Abstract
Cancer stem cells or cancer initiating cells are believed to contribute to cancer recurrence after therapy. MicroRNAs (miRNAs) are short RNA molecules with fundamental roles in gene regulation. The role of miRNAs in cancer stem cells is only poorly understood. Here, we report miRNA expression profiles of glioblastoma stem cell-containing CD133+ cell populations. We find that miR-9, miR-9* (referred to as miR-9/9*), miR-17 and miR-106b are highly abundant in CD133+ cells. Furthermore, inhibition of miR-9/9* or miR-17 leads to reduced neurosphere formation and stimulates cell differentiation. Calmodulin-binding transcription activator 1 (CAMTA1) is a putative transcription factor, which induces the expression of the anti-proliferative cardiac hormone natriuretic peptide A (NPPA). We identify CAMTA1 as an miR-9/9* and miR-17 target. CAMTA1 expression leads to reduced neurosphere formation and tumour growth in nude mice, suggesting that CAMTA1 can function as tumour suppressor. Consistently, CAMTA1 and NPPA expression correlate with patient survival. Our findings could provide a basis for novel strategies of glioblastoma therapy.
Keywords: cancer stem cells, gene silencing, glioblastoma, microRNAs, tumour suppressor
Introduction
MicroRNAs (miRNAs) are fundamental regulators of gene expression that direct processes as diverse as cell metabolism, lineage specification or cell differentiation (Bushati and Cohen, 2007; Bartel, 2009). MiRNAs are small RNA molecules with a size of about 18–25 nucleotides (nt). MiRNA genes are transcribed to primary miRNA transcripts, which are processed to miRNA precursors (pre-miRNAs) by the microprocessor complex in the nucleus. Pre-miRNAs fold into characteristic hairpin structures and are transported into the cytoplasm, where Dicer processes pre-miRNAs and generates a short double-stranded RNA. In further steps, one strand directly interacts with a member of the Argonaute (Ago) protein family (Peters and Meister, 2007; Hutvagner and Simard, 2008) and is incorporated into a miRNA–protein complex referred to as miRNP (Carthew and Sontheimer, 2009; Kim et al, 2009; Krol et al, 2010). The other strand, referred to as miRNA* (miRNA star), is degraded. In rare cases, however, both strands can give rise to functional miRNAs. One example for such a bifunctional miRNA is the miR-9/9* pair (Packer et al, 2008). MiRNAs guide miRNPs to partially complementary target sites on mRNAs and the miRNP–mRNA interaction leads to inhibition of translation or mRNA degradation (Huntzinger and Izaurralde, 2011).
MiRNAs are fundamental regulators of basic cellular processes and are frequently deregulated in tumours (Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Croce, 2009; Garzon et al, 2009). Numerous miRNA-profiling studies revealed that miRNA expression is altered in almost all types of cancer. Depending on the target mRNAs they regulate, miRNAs can be classified as tumour suppressors or oncogenes. The miRNAs miR-15 and miR-16 (chronic lymphocytic leukaemia), the let-7 family (lung and breast cancer) as well as miR-34 (pancreatic, colon and breast cancer) are well-characterized tumour suppressors (Calin et al, 2002; Johnson et al, 2005; He et al, 2007; Tarasov et al, 2007). On the other hand, miRNAs such as miR-155 (lymphomas), the miR-17-92 (lymphomas) cluster or miR-21 (variety of different cancers including glioblastomas) have been characterized as oncogenes (Chan et al, 2005; He et al, 2005; Costinean et al, 2006).
Cell populations with stem cell-like properties including self-renewal and differentiation have been identified in a variety of tumours (Lobo et al, 2007). These cancer stem cells (also referred to as tumour initiating cells) can initiate tumour growth, whereas other tumour cells fail to form new tumours when injected into nude mice. Cancer stem cells can be enriched in cell fractions that express specific surface proteins, such as CD44 in breast cancer or CD133 in colorectal cancer and a subgroup of primary astrocytic glioblastoma (Gilbertson and Rich, 2007; Lobo et al, 2007; Visvader and Lindeman, 2008; Tabatabai and Weller, 2011). According to the cancer stem cell hypothesis, cancer stem cells are believed to be the cause of relapse after therapy and contribute to treatment resistance (Reya et al, 2001). MiRNA expression profiling has been performed in several different tumour stem cell populations. In breast cancer cells, for example, it has been shown that let-7 regulates self-renewal and tumourigenicity of cancer initiating cells (Yu et al, 2007). Moreover, miR-34a is required for prostate cancer stem cell function and inhibition of miR-34a leads to reduced tumour growth (Liu et al, 2011). A detailed characterization of miRNA expression in glioblastoma stem cells has not yet been performed.
Here, we report the miRNA expression profile of CD133+ glioblastoma cell populations. We find that miR-9, miR-9*, miR-106b and miR-17 are highly abundant in glioblastoma stem cells. We further find that inhibition of miR-9/9* promotes neuronal differentiation, suggesting that miR-9/9* inhibit differentiation of glioblastoma stem cells and maintain their stemness. We identify the calmodulin-binding transcription activator 1 (CAMTA1) as miR-9/9* target. CAMTA1 overexpression substantially reduces colony formation, demonstrating that CAMTA1 is a novel tumour suppressor in glioblastoma. Consistently, we find that CAMTA1 expression correlates with glioblastoma patient survival. CAMTA1 is a putative transcription factor and we show that CAMTA1 regulates the expression of the natriuretic peptide A (NPPA, also referred to as atrial natriuretic factor (ANF)), which gives rise to peptide hormones with anti-proliferative effects.
Results
miRNA expression profiling of CD133+ glioblastoma stem cells
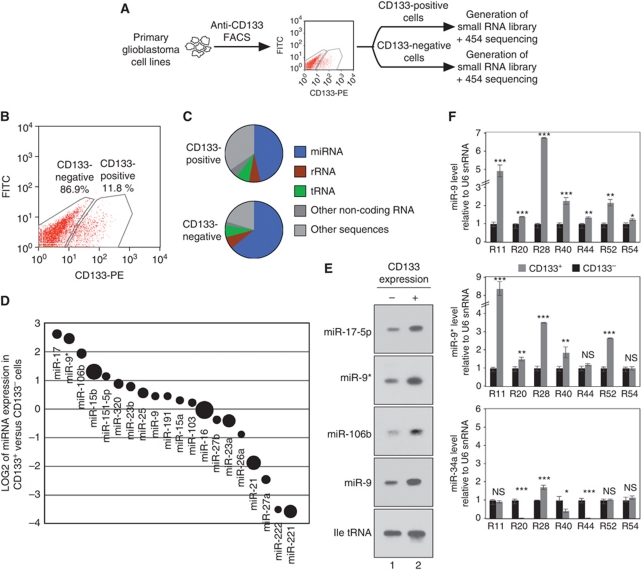
In order to analyse the contribution of miRNAs to the biology of glioblastoma stem cells, we isolated stem cell-containing CD133+ cells from the primary glioblastoma cell line R11 (Beier et al, 2007, 2008) by fluorescence-activated cell sorting (FACS, Figure 1A and B). Total RNA was extracted from CD133+ and CD133− cells and small RNA libraries were generated and sequenced (Figure 1C; Supplementary Tables 1 and 2). A detailed analysis of the individual miRNAs revealed that miR-9*, miR-17-5p, miR-106b and miR-15b were highly abundant in CD133+ cells, whereas miR-221, miR-222, miR-27a and miR-21 were more specific to CD133− cells (Figure 1D). The sequencing data were further validated by northern blotting. Signals for miR-17-5p, miR-9*, miR-106b and also miR-9 were much stronger in CD133+ cells (Figure 1E), demonstrating that these miRNAs are indeed differentially expressed between CD133+ and CD133− cells. Since the miR-9/9* pair was highly abundant in CD133+ R11 cells, we next confirmed the differential miR-9/9* expression in CD133+ cells in additional primary glioblastoma cell lines by qPCR (Beier et al, 2007; Figure 1F). Consistent with the data obtained from R11 cells, miR-9/9* expression is increased in CD133+ cell populations of many glioblastoma cell lines, whereas miR-34a was either not changed or less abundant in CD133+ cells. Although miR-9/9* expression is always stronger in CD133+ cells (with the exception of miR-9* in R54 cells, middle panel), some cell lines show only mild differences in miR-9/9* expression in CD133+ cells compared with CD133− cells. This might be due to the different tumour origin of the cell lines.
Figure 1.
miRNA expression profile of CD133+ versus CD133− glioblastoma cells. (A, B) Glioblastoma stem cells were sorted from primary glioblastoma cells (R11) and miRNA expression was analysed by cloning and deep sequencing. (C) RNA composition of the libraries. (D) Comparison of the CD133+ (positive value) and CD133− (negative value) miRNA expression profiles. The circle sizes indicate the abundance of the miRNA. (E) Validation of miR-17-5p, miR-9*, miR-106b and miR-9 in CD133+ glioblastoma cells (R11) by northern blotting. Ile tRNA served as loading control. (F) Validation of the differential expression of miR-9 (upper panel), miR-9* (middle panel) and miR-34a (control, lower panel) in different glioblastoma cell lines. In F, significance was assessed by two-sided Student's t-test (*P<0.05, **P<0.01, ***P<0.001, NS=not significant).
miR-9/9* are required for neurosphere formation and glioblastoma stem cell maintenance
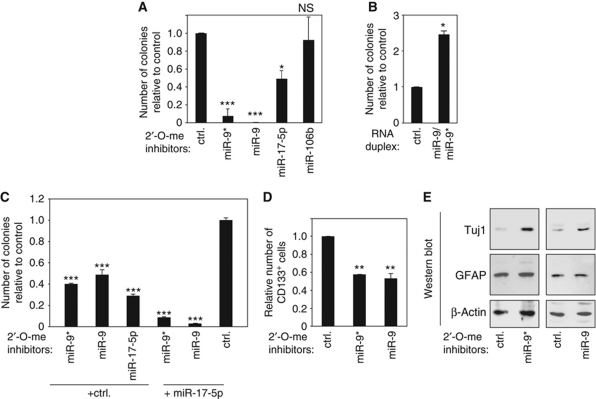
CD133+ glioblastoma cell populations show a neurosphere-like growth in tissue culture-based colony formation assays. Therefore, we analysed the impact of CD133+-specific miRNAs on neurosphere formation. Primary glioblastoma cells were transfected with 2′-O-methylated antisense inhibitors against miR-9, miR-9*, miR-17-5p or miR-106b and neurosphere growth was analysed (Figure 2A and Supplementary Figure S1 for validation of miRNA inhibition). Inhibition of miR-9 as well as miR-9* led to strongly reduced neurosphere formation. Inhibition of miR-17-5p reduced colony formation significantly whereas miR-106b inhibition had no effect (Figure 2A). Conversely, transfection of a miR-9/9* mimic increased neurosphere formation of primary glioblastoma cells (Figure 2B).
Figure 2.
miR-9/9* affect neurosphere formation and neuronal differentiation of glioblastoma stem cells. (A) miR-9*, miR-9, miR-17-5p and miR-106b were inhibited using 2′-O-methylated antisense oligonucleotides and neurosphere formation was analysed in R11 cells. (B) An miR-9/9* mimic was transfected into primary glioblastoma cells (R11) and colony formation was analysed. (C) Lower concentrations of inhibitors against miR-9/9* and miR-17 were co-transfected into R11 cells together with a control inhibitor or an miR-17 inhibitor and neurosphere formation was analysed. (D) miR-9/9* were inhibited by antisense inhibitors in primary glioblastoma cells (R11) and CD133+ cells were analysed by FACS. (E) miR-9/9* were inhibited as described above and the neuronal differentiation marker Tuj1 (upper panels), the glial differentiation marker GFAP (middle panels) were analysed by western blotting. β-Actin (lower panels) was analysed as loading control. In A–E, significance was assessed by two-sided Student's t-test (*P<0.05, **P<0.01, ***P<0.001, NS=not significant).
Since inhibition of both miR-9/9* and miR-17-5p affected neurosphere formation, we analysed whether the simultaneous inhibition of two miRNAs caused additive effects (Figure 2C). Indeed, the effects of miR-9 or miR-9* inhibition were significantly stronger, when miR-17-5p was co-inhibited, suggesting that both miRNAs function independently of each other and contribute to neurosphere formation. Of note, much lower inhibitor concentrations were used for the double-inhibition experiments; and therefore, the overall numbers differ from the experiments shown in Figure 1A.
Because miR-9/9* are highly abundant in CD133+ cells, we asked if inhibition of these miRNAs may influence the CD133+ cell population. Primary glioblastoma cells were transfected with inhibitors against miR-9 or miR-9* and CD133+ cells were sorted (Figure 2D). Indeed, inhibition of miR-9 or miR-9* led to a reduction of the CD133+ cell population, suggesting that both miRNAs are required for CD133+ glioblastoma stem cell maintenance. Reduced stem cell maintenance might lead to increased cell differentiation of primary glioblastoma cells. Thus, miR-9 or miR-9* was blocked and the neuronal differentiation marker Tuj1 or the glial marker GFAP were analysed by western blotting. Strikingly, CD133+ cell-depleted populations exhibited induced neuronal differentiation as evident from increased Tuj1 expression, whereas glial differentiation appeared not to be affected by miR-9/9* inhibition (Figure 2E). In summary, our data suggest that miR-9/9* help maintaining CD133+ cells probably by preventing differentiation.
CD133+ cancer stem cell miRNAs regulate the putative transcription factor CAMTA1
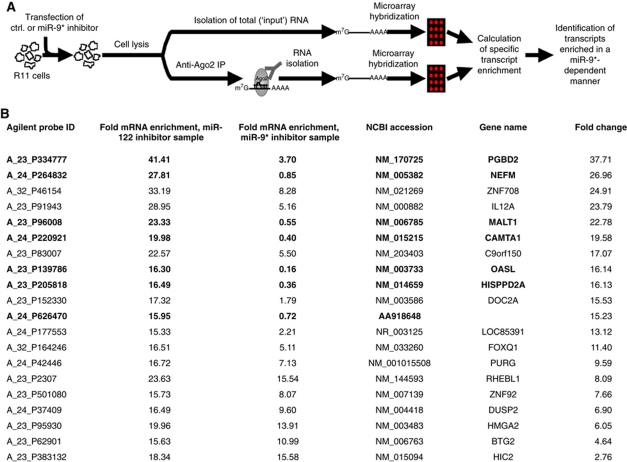
In order to understand the role of miR-9/9* in glioblastoma stem cells, it is important to know the identity of their individual target mRNAs. To find miR-9* targets in primary glioblastoma cells, we transfected inhibitors against miR-9* or control inhibitors and immunoprecipitated Ago2-bound mRNAs (Figure 3A; Beitzinger et al, 2007; Easow et al, 2007; Karginov et al, 2007). The miRNA inhibitor prevents mRNA binding and therefore miR-9* targets are selectively lost in the immunoprecipitates compared with control transfections. Using this approach, we identified a number of mRNAs that are strongly reduced in anti-Ago2 immunoprecipitates when miR-9* was inhibited (Figure 3B). We focused on CAMTA1, because it is expressed from the 1p36 locus that is frequently deleted in a subset of gliomas (Finkler et al, 2007). Strikingly, a minimal deletion comprising only the CAMTA1 gene has been identified, suggesting that CAMTA1 is indeed important for glioma formation (Barbashina et al, 2005).
Figure 3.
Identification of miR-9* target mRNAs. (A) Strategy to identify miR-9* target mRNAs. (B) List of most strongly Ago2-associated transcripts in primary glioblastoma cell line R11 in the presence of a miR-9* or miR-122 control inhibitor. miR-9* target mRNA candidates that were >10-fold depleted in the miR-9* inhibitor sample compared with the control are displayed in bold letters.
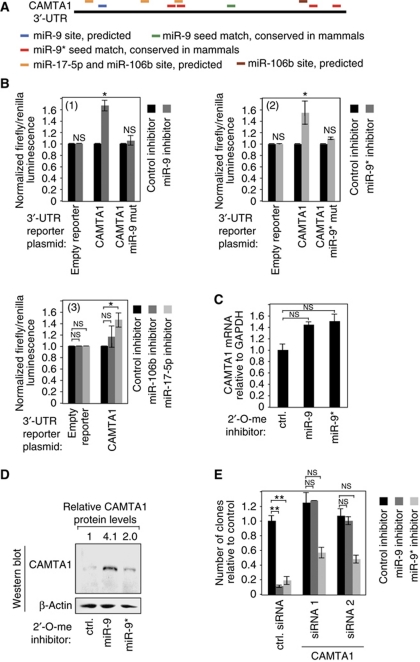
We found that the CAMTA1 3′-untranslated region (UTR) not only contains binding sites for miR-9* but also for miR-9, miR-106b and miR-17-5p, which are also highly abundant in CD133+ cell populations (Figure 4A). The CAMTA1 3′-UTR was fused to firefly luciferase and co-transfected together with inhibitors against miR-9/9* (Figure 4B, panels 1 and 2), miR-106b (panel 3) or miR-17-5p (panel 3) into primary glioblastoma cells. In all cases, firefly expression was elevated upon miRNA inhibition. Increased firefly activity was not observed, when reporters with mutated miR-9 or miR-9* binding sites were transfected. Furthermore, endogenous CAMTA1 mRNA as well as protein levels were elevated, when miR-9 or miR-9* was inhibited (Figure 4C and D). Of note, protein levels were much stronger increased than mRNA levels, suggesting that miR-9/9* may preferentially inhibit CAMTA1 translation. Since miR-9 and miR-9* inhibition blocked neurosphere formation, we hypothesized that this effect could be mediated through the induction of CAMTA1. Therefore, CAMTA1 was depleted by RNAi in primary glioblastoma cells (Supplementary Figure S2) and after 2 days, miR-9 or miR-9* was inactivated with antisense oligonucleotides (Figure 4E). Indeed, miR-9 inhibition effects on colony formation were rescued by CAMTA1 depletion. We also observed a significant rescue of miR-9* inhibition, although not as strong as observed for miR-9.
Figure 4.
miR-9/9* regulate CAMTA1 expression. (A) Location of miR-9 (blue/green), miR-9* (red), miR-17-5p (yellow) and miR-106b (brown) on the 3′-UTR of CAMTA1. (B) The CAMTA1 3′-UTR or variants with mutated miR-9 (1) or miR-9* (2) binding sites were fused to luciferase and luciferase expression was investigated in the presence or absence of miR-9 (1) or miR-9* (2) inhibitors in T98G cells. Panel (3) shows inhibitory effects of miR-106b and miR-17-5p on the wild-type CAMTA1 3′-UTR. (C) mRNA levels of CAMTA1 were analysed by qPCR upon miR-9 or miR-9* inhibition by antisense oligonucleotides in R11 cells. (D) CAMTA1 protein levels were analysed upon miR-9 or miR-9* inhibition in R11 cells. Western blotting with an anti-β-actin antibody was used as loading control. (E) Primary glioblastoma cells (R11) were pre-treated with two different siRNAs against CAMTA1 for CAMTA1 depletion. Subsequently, neurosphere formation assays were performed as described in Figure 2A using miR-9 or miR-9* inhibitors. In B, C and E, significance was assessed by two-sided Student's t-test (*P<0.05, **P<0.01, NS=not significant).
CAMTA1 functions as tumour suppressor in glioblastoma cells
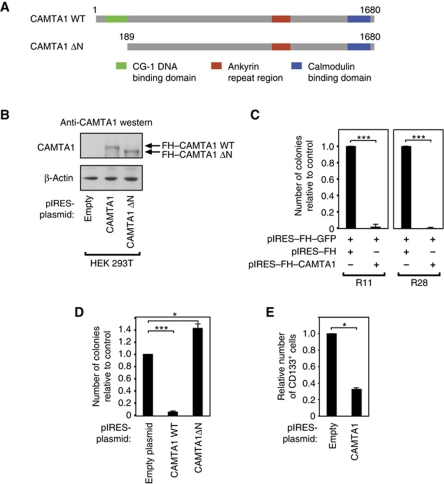
It has been suggested that CAMTA1 functions as tumour suppressor in neuroblastoma (Finkler et al, 2007; Henrich et al, 2011). However, a link between CAMTA1 function and glioblastoma has not been reported so far. To address this question, we cloned the CAMTA1 cDNA and transfected it into primary glioblastoma cells (Figure 5A–C). Strikingly, overexpression of CAMTA1 led to strongly reduced neurosphere formation in both R11 and R28 cells. CAMTA1 is a putative transcription factor that contains an N-terminal DNA binding domain (Figure 5A). We deleted the DNA binding domain (Figure 5D), transfected the mutated CAMTA1 into primary glioblastoma cells and again analysed neurosphere formation. Interestingly, the ΔN mutant that cannot bind DNA has no inhibitory effect on colony formation, indicating that overexpression of functional CAMTA1 inhibits neurosphere formation. Since miR-9/9* negatively regulate CAMTA1 expression, we hypothesized that CAMTA1 overexpression should have a similar effect on the CD133+ cell compartment as miR-9/9* inhibition (see Figure 2D). Indeed, overexpression of CAMTA1 reduced the number of CD133+ cells, suggesting that the miR-9/9* effect is at least in part due to CAMTA1 inhibition (Figure 5E).
Figure 5.
CAMTA1 has tumour suppressor activity in vitro. (A) Domain organization of CAMTA1 and the CAMTA1ΔN, where the DNA-binding domain has been deleted. (B) Flag/HA-tagged CAMTA1 (FH–CAMTA1) as well as FH–CAMTA1ΔN was expressed in HEK 293T cells and protein expression was analysed by western blotting using an anti-CAMTA1 antibody. (C) FH–CAMTA1 was expressed in two different primary glioblastoma cells (R11, left panel and R28, right panel) and neurosphere formation was analysed. (D) CAMTA1 and CAMTA1ΔN were expressed in R11 cells and neurosphere formation was analysed. (E) CAMTA1 was expressed in primary glioblastoma cells (R11) and the CD133+ cell population was analysed by flow cytometry. In C–E, significance was assessed by two-sided Student's t-test (*P<0.05, ***P<0.001).
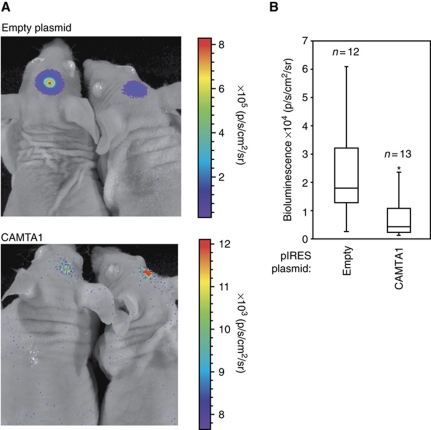
To further analyse the observed tumour suppressor activity of CAMTA1 in vivo, an R28 cell line stably expressing luciferase was transfected either with an empty plasmid or with a plasmid containing wild-type (wt) CAMTA1 (Figure 6). After transfection, cells were injected into nude mice brains and tumour growth was analysed 15 days after transfection by measuring luciferase activity. In agreement with the in vitro data, cells transfected with wt CAMTA1 showed strongly decreased tumour growth, whereas control cells formed tumours rapidly (Figure 6A and B). In summary, we have shown that CAMTA1 functions as tumour suppressor both in vitro and in vivo.
Figure 6.
CAMTA1 regulates tumour growth in an in vivo xenograft model. R28 cells, stably transfected with luciferase, were transfected with pIRES or pIRES-CAMTA1. Cells were then intracranially injected into mice brains. Tumour growth was quantified by measurement of luciferase activity 15 days after implantation. (A) Dorsal views of two representative mice are depicted from pIRES and pIRES-CAMTA1 group. The colour scales indicate the signal intensity at the surface of the animals recorded as total photons per second, square centimetre and steradian (p/s/cm2/sr). (B) Box plots of the quantified dorsal signals from both groups. Statistical significance was calculated with two-sided Student's t-test (*P<0.05).
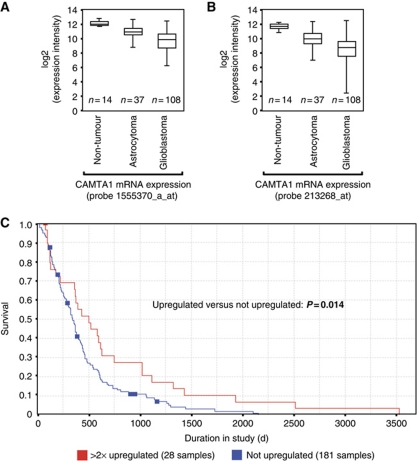
The striking effect of CAMTA1 overexpression on colony formation and tumour growth prompted us to ask whether CAMTA1 expression also correlates with survival of patients suffering from astrocytoma or glioblastoma. We analysed mRNA expression data sets from large patient cohorts using the Repository of Molecular Brain Neoplasia Data (REMBRANDT) and The Cancer Genome Atlas (TCGA) (Figure 7A and B). CAMTA1 mRNA expression was strongly decreased in astrocytoma as well as in glioblastoma patients compared with healthy individuals. Notably, grade IV glioblastomas show lower CAMTA1 expression than the less malignant astrocytomas. These results are in line with our previous results indicating a significant downregulation of CAMTA1 mRNA in cancer stem cells compared with neural stem cells (Lottaz et al, 2010). Finally, we correlated glioblastoma patient survival with low or intermediate (blue) and high (red) CAMTA1 expression (Figure 7C). Strikingly, glioblastoma patients with high CAMTA1 levels survived much longer than patients with lower or intermediate CAMTA1 expression, indicating that CAMTA1 is a tumour suppressor with a high prognostic value.
Figure 7.
CAMTA1 expression correlates with patient survival. (A, B) CAMTA1 mRNA expression was analysed in non-tumour brain tissue, WHO grade I–III astrocytoma and glioblastoma using the indicated affymetrix probes on a U-133A microarray. (C) Kaplan–Meier survival plot for glioblastoma patients with high, low and intermediate CAMTA1 mRNA expression, as determined by affymetrix probe 213268 on a U-133A microarray. Data were obtained from the REMBRANDT and the TCGA databases (see Materials and methods).
CAMTA1 stimulates the expression of the anti-proliferative peptide NPPA
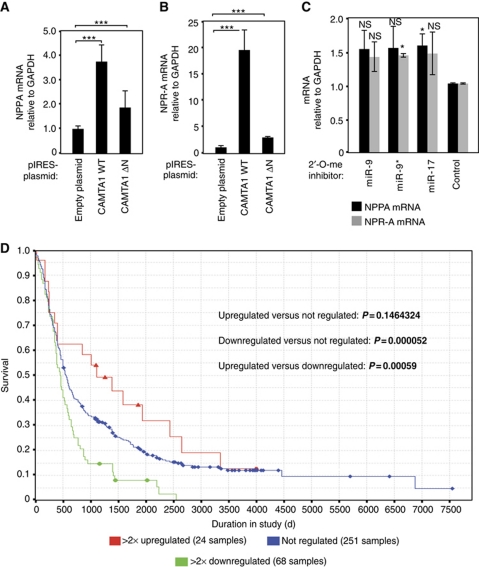
It has been shown that CAMTA2, which is highly homologous to CAMTA1, activates transcription of NPPA in the heart (Song et al, 2006). Interestingly, it has been demonstrated that NPPA has an anti-proliferative effect on glioblastoma cells in vitro (Vesely et al, 2007). We analysed whether CAMTA1 also activates the expression of NPPA in glioblastoma cells. A plasmid expressing wt CAMTA1 or a mutant lacking the DNA-binding domain was transfected into LNT-229 cells and NPPA expression was analysed by qPCR (Figure 8A). Wild-type CAMTA1 expression led to an increase of NPPA expression, while transfection of the mutated CAMTA1 had a much weaker effect. NPPA is a short secreted peptide, which is taken up by the natriuretic peptide receptor A (NPR-A). Therefore, we tested NPR-A expression and found that the receptor for NPPA is upregulated by CAMTA1 expression as well (Figure 8B). Finally, since miR-9, miR-9* and miR-17 regulate CAMTA1 expression in CD133+ cells, we tested whether inhibition of these miRNAs induces NPPA or NPR-A expression. MiRNAs were inhibited using 2′-O-methylated inhibitors and NPPA or NPR-A expression was subsequently analysed by qPCR (Figure 8C). Strikingly, inhibition of miR-9/9* or miR-17 increased NPPA and NPR-A expression presumably by inducing CAMTA1 expression.
Figure 8.
CAMTA1 regulates the peptide hormone NPPA and its receptor NPR-A. (A, B) CAMTA1 overexpression increases NPPA (A) and NPR-A mRNA levels (B). LNT-229 cells were transfected with indicated plasmids and NPPA or NPR-A mRNA levels were determined by qRT–PCR. Data were normalized to GAPDH mRNA levels as well as to the empty pIRES plasmid. (C) Inhibition of miR-9 and miR-9* increases NPPA and NPR-A mRNA levels. LNT-229 cells were transfected with indicated 2′-O-methyl antisense oligonucleotides and NPPA or NPR-A mRNA levels were determined by qRT–PCR. Data were normalized to GAPDH mRNA levels and control inhibitor-transfected samples. (D) NPPA expression correlates with glioma outcome. Kaplan–Meier survival plot for glioma patients (glioblastoma multiforme, oligodendroglioma and astrocytoma) with high, low and intermediate NPPA mRNA expression. Data were generated to Affymetrix probe 209957 on a U-133A microarray. In A–C, significance was assessed by two-sided Student's t-test (*P<0.05, ***P<0.001, NS=not significant).
NPPA has a strong anti-proliferative effect on glioblastoma cells in vitro; and therefore, we analysed whether NPPA expression correlates with patient survival (Figure 8D). We used the REMBRANDT database for our investigations. Consistent with our experimental data, we found that expression of NPPA correlates with patient survival. Patients with high NPPA levels (red) survived much longer than patients with intermediate (blue) and low (green) NPPA levels.
Taken together, we have found that miR-9/9* and miR-17 regulate the expression of the novel tumour suppressor CAMTA1 in CD133+ glioblastoma cells and CAMTA1 itself stimulates the expression of the anti-proliferative peptide NPPA.
Discussion
MiRNAs have been implicated in almost all types of cancer (Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Garzon et al, 2009). However, only a few studies have analysed the function of miRNAs in cancer stem cells (Yu et al, 2007; Ji et al, 2009; Shimono et al, 2009; Wong et al, 2010). We found that miR-9 and its corresponding miR-9* are highly expressed in cancer stem cell populations obtained from a subgroup of primary astrocytic glioblastomas. Both miRNAs function as oncogenes by repressing the novel tumour suppressor CAMTA1. Consistently, it has been shown that miR-9/9* are highly expressed in primary brain tumours (Huse et al, 2009; Nass et al, 2009). A recent study has analysed miRNA expression in 261 glioblastoma samples (Kim et al, 2011). According to their miRNA and mRNA expression profiles, glioblastomas were separated into five classes: glioblastomas with neural precursors, with oligoneural precursors, with multipotent precursors, with astrocytic precursors and with neuromesenchymal precursors. Interestingly, miR-9/9*, miR-17 or miR-106b were only found in glioblastomas with oligoneural precursors (Kim et al, 2011). It is thus unlikely that miR-9/9* or miR-17 inhibition would affect all glioblastomas. It is more likely that only those tumours with oligoneural precursors are affected. These findings have also impact on possible therapeutic approaches. Glioblastomas need to be classified by miRNA expression profiling first before miRNA inhibition could be used as potential treatment. However, it is important to note that glioblastomas have been classified using various approaches and the obtained classes very often differ significantly (Phillips et al, 2006; Lottaz et al, 2010; Verhaak et al, 2010; Huse et al, 2011).
MiR-9 has been implicated in cancer before. It is involved in cancer metastasis in breast cancer cells (Ma et al, 2010) or in colorectal cancer (Zhu et al, 2011). Interestingly, miR-9 expression is stimulated by MYCN in breast cancer and MYCN is closely related to the frequent 1p36 deletion (Ma et al, 2010; Mestdagh et al, 2010). It is tempting to speculate that MYCN might contribute to miR-9/9* expression in glioblastoma as well. Very recently, it has been demonstrated that among others, expression levels of miR-9 and miR-17 are correlated with malignant progression of gliomas (Malzkorn et al, 2010). In addition, miR-9 has been implicated in oligodendroglioma (Nelson et al, 2006). This supports our finding that these miRNAs are important for glioma pathogenesis. Furthermore, miR-9 is also important for neural development, for neuronal stem cell fate determination and for proliferation and migration of neural progenitors (Yoo et al, 2009; Zhao et al, 2009; Delaloy et al, 2010), supporting our model that miR-9 is important for glioblastoma stem cell function. Our data provide evidence that the bifunctional miR-9/9* inhibit CAMTA1 expression in glioblastoma stem cells and thereby contribute to robust cancer stem cell survival. We have demonstrated that expression of CAMTA1 in glioblastoma cells causes a strong reduction of colony formation both in vitro and in a xenograft model system. Therefore, we suggest that CAMTA1 is a novel tumour suppressor gene functioning in glioblastoma. Strikingly, it has been shown very recently that CAMTA1 has tumour suppressor activity in neuroblastoma cells supporting the idea that CAMTA1 is a tumour suppressor in a larger variety of neural tumours (Henrich et al, 2011).
The molecular functions of CAMTA1 have not been studied in detail yet. It has been shown in flies that CAMTA functions as transcription activator and is involved in the function of Rhodopsin, a G protein coupled light receptor (Han et al, 2006). In mammals, at least two CAMTA genes exist. The loss of CAMTA2 promotes cardiomyocyte hypertrophy (Song et al, 2006). Since CAMTA proteins can activate the expression of NPPA in the heart, we asked whether NPPA might also play a role in glioblastoma. Indeed, it has been demonstrated that NPPA, when incubated with glioblastoma and also other cancer cells, has a strong anti-proliferative effect in vitro (Vesely et al, 2005, 2007). Strikingly, CAMTA1 activates NPPA expression in glioblastoma cells as well. Moreover, CAMTA1 also stimulates the expression of the NPPA receptor, which might explain the tumour suppressor function of CAMTA1. Additionally, the NPPA gene is located in the 1p36 locus, and it is indeed deleted in a number of gliomas (Ichimura et al, 2008).
Based on our data, we propose the following model. In CD133+ cells, miR-9/9* and miR-17 expression is high compared with CD133− cells, which results in low CAMTA1 expression. Of note, it is likely that these miRNAs regulate several other target mRNAs, which might contribute to the observed phenotype as well. CAMTA1 inhibition also represses the expression of the secreted peptide NPPA. Upon CAMTA1 activation, NPPA expression is induced and the peptide is secreted. In parallel, CAMTA1 also stimulates the expression of the NPPA receptor NPR-A in order to allow cells to respond to NPPA. Since CAMTA1 expression is particularly low in the CD133+ fraction of many of our analysed glioblastoma cell lines (Supplementary Figure S3), it is tempting to speculate that the expression and also the sensitivity to the anti-proliferative peptide NPPA is repressed by miR-9/9* and miR-17 regulation of CAMTA1.
Further analysis of the cellular functions of the tumour suppressor CAMTA1 as well as its regulators miR-9/9* may finally lead to a better understanding of glioblastoma pathogenesis and ultimately to more efficient therapies that might be based on miR-9 and miR-9* inhibitors.
Materials and methods
Isolation of Ago2-associated RNAs
R11 cells were reverse transfected in four 10 cm plates per sample with miR-122 (control) or miR-9* 2′-O-methylated antisense oligonucleotide inhibitors for 2 days. Cells were lysed in 500 μl lysis buffer (150 mM KCl/25 mM Tris–HCl pH 7.5/2 mM EDTA/1 mM NaF/0.5% NP-40/0.5 mM DTT/0.5 mM AEBSF) per plate. Ribolock (Fermentas, 1 μl/ml of lysis buffer) was added before lysis. Lysates were cleared by centrifugation at 16 000 g for 10 min. For immunoprecipitation (IP) of endogenous Ago2, 3 ml of monoclonal anti-Ago2 11A9 hybridoma supernatant was coupled to 100 μl protein G-Sepharose (GE Healthcare) for 10 h at 4°C. Coupled beads were washed twice with PBS and subsequently incubated with cell lysate for 4 h at 4°C. All IP samples were washed three times with IP wash buffer (300 mM NaCl/50 mM Tris pH 7.5/1 mM NaF, 0.01% NP-40/5 mM MgCl2) and once with PBS. IP samples and corresponding samples containing 10% of input lysate were proteinase K digested, followed by phenol/chloroform/isopropyl alcohol extraction and precipitation of RNA in 80% ethanol at −20°C. RNA was pelleted, dried and treated with DNaseI (Fermentas) for 45 min at 37°C, followed by thermal inactivation of DNaseI. RNA integrity was assessed via Agilent 2100 Bioanalyzer (Agilent Technologies) prior to microarray hybridization.
Microarray experiments and data analysis
RNA from anti-Ago2 IP and input samples was processed using the SuperAmp RNA amplification protocol (Miltenyi Biotec). cDNA integrity was checked via Bioanalyzer platform. In all, 250 ng of each cDNA was labelled with Cy3 dye (Miltenyi Biotec) according to the manufacturer's protocol. Samples were hybridized to Agilent Whole Human Genome 4 × 44 K Oligo Microarrays. Fluorescence signals of the hybridized Agilent Microarrays were detected using Microarray Scanner System (Agilent Technologies).
Microarray data were analysed using Agilent Genespring software. Expression values below 0.01 were set to 0.01. Each measurement was divided by the 50th percentile of all measurements in that sample. All IP samples were normalized to the corresponding input RNA samples. The IP sample from control antagomir-transfected cells was normalized against the median of the corresponding input RNA sample and the IP sample from miR-9* antagomir-transfected cells was normalized against the median of the corresponding input RNA sample. For normalization, each measurement for each gene in the IP samples was divided by the median of that gene's measurements in the corresponding input RNA samples. IP to input ratios from miR-9*-transfected samples were then divided by the IP to input ratios from control-transfected sample.
Using this normalization procedure, the normalized expression value of each transcript in IP samples directly reflects its fold enrichment in the immunoprecipitated transcript pool relative to the input RNA pool. To filter for potential miRNA target mRNAs bound by Ago2, all transcripts that were >5-fold enriched in IPs from control antagomir-transfected cells were identified. Transcripts where the enrichment in miR-9* antagomir-transfected cells was >10-fold lower than in control-transfected cells were considered to be potential targets of miR-9*.
Cell culture and transfection
R11, R20, R28, R40, R44 and R52 cells were cultured at 37°C in DMEM-F12 medium supplemented with 20 ng/ml of each human recombinant epidermal growth factor, human recombinant basic fibroblast growth factor (both from R&D Systems), and human leukaemia inhibitory factor (Millipore), 2% B27 supplement (Invitrogen), 1% penicillin/streptomycin solution (PAA), and 1% MEM vitamins solution (Invitrogen). Cells were passaged every 7–10 days by trypsinization or by detaching with a pipette. In all, 50% of the medium was substituted twice weekly. HEK 293T, T98G and LNT-229 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin solution. Typically, cells were passaged every 3 days by trypsinization in a 1:10 ratio.
R11 cells (1 × 105 cells/well) were reverse transfected with 100 nM 2′-O-methyl oligonucleotides or 40 nM siRNAs in 6-well plates with 5 μl/well Lipofectamine 2000 (Invitrogen). For co-transfection of two 2′-O-methyl antisense oligonucleotides, each oligo was added to 50 nM final concentration, to give an overall concentration of 100 nM. The transfection mix was removed 24 h post transfection and fresh medium was added. T98G cells were transfected with Lipofectamine 2000 12 h after seeding according to the manufacturer's instructions, using 40 nM siRNAs or 80 nM 2′-O-methyl oligonucleotides.
For plasmid transfections, R11, R28 and R28-luc cells were electroporated using the Nucleofector Kit for mouse neural stem cells (Lonza), according to the manufacturer's instructions and the Nucleofector Device II (Lonza). In short, cells were detached by pipetting, washed once with DMEM/F12 and 3 × 106 cells were transfected with 5 μg of plasmid DNA using Program A-033. Directly after electroporation, cells were transferred into one well of a 6-well plate. Medium was replaced 24 h post transfection. For electroporation of LNT-229, the cell line nucleofector Kit R (Lonza) was used as described above.
Western blotting and northern blotting
To analyse levels of proteins other than CAMTA1, cells were lysed as previously described (Weinmann et al, 2009). For the analysis of CAMTA1 protein levels or corresponding loading controls, cells were lysed in nuclear lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris pH 8, 0.1% sodium deoxycholate, 2 mM 4-(2-Aminoethyl) benzenesulphonyl fluoride hydrochloride, 5 units/ml DNAaseI (Fermentas)) for 20 min and sonicated for 10 s on a Bandelin Sonopuls HD2070 sonifier. Lysates were cleared by centrifugation. Western blotting was performed as previously described (Hock et al, 2007). The following antibodies (dilutions) were used: Anti-mouse-HRP (1:5000), anti-rabbit-HRP (1:5000), mouse-anti-β-Actin AC15 (1:10 000), mouse-anti-Tuj1 (1:1000), rabbit-anti-GFAP (1:2000) and rabbit-anti-CAMTA1 (1:200). Northern blotting was performed as previously described (Lagos-Quintana et al, 2001) using either 2′-O-methyl oligonucleotide or DNA probes antisense to the miRNA of interest. The sequence of Ile tRNA probe was 5′-TGCTCCAGGTGAGGATCGAAC-3′.
Clonogenicity assays
1 × 105 R11 cells were transfected in 6-well plates as indicated. 2′-O-methylated antisense oligonucleotides and synthetic miRNAs were transfected twice with an interval of 7 days. Seven days after second transfection, cells were counted and ∼1000 cells were transferred into each well of a 48-well plate. Neurosphere-like clones were counted 4 weeks after plating. The number of clones per well were summed up for each sample and normalized to the number of clones obtained for control-transfected samples. Results of the described clonogenicity assays were reproduced by limited dilution assays with one cell per well. Therefore, 1 × 105 cells were transfected as indicated above. Seven days after second transfection, one cell was seeded into each well of a 96-well plate and neurosphere-like clones were counted 4 weeks after plating. Data for limited dilution assays are not shown.
In vivo tumour model and bioluminescence imaging
Intracranial glioblastoma xenografts were established in 10-week-old male NMRI:nu/nu mice (Charles River) as described previously (Beier et al, 2007). In brief, tumour cells were treated as indicated. Three hours after transfection, 1.4 × 105 viable cells (as determined by trypan blue exclusion) were injected 2 mm lateral to the midline and 4 mm anterior to bregma to a depth of 3 mm using a Hamilton syringe. The bioluminescence of implanted tumour cells was determined 15 days after implantation. One minute after injection of 150 mg/kg D-luciferin (Biosynth), mice were anaesthetized and emitted photons were registered for 5 min using Xenogen IVIS Lumina Imaging System (Caliper Life Sciences). The signal was normalized to background signal. Mice without detectable tumours were excluded from the analysis.
Lentiviral transduction of R28 cells
Glioblastoma stem cells were stably transduced using a lentiviral vector expressing firefly luciferase under control of the constitutive spleen focus forming virus LTR promoter.
Third-generation packaging, VSV-pseudotyped, self-inactivating lentiviral vectors were produced by transient transfection of HEK 293T cells using standard protocols (Wubbenhorst et al, 2010). Medium was changed to stem cell medium after 24 h. Supernatants were filtered through 0.45 μm filter and used for spin infection (Leisegang et al, 2008). Individual wells of a 24-well plate were coated with 400 μl RetroNectin (Takara Bio Europe SAS; final concentration 12.5 μg/ml) for 2 h at room temperature, subsequently incubated for 30 min at 37°C with bovine serum albumine (2% in PBS) and washed with PBS prior to addition of cells. 1 × 105 cells/ml suspension cells were added onto RetroNectin-coated wells. One millilitre virus containing supernatant supplemented with protamine sulphate (final concentration 4 μg/ml) was added to each well and infection was enhanced by 90 min centrifugation at 32°C and 800 g. Cells were incubated at 37°C. The next day, cells were washed off the wells, spun, resuspended in 5 ml fresh complete medium and further cultivated.
Cell lines
The generation of R11, R20, R28, R40, R44 and R52 cell lines from glioblastoma samples was previously described (Beier et al, 2007).
Antibodies
The following antibodies were used: Rat-anti-hsAgo2 (11A9; Rudel et al, 2008), mouse-anti-Tuj1 MMS-435p (Covance), mouse-anti-β-Actin AC15 (Abcam), rabbit-anti-GFAP (DAKO), anti-CD133-2 293C3-PE (Miltenyi), anti-rabbit-HRP, anti-mouse-HRP (both from Sigma). The polyclonal antibody to CAMTA1 was generated as follows: a GST-tagged fragment containing aa 294–864 of CAMTA1 was expressed in E. coli and used for the immunization of rabbits. Polyclonal antiserum was purified and used for western blotting.
Oligonucleotides
2′-O-methyl antisense oligonucleotides and siRNAs/miRNAs were chemically synthesized using RNA phosphoramidites (Pierce) on an Äkta Oligopilot 10 DNA/RNA synthesizer (GE Healthcare), according to the manufacturer's protocol. The sequences of 2′-O-methyl oligonucleotides were miR-9* antisense, 5′-ACUUUCGGUUAUCUAGCUUUAT-3′; miR-17-5p antisense, 5′-ACUACCUGCACUGUAAGCACUUUGT-3′; miR-106b antisense, 5′-AUCUGCACUGUCAGCACUUUAT-3′; miR-9 antisense, 5′-UCAUACAGCUAGAUAACCAAAGAT-3′; miR-122 antisense, 5′-ACAAACACCAUUGUCACACUCCAT-3′; miR-301 antisense, 5′-GCUUUGACAAUACUAUUGCACUGT-3′ and miR-330 antisense, 5′-GCCUAAGACACAGGCCCAGAGAT-3′. The siRNA sequences (sense and antisense) were CAMTA1 siRNA 1, 5′-CTACCGAAGTTATAAGAAAUT-3′, 5′-TTTCTTATAACTTCGGTAGUT-3′; CAMTA1 siRNA 2, 5′-GAAUCAAGCAGGAGAAUUUUT-3′, 5′-AAAUUCUCCUGCUUGAUUCGT-3′; control siRNA 1, 5′-UUGUCUUGCAUUCGACUAAUT-3′, 5′-UUAGUCGAAUGCAAGACAAUT-3′ and control siRNA 2, 5′-UCGAAGUAUUCCGCGUACGUT-3′, 5′-CGUACGCGGAAUACUUCGAUT-3′. The sequences of synthetic miRNAs were miR-9, 5′-UCUUUGGUUAUCUAGCUGUAUGAT-3′; miR-9*, 5′-AUAAAGCUAGAUAACCGAAAGUT-3′; miR-122, 5′-UGGAGUGUGACAAUGGUGUUUGT-3′ and miR-122*, 5′-AACGCCAUUAUCACACUAAAUAT-3′.
Flow cytometry
Cells were trypsinized and washed with DMEM-F12 and FACS buffer (PBS containing 1% BSA). 107 cells were resuspended in 80 μl FACS buffer containing 10% FcR blocking reagent (Miltenyi) and incubated for 5 min on ice. In all, 10 μl Anti-CD133-PE was added, and cells were incubated for 10 min on ice in the dark. Cells were pelleted and washed once with FACS buffer. Stained cells were sorted on an FACS Aria system (Becton Dickinson). Cell debris were gated out using a forward scatter/sideward scatter dot plot. CD133-negative and CD133-positive cell populations were identified using unstained cells as control.
RNA isolation
Total RNA for mRNA analyses was isolated using the Prep Ease kit (USB), according to the manufacturer's instructions. For small RNA detection, RNA was isolated using Trifast (Peqlab).
cDNA synthesis
cDNA for mRNA analysis was synthesized with random hexamer primers from 2 μg of total RNA using the First Strand cDNA synthesis kit (Fermentas), according to the manufacturer's protocol. To quantify miRNAs, RNA samples were treated with DNaseI (Fermentas), poly(A)-tailed using the poly(A)-tailing kit (Ambion) and reverse transcribed using the First Strand cDNA synthesis kit and the URT primer 5′-AACGAGACGACGACAGACTTTTTTTTTTTTTTTV-3′ (Hurteau et al, 2006).
qPCR
qPCR was performed on a MyiQ cycler (BioRad) using the Mesa Green qPCR mastermix (Eurogentec). The primers were GAPDH, 5′-TGGTATCGTGGAAGGACTCATGAC-3′, 5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′; β-actin, 5′-CTGGAGAAGAGCTACGAGCTG-3′, 5′-TTGAAGGTAGTTTCGTGGATG-3′; CAMTA1, 5′-ATCCTTATCCAGAGCAAATTCC-3′, 5′-AGTTTCTGTTGTACAATCACAG-3′; NPPA, 5′-CAGGATGGACAGGATTGGA-3′, 5′-TCTTCAGTACCGGAAGCTGTT-3′; NPR-A 5′-TCGAAACCACCAAACTCCTC-3′, 5′-AGTGGTGGGACTGAAGATGC-3′; hsa-miR-9, 5′-TCTTTGGTTATCTAGCTGTATG-3′; hsa-miR-9*, 5′-ATAAAGCTAGATAACCGAAAG-3′; hsa-miR-34a; 5′-TGGCAGTGTCTTAGCTGGTTG-3′; U6 snRNA, 5′-GATGACACGCAAATTCGTGAAG-3′ and universal RT primer for miRNA and U6 snRNA detection, 5′-AACGAGACGACGACAGACTTT-3′. Data were evaluated using the ddCt method with GAPDH or β-actin as reference mRNAs. Error bars were obtained from triplicate PCR samples by propagating the ddCt standard error of the mean through the exponential term as previously described (Livak and Schmittgen, 2001).
Generation and sequencing of small RNA libraries
Small RNA libraries were generated by Vertis Biotechnology AG and sequenced by 454 pyrosequencing as previously described (Tarasov et al, 2007).
Analysis of the sequencing results
Known miRNAs were identified by comparing the sequencing results with annotated miRNAs from the H. sapiens miRNA database (miRBase) using the Microsoft Excel software. Several miRNA reads were found to contain sequencing errors, typically starting from nucleotides 18 to 25 that were possibly due to the procedure of library preparation and/or pyrosequencing. Most errors were poly(A) insertions at the 3′-end of the reads. Therefore, those reads that were fully complementary to a known miRNA from nucleotides 1 to 18 but had additional (A) insertions at the 3′-end were re-annotated as miRNAs.
Read numbers for each miRNA were normalized to the total number of reads of the corresponding library. For calculation of miRNA expression, the normalized reads numbers from the CD133-negative cell library were divided by the normalized read numbers from the CD133-negative library.
Luciferase assays
To investigate miRNA effects on reporter constructs, T98G were used for high transfection efficiency and high firefly and renilla expression levels. T98G cells were co-transfected with 2′-O-methylated antisense oligonucleotides (80 nM final concentration) and pMIR-RL constructs (100 ng/well) in 48-well plates, using Lipofectamine 2000. Twenty-four hours after transfection, cells were lysed in passive lysis buffer (Promega). Luciferase activities were measured on a Mithras LB 940 luminometer (Berthold Technologies). Luciferase substrate reagents were purchased from PJK cryosystems. All samples were assayed in 4–6 replicates. Firefly/renilla luminescence ratios for individual samples were normalized to corresponding ratios of the empty pMIR-RL plasmid and control inhibitor-transfected samples.
Plasmids
The pMIR-RL dual luciferase vector was previously described (Beitzinger et al, 2007). The 3′-UTR of CAMTA1 mRNA was PCR amplified from R28 genomic DNA using the primers 5′-ATACGAGCTCAGACATACAGCAGCATCCCTTAGCAATGTG-3′ (forward), 5′-ATACGCCGGCGGAAATTTTCTTCATTTTTAATTTACAGCAG-3′ (reverse), digested with SacI and NaeI and ligated into pMIR-RL. For the analysis of miR-9 and miR-9* binding sites, all sites predicted by TargetScan 5.0 and all seed matches that are conserved in mammals and indicated in Figure 3A were mutated by PCR-based mutagenesis as follows: the nucleotides CAAA in miR-9* seed matches were replaced by GTTT and the nucleotides CTTT in miR-9* seed matches were replaced by GAAA. A cDNA fragment encoding CAMTA1 was PCR amplified from Marathon whole human brain cDNA library (BD Bioscience). CAMTA1 orf was re-amplified by using the primers 5′-ATACGATATCATGTGGCGCGCGGAGGGGAAATG-3′ (forward) and 5′-ATATGCGGCCGCTCAAGTTCCTTGGCCTTTTTCAATTCTTTCACTC-3′ (reverse) bearing EcoRV and NotI restriction sites, respectively, followed by restriction digestion and ligation into pIRESneo. Note that all CAMTA1 clones that were obtained contained an additional exon of 21 nt in size (AGCTGACATGGATAGCCTTGA, data not shown) compared with the Refseq sequence (NM_015215.1). The additional nucleotides are inserted after nt 4687 of NM_015215.1 and encode for seven additional amino acids which localize to the predicted Calmodulin binding domain. An N-terminal CAMTA1 deletion mutant lacking the amino acids 1–188 (nt 1–564) was cloned into pIRESneo and used as a control. All constructs were verified by sequencing.
Analysis of gene expression in glioma patients and correlation with survival
The results published here are in part based upon data generated by the REMBRANDT, a project led by the National Cancer Institute (NCI) (The Cancer Genome Atlas, 2008; Madhavan et al, 2009). Information about the REMBRANDT database can be found at https://caintegrator.nci.nih.gov/rembrandt/. TCGA project is a joined effort of the NCI and the National Human Genome Research Institute (The Cancer Genome Atlas, 2008). Information about TCGA can be found at http://cancergenome.nih.gov.
Statistical analysis
Experiments were performed in three biological replicates, unless stated otherwise. Mean values and standard error of the mean (s.e.m.) were calculated from all biological replicates. Error bars display ±s.e.m. Significance was assessed from biological replicates using two-sided Student's t-tests for unequal sample variance. P-values <0.05 were considered as significant.
Supplementary Material
Acknowledgments
We thank Sabine Rottmüller, Marion-Stefanie Roller, Dr Theo Thepen, Dr Christoph Jan Wruck and Bernd Haas for technical assistance. This work was supported by the Max-Planck-Society, in part by the Behrens-Weise-Foundation, the European-Research-Council (ERC, grant ‘sRNAs’), the NGFN+ Brain-Tumor-Network (No. 01GS0887), the Deutsche Forschungsgemeinschaft (DFG, SFB 773-project A6 (to GT) and Me 2064/2-2 to GM), NCCR Neural plasticity and repair (P4), the START program of the University of Aachen (to CB and DB) and the Bavarian Ministry of Education and Science (BayGene to GM). LW received a fellowship of the Boehringer Ingelheim Fonds. DB is supported by a career award for young female scientists.
Author contributions: DS and LW designed and performed experiments; analysed data and wrote parts of the manuscript. DB performed experiments and analysed data. GT designed experiments and analysed data. AE performed experiments. JYZ analysed data. MA performed experiments. MS performed experiments. MW analysed data and designed experiments. CB designed experiments and analysed data. GM supervised the project, designed experiments, did experimental analysis and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barbashina V, Salazar P, Holland EC, Rosenblum MK, Ladanyi M (2005) Allelic losses at 1p36 and 19q13 in gliomas: correlation with histologic classification, definition of a 150-kb minimal deleted region on 1p36, and evaluation of CAMTA1 as a candidate tumor suppressor gene. Clin Cancer Res 11: 1119–1128 [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP (2007) CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res 67: 4010–4015 [DOI] [PubMed] [Google Scholar]
- Beier D, Wischhusen J, Dietmaier W, Hau P, Proescholdt M, Brawanski A, Bogdahn U, Beier CP (2008) CD133 expression and cancer stem cells predict prognosis in high-grade oligodendroglial tumors. Brain Pathol 18: 370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G (2007) Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol 4: 76–84 [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM (2007) MicroRNA functions. Annu Rev Cell Dev Biol 23: 175–205 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6: 857–866 [DOI] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of microRNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136: 642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65: 6029–6033 [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 103: 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM (2009) Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 10: 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaloy C, Liu L, Lee JA, Su H, Shen F, Yang GY, Young WL, Ivey KN, Gao FB (2010) MicroRNA-9 coordinates proliferation and migration of human embryonic stem cell-derived neural progenitors. Cell Stem Cell 6: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easow G, Teleman AA, Cohen SM (2007) Isolation of microRNA targets by miRNP immunopurification. RNA 13: 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269 [DOI] [PubMed] [Google Scholar]
- Finkler A, Ashery-Padan R, Fromm H (2007) CAMTAs: calmodulin-binding transcription activators from plants to human. FEBS Lett 581: 3893–3898 [DOI] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM (2009) MicroRNAs in cancer. Annu Rev Med 60: 167–179 [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Rich JN (2007) Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer 7: 733–736 [DOI] [PubMed] [Google Scholar]
- Han J, Gong P, Reddig K, Mitra M, Guo P, Li HS (2006) The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 127: 847–858 [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich KO, Bauer T, Schulte J, Ehemann V, Deubzer H, Gogolin S, Muth D, Fischer M, Benner A, Konig R, Schwab M, Westermann F (2011) CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res 71: 3142–3151 [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G (2007) Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110 [DOI] [PubMed] [Google Scholar]
- Hurteau GJ, Spivack SD, Brock GJ (2006) Potential mRNA degradation targets of hsa-miR-200c, identified using informatics and qRT-PCR. Cell Cycle 5: 1951–1956 [DOI] [PubMed] [Google Scholar]
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, Holland EC (2009) The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 23: 1327–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse JT, Phillips HS, Brennan CW (2011) Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia 59: 1190–1199 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9: 22–32 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Vogazianou AP, Liu L, Pearson DM, Backlund LM, Plant K, Baird K, Langford CF, Gregory SG, Collins VP (2008) 1p36 is a preferential target of chromosome 1 deletions in astrocytic tumours and homozygously deleted in a subset of glioblastomas. Oncogene 27: 2097–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L (2009) MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS ONE 4: e6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ (2007) A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA 104: 19291–19296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Huang W, Park R, Park PJ, Johnson MD (2011) A developmental taxonomy of glioblastoma defined and maintained by microRNAs. Cancer Res 71: 3387–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W (2010) The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Leisegang M, Engels B, Meyerhuber P, Kieback E, Sommermeyer D, Xue SA, Reuss S, Stauss H, Uckert W (2008) Enhanced functionality of T cell receptor-redirected T cells is defined by the transgene cassette. J Mol Med 86: 573–583 [DOI] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, Wiggins JF, Bader AG, Fagin R, Brown D, Tang DG (2011) The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med 17: 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lobo NA, Shimono Y, Qian D, Clarke MF (2007) The biology of cancer stem cells. Annu Rev Cell Dev Biol 23: 675–699 [DOI] [PubMed] [Google Scholar]
- Lottaz C, Beier D, Meyer K, Kumar P, Hermann A, Schwarz J, Junker M, Oefner PJ, Bogdahn U, Wischhusen J, Spang R, Storch A, Beier CP (2010) Transcriptional profiles of CD133+ and CD133− glioblastoma-derived cancer stem cell lines suggest different cells of origin. Cancer Res 70: 2030–2040 [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, Westermann F, Speleman F, Vandesompele J, Weinberg RA (2010) miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol 12: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K (2009) Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 7: 157–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G (2010) Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol 20: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Fredlund E, Pattyn F, Schulte JH, Muth D, Vermeulen J, Kumps C, Schlierf S, De Preter K, Van Roy N, Noguera R, Laureys G, Schramm A, Eggert A, Westermann F, Speleman F, Vandesompele J (2010) MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene 29: 1394–1404 [DOI] [PubMed] [Google Scholar]
- Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H, Schlosberg A, Kuker H, Sion-Vardy N, Tobar A, Kharenko O, Sitbon E, Lithwick Yanai G, Elyakim E, Cholakh H, Gibori H, Spector Y, Bentwich Z, Barshack I, Rosenfeld N (2009) MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol 19: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Baldwin DA, Kloosterman WP, Kauppinen S, Plasterk RH, Mourelatos Z (2006) RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 12: 187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 28: 14341–14346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters L, Meister G (2007) Argonaute proteins: mediators of RNA silencing. Mol Cell 26: 611–623 [DOI] [PubMed] [Google Scholar]
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9: 157–173 [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414: 105–111 [DOI] [PubMed] [Google Scholar]
- Rudel S, Flatley A, Weinmann L, Kremmer E, Meister G (2008) A multifunctional human Argonaute2-specific monoclonal antibody. RNA 14: 1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF (2009) Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN (2006) The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125: 453–466 [DOI] [PubMed] [Google Scholar]
- Tabatabai G, Weller M (2011) Glioblastoma stem cells. Cell Tissue Res 343: 459–465 [DOI] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas (TCGA) Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS et al. (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17: 98–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely BA, Eichelbaum EJ, Alli AA, Sun Y, Gower WR Jr, Vesely DL (2007) Four cardiac hormones eliminate 4-fold more human glioblastoma cells than the green mamba snake peptide. Cancer Lett 254: 94–101 [DOI] [PubMed] [Google Scholar]
- Vesely BA, Song S, Sanchez-Ramos J, Fitz SR, Solivan SM, Gower WR Jr, Vesely DL (2005) Four peptide hormones decrease the number of human breast adenocarcinoma cells. Eur J Clin Invest 35: 60–69 [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 8: 755–768 [DOI] [PubMed] [Google Scholar]
- Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G (2009) Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell 136: 496–507 [DOI] [PubMed] [Google Scholar]
- Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, Chen CZ, Cleary ML (2010) The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res 70: 3833–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wubbenhorst D, Dumler K, Wagner B, Wexel G, Imhoff A, Gansbacher B, Vogt S, Anton M (2010) Tetracycline-regulated bone morphogenetic protein 2 gene expression in lentivirally transduced primary rabbit chondrocytes for treatment of cartilage defects. Arthritis Rheum 62: 2037–2046 [DOI] [PubMed] [Google Scholar]
- Yoo AS, Staahl BT, Chen L, Crabtree GR (2009) MicroRNA-mediated switching of chromatin-remodelling complexes in neural development. Nature 460: 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131: 1109–1123 [DOI] [PubMed] [Google Scholar]
- Zhao C, Sun G, Li S, Shi Y (2009) A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat Struct Mol Biol 16: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S, Ge W (2011) MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol (DOI:10.1007/s12032-011-9975-z) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.