacity to interact with ΔNHAB1 in the absence of ABA while dimeric receptors do not. This observation is in agreement with the strong ABA dependence of PYR1–HAB1 and PYL1–HAB1 interactions observed previously in a yeast two-hybrid (Y2H) assay system and the ABA-independent interactions observed for PYL5, PYL6 and PYL8 (Park et al, 2009; Santiago et al, 2009b). We took advantage of this assay to identify PYR1 mutations leading to altered PYR1–PP2C interactions. Using a previously described error-prone PCR mutagenized PYR1 library (Peterson et al, 2010), we screened yeast cells for PYR1 mutants that enable interaction with HAB1 in the absence of ABA, which led to the identification of a missense mutant in which histidine 60 was replaced by proline (Figure 3A).
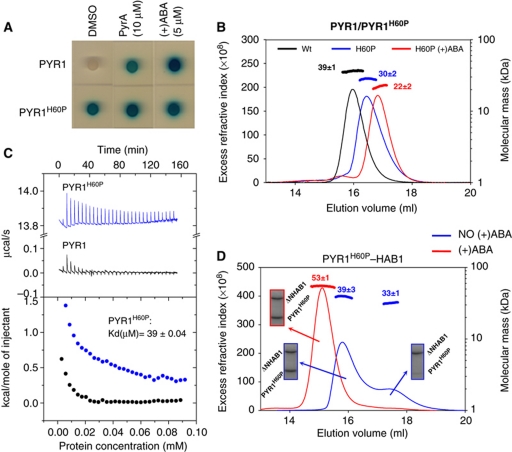
Figure 3.
The PYR1 His60Pro mutation favours both dimer dissociation and constitutive interaction with ΔNHAB1. (A) Y2H assay showing constitutive interaction between PYR1H60P and ΔNHAB1. This interaction is strictly dependent on ABA or its analogue Pyrabactin for wt PYR1. (B) SEC-MALLS analysis of wt PYR1 (black) and PYR1H60P in the absence (blue) and presence (red) of 1 mM (+)ABA. The shift of PYR1H60P towards lower apparent molecular size in SEC and MALLS in the absence of ABA is indicative of equilibrium between monomeric and dimeric forms. PYR1H60P dissociates completely into monomeric species in the presence of 1 mM ABA (red curve). (C) Comparison of the dissociation rates of PYR1 and PYR1H60P measured by ITC. Concentrated solutions of both proteins were repeatedly injected into the ITC cell filled with buffer and dissociation thermograms were recorded. The estimated dimer dissociation constant for PYR1H60P is indicated. (D) SEC-MALLS analysis showing constitutive interactions between ΔNHAB1 and PYR1H60P. Experiments were conducted by mixing equimolecular concentrations of both proteins before injection, in the absence (blue) and in the presence (red) of 1 mM (+)ABA. The composition of the SEC peaks was analysed by SDS–PAGE (boxes). PYR1H60P shows a weak interaction with ΔNHAB1 in the absence of ABA.
The SEC-MALLS analysis of the H60P mutant produced a single elution peak with an apparent mass of 30 kDa (Figure 3B), which is between those expected for the dimer and monomer species. This elution peak is wide and asymmetric, indicative of polydispersity. In contrast, in the presence of ABA, PYR1H60P elutes as a single symmetric peak with an apparent mass of 22 kDa, as expected for the monomeric form. This indicates that in the absence of ABA, PYR1H60P behaves as a mixture of dimeric and monomeric species in rapid exchange. In order to confirm this, we measured dimer dissociation rates by ITC with wt and mutant PYR1. For this purpose, concentrated solutions of either protein were injected into the ITC cell filled with buffer. In this way, the sudden dilution of the protein promotes dissociation and the associated heat can be measured. As can be observed (Figure 3C), the thermogram corresponding to PYR1H60P shows a profile typical of protein dissociation, with endothermic peaks whose area decrease as the protein concentration in the ITC cell increases. The calculated dissociation constant is 39 μM. In contrast, wt PYR1 produced much weaker signals that disappeared after a few injections, indicating that limited dissociation occurs only in extremely diluted conditions. We next tested whether the PYR1H60P mutant could interact in vitro with ΔNHAB1 in the absence of ABA. For this purpose, PYR1H60P and ΔNHAB1 were mixed in equimolar amounts and analysed by SEC-MALLS. As shown in Figures 2 and 3A, wt PYR1 does not interact with ΔNHAB1 in the absence of ABA. However, PYR1H60P shows partial interaction with ΔNHAB1, similar to monomeric receptors (Figure 2), and confirms the constitutive interactions detected by the Y2H assay (Figure 3A).
We measured the apparent affinity and enthalpy for ABA binding of the PYR1H60P mutant and compared it with that of wt PYR1 (Figure 4A and B). The introduction of this mutation leads to a significant increase in apparent ABA-binding affinity with a dissociation constant of 3 μM and a negative enthalpy, similar to that of monomeric receptors. This enhanced ABA-binding affinity was confirmed in a functional assay based on the in vitro reconstitution of the ABA signalling cascade (Fujii et al, 2009). For this purpose, wt PYR1 or PYR1H60P was combined in a test tube with ΔNHAB1, SnRK2.6/OST1 and a C-terminal deletion of the ABF2 transcription factor (ΔCABF2, amino acids 1–173), as described in Materials and methods. In this assay, OST1 activity is measured as autophosphorylation as well as trans-phosphorylation of its natural substrate, ΔCABF2. In the absence of ABA, OST1 activity is inhibited by ΔNHAB1-mediated dephosphorylation, while addition of ABA leads to the receptor-mediated inactivation of the phosphatase and increased levels of OST1 activity (Fujii et al, 2009; Dupeux et al, 2011). As can be observed in Figure 4C, PYR1H60P was able to recover OST1 activity at lower ABA concentrations than wt PYR1. This result is in agreement with the measured increase in ABA-binding affinity and indicates that a monomeric variant of PYR1 is more effective than the dimeric PYR1 for the inhibition of the phosphatase activity of ΔNHAB1 at lower ABA concentrations.
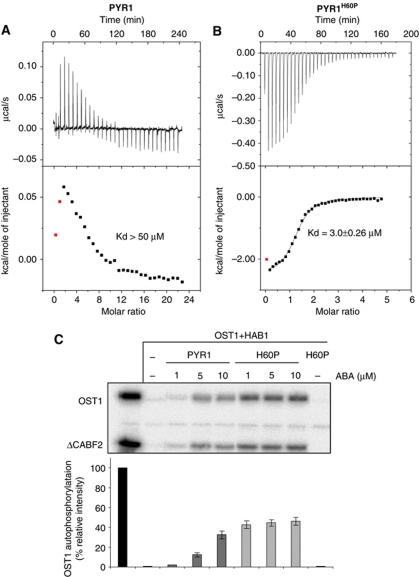
Figure 4.
The PYR1 His60Pro mutation leads to increased affinity for ABA. Typical thermograms resulting from isothermal titration calorimetry experiments with wt PYR1 (A) and PYR1H60P (B) (the concentration of PYR1 is 8 times higher than that of PYR1H60P). The estimated Kds for ABA binding are indicated. The increase in affinity and the change in enthalpy, endothermic for PYR1 and exothermic PYRH60P can be appreciated. (C) In vitro OST1 kinase activity assays in the presence of ΔNHAB1, wt PYR1 and the mutant PYR1H60P. The experiment shows an enhanced recovery of OST1 activity in the presence of PYR1H60P at low ABA concentrations, as compared with wt PYR1. The top panel shows a representative autoradiogram from a protein kinase assay. The bottom panel shows the quantification of the activity based on relative intensity of bands corresponding to OST1 phosphorylation products, using a phosphoimager Image Gauge V.4.0. Quantification of ΔCABF2 was also performed obtaining similar results. Standard error measurements are shown (n=3).
Taken together, these results demonstrate that the His60Pro mutation produces a destablization of the PYR1 dimer and that this renders the mutant PYR1 capable of establishing weak, but clearly detectable, interactions with ΔNHAB1 in the absence of the hormone, supporting the notion that dimerization contributes to prevent interaction between receptor and phosphatase in the absence of ABA by occluding the phosphatase docking region on the surface of the receptor. Moreover, they demonstrate that the differences in apparent ABA-binding affinity between monomeric and dimeric receptors are produced by a negative contribution of dimerization to the receptor activation process.
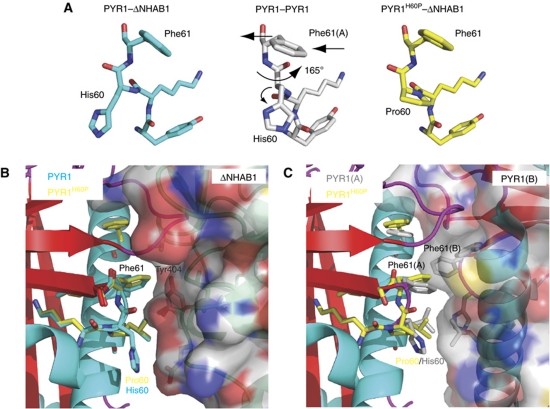
A proline at position 60 of PYR1 hinders peptide backbone rearrangements required for the formation of the receptor dimer
The structure of the ternary complex formed by PYR1H60P, ABA and the catalytic core of HAB1 (ΔNHAB1) was obtained by X-ray crystallography (see Table II for crystallographic data collection and refinement statistics). The electron density around position 60 of PYR1 confirmed the substitution of histidine for proline (see Supplementary Figure S3). The mutant complex is remarkably similar to that formed by wt PYR1 and ΔNHAB1 (Dupeux et al, 2011). However, the comparison of these two complexes with the structure of dimeric PYR1 indicates a major rearrangement of the peptide backbone around residue 60, with a small change in the Phi angle and a rotation of the Psi torsion angle of 165° (Figure 5A). This conformational change influences the position of the side chain of the neighbouring residue, Phe61 at the core of the dimerization interface. As can be observed in Figure 5B, in both the wt and mutant ternary complexes Phe61 of PYR1 occupies a hydrophobic pocket at the centre of the receptor–phosphatase interface. In dimeric PYR1, this residue occupies an equivalent position, but this time at the interface between the two PYR1 monomers. However, in this case, the space available to Phe61 is restricted by the equivalent Phe61 residue from the second molecule in the dimer, whose side chain protrudes into this hydrophobic pocket (Figure 5C). The modification of the backbone torsion angles around residue 60 in wt PYR1 produces an outward displacement of about 1 Å in the position of the Cα and the side chain of Phe61 in each molecule of the dimer (Figure 5A). This rearrangement is necessary to prevent a steric clash between the two Phe61 residues at the dimer interface. While introduction of a proline at position 60 is compatible with the formation of a phosphatase complex, as demonstrated here (Figure 5B), this residue limits the flexibility of the peptide backbone at this critical position, hindering in turn the outward movement of Phe61 and leading to a steric clash at the dimer interface, explaining the decreased tendency of this receptor variant to form dimers.
Table 2. Crystallographic data collection and refinement statistics of the PYR1H60P–ABA–ΔNHAB1 complex (brackets indicated values for highest resolution shell).
| Data collection | |
| Space group | P212121 |
| Unit cell a, b, c | 45.4 65.9 172.00 |
| α, β, γ | 90.0 90.0 90.0 |
| Wavelength | 0.979240 |
| Resolution | 25.0–2.10 (2.20-2.21) |
| Rmerge (%) | 9.0 (44.3) |
| I/σI | 19.70 (4.42) |
| Completeness | 99.7% (99.9) |
| Redundancy | 7.14 (7.3) |
| No. of reflections | 222 258 |
| No. of unique refl. | 31 132 |
| Refinement | |
| Resolution range (Å) | 24.29–2.1 |
| No. of unique refl. | 31132 |
| No. of refl in test set | 1557 |
| Rwork (%) | 18.0 |
| Rfree (%) | 23.6 |
| No. of atoms | 3951 |
| Protein | 3731 |
| Ligand | 29 |
| Solvent | 206 |
| B-factors | 35.807 |
| R.m.s. deviations | |
| Bond length | 0.02 |
| Angles | 1.74 |
Figure 5.
Structure of the PYR1H60P mutant and its effect on the dimerization interface. The structure of PYR1H60P in complex with ΔNHAB1 and (+)ABA was obtained by X-ray crystallography (3ZVU). (A) Detail of the conformation of peptide backbone at position 60 and its effect on Phe61 in the wt PYR1–ΔNHAB1 complex (3QN1) (left, blue), the PYR1 dimer (3K90) (centre, grey) and the PYR1H60P–ΔNHAB1 (right, yellow). Arrows indicate the rearrangement of the peptide backbone around His60 and the displacement of Phe61 in dimeric PYR1. (B) Detail of the receptor–PP2C interface in the wt PYR1–ΔNHAB1 and the PYR1H60P–ΔNHAB1 complexes. Wt PYR1 is shown as in Figure 1 with relevant residues at the α4/β2 loop in blue. The corresponding residues of PYR1H60P are shown in yellow. The backbone of ΔNHAB1 is shown in green with semitransparent molecular surface. As can be appreciated except for the His/Pro substitution, both PYR1 proteins form virtually identical interfaces with Phe61 occupying a pocket at the centre of the receptor–phosphatase interface. (C) The structure of PYR1H60P (yellow) from the model above was overlaid on subunit A of the PYR1 dimer (backbone as in Figure 1 and side chains in grey). The structure of the second subunit in the PYR1 dimer is shown with semitransparent molecular surface and critical side chains in grey. The modifications in the conformation of the α4/β2 loop and the outward displacement of Phe61 in dimeric PYR1 leave space to accommodate the side chain of Phe61(B) of the second subunit in the dimer, which occupies the centre of the dimerization interface. The position of Phe61 from PYR1H60P (yellow) partially overlaps with that of Phe61(B). The modification of the backbone torsion angles at position 60 in the PYR1 dimer (A) produces an outward displacement of Phe61 preventing a steric clash between the two Phe61 residues at the dimer interface. The introduction of a proline at position 60 limits backbone flexibility hindering this rearrangement.
Competition for ABA binding could lead to preferential activation of monomeric receptors
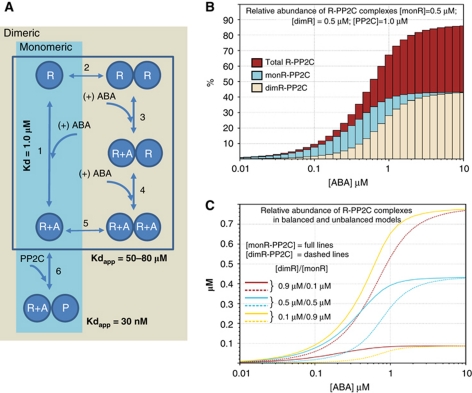
Both monomeric and dimeric receptors can form high-affinity complexes with PP2Cs, and once they are formed, both complexes have a similar stability (Ma et al, 2009; Santiago et al, 2009b). However, our results indicate that dimeric receptors (i) need to dissociate into monomers before they can interact with and inhibit a PP2C and (ii) have a lower intrinsic affinity for ABA. We reasoned that if both monomeric and dimeric receptors are expressed in the same cell, then the monomeric receptors would have a competitive advantage over dimeric ones for ABA binding and the formation of ternary complexes, especially in situations when ABA concentrations would be limiting. In order to explore this concept, we performed in silico modelling of the receptor activation process. We considered a model consisting of generic monomeric and dimeric receptors that compete for binding to the same pool of ABA and PP2C molecules. The reaction diagram used in this model is shown in Figure 6A. Dimeric receptors exist in a monomer–dimer equilibrium (reaction 2) and can form a ternary complex in two different ways. A high-affinity pathway involving binding of ABA to the monomeric form with an equilibrium constant similar to that of monomeric receptors (reactions 1 and 6) and a low-affinity pathway involving binding of ABA to dimeric receptors and dissociation of the receptor dimer (reactions 3–6). The apparent Kds for the global reactions leading to the formation of receptor–ABA and receptor–ABA–phosphatase complexes have been experimentally determined (highlighted in Figure 6A) and were used to estimate the equilibrium constants for the individual reactions in the model.
Figure 6.
Model of the abscisic acid receptor activation process. (A) The reaction equilibria leading to the formation of receptor–ABA–PP2C ternary complexes for monomeric and dimeric PYR/PYL/RCAR proteins considered in the modelling study are indicated. The apparent Kds for the reactions measured experimentally (formation of receptor–ABA and receptor–ABA–phosphatase complexes) are indicated, and were used to parameterize the model. This model was used to explore the response to ABA when both monomeric and dimeric PYR/PYL/RCAR proteins compete for the same pool of ABA and PP2C molecules. (B) Results corresponding to a balanced model with dimer dissociation constant of 0.05 μM. The amount of total receptor–ABA–PP2C ternary complexes (red) as well as the proportion of ternary complexes formed with monomeric (monR-PP2C, cyan) and dimeric receptors (dimR-PP2C, brown) are indicated. (C) Activation of dimeric and monomeric receptors in balanced and unbalanced models. The concentration of ternary complexes formed by dimeric (dimR-PP2C dashed lines) and monomeric (monR-PP2C full lines) receptors at different ABA concentrations is indicated for situations involving different proportions of monomeric and dimeric receptors (as indicated). The dimer dissociation constant and the total receptor and PP2C concentrations were as in (B). As can be appreciated, these results indicate that overexpression of dimeric receptors can compensate for their thermodynamic disadvantage, and that the relative abundance of monomeric receptors has a strong influence on the concentration of ABA at which activation of dimeric receptors commences.
The equilibrium constant for dimer dissociation (reaction 2) is an important parameter in the model, since it determines the proportion of monomeric and dimeric forms of the receptor. Due to the stability of the PYR1 dimers, it was not possible to determine this value experimentally. A series of models were generated with values of this dissociation constant in a range between 0.3 and 0.05 μM. Values above this range would have been easily detected by the ITC dissociation experiment with wt PYR1, while values much lower than 0.05 μM would result in a very stable receptor dimer, which would prevent formation of the ternary complex. The concentrations of receptor and PP2C molecules were chosen so that the proportion between receptors (both types combined) and PP2C molecules was always 1:1. However, three different situations were considered: monomeric and dimeric receptors in equal amounts (balanced model) and concentrations of dimeric receptors either exceeding or significantly below that of monomeric receptor (unbalanced models). The steady-state concentrations of the different species were determined for a range of protein and ABA concentrations.
Figure 6B shows the contribution of monomeric and dimeric receptors to the formation of ternary complexes at ABA concentrations ranging from 10 nM to 10 μM for PP2C and receptor concentrations of 1 μM and with a value of the dimerization constant of 0.05 μM. As can be appreciated, at 0.6 μM ABA, >50% of the PP2C is forming ternary complexes, which represents the inactive form of the phosphatase. However, two thirds of these ternary complexes are formed by monomeric receptors and only one third by dimeric ones. The competitive advantage of monomeric receptors is stronger at lower ABA concentrations. For example, at 0.2 μM ABA, 80% of the ternary complexes are formed by monomeric receptors while at 1 μM ABA, they represent 60%. When the system reaches saturation, at ABA concentrations between 2 and 10 μM, both receptor types contribute similarly to the inhibition of the phosphatase activity. The behaviour of the system is strongly influenced by the relative abundance of monomeric and dimeric receptors and the competitive disadvantage of dimeric receptors can be compensated if they are more abundant than monomeric ones. For example, when the concentration of dimeric receptors is nine-fold higher than that of monomeric ones, dimeric receptors contribute two thirds of the ternary complexes at 0.2 μM ABA and >85% at 1 μM ABA (Figure 6C). If monomeric receptors are more abundant their competitive advantage is exacerbated.
The competitive advantage of monomeric receptors decreases with increasing values of the dimerization constant and at lower receptor concentrations, both conditions that would promote the dissociation of dimeric receptors. However, an advantage towards the activation of monomeric receptors is always observed for all the balanced models, even when protein concentrations are below the value of the dimer dissociation constant. Finally, the absolute PP2C and receptor concentrations determine the concentration of ABA required to attaining substantial inhibition of the phosphatase activity.
Discussion
Recent structural studies have contributed significantly to the understanding of the process of ABA perception and the activation of the plant ABA signalling pathway (Melcher et al, 2009, 2010a; Miyazono et al, 2009; Nishimura et al, 2009; Santiago et al, 2009a; Yin et al, 2009; Peterson et al, 2010; Dupeux et al, 2011). The data presented here provide novel mechanistic insight into the process of activation of PYR/PYL/RCAR abscisic acid receptors and show how the capacity of the apo form of the receptor to dimerize or not divides this protein family in two groups with distinctive properties. These differential properties may have important consequences for the activation of the ABA response pathway when they are considered in the cellular context.
Our analysis indicates that PYR/PYL/RCAR proteins can be separated into two distinct subclasses: one corresponding to dimeric receptors, to which PYR1, PYL1 and PYL2 belong, and another corresponding to monomeric receptors, including at least PYL5, PYL6 and PYL8. Dimeric receptors display lower apparent affinities for ABA and have a strict requirement on ABA for interaction with PP2Cs, while monomeric receptors have a significantly higher intrinsic affinity for ABA and can form low-affinity complexes with PP2Cs in the absence of ABA (Table I; Figure 2; Supplementary Figure S1). These distinctive properties do not result from differences in the ABA-binding pocket. Instead, they are the consequence of the oligomeric state of the apo form of the receptor. In dimeric receptors, the PP2C-interaction surface is occluded in the absence of ABA (Figure 1), and ABA binding is required both to promote the closed conformation of the gating loops and to induce dimer dissociation. This releases the PP2C-interaction region on the surface of the receptor, making it competent for interaction with a phosphatase. The lower ABA affinity characteristic of dimeric receptors is the consequence of the thermodynamic penalty imposed by dimer dissociation on the receptor activation process. For dimeric receptors, the heat released by hormone binding is reabsorbed by the process of dimer dissociation, which is endothermic (Figure 3C), resulting in net positive enthalpies and a less favourable ΔG for the overall receptor activation process, decreasing in turn the apparent ABA binding affinity (Figure 4A). The activation of monomeric receptors does not involve this unfavourable energetic contribution, which explains their higher intrinsic affinity for ABA, but on the other hand, in their ABA-free form, the PP2C-interaction region is not occluded, leading to a certain capacity to establish weak interactions with phosphatases (Figure 2; Supplementary Figure S1). The distinctive properties of monomeric and dimeric receptors are particularly well illustrated by the wt and PYR1H60P proteins. Even though their ABA-binding pockets and PP2C-interaction regions are virtually identical (Nishimura et al, 2009; Santiago et al, 2009a; Melcher et al, 2010a; Peterson et al, 2010; Dupeux et al, 2011; and this work), the different oligomeric state of their apo forms entails different ABA sensitivity, thermodynamic properties and ability to activate OST1.
The analysis of the ABA binding regions in the PYR/PYL/RCAR family indicates that, for dimeric receptors like PYR1, PYL1 and PYL2, the residues involved in dimerization are surrounded or adjacent to residues involved in ABA binding (Melcher et al, 2009, 2010a; Miyazono et al, 2009; Nishimura et al, 2009; Santiago et al, 2009a; Yin et al, 2009; Peterson et al, 2010; Dupeux et al, 2011; see Supplementary Figure S4). This configuration is poised to directly transmit and convert ligand binding into dimer disruption. Indeed, by comparing the structures of apo and ABA-bound PLY2, Yin et al (2009) had already noted a significant weakening of the dimer interface upon ABA binding. However, its biological significance was not interpreted. The weakening of the dimerization interface is mostly due to the closing of the gating loops induced by ABA binding, which in the apo form are open and establish both polar and hydrophobic contacts with the neighbouring subunit, and to a slight change in the relative orientation of the two protomers, all of which results in a reduction in the number of Van der Waals contacts and hydrogen bonds between the two subunits and a reduction of the dimer interaction surface (Yin et al, 2009). Similar differences can be observed between the ABA bound and free forms of PYL1, PYL2 and PYR1.
The thermodynamic disadvantage for the activation of dimeric receptors will be offset by the formation of the new receptor–phosphatase interface in the ternary complex, with a favourable enthalpic contribution, which explains why both monomeric and dimeric receptors show increased affinities for ABA in the presence of clade A PP2Cs (Ma et al, 2009; Melcher et al, 2009; Miyazono et al, 2009; Santiago et al, 2009b; Yin et al, 2009). However, our results indicate that if both types of receptors are equally expressed in a single cell, then the monomeric receptors would contribute more significantly than dimeric ones to the inhibition of the PP2Cs at low and mid-range ABA concentrations. For example, during the early stages of the stress response, or when intracellular ABA levels are low, monomeric receptors are likely to play a major role in the control of the activity of clade A PP2Cs, while dimeric receptors might only be fully activated under more severe stress conditions or at a later stage in the response. This may explain the long time observed ability of ABA to elicit responses over a wide range of concentrations (Trewavas, 1991). For instance, at nanomolar concentrations, ABA triggers changes in stomatal aperture, at submicromolar concentrations it delays or inhibits seed germination and yet higher ABA concentrations are required to inhibit root growth (Rubio et al, 2009). Additionally, in silico modelling suggests that the relative abundance of monomeric and dimeric receptors and PP2Cs is likely to play a critical role in tuning the response of the system. For example, changes in the expression profile of PYR/PYL/RCAR proteins might influence the ABA response, offering a potential mechanism through which the plant could differentially modulate the response to stress in specific organs or tissues, or under particular physiological conditions. Intriguingly, it is apparent from many microarray experiments that both the mRNAs for receptors and clade A PP2Cs are regulated transcriptionally by ABA (Huang et al, 2007). This highlights the importance of efforts to obtaining quantitative proteomic data on components of the ABA pathway in well-defined cell types (Kline et al, 2010).
The PYR/PYL/RCAR family contains 14 members in Arabidopsis (Ma et al, 2009; Park et al, 2009; Nishimura et al, 2010), only a number of which have been characterized so far. The model of activation emerging from this work can be used to predict the properties of yet uncharacterized receptors. For instance, the affinity for ABA of the receptors, in the absence of a PP2C, could be used to predict their oligomeric state. On the other hand, the structural analysis of the PYRH60P mutant indicates that a proline at position 60 is unfavourable for homodimerization, since it hinders conformational changes at the peptide backbone required to accommodate Phe61 at the dimer interface. Phe61 is strictly conserved in the PYR/PYL/RCAR family (see Supplementary Figure S4), and similar conformational changes are found at equivalent positions in the PYL1 and PYL2 dimers. This suggest that proteins like PYL7, PYL9 and PYL10, which contain a proline at the position equivalent to that of His60 in PYR1, are likely to be monomeric and show higher ABA sensitivity. This is indeed the case for PYL8 as we show here. Other proteins, like PYL5 and PYL6 seem to acquire a monomeric structure through a different mechanism involving other amino-acid variations at the dimerization interface. Further structural characterization of members of the PYRL/PYL/RCAR family should contribute to understand these mechanisms.
Finally, our results indicate that monomeric ABA receptors are able to interact with the catalytic core of the HAB1 phosphatase in the absence of ABA in vitro. This suggests that, although the hormone plays a critical role in the stabilization of the ternary complex, a less stable complex can be formed between the phosphatase and monomeric receptors. Constitutive interactions between monomeric PYR/PYL/RCAR receptors and PP2Cs have been observed in yeast two hybrid assays (Ma et al, 2009; Park et al, 2009; Santiago et al, 2009b) and also in plants (Nishimura et al, 2010). However, under such experimental conditions it is difficult to exclude completely the presence of small amounts of ABA. The protein concentrations used in our SEC-MALLS assays are probably higher than those found under physiological conditions, but they allow to completely rule out the presence of ABA, lending weight to previous observations of constitutive interactions (Ma et al, 2009; Park et al, 2009; Santiago et al, 2009b; Nishimura et al, 2010). These interactions might contribute a certain basal activation of the components of the ABA pathway in the plant. Moreover, monomeric receptors would efficiently compete with dimeric receptors for low endogenous ABA levels that might be necessary for the regulation of certain developmental processes not related to stress. Indeed, both ABA-deficient and ABA-insensitive mutants are severely impaired in growth and reproduction even under high humidity conditions (Cheng et al, 2002; Barrero et al, 2005; Fujii and Zhu, 2009). Alternatively, the activation of the apo form of some monomeric receptors might be modulated through interaction with yet unidentified non-receptor proteins, preventing unliganded interactions with PP2Cs.
The results presented here reveal an unexpected level of complexity in the molecular mechanisms governing the activation of the PYR/PYL/RCAR receptors and provide a novel framework for the understanding of the ABA signalling pathway and the activation of the stress response in plants. More generally, this work illustrates how receptor oligomerization can modulate ligand binding affinity and the level of activation of a signalling pathway by influencing the thermodynamics of the overall receptor activation reaction. This implies that a high degree of amino-acid sequence conservation at a ligand binding site does not necessarily lead to similar binding properties.
Materials and methods
Construction of plasmids, expression and purification of 6 × HIS fusion proteins
Plasmids pETM11 or pET28a were used to generate N-terminal His6-tagged recombinant proteins. The cloning of 6 × his–ΔNHAB1 (lacking residues 1–178), PYR1 and PYL5 constructs were previously described (Santiago et al, 2009a, 2009b). Full-length PYL1, PYL6 and PYL8 were cloned in pETM11. PYR1Y120A mutant was produced using the Overlap extension procedure (Ho et al, 1989) and cloned into pETM11. The coding sequence of OST1 and a C-terminal deletion of ABF2 (ΔCABF2, amino acids 1–173) were cloned into pET28a.
The H60P mutation was obtained in a screen designed to identify altered PYR1–PP2C interactions, the coding sequence for PYR1 was mutated by error-prone PCR and the mutagenized library was transformed into S. cerevisiae strain Y190 containing pAD-HAB1 and 1 × 106 colonies were screened. Y190 contains a galactose-inducible HIS3 nutritional reporter that allows positive selection for PYR1–HAB1 interactions in the absence of exogenously added ABA by growth on media containing 3-amino triazole. Once identified, the H60P mutation was introduced into PYR1 using the QuickChange site directed mutagenesis protocol.
Protein expression and purification
BL21(DE3) cells transformed with the corresponding constructs in pETM11 or pET28a vectors were grown in LB medium to an OD600 of 0.6–0.8. At this point, 1 mM IPTG was added and the cells were harvested after overnight incubation at 20°C. Proteins were purified as described by Santiago et al (2009b). Briefly, cell pellets were resuspended in lysis buffer (30 mM Tris pH 7.5, 500 mM NaCl, 15 mM Imidazole, 1 mM MnCl2, 1 mM β-mercaptoethanol and Protease cocktail inhibitor), lysed with a microfluidizer (Microfuildics) and purified by Ni-affinity. For proteins produced from pETM11 vectors, the 6 × his tag was excised by digestion with the TEV protease. In the case of PYR1, PYL1, PYL6 and PYL8, an additional step of gel filtration using an S200 column (GE) was carried out.
Crystallization and structure solution
The PYR1H60P–ABA–ΔNHAB1 ternary complex was prepared by mixing PYR1H60P, ΔNHAB1 and ABA to final concentrations of 3, 5 mg/ml and 1 mM, respectively, in 20 mM Tris pH 7.5, 150 mM NaCl, 1 mM MnCl2 and 1 mM β-mercaptoethanol. Crystallization conditions for the complex were identified at the High Throughput crystallization Laboratory of EMBL Grenoble Outstation (https://embl.fr/htxlab) as described in Marquez et al (2007). The crystals used for data collection were obtained by the vapour diffusion method in 0.25 M NaCl, 19% Peg 3350 at 20°C. X-ray diffraction data were collected at the ID23-1 beamline of the ESRF to 2.1 Å resolution. Initial phases were obtained by the molecular replacement with the program Phaser (McCoy et al, 2007) using the wt PYR1–ΔNHAB1 complex as search model (3QN1) (Dupeux et al, 2011). Successive rounds of automatic refinement and manual building were carried out with RefMac5 (Murshudov et al, 1997) and Coot (Emsley and Cowtan, 2004).
Size-Exclusion Chromatography-Multiple Angle Laser Light Scattering
SEC was performed using an S200 Superdex column (GE Healthcare) equilibrated with 20 mM Tris pH 7.5, 150 mM NaCl and 1 mM β-mercaptoethanol. For the experiments with ABA, either 1 or 5 mM ABA was included in the equilibration buffer. Receptor proteins were injected at a concentration of 80 μM. For the analysis of receptor–PP2C complexes receptor molecules were mixed prior to injection with equimolecular amounts of ΔNHAB1 (at a final concentration of 80 μM each protein) in the presence or absence of 1 mM (+)ABA. All separations were performed at 20°C with a flow rate of 0.5 ml/min. On-line MALLS detection was performed with a DAWN-EOS detector (Wyatt Technology Corp., Santa Barbara, CA) using a laser emitting at 690 nm. Data were analysed and weight-averaged molar masses (Mw) were calculated using the ASTRA software (Wyatt Technology Corp.) as described previously (Gerard et al, 2007).
Determination of ABA-binding affinity and dimer dissociation constants
ITC experiments were performed using a VP-ITC calorimeter equipped with the control, data acquisition and analysis software ORIGIN 7. All solutions were centrifuged, degassed to avoid formation of bubbles and equilibrated to the corresponding temperature prior to each experiment. Experiments were carried out at pH 7.5 and 25°C. In the binding experiments, protein solution in the calorimetric cell was titrated with the (+)ABA ligand dissolved in the dialysis buffer (20 mM Tris, 150 mM NaCl, 1 mM and 1 mM β-mercaptoethanol). PYL6 and His6–PYR1(H60P) were assayed at a protein concentration of 20 μM, and the concentration of (+)ABA stock solution in the injection syringe was 0.8 mM. PYR1 was assayed at protein concentrations from 50 to 200 μM, and the concentration of the (+)ABA stock solutions varied from 0.8 to 2 mM. The titrations were carried out within a series of 30 injections of 7 μl each one. The heat evolved after each ligand injection was obtained from the integral of the calorimetric signal. The heat due to the binding reaction between protein and the ligand was obtained as the difference between the heat of reaction and the corresponding heat of dilution obtained from independent titration experiments of (+)ABA into buffer. The resulting binding isotherms were analysed by non-linear least squares fitting of the experimental data to an one-set of sites model with the Microcal Origin (OriginLab Corp., Northapton, MA) software. Data shown are the average of three independent experiments±s.d.
PYR1 dissociation experiments were performed with sequences of 30 injections of 6 μl each. The cell contained 20 mM Hepes pH 7.5, 150 mM NaCl and 1 mM β-mercaptoethanol. The concentration of PYR1 or PYR1H60P stock solution in the injection syringe was 0.7 mM. The injections were performed over a period of 6 s, with 5 min intervals between injections, reaching a final protein concentration of 75 μM in the cell. Data were analysed and fitted using the dissociation fitting model provided by Microcal Origin software, implemented with corrected DeltaH.ogs and Dissociation.fdf files kindly provided by Professor Alan Cooper (Chemistry Department, University of Glasgow).
For wt PYR1, an independent determination of the ABA-binding affinity was carried out by NMR. Experiments were done at 37°C on a sample of 15N-labelled PYR1 containing 150 mM NaCl and 20 mM Tris pH 7.5. 1H-15N HSQC spectra were acquired at a 1H resonance frequency of 600 or 800 MHz. An estimation of the dissociation constant, Kd, of the PYR1:ABA complex was obtained by titration of ABA into an 15N-labelled sample of PYR1. Measurements were done at 10 different ABA/PYR1 concentration ratios ranging from 0 to 2.59. For each step of the titration, a 1H-15N HSQC was recorded. Measurements of the dimer dissociation constants were carried out by ITC through serial injections of protein solutions of either wt or His6PYR1(H60P) into the cell containing the same buffer. Binding isotherms were analysed by non-linear least squares fitting with the ORIGIN 7 software (OriginLab Corp.). The dissociation data were analysed with the dissociation model provided by Dr Alan Cooper's (http://www.chem.gla.ac.uk/staff/alanc/service1.htm). Data shown are the average of three independent experiments±s.d.
OST1 in vitro activity assays
OST1 phosphorylation assays were done as described previously (Belin et al, 2006; Vlad et al, 2009). Inactivation of OST1 was achieved by incubation with the protein phosphatase ΔNHAB1. Assays to test recovery of OST1 activity were done by previous incubation during 10 min of ΔNHAB1 with either wt PYR1 or PYR1H60P proteins in 30 μl of kinase buffer (20 mM Tris–HCl pH 7.8, 20 mM MgCl2, 2 mM MnCl2) in the presence of the indicated concentration of (+)ABA. Then, ΔCABF2 recombinant protein (100 ng) and 3.5 μCi of γ-32ATP (3000 Ci/mmol) were added to the mix and the reaction was incubated for 50 min. The reaction was stopped by adding Laemmli buffer. After the reaction, proteins were separated by SDS–PAGE using an 8% acrylamide gel and transferred onto an Immobilon-P membrane (Millipore). Radioactivity was detected using a Phosphoimage system (FLA5100, Fujifilm). Quantification of activity based on relative intensity of phosphorylated bands was performed using a phosphoimager Image Gauge V.4.0. The data presented are averages of at least three independent experiments.
In silico modelling of ABA pathway activation
Computational modelling was performed on the Infobiotics Workbench executable biology platform (http://www.infobiotics.org/) using a stochastic P system framework (Twycross et al, 2010). A multi-compartment stochastic simulation algorithm (Romero-Campero et al, 2009) was used for simulations. Two separate models were initially considered, corresponding to monomeric and dimeric receptors. The model for dimeric receptors included all reactions in Figure 6A while that of monomeric receptors included only reactions 1 and 6. Those reaction rates that were not experimentally available were estimated using the CMA-ES optimization algorithm (Hansen and Ostermeier, 2001). The values of the estimated parameters were progressively adjusted through this algorithm so that the individual models reproduce the apparent ABA binding constants of the experimentally measured reactions. These reactions include the formation of ternary receptor–ABA–PP2C complexes (a Kd value of 30 nM was chosen as representative of generic monomeric and dimeric receptors), binding of ABA to a monomeric receptor or to the monomeric form of a dimeric receptor (Kd of 1.0 μM) and binding of ABA to dimeric receptors (with Kds of 52, 59 and 97 μM for PYL1, PYL2 and PYR1, respectively, a value of 60 μM was chosen). Once the individual models were parameterized, a combined model was generated simulating the response of monomeric and dimeric receptors sharing a common pool of ABA and PP2C molecules. This combined model was studied over a range of ABA (1 nM–40 μM) and PP2C (0.1 nM–10 μM) concentrations for situations corresponding to balanced or unbalanced receptor concentrations (see main text). All models were encoded in SBML version 4 level 2 (http://sbml.org/). SBML files will be made publicly available on the Biomodels database (http://www.ebi.ac.uk/biomodels/).
Accession codes
Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank under accession code 3ZVU.
Supplementary Material
Acknowledgments
We are grateful to the European Synchrotron Radiation Facility (ESRF) and the EMBL for access to macromolecular crystallography beam lines. This work was supported by Ministerio de Educación y Ciencia, Fondo Europeo de Desarrollo Regional, Consejo Superior de Investigaciones Científicas (grant BIO2008-00221 to PLR; fellowships to JS and LR; Juan de la Cierva contract to MGG) and by the BBSRC/EPSRC grant BB/D019613/1 to CPIB (MH, JT and NK). Access to the High Throughput Crystallization facility of the Partnership for Structural Biology in Grenoble (PSB) (https://embl.fr/htxlab) was supported by the P-CUBE project funded by the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement no 227764.
Author contributions: FD and JS contributed with the cloning, protein purification, ITC and MALLS analysis. KB contributed crystallization and structure solution. JT, NK and MH performed in silico modelling of ABA pathway activation. LR and MGG produced PYL proteins, generated the PYR1Y120 mutant and carried out phosphorylation experiments. SYP and SC obtained the PYR1H60P mutant. MRJ and MB carried out the NMR experiments. SC and PLR contributed to experimental design, discussion and writing of MS. JAM coordinated the work and discussions and co-wrote the MS.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barrero JM, Piqueras P, Gonzalez-Guzman M, Serrano R, Rodriguez PL, Ponce MR, Micol JL (2005) A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. J Exp Bot 56: 2071–2083 [DOI] [PubMed] [Google Scholar]
- Belin C, de Franco PO, Bourbousse C, Chaignepain S, Schmitter JM, Vavasseur A, Giraudat J, Barbier-Brygoo H, Thomine S (2006) Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol 141: 1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Wilhelm G, Russell LJ (eds). (2000) Biochemistry & Molecular Biology of Plants. Somerset, NJ: John Wiley & Sons Ltd [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M, Koshiba T, Sheen J (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14: 2723–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, Santiago J, Gonzalez-Guzman M, Rodriguez L, Rubio S, Park SY, Cutler S, Rodriguez PL, Marquez J (2011) Modulation of ABA signaling in vivo by an engineered receptor-insensitive PP2C allele. Plant Physiol 156: 106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol 50: 2123–2132 [DOI] [PubMed] [Google Scholar]
- Geiger D, Maierhofer T, Al-Rasheid KA, Scherzer S, Mumm P, Liese A, Ache P, Wellmann C, Marten I, Grill E, Romeis T, Hedrich R (2011) Stomatal closure by fast Abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor RCAR1. Sci Signal 4: ra32. [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, Romeis T, Hedrich R (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard FC, Ribeiro Ede A Jr, Albertini AA, Gutsche I, Zaccai G, Ruigrok RW, Jamin M (2007) Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46: 10328–10338 [DOI] [PubMed] [Google Scholar]
- Hansen N, Ostermeier A (2001) Completely derandomized self-adaptation in evolution strategies. Evol Comput 9: 159–195 [DOI] [PubMed] [Google Scholar]
- Harris MJ, Outlaw WH, Mertens R, Weiler EW (1988) Water-stress-induced changes in the abscisic acid content of guard cells and other cells of Vicia faba L. leaves as determined by enzyme-amplified immunoassay. Proc Natl Acad Sci USA 85: 2584–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Huang D, Jaradat MR, Wu W, Ambrose SJ, Ross AR, Abrams SR, Cutler AJ (2007) Structural analogs of ABA reveal novel features of ABA perception and signaling in Arabidopsis. Plant J 50: 414–428 [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lee SC, Lan W, Buchanan BB, Luan S (2009) A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc Natl Acad Sci USA 106: 21419–21424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448–1452 [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068 [DOI] [PubMed] [Google Scholar]
- Marquez JA, Galfre E, Dupeux F, Flot D, Moran O, Dimasi N (2007) The crystal structure of the extracellular domain of the inhibitor receptor expressed on myeloid cells IREM-1. J Mol Biol 367: 310–318 [DOI] [PubMed] [Google Scholar]
- McCourt P, Creelman R (2008) The ABA receptors—we report you decide. Curr Opin Plant Biol 11: 474–478 [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, Kovach A, Li J, Wang Y, Peterson FC, Jensen DR, Yong EL, Volkman BF, Cutler SR, Zhu JK, Xu HE (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Xu Y, Ng LM, Zhou XE, Soon FF, Chinnusamy V, Suino-Powell KM, Kovach A, Tham FS, Cutler SR, Li J, Yong EL, Zhu JK, Xu HE (2010a) Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol 17: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Zhou XE, Xu HE (2010b) Thirsty plants and beyond: structural mechanisms of abscisic acid perception and signaling. Curr Opin Struct Biol 20: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452–1455 [DOI] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang HJ, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, Yoshida T, Kodaira KS, Yamaguchi-Shinozaki K, Tanokura M (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Sarkeshik A, Nito K, Park SY, Wang A, Carvalho PC, Lee S, Caddell DF, Cutler SR, Chory J, Yates JR, Schroeder JI (2010) PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J 61: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, Alfred SE, Bonetta D, Finkelstein R, Provart NJ, Desveaux D, Rodriguez PL, McCourt P, Zhu JK, Schroeder JI, Volkman BF et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson FC, Burgie ES, Park SY, Jensen DR, Weiner JJ, Bingman CA, Chang CE, Cutler SR, Phillips GN Jr, Volkman BF (2010) Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol 17: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Campero FJ, Twycross J, Camara M, Bennett M, Gheorghe M, Krasnogor N (2009) Modular assembly of cell systems biology models using P systems. Int J Found Comput Sci 20: 427–442 [Google Scholar]
- Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL (2009) Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol 150: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL (2004) Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J 37: 354–369 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Marquez JA (2009a) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL (2009b) Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J 60: 575–588 [DOI] [PubMed] [Google Scholar]
- Szostkiewicz I, Richter K, Kepka M, Demmel S, Ma Y, Korte A, Assaad FF, Christmann A, Grill E (2010) Closely related receptor complexes differ in their ABA selectivity and sensitivity. Plant J 61: 25–35 [DOI] [PubMed] [Google Scholar]
- Trewavas A (1991) How do plant growth substances work? II. Plant Cell Environ 14: 1–12 [Google Scholar]
- Twycross J, Band LR, Bennett MJ, King JR, Krasnogor N (2010) Stochastic and deterministic multiscale models for systems biology: an auxin-transport case study. BMC Syst Biol 4: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues PE, Zhu JK (2007) New developments in abscisic acid perception and metabolism. Curr Opin Plant Biol 10: 447–452 [DOI] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Lauriere C, Merlot S (2009) Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21: 3170–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt PJ (1998) Submicrometer particle sizing by multiangle light scattering following fractionation. J Colloid Interface Sci 197: 9–20 [DOI] [PubMed] [Google Scholar]
- Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N (2009) Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol 16: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Yoshida R, Hobo T, Ichimura K, Mizoguchi T, Takahashi F, Aronso J, Ecker JR, Shinozaki K (2002) ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol 43: 1473–1483 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.