Abstract
Dicer proteins function in RNA interference (RNAi) pathways by generating small RNAs (sRNAs). Here, we report the solution structure of the C-terminal domain of Schizosaccharomyces pombe Dicer (Dcr1). The structure reveals an unusual double-stranded RNA binding domain (dsRBD) fold embedding a novel zinc-binding motif that is conserved among dicers in yeast. Although the C-terminal domain of Dcr1 still binds nucleic acids, this property is dispensable for proper functioning of Dcr1. In contrast, disruption of zinc coordination renders Dcr1 mainly cytoplasmic and leads to remarkable changes in gene expression and loss of heterochromatin assembly. In summary, our results reveal novel insights into the mechanism of nuclear retention of Dcr1 and raise the possibility that this new class of dsRBDs might generally function in nucleocytoplasmic trafficking and not substrate binding. The C-terminal domain of Dcr1 constitutes a novel regulatory module that might represent a potential target for therapeutic intervention with fungal diseases.
Keywords: dicer, dsRBD, heterochromatin, RNA interference, zinc-binding domain
Introduction
RNA interference (RNAi) designates a set of conserved pathways found in many eukaryotes. These pathways are involved in various processes ranging from the control of gene expression to silencing of mobile genetic elements, combating viruses and maintaining genomic integrity (Grishok et al, 2001; Baulcombe, 2004; Huisinga and Elgin, 2009; van Wolfswinkel and Ketting, 2011). Additionally, RNAi is also being pursued as a promising tool for the treatment of human diseases. Common to all RNAi pathways is the association of small RNAs (sRNAs) with members of the Argonaute family of proteins, forming the core component of a diverse set of protein–RNA complexes called RNA-induced silencing complexes (RISCs; Carmell et al, 2002; Hutvagner and Simard, 2008). To date, at least three classes of RISC-associated sRNAs have been identified (Siomi and Siomi, 2009): short interfering RNAs (siRNAs), microRNAs (miRNAs) and PIWI-interacting RNAs (piRNAs). These sRNAs guide the RISCs through base-pairing interactions to homologous sequences, which usually results in reduced activity of the target sequence (Ghildiyal and Zamore, 2009).
In Schizosaccharomyces pombe (S. pombe), the RNAi pathway functions in association with chromatin to trigger co-transcriptional gene silencing (CTGS) and the formation of heterochromatin. Deletions of any of the three genes encoding the RNAi proteins Dicer, Argonaute and RNA-dependent RNA polymerase (dcr1+, ago1+ and rdp1+, respectively) result in loss of CTGS, chromosome segregation defects, and greatly reduced levels of H3K9 methylation (H3K9me) at centromeric repeats (Volpe et al, 2002, 2003; Woolcock et al, 2011). siRNAs corresponding to heterochromatic regions are loaded into an Argonaute-containing complex referred to as RNA-induced transcriptional silencing (RITS) complex in S. pombe (Reinhart and Bartel, 2002; Verdel et al, 2004). Importantly, the RITS complex is physically linked to heterochromatin and this interaction, as well as siRNA generation, depends on the histone H3K9 methyltransferase Clr4 (Motamedi et al, 2004; Cam et al, 2005; Sugiyama et al, 2005). These and other observations have led to a model in which the association of the RITS complex with chromatin is proposed to involve base pairing between siRNAs and the nascent RNA polymerase II (RNApII) transcripts. Subsequently, RITS would recruit histone-modifying enzymes such as Clr4, leading to the generation and spreading of heterochromatin (Buhler et al, 2006; Moazed, 2009).
The biogenesis of siRNAs is mediated by Dcr1, which processes double-stranded RNA (dsRNA) precursors endonucleolytically. Resembling canonical Dicers in animals, S. pombe Dcr1 contains an N-terminal helicase/ATPase domain, followed by a DUF283 domain, a Platform domain (Macrae et al, 2006) and a PAZ-like domain, two RNase III domains, and a long C-terminal domain that bears a divergent double-stranded RNA binding domain (dsRBD) and a short motif referred to as C33 (Figure 1A). Notably, the C-terminus of Dcr1, comprising the dsRBD and C33, is dispensable for processing dsRNA in vitro but not in vivo, where it functions to control the subcellular localization of Dcr1 (Colmenares et al, 2007; Emmerth et al, 2010). We have shown previously that C33 functions to counteract dsRBD-mediated export of Dcr1 to the cytoplasm, resulting in nuclear accumulation of Dcr1. This is crucial for a functional RNAi pathway and establishment of heterochromatin in S. pombe, as mutations rendering Dcr1 cytoplasmic abolish siRNA generation and the methylation of H3K9 (Emmerth et al, 2010).
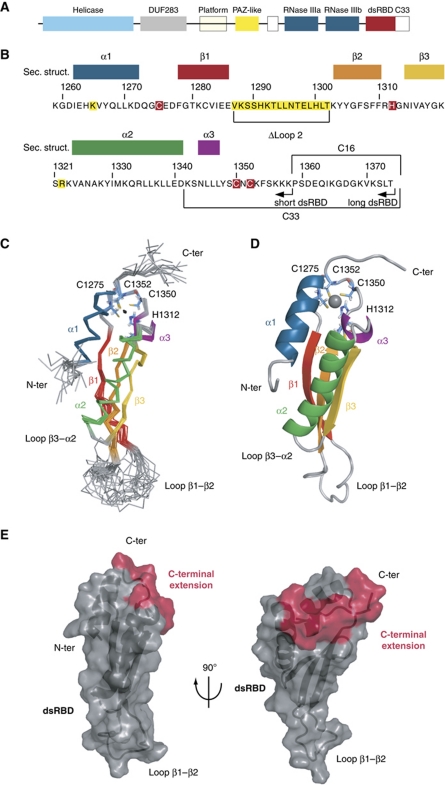
Figure 1.
NMR solution structure of the Dcr1 C-terminus. (A) Domain architecture of S. pombe Dcr1 (not drawn to scale). (B) Sequence of the C-terminus (dsRBD+C33) with the corresponding secondary structures indicated (colour code is the same as in C, D). The CHCC zinc coordination motif is highlighted in red. Arrows indicate the C-terminal ends of the different constructs used for structure determination. The residues highlighted in yellow are involved in dsRNA binding. Amino-acid numbers refer to the S. pombe Dcr1 protein (UniprotKB Q09884). (C) NMR ensemble. Overlay of the 20 final structures with colour-coded secondary structure elements: α1 in blue, β-strands 1–3 in red-orange-yellow, α2 in green and α3 in purple. The four zinc ligands (C1275, H1312, C1350 and C1352) are represented as sticks in light blue. The zinc ion is represented as a black dot. (D) Cartoon representation of the lowest energy structure. The same colour code is used for secondary structure elements. The zinc ion is represented as a grey sphere. (E) Visualization of the extended dsRBD fold on the protein surface. The canonical elements of the dsRBD are represented in grey and the C-terminal extension in red. Left panel: Same orientation of the domain as in (C) and (D). Right panel: Side view.
That dsRBDs can mediate nucleocytoplasmic trafficking has been demonstrated for other proteins (Strehblow et al, 2002; Chen et al, 2004; Gwizdek et al, 2004; Macchi et al, 2004). However, mechanistically this process is poorly understood. For instance, it is not clear to what extent binding to dsRNA contributes to this rather unexpected function of dsRBDs. Similarly, how C33 of Dcr1 contributes to the inhibition of the export-promoting activity of the dsRBD remains unknown. To address these questions, we determined the NMR solution structure of the C-terminal domain of Dcr1. Although the structure reveals a typical dsRBD fold, additional structural elements are present. This includes a novel zinc-binding motif, formed by four residues that are encoded in both the dsRBD and C33. We demonstrate that zinc coordination by this motif is required for the formation of a protein–protein interaction surface that is required for proper Dcr1 localization. Surprisingly, although the dsRBD binds to dsRNA strongly, this property is dispensable for proper functioning of Dcr1 in the S. pombe RNAi pathway. This raises the possibility that dicer dsRBDs might generally function in nucleocytoplasmic transport and not necessarily in substrate binding.
Results
The dsRBD of Dcr1 embeds an unexpected zinc-binding motif
To obtain novel insights into the role of C33 in promoting nuclear accumulation of Dcr1, we studied the Dcr1 C-terminal domain with NMR spectroscopy (1259–1374, long-dsRBD construct; Figure 1B). We assigned backbone and side-chain NMR chemical shifts using standard triple-resonance experiments as described in Materials and methods and could achieve 80.1% of proton assignment. NOE distance restraints were obtained from four NOESY spectra and used to run initial structure calculations. As expected, the preliminary folds revealed a typical dsRBD fold for residues 1262–1341. More surprisingly, the first half of C33 appeared to contact the dsRBD. Strikingly, we observed that four side chains with two residues from the dsRBD and two from C33 (in total three cysteines and one histidine; highlighted in red in Figure 1B) appear in close proximity in this preliminary structure, strongly suggesting that these four side chains might be coordinating a zinc ion. Subsequent atomic absorption spectroscopy (AAS) and mass spectrometry measurements confirmed that the Dcr1 C-terminal domain containing the dsRBD and the C33 indeed include one zinc ion per molecule (Supplementary Figure S1; Table I). 15N{1H}-NOE measurement (Supplementary Figure S2) further confirmed that the first half of C33 fragment is structured and participates in the overall fold of the domain, while the last 16 residues (1359–1374) are flexible. To reduce signal overlap and therefore improve the precision of the structure, we decided to investigate a construct lacking the last 16 residues of Dcr1 (1259–1358, short-dsRBD construct; Figure 1B). Also, ZnCl2 was added during the protein preparation, which helped to stabilize the recombinant short-dsRBD construct and improved the overall quality of the NMR spectra.
Table 1. Zinc binding determination by ESI-MS and atomic absorption spectroscopya.
| Observed apoprotein mass (Da) | Observed holoprotein mass (Da) | Expected apoprotein mass (Da) | Expected holoprotein mass with 1 Zn2+ (Da) | Atomic absorption spectroscopy (zinc/protein ratio) |
|---|---|---|---|---|
| 15 499.0±0.5 | 15 562.0±0.5 | 15 498.8 | 15 562.2 | 1.2±0.1 |
| aThe long-dsRBD protein construct was analysed for bound metal by ESI-MS in 10 mM ammonium acetate at pH 7.0 to observe the holo form of the protein and under denaturing conditions (0.1% formic acid and 50% acetonitrile) to observe the apo form of the protein. The expected neutral mass of the holoprotein is calculated according to holo=apo+Zn2+–2H+, where MZn=65.4 Da (Lei et al, 1998). For AAS, the protein concentration was determined by UV absorbance at 280 nm taking a theoretical extinction coefficient ε=8940M−1 cm−1. | ||||
Final protein structure determination
The NMR solution structure of the Dcr1 C-terminal domain (1259–1358, short-dsRBD construct) was determined using 2308 NOE-derived constraints. The structure is very precise with a backbone r.m.s.d. of 0.46±0.12 Å for the NMR ensemble of conformers. Hydrogen-bonded amides were established as slowly exchanging protons in the presence of D2O. Their hydrogen-bond acceptors were identified from preliminary structures as well as from analysis of the characteristic NOE pattern found in α-helices and β-sheets. Characteristic NOE contacts between the ligands of the zinc ion further confirmed the composition of the coordination sphere that involves C1275 and H1312 from the dsRBD, and C1350 and C1352 from C33 (Supplementary Figure S3). In addition, long-range 1H-15N HSQC shows a characteristic pattern of cross-peaks that allowed us to unambiguously identify H1312 as coordinating the zinc ion via its Nε (Supplementary Figure S4; Legge et al, 2004; Pelton et al, 1993). Therefore, we added distance constraints to the four zinc-binding residues to impose a tetrahedral coordination around the zinc ion. NMR experimental constraints and refinement statistics are presented in Table II.
Table 2. NMR experimental restraints and structural statistics.
| Distance restraints | |
| Total NOE | 2308 |
| Intraresidue | 520 |
| Sequential | 600 |
| Medium range (∣i–j∣<5 residues) | 521 |
| Long range (∣i–j∣⩾5 residues) | 667 |
| Hydrogen bond | 41 |
| Zinc coordination restraints | |
| Distance restraints | 4 |
| Bond angles | 10 |
| Dihedral angles | 1 |
| Structure statistics | |
| NOE violations (mean±s.d.) | |
| Number of NOE violations >0.2 Å | 4.7±1.8 |
| Maximum NOE violation (Å) | 0.30±0.05 |
| R.m.s.d. from average structurea (Å) | |
| Backbone | 0.46±0.12 Å |
| Heavy atoms | 0.95±0.14 Å |
| Mean deviation from ideal covalent geometry | |
| Bond length (Å) | 0.007 |
| Bond angles (deg) | 0.9 |
| Ramachandran analysis | |
| Most favoured region | 79.6% |
| Allowed region | 20.2% |
| Disallowed region | 0.2% |
| aProtein r.m.s.d. was calculated using residues 1262–1286, 1303–1353 for the ensemble of 20 refined structures. | |
The Dcr1 C-terminal domain adopts an extended dsRBD fold due to a novel zinc-binding motif
The residues ranging from I1262 to D1341 form the dsRBD core domain with the canonical αβββα topology (Bycroft et al, 1995; Kharrat et al, 1995), with the two α-helices packed against the 3-stranded antiparallel β-sheet (Figure 1C and D). The residues of C33 that immediately follow helix α2 are well structured (Figure 1C and D; Supplementary Figure S2) and fold back on the dsRBD. N1344 to L1347 fold in a short α-helical turn (helix 3) that is followed by C1350 and C1352 coordinating zinc with C1275 and H1312 from loop 1 and loop 3 of the dsRBD, respectively (Figure 1C and D). This zinc-binding site is unusual because of the long spacing between the first and the last residue coordinating the zinc atoms (76 residues) as well as the spacing between the individual coordinating side chains (C-X36-H-X37-C-X-C). Typically, zinc-binding motifs are found within 20–40 residues (Auld, 2001) with the most abundant form being the C2H2 zinc fingers that adopt a ββα fold (Laity et al, 2001). To our knowledge, the present ligand arrangement in the Dcr1 C-terminal domain is unprecedented among zinc-binding domains. Rather than being an independent fold of the dsRBD, this novel zinc-binding domain is formed jointly by dsRBD and C33, and therefore constitutes an extended dsRBD fold (Figure 1E).
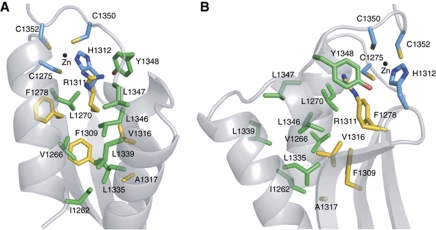
Binding of zinc to a protein is unlikely to sustain the large entropy changes that would result from the ordering of a very mobile peptide chain (Auld, 2001). Thus, in addition to the CHCC zinc-binding motif, other structural elements are required to maintain proper folding of the C-terminus. Indeed, the short segment joining helix 2 to the C-X-C part of the zinc-binding motif contains three hydrophobic residues (L1346, L1347 and Y1348) that form hydrophobic interactions with residues from the dsRBD (helix 1, helix 2 and the β-sheet; Figure 2). The two leucines interact with other leucine side chains, constituting most of the hydrophobic core of the protein. Importantly, five leucine residues distributed along the three helices together with other hydrophobic residues of helix 1 (I1262 and V1266) bring the three α-helices, as well as the hydrophobic surface of the β-sheet, which is mainly composed of aromatic residues, in close contact (Figure 2). Most surprisingly, the side chain of R1311 is also buried in the hydrophobic core of the protein, in close proximity to the zinc coordination site (Figure 2). The vicinity of this positively charged arginine to the CHCC motif would counterbalance the overall −1 net charge of a zinc ion coordinated by three deprotonated cysteines.
Figure 2.
Close-up view of the hydrophobic core of the domain. (A) Front view. (B) Side view. Side chains belonging to the three α-helices and to the β-sheet are shown as green and yellow sticks, respectively. The four zinc ligands are coloured in light blue. The zinc ion is represented as a black dot.
Zinc coordination is required for nuclear localization of Dcr1
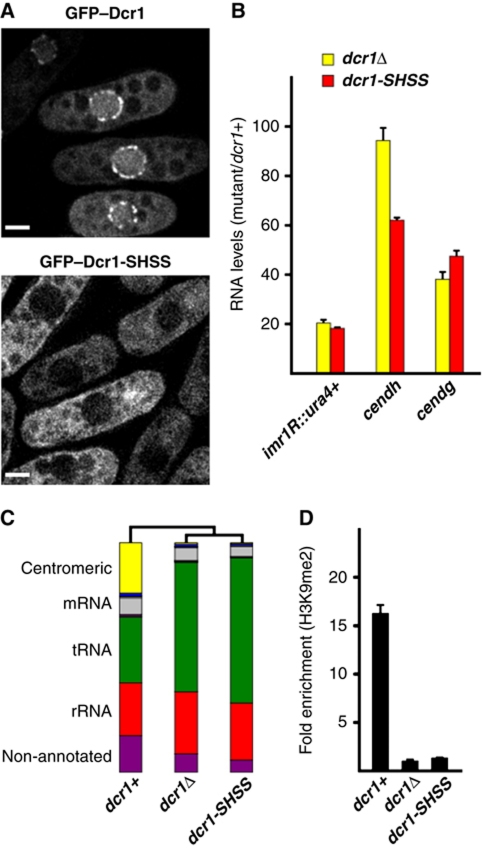
We have previously demonstrated that C33 functions to counteract dsRBD-mediated export of Dcr1 to the cytoplasm, resulting in nuclear accumulation of Dcr1 (Emmerth et al, 2010). This could be achieved by binding of C33 to a putative nuclear retention factor. However, our structural analysis strongly suggests that C33 rather contributes to the overall folding of the C-terminus of Dcr1 by providing two cysteines to form the newly identified zinc-binding domain. To test whether disruption of zinc coordination would result in a similar phenotype as deleting C33, we mutated the three cysteines in the CHCC motif of endogenous Dcr1 into serines (Dcr1-CHCC to Dcr1-SHSS). This resulted in mainly cytoplasmic localization of the protein without changing the amount of protein, similar to what we observed previously when C33 was deleted entirely (Figure 3A; Supplementary Figure S5A). Therefore, it appears that it is the extended dsRBD structure formed upon zinc coordination, rather than the amino-acid sequence of C33 per se, that is responsible for preventing nuclear export of Dcr1. This is further supported by our observation that deletion of the last 16 amino acids of Dcr1 did not lead to a change in localization of the protein (Supplementary Figure S5B).
Figure 3.
Structuring of the C-terminus of Dcr1 via coordination of zinc is essential for RNAi-mediated heterochromatin formation. (A) Live-cell imaging of wild-type GFP–Dcr1 and the zinc motif mutant GFP–Dcr1-SHSS. Scale bars=2 μm. (B) Quantitative real-time RT–PCR showing that dcr1-SHSS cannot silence centromeric repeats. cendg, cendh and imr1R∷ura4+ RNA levels were normalized to act1+ RNA and are shown in relation to dcr1+ cells. Error bars represent standard deviations (s.d.). (C) Total small RNA profiles of wild-type, dcr1Δ and dcr1-SHSS cells as determined by Illumina Sequencing. Major small RNA classes relative to the total amount of small RNAs sequenced are indicated. See Supplementary Table I for read numbers. (D) ChIP experiment showing that mutations in the zinc-coordinating motif of Dcr1 (Dcr1-SHSS) abolish H3K9 methylation at a centromeric ura4+ transgene (imr1R∷ura4+). Fold-enrichment values from one representative experiment, normalized to act1+, are shown. The value for dcr1Δ cells was set to 1. Error bars represent standard deviations (s.d.).
To test whether disrupting the coordination of zinc has the same functional consequences as deleting C33, we investigated heterochromatic gene silencing and the generation of centromeric siRNAs by quantitative RT–PCR and sRNA deep sequencing, respectively. Consistent with a failure to retain Dcr1 in the nucleus, silencing of centromeric heterochromatin and the generation of centromeric siRNAs were lost in cells expressing Dcr1-SHSS (Figure 3B and C; Supplementary Figure 6; Supplementary Table I). Concomitant with the observed loss of silencing, H3K9 methylation was lost in Dcr1-SHSS expressing cells (Figure 3D). In conclusion, coordination of zinc by the C-terminus of Dcr1 is required for its nuclear retention and thus RNAi-mediated heterochromatin assembly.
The Dcr1 dsRBD binds dsRNA and dsDNA
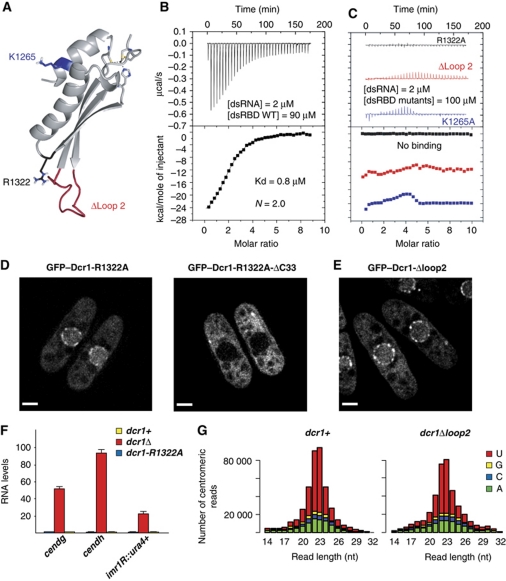
It has been demonstrated for other dsRBDs that they can mediate protein import and/or export (Strehblow et al, 2002; Chen et al, 2004; Gwizdek et al, 2004; Macchi et al, 2004), and it has been discussed that RNA may regulate this process (Fritz et al, 2009). In canonical dsRBDs, both helix 1 and the loop between β1 and β2 contact the minor groove of a dsRNA helix (Supplementary Figure S7A; Ryter and Schultz, 1998; Ramos et al, 2000; Wu et al, 2004; Stefl et al, 2010). Interestingly, the dsRBD of Dcr1 has an unusually long loop between β1 and β2 (14 residues compared with 4–6 residues in most dsRBDs), raising the question as to whether the C-terminal domain of Dcr1 would bind dsRNA at all. We, thus, investigated the nucleic acid binding properties of the extended dsRBD by isothermal titration calorimetry (ITC). Similar to canonical dsRBDs (St Johnston et al, 1992; Bass et al, 1994), the isolated domain (Figure 4A) binds rather strongly to a regular A-form RNA helix (Kd=0.8 μM, with two dsRBDs bound per 24 bp dsRNA helix) (Figure 4B), but binds only weakly to ssRNA or ssDNA (Supplementary Figure S7B). The absence of binding to a regular B-form DNA helix has been reported for various dsRBDs (Burd and Dreyfuss, 1994; Fierro-Monti and Mathews, 2000; Saunders and Barber, 2003), although competition binding studies revealed that some dsRBDs can also bind to dsDNA, albeit with weaker affinity (Bass et al, 1994). Surprisingly, the extended dsRBD of Dcr1 showed a fairly strong affinity to a regular B-form DNA helix (Kd=1.5 μM), although only one molecule is bound per dsDNA (Supplementary Figure S7B). This raises the question of how this dsRBD can adapt to bind both a B-form DNA helix and an A-form RNA helix. We speculate that the unusually long β1–β2 loop and the unusual position of helix 1 relative to the rest of the domain (see Discussion) might be responsible for this dual binding capacity.
Figure 4.
Key residues in the dsRBD that are required for dsRNA binding are dispensable for proper functioning of Dcr1 in the RNAi pathway. (A) Location of residues important for the binding of dsRNA. (B) Affinity of the Dcr1 dsRBD for dsRNA as determined by ITC. (C) ITC measurements with dsRNA and Dcr1 dsRBD mutants (R1322A, Δloop 2 and K1265A). Data for Δloop 2 and K1265A are shifted on the y axis for clarity. (D) Live-cell imaging of GFP–Dcr1-R1322A and GFP–Dcr1-R1322A-ΔC33. Scale bars=2 μm. (E) Live-cell imaging of GFP–Dcr1-Δloop2. Scale bars=2 μm. (F) Quantitative real-time RT–PCR showing that heterochromatic gene silencing is not affected in dsRBD mutants that cannot bind dsRNA. cendg, cendh and imr1R∷ura4+ RNA levels are shown relative to dcr1+ cells and were normalized to act1+ RNA. Error bars represent standard deviations (s.d.). (G) Size distribution and the 5′-most nucleotide of centromeric sRNAs from dcr1+ and dcr1-Δloop2 cells.
RNA binding by the extended dsRBD is dispensable for proper functioning of Dcr1
Having shown that the extended dsRBD of Dcr1 binds dsRNA with high affinity, we set out to test if RNA binding is important for import/export-promoting activity of the dsRBD of Dcr1. We created mutations in three regions of the dsRBD (K1265A in helix 1, Δloop 2 and R1322A in loop 4, Figure 4A and Figure 1B, Supplementary Figure S5A for stability) that can be predicted to be involved in RNA binding based on previous structures of dsRBDs in complex with dsRNA (Ryter and Schultz, 1998; Ramos et al, 2000; Wu et al, 2004; Stefl et al, 2006, 2010). As expected, none of these dsRBD mutants could bind dsRNA in vitro (Figure 4C), despite being properly folded (as shown by 1D 1H NMR spectra). This further demonstrates that the extended dsRBD of Dcr1 binds RNA in a canonical mode despite its rather unusual structural features. Moreover, binding to dsDNA is also lost in these three mutants (Supplementary Figure S7C). This allowed us to generate mutant strains expressing Dcr1 that has lost its ability to bind dsRNA via the extended dsRBD.
Interestingly, nuclear localization of Dcr1 was not affected in any of the three mutant strains GFP–Dcr1-R1322A (Figure 4D), GFP–Dcr1-K1265A or GFP–Dcr1-Δloop 2 (Figure 4E), demonstrating that nuclear import of Dcr1 is unlikely to be dependent on binding of dsRNA via the extended dsRBD. Instead, one might speculate that dsRNA binding could be important to mediate the rapid export observed in the absence of C33 or of a functional CHCC motif. However, this is also highly unlikely because we assume that a dsRBD lacking C33 or the zinc-binding motif is unstructured, and therefore cannot bind RNA. Most importantly, nuclear localization of the mainly cytoplasmic Dcr1ΔC33 could not be restored by mutating R1322 to alanine (Figure 4D; Supplementary Figure S5A for stability). Thus, the dsRBD of Dcr1 functions in nucleocytoplasmic trafficking independently of its affinity for dsRNA.
Surprisingly, we found that dsRNA binding through the dsRBD is also dispensable for heterochromatin silencing and siRNA generation (Figure 4F and G; Supplementary Figure S6). Consistent with this observation, in vitro processing of dsRNA into siRNAs by Dcr1 has previously been shown not to require the extended dsRBD (Colmenares et al, 2007). Therefore, rather than substrate binding and processing, the main function of the unusual and elongated dsRBD of Dcr1 appears to be to regulate the subcellular localization of Dcr1 independently of its affinity for dsRNA.
The extended dsRBD of Dcr1 exposes a protein interaction surface that functions in nuclear retention
Our finding that mutating the zinc coordination motif has the same functional consequences as deleting C33, together with the fact that mutations in the zinc-binding motif render the protein insoluble in vitro, strongly suggested that coordination of zinc is required for the proper folding of the protein. Because nuclear retention of Dcr1 is lost in both C33 and CHCC mutants, and loss of RNA binding does not affect nuclear localization, we speculated that an exposed protein surface unique to the extended dsRBD might be crucial for this nuclear retention.
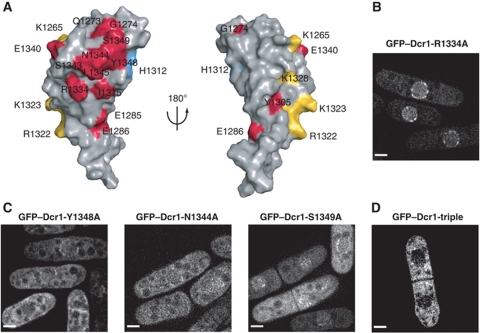
In order to identify such a putative protein interaction surface, we aligned the sequence of the C-terminal domain of S. pombe Dcr1 with the two closest annotated proteins (S. octosporus Dcr1 and S. cryophilus Dcr1; 31 and 33% sequence identity, respectively; see Supplementary Figure S8 for the sequence alignment). This revealed a patch of conserved residues (shown in red in Figure 5A) that cluster on one side of the domain around the third most N-terminal residue of C33 (N1344). In order to test whether this conserved surface is important for Dcr1 localization, we mutated residues in the centre of the surface (N1344A, Y1348A, S1349A; belonging to C33) or at its periphery (R1334A; belonging to α-helix 2). Whereas mutating R1334 to A did not affect Dcr1 localization (Figure 5B), the three residues in the middle of the putative protein–protein interaction surface turned out to be crucial for proper Dcr1 localization, resulting in reduced nuclear localization when mutated singly and in a complete loss of nuclear localization when mutated together (Figure 5C and D).
Figure 5.
A cluster of conserved residues is required for proper localization of Dcr1. (A) Solvent exposed residues with conserved biochemical properties among S. pombe, S. octosporus and S. cryophilus are displayed in colour on the surface of the domain. Residues that might be conserved for dsRNA binding are shown in yellow and H1312 that is conserved for zinc coordination is coloured in blue. All other residues are shown in red. Left: Front view. Right: Back view. (B) Live-cell imaging of GFP–Dcr1-R1334A. Scale bars=2 μm. (C) Live-cell imaging of GFP–Dcr1-Y1348A, GFP–Dcr1-N1344A and GFP–Dcr1-S1349A. Scale bars=2 μm. (D) Live-cell imaging of a strain expressing GFP–Dcr1 containing the mutations Y1348A, N1344A and S1349A (GFP–Dcr1-triple). Scale bars=2 μm.
These results are consistent with the idea that nuclear accumulation of Dcr1 is mediated by binding to a putative nuclear retention factor, which is likely to be mediated by this conserved surface. Importantly, C33 is required to establish the Dcr1 protein–protein interaction surface by contributing residues that are either important for the folding of the extended dsRBD or that belong to the exposed protein–protein interaction surface itself.
The novel zinc-binding motif is conserved in yeast dicers
In contrast to S. pombe Dcr1, human Dicer (DICER1) localizes mainly to the cytoplasm (Billy et al, 2001; Provost et al, 2002). Strikingly, only Dcr1 but not DICER1 has an extended C-terminus and we have therefore previously attempted to restrict DICER1 to the nucleus by the addition of C33 to its C-terminus. However, this had no effect on DICER1 localization, neither in human nor in S. pombe cells (SE and MB, unpublished observation), further corroborating our conclusion that C33 is unlikely to bind a nuclear retention factor on its own. With the structure of the Dcr1 dsRBD solved, our failure to restrict DICER1 to the nucleus can be explained. Because C1275 and H1312 in the dsRBD of Dcr1 are not conserved in the dsRBD of DICER1, addition of C33 is not sufficient to coordinate zinc. Thus, DICER1+C33 cannot fold into an extended dsRBD as observed for the C-terminus of Dcr1.
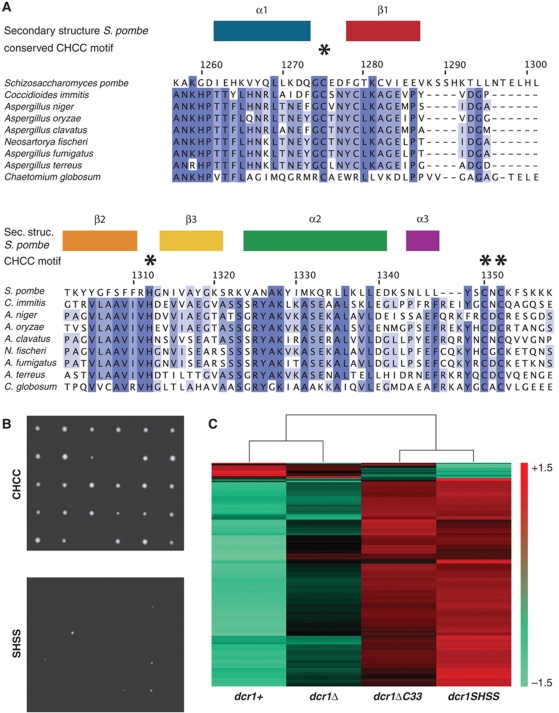
Intrigued by this difference between the two proteins, we performed a refined search for proteins with at least one RNase III domain, followed by a dsRBD and a C-terminal extension of at least 15 amino acids, and looked for the presence of a potential zinc coordination motif similar to the one found in S. pombe Dcr1. This search revealed 39 proteins of the dicer family in which the CHCC motif is found. Surprisingly, except for the CHCC motif, most amino acids of the dsRBD or C33 differ between S. pombe and other dicer proteins (Supplementary Figure S9). Strikingly, many of the dicers in which the four zinc-binding ligands are present belong to human or plant pathogens (Figure 6A; Supplementary Table II). This is particularly interesting because an S. pombe mutant strain that fails to coordinate zinc grows much more slowly and is less viable than wild-type or dcr1Δ cells (Figure 6B). Concomitantly, dramatic changes in gene expression can be observed upon mutating the zinc-coordinating residues but not upon deletion of the dcr1+ gene (Figure 6C), in accordance with our previous observation that a functional C33 prevents Dcr1 from acting promiscuously (Emmerth et al, 2010). Thus, if growth would be similarly reduced in pathogenic yeasts, disruption of zinc coordination by Dcr1 in these organisms might represent an appealing strategy to fight pathogenic fungi.
Figure 6.
Mutations in the conserved zinc coordination motif of Dcr1 are toxic in S. pombe. (A) Conservation of the Dcr1 CHCC motif in dicer proteins of human pathogenic yeasts. The highly conserved residues are indicated by asterisks. Amino-acid numbers refer to the S. pombe Dcr1 protein (UniprotKB Q09884). (B) Single wild-type (CHCC) and zinc coordination motif mutant (SHSS) cells were dissected onto a 5 × 6 matrix on EMMc-agar plates and grown at 30°C for ∼5–6 days. (C) Heatmap displaying the genes that were upregulated or downregulated at least 1.5-fold (P=0.05) in dcr1SHSS cells compared with wild type on S. pombe tiling arrays. Artificially scaled expression values are shown for the strains indicated. Three biological replicates were performed.
Discussion
In this study, we determined the solution structure of the C-terminal domain of S. pombe Dcr1 which revealed an extended dsRBD embedding a novel zinc-binding motif that is conserved among dicers in yeast. This zinc-binding motif is formed jointly by two side chains of the dsRBD and two from C33, and could represent a potential drug target for therapeutic intervention with fungal diseases. The insights gained from this study have important implications for our mechanistic understanding of RNAi and will be discussed below.
Dcr1's dsRBD is an elongated, non-canonical dsRBD
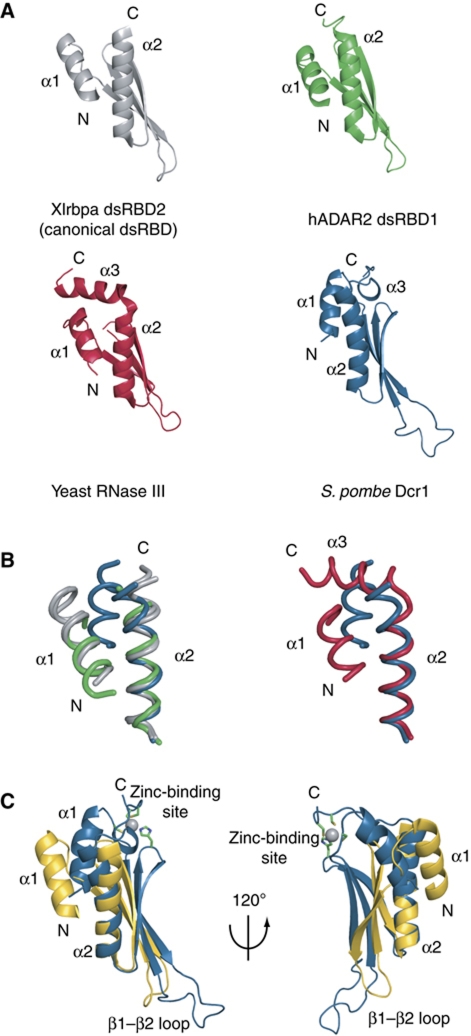
In this study, we found that the Dcr1 C-terminal domain adopts an extended dsRBD structure. Similarly, the dsRBD of the budding yeast RNase III protein Rnt1p has an extended fold with the region immediately C-terminal to the dsRBD forming a long C-terminal helix (α3, Figure 7A). This additional α-helix 3 is indispensable for the dsRBD fold and was proposed to contribute indirectly to RNA binding by helping position helix α1, which is the primary determinant of RNA recognition by Rnt1p (Leulliot et al, 2004; Wu et al, 2004). Despite this additional element, the dsRBD of Rnt1p adopts a very similar dsRBD fold compared with other dsRBDs (Figure 7B) like dsRBD2 of Xlrbpa (Ryter and Schultz, 1998) or the two dsRBDs of ADAR2 (Stefl et al, 2006). This is not what we observe in the structure of the dsRBD of S. pombe Dcr1. By comparing the Dcr1 extended dsRBD with others, one can see the different position of α-helix 1 relative to the rest of the domain (Figure 7B and C). Furthermore, the β-sheet of the Dcr1 dsRBD adopts a different curvature compared with other dsRBDs due to the involvement in the β2–β3 loop of H1312 in zinc coordination (Figure 7C). In summary, the Dcr1 dsRBD is not only extended by C33 and zinc coordination but also adopts a very distinct fold compared with other dsRBDs. Remarkably, the Dcr1 dsRBD still binds dsRNA strongly, despite an extended fold, an unusual position of α-helix 1, and an unusually long β1–β2 loop. These unique features might explain why the Dcr1 dsRBD also binds dsDNA. Finally, we note that it is possible that a regular dsRNA might not be the optimal RNA target of the Dcr1 dsRBD. Similar to the dsRBD of Rnt1p that recognizes AGNN tetraloops (Leulliot et al, 2004; Wu et al, 2004), or dsRBDs of ADAR2 that have been shown to be partly sequence specific (Stefl et al, 2010), the dsRBD of Dcr1 might preferentially recognize a particular RNA motif. It will be interesting to investigate such RNA sequence and/or structural preferences of the Dcr1 dsRBD in future studies.
Figure 7.
Structural comparison with other dsRBDs. (A) Structures of Xlrbpa dsRBD2 in grey (PDB code 1DI2), human ADAR2 dsRBD1 in green (PDB code 2L3C) and yeast RNase III Rnt1p in red (PDB code 1T4O) were superimposed over Cα atoms over the entire domain. S. pombe Dcr1 dsRBD in blue was superimposed with the other structures over Cα atoms of the C-terminal part of β-strand 3, β3–α2 loop and helix 2 in order to emphasize the differences in the position and the relative orientation of helix 1 and the β-sheet surface. (B) Relative position of helix 1 and helix 2. Only helices are displayed as ribbon. Left panel: Xlrbpa dsRBD2 and A. aeolicus RNase III (PDB code 1RC7) are displayed in grey, human ADAR2 dsRBD1 in green and S. pombe Dcr1 dsRBD in blue. Right panel: Yeast RNase III Rnt1p is displayed in red and S. pombe Dcr1 dsRBD in blue. (C) Superposition of Xlrbpa dsRBD2 in yellow and S. pombe Dcr1 dsRBD in blue over Cα atoms of the C-terminal part of β-strand 3, β3–α2 loop and helix 2, showing the unusual position of helix 1 and the β-sheet relative to helix 2.
Mechanism of nuclear retention
In our previous study, we demonstrated that the C-terminus of Dcr1 functions to control the subcellular localization of Dcr1. We have shown that the dsRBD can function as a strong nuclear export signal, which is under negative control by C33 (Emmerth et al, 2010). The C-terminal structure of Dcr1 determined in this study provides an elegant explanation for why our previous attempts to use C33 as a nuclear retention signal on heterologous proteins have failed. Rather than interacting with a nuclear retention factor directly, C33 contributes to the overall fold of the C-terminus by providing two of the four zinc-binding residues. These results in the formation of a putative protein–protein interaction surface that is characterized by a patch of conserved residues that cluster around the side chain of N1344 at the beginning of helix 3. This protein interaction surface would allow the attachment of Dcr1 to an as yet unidentified nuclear protein, resulting in nuclear accumulation of Dcr1. Even though we favour such a model, we cannot rule out the possibility that export-promoting features of the dsRBD are buried in the overall fold of the C-terminus and that this also contributes to nuclear retention of Dcr1. Only if zinc coordination was disrupted or C33 was completely missing, the export signal would become exposed and Dcr1 rapidly exported. The former model is particularly appealing to us because it would allow regulation of nucleocytoplasmic localization of Dcr1 via modulating the interaction between Dcr1 and the nuclear retention factor. This is an intriguing possibility, which we will investigate more closely in our future studies.
The role of dicer dsRBDs
The solution structure of the Dcr1 dsRBD allowed us to design specific mutations that interfere with binding to dsRNA without disturbing the overall structure of the domain. Intriguingly, we found that key residues in the dsRBD that are required for dsRNA binding are dispensable for proper functioning of Dcr1 in the RNAi pathway. This is surprising because one would assume that dicer proteins contain dsRBDs to get a hold on their dsRNA substrates. Thus, rather than substrate binding and processing, the main function of the Dcr1 dsRBD appears to be regulation of the subcellular localization of Dcr1 independently of its affinity for dsRNA. Interestingly, dicer proteins in animals are known to interact with other dsRBD-containing proteins, such as DICER1 with TRBP (Chendrimada et al, 2005; Haase et al, 2005), or Drosophila Dcr-1 and Dcr-2 with Loquacious and R2D2, respectively (Liu et al, 2003; Forstemann et al, 2005; Saito et al, 2005). We speculate that dicer dsRBDs might generally function in nucleocytoplasmic transport and not necessarily substrate binding. Dicers in some organisms might therefore have acquired auxiliary dsRBD proteins that would increase their specificity or efficiency in dsRNA processing.
Conservation of the zinc-binding motif
The unusual spacing between C and H residues in the zinc-binding motif makes it difficult to search for homologies. Therefore, we restricted our analysis to proteins with at least one RNase III domain, followed by a dsRBD and a C-terminal extension of at least 15 amino acids. This analysis revealed that the CHCC zinc-binding motif is highly conserved in the C-terminal domains of yeast dicers (Figure 6; Supplementary Figure S9).
Whereas the CHCC zinc-binding motif is highly conserved, the remaining sequences can be rather divergent even within the yeast group. The region between helix 2 and the C-X-C motif, as well as the sequence that follows the C-X-C motif, is well conserved neither in amino-acid composition nor in length (Supplementary Figure S9). For example, the region between helix 2 and the C-X-C motif in S. pombe Dcr1 is three amino acids shorter than in many other dicer sequences (Figure 6A; Supplementary Figure S9), suggesting the presence of an additional turn to helix 3 in other species. We also noted the presence of a buried arginine (R1311) side chain in the core of the S. pombe domain, whose positive charge would not be favourable in such a hydrophobic environment. However, we note that the vicinity of the CHCC zinc-binding site, with an overall −1 net charge, is likely to counterbalance this positive charge. Interestingly, in any other CHCC zinc-binding motif-containing dsRBD, this arginine is replaced by a hydrophobic residue (Figure 6A; Supplementary Figure S9), thus forming a more classical hydrophobic core.
Finally, the high degree of conservation and its mutational intolerance make the zinc-binding motif a prime antifungal target. Ejection of zinc from HIV type 1 nucleocapsid protein by disulphide benzamides or other molecules has been shown to have an antiviral effect (Rice et al, 1995; Pannecouque et al, 2010). Similarly, compounds reacting with the Dcr1 zinc-binding motif may offer new antifungal strategies to cope with the increasing incidence of invasive mycoses.
Materials and methods
Strains and plasmids
Fission yeast strains used in this study are listed in Supplementary Data and were grown at 30°C in YES. All strains were constructed following a standard PCR-based protocol (Bahler et al, 1998) and confirmed by sequencing. For protein expression, the different constructs, consisting of residues 1258–1374 (long dsRBD) or residues 1258–1358 (short dsRBD) of S. pombe Dcr1 (Figure 1B), were subcloned in E. coli expression vector pET28a+ between NdeI and XhoI cloning sites. The constructs hold an N-terminal tag whose sequence MGSSHHHHHHSSGLVPRGSHM includes a 6 histidine stretch used for protein purification. Mutagenesis was performed using the Quickchange Kit (Stratagene) following the manufacturer′s recommendations.
Protein expression and purification
Proteins were overexpressed in BL21(DE3) Codon-plus (RIL) cells in either LB media or M9 minimal media supplemented with 15NH4Cl and 13C-labelled glucose. All media were supplemented with 0.1 mM ZnCl2. The cells were grown at 37°C to OD600 ∼0.4, cooled down at 18°C and induced at OD600 ∼0.6 by adding isopropyl-β-D-thiogalactopyranoside to a final concentration of 0.25 mM. Cells were harvested 20 h after induction by centrifugation. The protein of interest was purified from both the soluble and insoluble fraction of E. coli. Cell pellets were resuspended in lysis buffer (Tris–HCl pH 7.5 50 mM, NaCl 500 mM, imidazole 20 mM, ZnCl2 0.1 mM, DTT 1 mM, Tween-20 0.2%), and lysed by sonication. Cell lysates were centrifuged for 30 min at 45 000 g. When the protein is expressed in LB media, about 40% remains in the soluble fraction, whereas it expresses almost exclusively in the inclusion bodies when expressed in M9 minimal media. On the one hand, the supernatant was loaded on an Ni-NTA column on a ÄKTA Prime purification system (Amersham Biosciences), and the protein of interest was eluted with an imidazole gradient. The fractions containing the protein were pooled, dialysed against the NMR buffer (NaPi pH 7.0 25 mM, KCl 75 mM, DTT 2 mM and ZnCl2 10 μM), and concentrated to ∼0.5 mM using Vivaspin 5000 MWCO (Sartorius Stedim Biotech). On the other hand, the pellet was dissolved in 6 M guanidinium chloride and DTT was added to a final concentration of 1 mM. The resuspended proteins from the insoluble fraction were then loaded on an Ni-NTA column equilibrated with Tris–HCl pH 7.5 50 mM, guanidinium chloride 4 M. The bound protein was then eluted with an imidazole gradient. The fractions containing the protein of interest were pooled, concentrated and subjected to a refolding protocol. Shortly, small amounts of purified protein were added step by step in a large amount of refolding buffer (Tris–HCl pH 7.5 50 mM, KCl 300 mM, 2-mercaptoethanol 1 mM and ZnCl2 0.1 mM) at 4°C under stirring. The protein solution was then dialysed against the NMR buffer and concentrated as for the protein purified from the soluble fraction. The protein purified from the insoluble fraction gives exactly the same 1H NMR spectrum as the one purified from the soluble fraction. This shows that both proteins adopt the same structure, and thus validate our refolding protocol.
NMR spectroscopy
All NMR spectra were recorded at 298 K in a buffer containing 25 mM NaPi pH 7.0, 75 mM KCl, 2 mM DTT, 10 μM ZnCl2 on Bruker AVIII-500 MHz, AVIII-600 MHz, AVIII-700 MHz and Avance-900 MHz spectrometers (all equipped with a cryoprobe). The data were processed using TOPSPIN 2.1 (Bruker) and analysed with Sparky (http://www.cgl.ucsf.edu/home/sparky/). Protein resonances were assigned using 2D (1H-15N)-HSQC, 2D (1H-13C)-HSQC, 3D HNCA, 3D HNCACB, 3D CBCA(CO)NH, 3D TOCSY-(1H-15N)-HSQC, 3D (H)CCH-TOCSY, 3D NOESY-(1H-15N)-HSQC and two 3D NOESY-(1H-13C)-HSQC optimized for the observation of protons attached to aliphatic carbons and to aromatic carbons, respectively. In addition, the assignment of aromatic protons was conducted using 2D (1H-1H)-TOCSY and 2D (1H-1H)-NOESY measured in D2O. We recorded all 3D NOESY spectra with a mixing time of 150 ms and the 2D NOESY spectra with a mixing time of 120 ms. Long-range 1H-15N HSQC spectrum was measured at 600 MHz and the delay during which 15N and 1H signals become antiphase was set to 22 ms to refocus magnetization arising from 1JNH couplings (Pelton et al, 1993).
Protein structure determination
Automated NOE cross-peak assignments (Herrmann et al, 2002a) and structure calculations with torsion-angle dynamics (Guntert et al, 1997) were performed using the macro noeassign of the software package CYANA 2.1. (Guntert, 2004). Peak lists of the four NOESY spectra were generated as input with the program ATNOS (Herrmann et al, 2002b) and manually cleaned to remove artefact peaks. The input also contained 41 hydrogen-bond restraints and restraints that defined the coordination geometry around the zinc. Hydrogen-bonded amides were identified as slowly exchanging protons in presence of D2O. Their bonding partner was identified from preliminary structures. We calculated 100 independent structures that we refined in a water shell with the program CNS 1.21 (Brunger et al, 1998; Brunger, 2007) using a water-refinement protocol successfully employed to refine a large set of NMR structures calculated with CYANA (Nederveen et al, 2005). The coordination geometry around the zinc has been defined with four distance restraints between the metal ion and the four ligand atoms that directly contact the metal centre (Cysteine Sγ and Histidine Nε), 10 bond angle and 1 dihedral angle restraints that defined a tetrahedral coordination sphere (Neuhaus et al, 1992). The 20 best energy structures were analysed using PROCHECK-NMR (Laskowski et al, 1996). Structures were visualized and figures were prepared using program MOLMOL (Koradi et al, 1996) and PYMOL (http://www.pymol.org).
Atomic absorption spectroscopy
Samples of unlabelled long dsRBD purified from the soluble protein fraction were extensively dialysed against ammonium acetate pH 7.0 100 mM, and analysed for Zn2+ content using an atomic absorption spectrometer from Varian (model spectrAA 220 FS). Zn2+ standards of 0–1.5 p.p.m. were prepared from a 1000 p.p.m. calibrated stock solution in the same buffer as the protein sample. Absorption measurement was repeated five times for standards and sample. The relative standard deviation for each sample was <1%.
Mass spectrometry
Samples for mass spectrometry were prepared either under denaturing conditions in aqueous 50% acetonitrile and 0.1% formic acid to a final pH of 2.0 to assess the mass of the apo form of the protein, or under native conditions in 10 mM ammonium acetate pH 7.0 to measure the mass of the holo form. Samples were analysed at the Functional Genomics Center Zurich on an electrospray time-of-flight mass spectrometer. Deconvoluted mass spectra were obtained using the MaxEnt1 software.
Nucleic acid samples used in ITC binding assays
To assess the nucleic acid binding properties of the dsRBD, we produced two complementary RNA fragments of arbitrary sequence by in vitro transcription with T7 polymerase (RNAfwd of sequence 5′-GGGAUCAAUAUGCUAAGCGAUCCC-3′ and RNArev being the reverse complement). RNA was purified by anion-exchange high-pressure liquid chromatography under denaturing conditions. The two complementary RNA strands were mixed to a 1:1 ratio, heated up at 95°C for 5 min and slowly cooled down to allow formation of dsRNA. DNA oligonucleotides of the same sequence (DNAfwd of sequence 5′-GGGATCAATATGCTAAGCGATCCC-3′ and DNArev being the reverse complement) were purchased from Microsynth AG (Switzerland) and purified by anion-exchange high-pressure liquid chromatography under denaturing conditions. The dsDNA duplex was formed as described for the dsRNA. ITC experiments with ssRNA and ssDNA were performed with RNAfwd and DNAfwd, respectively.
Isothermal titration calorimetry
ITC experiments were performed on a VP-ITC instrument (MicroCal) calibrated according to the manufacturer's instructions. The samples of protein and nucleic acids were prepared in and dialysed against the ITC buffer (NaPi pH 7.0 25 mM, KCl 75 mM, 2-mercaptoethanol 2 mM). The concentration of protein and nucleic acid was determined using OD absorbance at 280 and 260 nm, respectively. The sample cell (1.4 ml) was loaded with 2 μM of nucleic acid (dsRNA, dsDNA, ssRNA and ssDNA); dsRBD and mutant concentrations in the syringe were between 90 and 130 μM. Titration experiments were done at 25°C and typically consisted of 35–45 injections, each of 6–8 μl volume with a 5-min interval between additions. Stirring rate was 307 r.p.m. Raw data were integrated, corrected for non-specific heats, normalized for the molar concentration. Three parameters were fitted (the association constant Ka, the binding enthalpy ΔH and the number of site N) using the equation for 1:1 binding model.
Yeast live-cell fluorescence microscopy
S. pombe pre-cultures were grown in YES (sterile filtered components only) for 8 h at 30°C, diluted to 105 cells per ml, and grown for 14–16 h at 30°C to a concentration of about 5 × 106 cells per ml. Microscopy was performed on cells spread on agarose patches containing YES medium with 3% glucose. Images were captured on a Delta Vision built of an Olympus IX70 widefield microscope equipped with a CoolSNAP HQ2/ICX285 camera. Image stacks of 12–15 μm Z-distance were acquired with a Z-step size of 0.2 μm and deconvolved using the softworks (Delta Vision) software.
RNA analysis
RNA isolation, quantitative real-time RT–PCR, tiling arrays, as well as sRNA deep sequencing and analysis of the data were performed as described (Emmerth et al, 2010).
Multiple sequence alignment
All entries in UniProtKB (release 2010_10) were searched for Pfam domain PF00636 (RNase3). Sequences containing one or more RNase3 domains were cut after the (last) predicted RNase3 domain, and the Pfam domain PF00035 (double-stranded RNA binding motif) was aligned to the remaining C-terminal part. The fragments were further selected for the presence of at least 15 amino acids following the putative RNA binding motif and the presence of a Cys, followed by a His followed by Cys-X-Cys distributed over the whole length of the fragment. The distances between the first Cys and the His, and the His and the Cys-X-Cys in the remaining 58 fragments were manually compared with the numbers from S. pombe and S. japonicus. Thereby, another 15 proteins were eliminated. Only one protein was selected from proteins originating from the same species, but different strains. Proteins originating from the same species and the same strain, which were only different in five amino acids were reduced to one fragment as well. At this stage, C5WYA8 (a putative uncharacterized protein from Sorghum bicolor, the last non-fungi sequence) was removed, as it distorted the multiple sequence alignment created with Clustalw2 (Larkin et al, 2007). The multiple sequence alignment was manually adjusted and coloured (′Clustalx′) with Jalview (Waterhouse et al, 2009) and sequences to the left and the right of the dsRNA binding motif were removed as indicated in the figure. The UniProt accession for the S. pombe dicer protein is highlighted in red. Accessions in bold are proteins expressed in human or plant pathogenic yeast as listed in Supplementary Table II.
Accession numbers
The chemical shifts of Dcr1 dsRBD have been deposited in the BioMagResBank under accession number 17315. The coordinates of the structure have been deposited in the Protein Data Bank under accession code 2L6M.
Tiling array and deep sequencing data are deposited at GEO (http://www.ncbi.nlm.nih.gov/geo, accession numbers GSE30586 and GSE30549, respectively).
Supplementary Material
Acknowledgments
We are grateful to Serge Chesnov (Functional Genomics Center Zürich) for the measurements of ESI-MS spectrum; Daniel Fodor (ETH Zürich) for technical assistance with AAS; Heinz Gut and Jeremy Keusch (FMI Basel) for help with construct design and protein expression; and Irene Beusch (ETH Zürich) for her involvement in chemical shift assignment. We thank Helge Grosshans, Susan Gasser and Yvon Jaillais for critical reading of the manuscript and all the members of the Allain and Bühler laboratories for fruitful discussions. Research in the Bühler laboratory is supported by the Novartis Research Foundation, the Swiss National Science Foundation, the Gebert Rüf Foundation and the EMBO YIP program. Research in the Allain laboratory is supported by the Swiss National Science Foundation (Nr 310030-131031) and the SNF-NCCR structural biology. PB is supported by the Postdoctoral ETH Fellowship Program.
Author contributions: MB and FH-TA designed the project; PB prepared protein and RNA samples for structural studies and ITC, measured and analysed NMR data, and performed structure calculations and ITC measurements; PB and FH-TA analysed the structure; SE and YS generated mutant yeast strains; YS cloned Dcr1 fragments for recombinant protein production in E. coli and insect cells, and performed protein expression tests. SE performed all the functional analysis; H-RH performed homology searches. PB, SE, FH-TA and MB wrote the manuscript; all authors discussed the results and approved the manuscript.
Footnotes
Patent application (EP10191173.3) has been filed.
References
- Auld DS (2001) Zinc coordination sphere in biochemical zinc sites. Biometals 14: 271–313 [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Bass BL, Hurst SR, Singer JD (1994) Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Curr Biol 4: 301–314 [DOI] [PubMed] [Google Scholar]
- Baulcombe D (2004) RNA silencing in plants. Nature 431: 356–363 [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2: 2728–2733 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D (2006) Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell 125: 873–886 [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Bycroft M, Grunert S, Murzin AG, Proctor M, St Johnston D (1995) NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J 14: 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam HP, Sugiyama T, Chen ES, Chen X, FitzGerald PC, Grewal SI (2005) Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet 37: 809–819 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Chen T, Brownawell AM, Macara IG (2004) Nucleocytoplasmic shuttling of JAZ, a new cargo protein for exportin-5. Mol Cell Biol 24: 6608–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmenares SU, Buker SM, Buhler M, Dlakic M, Moazed D (2007) Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell 27: 449–461 [DOI] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M (2010) Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113 [DOI] [PubMed] [Google Scholar]
- Fierro-Monti I, Mathews MB (2000) Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem Sci 25: 241–246 [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P, Jantsch MF (2009) RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol Cell Biol 29: 1487–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet 10: 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Guntert P (2004) Automated NMR structure calculation with CYANA. Methods Mol Biol 278: 353–378 [DOI] [PubMed] [Google Scholar]
- Guntert P, Mumenthaler C, Wuthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273: 283–298 [DOI] [PubMed] [Google Scholar]
- Gwizdek C, Ossareh-Nazari B, Brownawell AM, Evers S, Macara IG, Dargemont C (2004) Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J Biol Chem 279: 884–891 [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W (2005) TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002a) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319: 209–227 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Guntert P, Wuthrich K (2002b) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR 24: 171–189 [DOI] [PubMed] [Google Scholar]
- Huisinga KL, Elgin SC (2009) Small RNA-directed heterochromatin formation in the context of development: what flies might learn from fission yeast. Biochim Biophys Acta 1789: 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Kharrat A, Macias MJ, Gibson TJ, Nilges M, Pastore A (1995) Structure of the dsRNA binding domain of E. coli RNase III. EMBO J 14: 3572–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14: 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
- Laity JH, Lee BM, Wright PE (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 11: 39–46 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Legge GB, Martinez-Yamout MA, Hambly DM, Trinh T, Lee BM, Dyson HJ, Wright PE (2004) ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J Mol Biol 343: 1081–1093 [DOI] [PubMed] [Google Scholar]
- Lei QP, Cui X, Kurtz DM Jr, Amster IJ, Chernushevich IV, Standing KG (1998) Electrospray mass spectrometry studies of non-heme iron-containing proteins. Anal Chem 70: 1838–1846 [DOI] [PubMed] [Google Scholar]
- Leulliot N, Quevillon-Cheruel S, Graille M, van Tilbeurgh H, Leeper TC, Godin KS, Edwards TE, Sigurdsson ST, Rozenkrants N, Nagel RJ, Ares M, Varani G (2004) A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III. EMBO J 23: 2468–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Macchi P, Brownawell AM, Grunewald B, DesGroseillers L, Macara IG, Kiebler MA (2004) The brain-specific double-stranded RNA-binding protein Staufen2: nucleolar accumulation and isoform-specific exportin-5-dependent export. J Biol Chem 279: 31440–31444 [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA (2006) Structural basis for double-stranded RNA processing by Dicer. Science 311: 195–198 [DOI] [PubMed] [Google Scholar]
- Moazed D (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature 457: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D (2004) Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119: 789–802 [DOI] [PubMed] [Google Scholar]
- Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Guntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM (2005) RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59: 662–672 [DOI] [PubMed] [Google Scholar]
- Neuhaus D, Nakaseko Y, Schwabe JW, Klug A (1992) Solution structures of two zinc-finger domains from SWI5 obtained using two-dimensional 1H nuclear magnetic resonance spectroscopy. A zinc-finger structure with a third strand of beta-sheet. J Mol Biol 228: 637–651 [DOI] [PubMed] [Google Scholar]
- Pannecouque C, Szafarowicz B, Volkova N, Bakulev V, Dehaen W, Mely Y, Daelemans D (2010) Inhibition of HIV-1 replication by a bis-thiadiazolbenzene-1,2-diamine that chelates zinc ions from retroviral nucleocapsid zinc fingers. Antimicrob Agents Chemother 54: 1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelton JG, Torchia DA, Meadow ND, Roseman S (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci 2: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 19: 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP (2002) Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Rice WG, Supko JG, Malspeis L, Buckheit RW Jr, Clanton D, Bu M, Graham L, Schaeffer CA, Turpin JA, Domagala J, Gogliotti R, Bader JP, Halliday SM, Coren L, Sowder RC II, Arthur LO, Henderson LE (1995) Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science 270: 1194–1197 [DOI] [PubMed] [Google Scholar]
- Ryter JM, Schultz SC (1998) Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J 17: 7505–7513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC (2005) Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol 3: e235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders LR, Barber GN (2003) The dsRNA binding protein family: critical roles, diverse cellular functions. FASEB J 17: 961–983 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457: 396–404 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Brown NH, Gall JG, Jantsch M (1992) A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA 89: 10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, Allain FH (2010) The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell 143: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH (2006) Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure 14: 345–355 [DOI] [PubMed] [Google Scholar]
- Strehblow A, Hallegger M, Jantsch MF (2002) Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell 13: 3822–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI (2005) RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA 102: 152–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wolfswinkel JC, Ketting RF (2011) The role of small non-coding RNAs in genome stability and chromatin organization. J Cell Sci 123: 1825–1839 [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, Allshire RC (2003) RNA interference is required for normal centromere function in fission yeast. Chromosome Res 11: 137–146 [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolcock KJ, Gaidatzis D, Punga T, Buhler M (2011) Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe. Nat Struct Mol Biol 18: 94–99 [DOI] [PubMed] [Google Scholar]
- Wu H, Henras A, Chanfreau G, Feigon J (2004) Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci USA 101: 8307–8312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.