Abstract
ADAR2 catalyses the deamination of adenosine to inosine at the GluR2 Q/R site in the pre-mRNA encoding the critical subunit of AMPA receptors. Among ADAR2 substrates this is the vital one as editing at this position is indispensable for normal brain function. However, the regulation of ADAR2 post-translationally remains to be elucidated. We demonstrate that the phosphorylation-dependent prolyl-isomerase Pin1 interacts with ADAR2 and is a positive regulator required for the nuclear localization and stability of ADAR2. Pin1−/− mouse embryonic fibroblasts show mislocalization of ADAR2 in the cytoplasm and reduced editing at the GluR2 Q/R and R/G sites. The E3 ubiquitin ligase WWP2 plays a negative role by binding to ADAR2 and catalysing its ubiquitination and subsequent degradation. Therefore, ADAR2 protein levels and catalytic activity are coordinately regulated in a positive manner by Pin1 and negatively by WWP2 and this may have downstream effects on the function of GluR2. Pin1 and WWP2 also regulate the large subunit of RNA Pol II, so these proteins may also coordinately regulate other key cellular proteins.
Keywords: ADAR2, GluR2, Pin1, RNA editing, WWP2
Introduction
The AMPA class of glutamate-gated ion channel receptors (GluR) are impermeable to calcium if a GluR2 subunit is present in the tetrameric receptor (Hollmann et al, 1991; Verdoorn et al, 1991). This impermeability to calcium results from RNA editing of the GluR2 transcript. The enzyme that catalyses this RNA editing event is a member of the family of adenosine deaminases that act on RNA (ADARs). ADAR2 specifically deaminates an adenosine residue in a glutamine (Q) codon to an inosine that is read as guanosine by reverse transcriptase and the translational machinery. ADAR2 converts the glutamine (Q) codon to an arginine (R) codon with 100% efficiency at the GluR2 Q/R site changing a key residue in the ion channel pore and rendering AMPA receptors assembled with this subunit impermeable to calcium (Sommer et al, 1991). The editing event also regulates AMPA receptor assembly, slowing the passage of the GluR2 subunit through the ER thus ensuring correct receptor assembly (Greger et al, 2003). Failure of RNA editing at this site can lead to neuronal cell death due to the influx of calcium (Higuchi et al, 2000). A decrease in editing at this site has been reported in sporadic ALS motor neurons (Kawahara et al, 2004) and in hippocampal neurons following transient forebrain ischaemia in a rat model of stroke (Peng et al, 2006).
Mice that are null mutants for ADAR2 are seizure-prone and die within 3 weeks after birth (Higuchi et al, 2000). Lethality in these Adar2−/− mice can be rescued by knocking-in the edited isoform of GluR2 (GluR2R). This experiment suggests that despite ADAR2 having other transcripts that it edits, the critical site is the Q/R site in GluR2 transcripts. These rescued mice have a normal phenotype, suggesting that the unedited GluR2 isoform does not have an essential biological function.
For this deamination event to occur, ADAR2 must recognize and bind to double-stranded (ds)RNA that is formed at the editing site between the edited exon and the downstream intron (Higuchi et al, 1993). Identified transcripts edited specifically by ADAR2 are mostly expressed in the CNS even though the protein is also expressed in other tissues. RNA editing occurs before splicing and ADAR2 localizes to the nucleus. In some cells, ADAR2 accumulates within the nucleolus (Desterro et al, 2003; Sansam et al, 2003); however, this localization is dynamic. When transcripts that can be edited are overexpressed in these cells, ADAR2 relocalizes to the nucleoplasm (Desterro et al, 2003).
Until now the only regulator found to influence ADAR2 expression is CREB, which can induce ADAR2 expression in hippocampal CA1 neurons in rat brain (Peng et al, 2006). In this study, we demonstrate that ADAR2 is dynamically regulated post-translationally by the phosphorylation-dependent peptidyl-prolyl cis/trans isomerase Pin1 (peptidyl-prolyl isomerase NIMA interacting protein 1). Pin1 binds to a phosphorylated serine or threonine residue preceding a proline residue and catalyses the cis/trans isomerization of the peptide bond (Lu et al, 1999). This conformational change can have a range of consequences on the function of target proteins, altering catalytic activity, stability or subcellular localization (for review see Lu and Zhou, 2007). Pin1 binds to the amino-terminus of ADAR2 in a phosphorylation-dependent manner. In the absence of Pin1, ADAR2 protein is more labile and is mislocalized to the cytoplasm, where it is unable to edit pre-mRNAs and there is a decrease in editing of the Q/R and R/G sites in endogenous GluR2 transcripts. Pin1 is therefore a positive regulator of ADAR2 editing activity.
We also identify a negative regulator of ADAR2 activity, which is WWP2; a HECT (homologous to the E6-AP C terminus) E3 ubiquitin ligase (Pirozzi et al, 1997). WWP2 binds to a conserved PPxY motif in ADAR2 and this interaction results in ubiquitination and subsequent degradation of ADAR2. An increase in the expression of WWP2 results in a decrease in ADAR2 protein level. This report of the post-translational regulation of ADAR2 demonstrates how RNA editing activity is controlled by coordinate action of two regulators.
Results
Phosphorylation sites near the N-terminus of ADAR2
When human ADAR2 was purified to homogeneity from HeLa cells, enzymatic activity was very labile (O'Connell et al, 1997). However, recombinant human ADAR2 protein purified after overexpression in the yeast Pichia pastoris is active and stable. To determine if the protein is regulated by post-translational modification, we performed mass spectrometry on recombinant ADAR2 purified from P. pastoris and identified two phosphorylated serines near the amino-terminus, serine (S) 26 and S31 (Supplementary Figure S1). Phosphorylation at S26 has been independently verified (Dephoure et al, 2008). The amino-terminal region of ADAR2 is of interest since it has been shown to be important for dimerization of the protein and autoinhibition of catalytic activity (Gallo et al, 2003; Macbeth et al, 2004).
ADAR2 interacts with Pin1
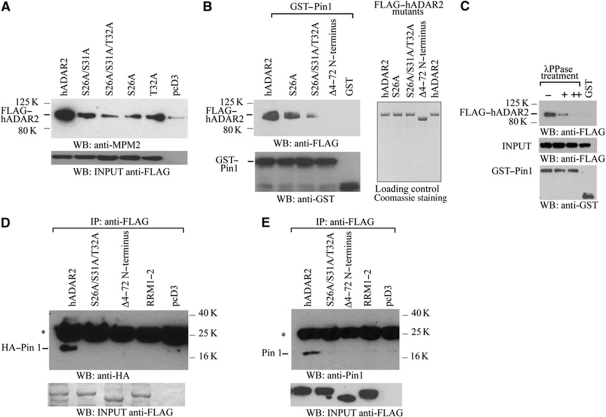
The phosphorylated residues near the N-terminus of ADAR2 are within potential recognition motifs (Ser/Thr-Pro) for the phosphorylation-dependent peptidyl-prolyl cis/trans isomerase Pin1, a well-conserved and extremely efficient enzyme for transducing post-translational modifications into conformational changes in key cellular proteins (Lu and Zhou, 2007). To determine whether Pin1 interacts with ADAR2, HEK293T cells were transiently transfected with a construct expressing ADAR2 bearing a FLAG epitope tag at the N-terminus and tetra-His tag at the C-terminus. After 24 h, the cells were harvested, whole cell protein extracts were immunoprecipitated with anti-FLAG monoclonal antibody and analysed by immunoblot detection of the immunoprecipitate with mouse anti-mitotic phosphoprotein monoclonal-2 (MPM-2) antibody (Davis et al, 1983), that recognizes the phosphorylated Pin1 motif (Ser/Thr-Pro) in proteins. As shown in Figure 1A, α-MPM-2 recognizes the FLAG-tagged ADAR2 protein. We mutated T32, as this is the residue that precedes the proline so it may be important for Pin1 binding. When the immunoprecipitation was repeated with alanine (A) substitutions for S26, S26/31 or T32 at the amino-terminus, the antibody recognized ADAR2 less efficiently and loss of binding of the MPM-2 antibody was particularly evident with the triple mutant ADAR2S26A/S31A/T32A (Figure 1A), suggesting that the amino-terminus of ADAR2 harbours phosphorylated S/T-P sites at the amino-terminus that are likely to bind Pin1.
Figure 1.
The amino-terminus of ADAR2 harbours a Pin1-binding site. (A) The anti-MPM-2 antibody recognizes potential Pin1 sites in ADAR2 purified after overexpression in P. pastoris. Immunoblot analysis with anti-MPM-2 antibody of anti-FLAG immunoprecipitates from lysates of HEK293T cells transfected with FLAG-tagged hADAR2, ADAR2S26A/S31A, ADAR2S26A/S31A/T32A, ADAR2S26A, ADAR2T32A or pcD3. The minor band in the lane with pcD3 is contamination from the neighbouring lane. ADAR input visualized with anti-FLAG antibody, lower panel. (B) Purified ADAR2 binds in vitro to Pin1 immobilized on beads. (Upper left panel) Immunoblot analysis with anti-FLAG antibody of the binding of FLAG-tagged ADAR2, ADAR2S26A, ADAR2S26A/S31A/T32A, ADAR2Δ4–72 bound to GST–Pin1 or GST on glutathione beads. (Lower panel) GST input visualized with anti-GST antibody. (Right panel) Purified ADAR proteins stained with Coomassie. (C) Binding of purified ADAR2 to Pin1 depends on phosphorylation of Pin1 sites on ADAR2. λ phosphatase treatment of lysate from HEK293T cells transfected with ADAR2 for 0 (−), 2 h (+), 3 h (++) prior to incubation with GST–Pin1. Immunoblot analysis of ADAR2 with anti-FLAG antibody. Middle and lower panels are input loading controls. (D) Pin1 binds to ADAR2 in HEK293T cells. Coimmunoprecipitation of ADAR2 and Pin1 performed with anti-FLAG antibody on HEK293T cell lysate cotransfected with HA–Pin1 and either FLAG-tagged ADAR2, ADAR2Δ4–72, ADAR2RRM1–2, ADAR2S26A/S31A/T32A or pcD3. HA–Pin1 was detected with anti-HA antibody. Asterisks represent IgG light chain. (Lower panel) Immunoblot of input proteins with anti-FLAG antibody. (E) Endogenous Pin1 detected with anti-Pin1 antibody after immunoprecipitation with anti-FLAG antibody from cell lysates of HEK293T cells transfected with FLAG-tagged ADAR2, ADAR2S26A/S31A/T32A, ADAR2Δ4–72, ADAR2RRM1–2 or pcD3. (Lower panel) Immunoblot of input proteins detected with anti-FLAG antibody. Asterisks represent IgG light chain.
The ability of ADAR2 to bind to Pin1 was next evaluated by in vitro binding assays with GST–Pin1 and recombinant ADAR2 purified from P. pastoris. As shown in Figure 1B (left panel), ADAR2 binds strongly to GST–Pin1 whereas ADAR2 did not interact with the GST beads alone. To map the interaction between Pin1 and ADAR2, ADAR2S26A, ADAR2S26A/S31A/T32A or an N-terminal deletion of ADAR2 from amino acid to 4–72 (Wong et al, 2003) were purified from P. pastoris (Figure 1B, right panel) and similarly tested for interaction with GST–Pin1. The interaction of GST–Pin1 with ADARS26A was slightly weaker than with wild-type ADAR2 and interaction was drastically decreased with the triple mutant ADAR2S26A/S31A/T32A, and totally absent with ADAR2Δ4–72 (Figure 1B, left panel). To determine if the interaction with Pin1 depends on ADAR2 phosphorylation, a transient transfection of ADAR2 into HEK293T cells was performed and the lysate was treated with λ phosphatase followed by a pull-down assay with GST–Pin1 beads. The interaction between ADAR2 and Pin1 was observed and this was abolished with a longer λ phosphatase treatment (Figure 1C).
As these experiments were performed in vitro, we then analysed the Pin1 ADAR2 interaction in HEK293T cells by transiently cotransfecting with constructs expressing FLAG-tagged ADAR2 and HA-tagged Pin1. The cells were harvested after 24 h and an immunoprecipitation of the lysate was performed with anti-FLAG monoclonal antibody and the precipitate was detected on an immunoblot with an anti-HA antibody (Figure 1D). Only the wild-type ADAR2 interacted with HA-tagged Pin1. Neither the triple alanine mutant nor ADAR2Δ4–72 interacted with Pin1. In addition, ADAR2 that has mutations in both RNA-binding domains and cannot bind to dsRNA (ADAR2RRM1–2) (Valente and Nishikura, 2007) does not interact with Pin1. Therefore, ADAR2 has to bind to RNA before it can interact with Pin1. In the in vitro binding assays with GST–Pin1 and recombinant ADAR2 purified from P. pastoris (Figure 1B), ADAR2 appears to interact with GST–Pin1 in the absence of dsRNA. However in our experience, it is difficult to eliminate all the dsRNA present in the purified protein fraction from yeast (Gallo et al, 2003) so therefore we presume that this in vitro reaction is also mediated by dsRNA. Similar results were obtained when HEK293T cells were transiently transfected with FLAG–ADAR2 followed by coimmunoprecipitation with endogenous Pin1 (Figure 1E). These results demonstrate that Pin1 binds to ADAR2 in a phosphorylation-dependent manner and that this interaction occurs at the amino-terminal of ADAR2 after it has bound to RNA.
Pin1 expression is required for optimal editing at the GluR2 Q/R site
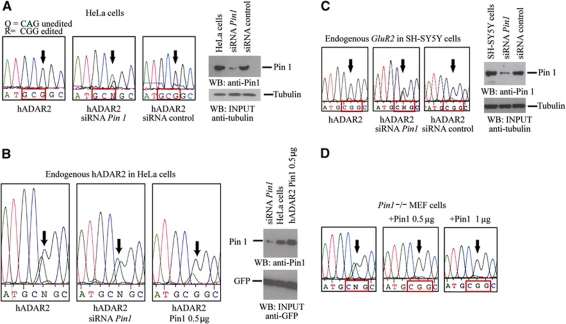
Since ADAR2 converts a glutamine (Q) codon to an arginine (R) codon with 100% efficiency at the GluR2 Q/R site in neurons, the important question is whether the interaction between Pin1 and ADAR2 affects editing activity at the critical GluR2 Q/R site. To address this point, we analysed editing of the GluR2 Q/R site in HeLa cells. To increase the level of editing at the Q/R site by ADAR2 in HeLa cells, we transiently cotransfected a plasmid encoding ADAR2 with the GluR2 B13 minigene. The level of editing rose to 100%. We then cotransfected an siRNA specific for Pin1 and editing fell to 53% (Figure 2A).
Figure 2.
Pin1 is required for efficient editing at the GluR2 Q/R site. (A) DNA sequence chromatograph of the RT–PCR product of the region encompassing the Q/R site (arrow) encoded by the GluR2 B13 minigene transiently cotransfected with ADAR2 (2 μg) in HeLa cells, editing is 100% (left chromatograph). Editing of the Q/R site drops to 53% when an siRNA specific for Pin1 is cotransfected together with plasmids encoding both ADAR2 and the GluR2 B13 minigene (middle chromatograph). Editing is 100% at the GluR2 Q/R site when a control siRNA is cotransfected (right chromatograph). Immunoblot analysis of cell lysate from HeLa cells with either anti-Pin1 or anti-tubulin antibodies (right panel). (B) (Left panel) Sequencing chromatogram of editing by endogenous ADAR2 at the Q/R site of RT–PCR product pools from the GluR2 B13 minigene transcript that has been transiently transfected into HeLa cells. Arrows indicate Q/R editing site in all panels. Immunoblot analysis with anti-Pin1 antibody of HeLa cell extracts that have been cotransfected with GFP in the presence of either Pin1-specific siRNA, no siRNA or HA–Pin1 construct (0.5 μg). Proteins are detected with anti-Pin1 antibody and anti-GFP antibody as a loading control (right panel). (C) Chromatograph of editing of endogenous GluR2 transcript at the Q/R site in neuroblastoma SH-SY5Y cells transfected with ADAR2 (2 μg). Editing is 100% at the Q/R site (left chromatograph). A decrease in editing is observed when an siRNA specific for Pin1 was cotransfected (middle chromatograph). A control siRNA does not affect editing when transfected (right chromatograph). Arrows indicate the Q/R site. Cell lysates of SH-SY5Y cells were analysed by immunoblot with either anti-Pin1 or anti-tubulin (right panel). (D) Chromatograph of editing at the Q/R site of GluR2 B13 minigene transcript in Pin1−/− MEF cells transfected with ADAR2 (2 μg). Editing increased to 100% when Pin1−/− MEF cells were cotransfected with either 0.5 or 1 μg of a construct expressing Pin1. An arrow marks the Q/R editing site in all the chromatographs.
We also analysed editing at the Q/R site in the GluR2 B13 minigene transcript (Higuchi et al, 1993) by endogenous ADAR2 and observed it was 60% efficient in this cell line; however, when siRNA specific for Pin1 was cotransfected, the level of editing fell to 46% (Figure 2B). Editing was restored to 74% when Pin1 was overexpressed in HeLa cells.
We also examined the effect of reducing Pin1 expression on editing of endogenously expressed GluR2 transcript in a neuroblastoma cell line, SH-SY5Y (Figure 2C). In this cell line, ADAR2 was cotransfected with either a Pin1-specific siRNA or a control siRNA and editing of the endogenous GluR2 transcript was analysed. Again the level of editing dropped from 100% to ∼60% at the Q/R site when there was a reduction in Pin1 expression. We also analysed editing at the R/G site in the GluR2 transcript and found it was 69% but dropped to 45% when siRNA specific for Pin1 was cotransfected whereas editing was 73% when a control siRNA was cotransfected. The reduction in Pin1 expression for this experiment is shown in Supplementary Figure S2.
To examine the effect of complete Pin1 elimination, we cotransfected constructs expressing the GluR2 B13 minigene and ADAR2 into an immortalized mouse fibroblast cell line derived from Pin1−/− mice (Figure 2D) (Fujimori et al, 1999). The editing activity at the Q/R site was ∼50% and increased to 100% when a construct expressing Pin1 was reintroduced in these cells. All these experiments strongly suggest that ADAR2 requires Pin1 for maximal editing of the critical Q/Rand R/G sites in GluR2 transcripts.
Pin1 has a role in the nuclear localization of ADAR2
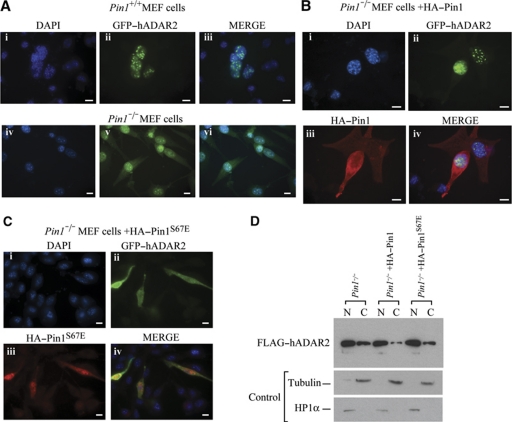
Pin1 has many diverse activities within the cell and it can alter the cellular localization of its substrate, as occurs with β-catenin (Ryo et al, 2001). Although ADAR2 has been documented as nuclear, recent evidence demonstrated that in human motor neurons in spinal cord sections, ADAR2 is both nuclear and cytoplasmic (Aizawa et al, 2010). Interestingly, a deletion of the amino-terminal residues 4–72 renders ADAR2 cytoplasmic (Wong et al, 2003) and it has also been demonstrated that this region is required for nuclear localization as it contains a non-canonical NLS within the first 64 amino acids (Desterro et al, 2003). As this deletion removes the Pin1-binding site, we wondered if preventing Pin1 binding also leads to mislocalization of ADAR2. To elucidate this we transiently transfected GFP-tagged ADAR2 into Pin1+/+ and Pin1−/− MEF cells and performed immunofluorescence detection of ADAR2 (Figure 3A). In the absence of Pin1, wild-type ADAR2 is mislocalized in the cytoplasm (Figure 3A, lower panel). Mislocalization of ADAR2 is confirmed when nuclear and cytoplasmic fractionation is performed on Pin1−/− MEF cells transiently transfected with FLAG-tagged ADAR2 (Figure 3D). When Pin1 was reintroduced into these cells, the level of ADAR2 in the cytoplasm was significantly reduced (Figure 3B and D). This effect of Pin1 on the localization of ADAR2 requires Pin1 prolyl-isomerase enzymatic activity as a Pin1S67E mutant that is catalytically inactive was unable to restore ADAR2 localization to the nucleus (Figure 3C). GFP–ADAR2 is localized to the nucleus when Pin1 is present; however, cytoplasmic localization of GFP–ADAR2 increases following cotransfection with catalytically inactive Pin1. Increased FLAG–ADAR2 is also observed in the cytoplasmic fraction of Pin1−/− MEF cells transiently transfected with FLAG-tagged ADAR2 (Figure 3D).
Figure 3.
Pin1 is required for nuclear localization of ADAR2. (A) ADAR2 is mislocalized from the nucleus to the cytoplasm in Pin1−/− MEF cells. GFP–ADAR2 immunofluorescence in Pin1+/+ and Pin1−/− MEF cells cotransfected with GluR2 B13 minigene and GFP–ADAR2. DAPI staining of nuclei (i, iv), GFP fluorescence of cell (ii, v) and merged (iii, vi). (B) Nuclear localization of ADAR2 is restored in Pin1−/− MEF cells by transfection of HA–Pin1. GFP–ADAR2 (green) direct and HA–Pin1 (red) indirect immunofluorescence detection in Pin1−/− MEF cells cotransfected with GluR2 B13 minigene and (ii) GFP–ADAR2 (green) and (iii) HA–Pin (red). (i) DAPI staining of nuclei. (iv) Merge of all three images. (C) Nuclear localization of ADAR2 depends on catalytic activity of Pin1. GFP–ADAR2 (green) and HA–Pin1S67E (red) in Pin1−/− MEF cells cotransfected with GluR2 B13 minigene and (ii) GFP–ADAR2 (green) and (iii) HA–Pin1S67E (red). (i) DAPI staining of nuclei. (iv) Merge of all three images. All photographs were taken at the same exposure. Scale bar, 10 μm. (D) Nucleo-cytoplasmic fractionation. Immunoblot analysis with anti-FLAG antibody of nuclear and cytoplasmic fractions of Pin1−/− MEF cells transfected with FLAG–ADAR2 (lanes 1 and 2). Pin1 was cotransfected with FLAG–ADAR2 in Pin1−/− MEF cells (lanes 3 and 4). HA–Pin1S67E was cotransfected in Pin1−/− MEF cells (lanes 5 and 6). (Lower panel) Immunoblot of fractionated Pin1−/− MEF cells with tubulin as a marker for cytoplasmic fraction and HP1α for nuclear fraction.
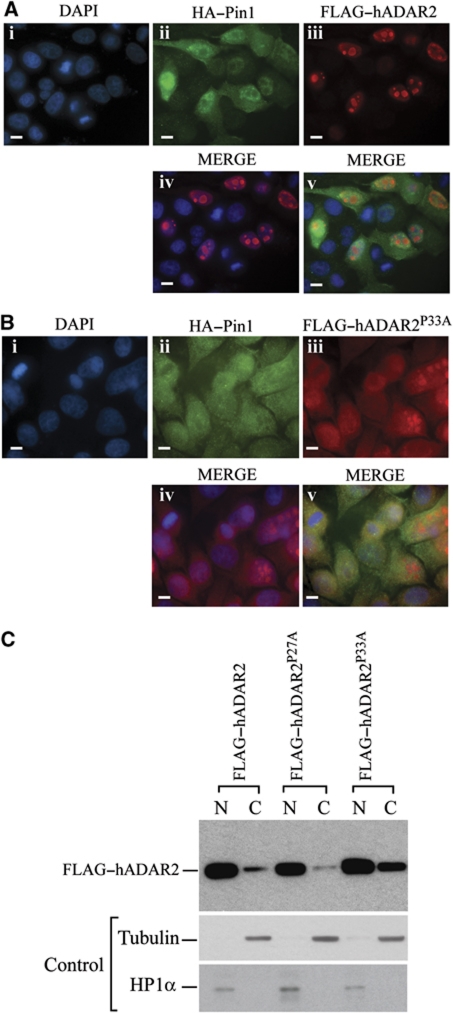
As Pin1 recognizes a phosphorylated serine or threonine preceding a proline, we replaced the phosphorylated amino acids as well as the prolines with alanine to determine if all were required for nuclear localization. As expected, the triple mutant FLAG–ADAR2S26/S31A/T32A was present in the cytoplasm (Supplementary Figure S3) and this appears slightly different to ADAR2Δ4–72 that is more localized around the nuclear periphery (Supplementary Figure S4). When the proline mutants were generated; FLAG–ADAR2P27A and FLAG–ADAR2P33A, were transiently transfected into HeLa cells together with HA–Pin1 (Figure 4), FLAG–ADAR2P33A was present in the cytoplasm as detected by immunofluorescence as well as by nuclear and cytoplasmic fractionation (Figure 4B and C) whereas FLAG–ADAR2P27A is nuclear. This implies that the second proline is the critical one, thus the phosphorylation of T32 may be the critical site for Pin1 binding and P33 for isomerization. Notably, this is also the most conserved Pin1 site in vertebrate ADAR2 sequences (Supplementary Figure S1).
Figure 4.
Proline 33 of ADAR2 is required for the Pin1 effect on ADAR2 nuclear localization. (A) Normal localization of ADAR2 and Pin1. Immunofluorescence of HeLa cells cotransfected with GluR2 B13 minigene, FLAG–ADAR2 and HA–Pin1 stained with (i) DAPI, (ii) anti-HA–Pin1 (green), (iii) anti-FLAG–ADAR2 (red), (iv) Merge of DAP1 and FLAG and (v) merge of all three images. (B) Nuclear localization of ADAR2 depends on Proline 33. Immunofluorescence of HeLa cells cotransfected with GluR2 B13 minigene, FLAG–ADAR2P33A and HA–Pin1 stained with (i) DAPI, (ii) anti-HA–Pin1 (green), (iii) anti-FLAG–ADAR2P33A (red), (iv) Merge of DAP1 and FLAG and (v) merge of all three images. All photographs were taken at the same exposure. Scale bar, 10 μm. (C) Nucleo-cytoplasmic fractionation of wild-type and ADARP27A and ADAR2P33A mutants. Immunoblot analysis with anti-FLAG antibody of nuclear and cytoplasmic fractions of HeLa cells transfected with FLAG–ADAR2 (lanes 1 and 2), ADARP27A (lanes 3 and 4), ADAR2P33A (lanes 5 and 6). (Lower panels) Immunoblot of fractionated HeLa cells with tubulin as a cytoplasmic marker and HP1α as a nuclear marker.
Pin1 stabilizes ADAR2
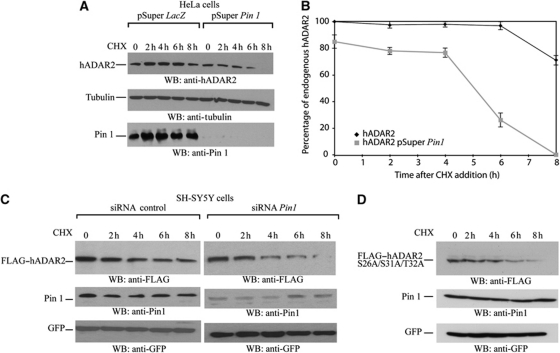
We wanted to elucidate if Pin1 had other effects on ADAR2. As Pin1 has been shown to influence the stability of proteins such as β-catenin (Ryo et al, 2001), NF-κB (Ryo et al, 2003) and p53 (Zacchi et al, 2002; Zheng et al, 2002), we wondered if Pin1 also influences the stability of ADAR2. The level of Pin1 was reduced in HeLa cells by transfecting either pSuper Pin1 siRNA (Rustighi et al, 2009) or a control pSuper LacZ siRNA so that the level of endogenous ADAR2 could be analysed (Figure 5A). A similar experiment was performed in the neuroblastoma cell line SH-SY5Y (Figure 5C). Twenty-four hours after transfection, cycloheximide was added to prevent further protein synthesis, a time course from 0 to 8 h was performed to chase the decay of ADAR2 protein and the samples were analysed by immunoblot analysis to determine ADAR2 levels. In both cell lines, the protein level of ADAR2 decreased when cycloheximide was added; however, the decrease was more dramatic when Pin1 expression was reduced (Figure 5A–C). The stability of the triple mutant ADAR2S26A/S31A/T32A was also analysed after cycloheximide treatment (Figure 5D). As predicted this mutant protein was unstable as it could no longer interact with Pin1. Therefore, Pin1 affects the stability of ADAR2.
Figure 5.
Pin1 contributes to stability of ADAR2 protein. (A) Knockdown of Pin1 in HeLa cells destabilizes ADAR2 in a cycloheximide time course. HeLa cells were transfected with either pSuper LacZ or pSuper Pin1. Cycloheximide (50 μg/μl) was added to both and a time course from 0 to 8 h was performed. Cell lysates were analysed by immunoblot and the antibodies used were anti-ADAR2 (top panel), anti-tubulin as a loading control (middle panel) and Pin1 (bottom panel). (B) Quantification of (A). (C) Pin1 knockdown destabilization of FLAG–ADAR2 in SH-SY5Y neuroblastoma cells. SH-SY5Y cells were cotransfected with FLAG-tagged ADAR2 and a control siRNA or Pin1-specific siRNA. Cycloheximide (50 μg/μl) was added to both and a time course from 0 to 8 h was performed. Cell lysates were analysed by immunoblot and the antibodies used were anti-FLAG (top panel), anti-Pin1 (middle panel) and GFP as loading control (bottom panel). (D) ADAR2 mutant in the Pin1-binding site is less stable. SH-SY5Y neuroblastoma cells were transfected with FLAG-tagged ADAR2S26A/S31A/T32A and cycloheximide (50 μg/μl) was added and a time course was performed from 0 to 8 h. Cell lysates were analysed by immunoblot and the antibodies used were anti-FLAG (top panel), anti-Pin1 (middle panel) and GFP as loading control (bottom panel).
The E3 ubiquitin ligase WWP2 interacts with ADAR2
Mass spectrometry was performed on the original samples of ADAR2 purified from large quantities of HeLa cell nuclear fractions (O'Connell et al, 1997) and one of the proteins that copurified with ADAR2 was WWP2, an E3 ubiquitin ligase containing four WW domains as well as a HECT domain. As ADAR2 was unstable in the absence of Pin1, we wondered if this E3 ligase was involved. WWP2 can bind directly to a PPxY motif within its substrate. Analysis of the ADAR2 amino-acid sequence revealed that this motif was present twice, at the amino-terminus and carboxyl-terminus of ADAR2 and was highly conserved (Supplementary Figure S5).
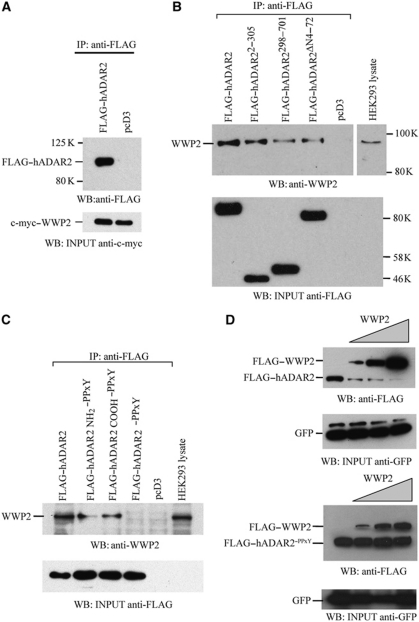
To demonstrate that ADAR2 and WWP2 interact, HEK293T cells were transiently cotransfected with constructs expressing FLAG–ADAR2 and WWP2 with a c-myc epitope tag at its amino-terminus. An immunoprecipitation was performed with anti-FLAG monoclonal antibody to precipitate FLAG–ADAR2 and c-myc–WWP2 was present in this precipitate (Figure 6A). To determine which motif in ADAR2 WWP2 binds to, transient transfections were performed in HEK293T cells with constructs expressing full-length and truncated forms of ADAR2 all with FLAG epitope at their amino-terminus. Immunoprecipitation with anti-FLAG monoclonal antibody followed by immunoblot detection with an anti-WWP2 antibody showed that endogenous WWP2 interacts best with full-length ADAR2 and with a truncated protein containing the amino-terminus of ADAR2 (Figure 6B). The interaction of WWP2 with ADAR2 appears to be weaker with the site in the deaminase domain or when the amino-terminus of ADAR2 was deleted. We repeated this immunoprecipitation with anti-FLAG with mutants of ADAR2 in which the binding site for WWP2 in the amino-terminus, carboxyl-terminus or a combination of both binding sites were mutated (Figure 6C). The single mutants decrease the interaction between ADAR2 and WWP2 whereas the interaction was completely abolished with the double mutant.
Figure 6.
WWP2 interacts with ADAR2. (A) Coimmunoprecipitation of ADAR2 and WWP2 performed with anti-FLAG antibody in HEK293T cell lysate cotransfected with FLAG–ADAR2 and c-myc–WWP2 or pcD3 empty vector. FLAG–ADAR2 was detected with FLAG antibody. WWP2 input visualized with anti-c-myc antibody (bottom panel). (B) Endogenous WWP2 detected with anti-WWP2 antibody after immunoprecipitation with anti-FLAG antibody from lysates of HEK293T cells transfected with FLAG-tagged ADAR2, ADAR22–305, ADAR2298–701, ADAR2Δ4–72 or pcD3. Immunoblot of input proteins detected with anti-FLAG antibody (bottom panel). (C) Immunoprecipitation of FLAG–ADAR2 and mutants with mutations in the amino, carboxyl binding site for WWP2 or a combination of both was performed with anti-FLAG antibody in HEK293 cells. Endogenous WWP2 detected with anti-WWP2 antibody. (Lower panel) Immunoblot of input proteins detected with anti-FLAG antibody. (D) Immunoblot analysis with anti-FLAG antibody of lysates from HeLa cells cotransfected with FLAG–ADAR2 and increasing amount of FLAG–WWP2 (0.5, 1, 2.5 μg) (upper panel) and this experiment was repeated with the mutant ADAR2PPxY that is unable to bind to WWP2 (lower panel). The efficiency of transfection was normalized to GFP expression. GFP input visualized with anti-GFP antibody is shown below both panels.
When a cotransfection was performed in HeLa cells with a constant amount of a plasmid encoding ADAR2 and an increase in the plasmid expressing WWP2, a drastic decrease in ADAR2 protein level was observed (Figure 6D). However, when this experiment was repeated with the double ADAR2–PPxY mutant then the level of the mutant protein did not change as it is no longer a substrate for WWP2. These experiments demonstrated that WWP2 can interact with ADAR2 via the PPxY motif present in ADAR2. An increased expression of WWP2 in HeLa cells resulted in a reciprocal decrease in ADAR2 levels; however, the protein level of the ADAR2–PPxY mutant unable to bind WWP2 remained stable, demonstrating that WWP2 can cause a decrease in ADAR2 protein levels.
WWP2 poly-ubiquitinates ADAR2
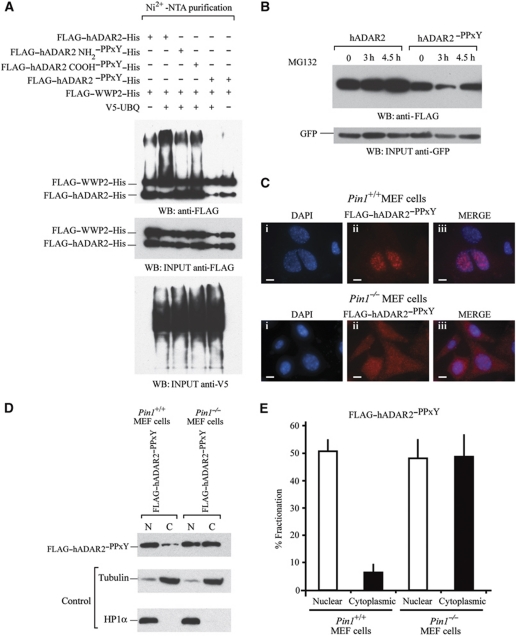
To verify that ADAR2 is indeed poly-ubiquinated by WWP2, we performed an ubiquitin assay with extract from HEK293T cells. The cells were transiently cotransfected with ADAR2 or a mutant where either single or both PPxY motifs in ADAR2 were mutated and a further construct expressing WWP2. Poly-ubiquitination of ADAR2 was detected in extracts from cells cotransfected with constructs expressing wild-type ADAR2 and WWP2 proteins but there was a decrease in poly-ubiquitination with both the ADAR2NH2–PPxY and ADAR2COOH–PPxY single mutants. Only in the presence of ADAR2–PPxY was there a complete loss of poly-ubiquitination (Figure 7A). Poly-ubiquitination was also observed in the absence of V5-UBQ as there was sufficient endogenous ubiquitin in the cell extract (Figure 7A, lane 1).
Figure 7.
WWP2 is required for ADAR2 ubiquitination and subsequent degradation in the cytoplasm. (A) In vivo ubiquitination assays. FLAG–ADAR2–His, FLAG–WWP2–His and V5-UBQ were transfected in HEK293T cells followed by purification of ubiquitination complexes from lysates with Ni2+-NTA. In lane 1, cotransfection was with FLAG–ADAR2–His, FLAG–WWP2–His. Lane 2, cotransfection was with FLAG–ADAR2–His, FLAG–WWP2–His and V5-UBQ. Lane 3, cotransfection of FLAG–ADAR2 NH2–PPxY–His, FLAG–WWP2–His and V5-UBQ. Lane 4, cotransfection of FLAG–ADAR2 COOH–PPxY–His, FLAG–WWP2–His and V5-UBQ. Lane 5, cotransfection of FLAG–ADAR2–PPxY–His (double mutant), FLAG–WWP2–His and V5-UBQ. Lane 6 is the same as lane 5 without the addition of V5-UBQ and is the negative control. (Middle panel) Immunoblot of input proteins detected with anti-FLAG antibody. (Lower panel) Immunoblot of V5-UBQ present in the purified complex detected with anti-V5 antibody. (B) Immunoblot at 24 h following transfection of FLAG–ADAR2 and FLAG–ADAR2–PPxY with 20 μM MG132 to inhibit protein degradation. The proteasomal inhibitor MG132 was added to both and a time course from 0 to 4.5 h was performed in HeLa cells. Cell lysates were normalized to GFP levels (lower panel). (C) Immunofluorescence of Pin1+/+and Pin1−/− MEF cells cotransfected with GluR2 and FLAG–ADAR2–PPxY (double mutant). (i) DAPI staining of nuclei. (ii) Anti-FLAG–ADAR2–PPxY (red). (iii) Merge of DAP1 and FLAG. Scale bar, 10 μm. All photographs were taken at the same exposure. (D) Nuclear and cytoplasmic fractionation of FLAG–ADAR2–PPxY in Pin1+/+and Pin1−/− MEF cells. (Lower panels) Immunoblot of fractionated MEF cells with tubulin as a cytoplasmic marker and HP1α as a nuclear marker. (E) Quantification of (D).
To demonstrate that the proteosome affected the stability of ADAR2, a time course was performed in the presence of the proteosome inhibitor MG132 (Figure 7B). An increase in the level of ADAR2 was observed; however, there was not a reciprocal increase in the levels of the ADAR2–PPxY mutant. This mutant could no longer bind WWP2 so the protein level could no longer be regulated by the proteasome, therefore, the proteosome inhibitor had no effect on its stability.
To determine if an increase in stability of ADAR2 would affect its localization, immunofluorescence was performed in Pin1+/+ and Pin1−/− MEF cells and were transiently transfected with ADAR2–PPxY. A cytoplasmic accumulation of ADAR2–PPxY was evident (Figure 7C). Previously it had been difficult to observe an accumulation of ADAR2 within the cytoplasm as the protein was being degraded by WWP2 (Figure 3A). However, as the binding of WWP2 is impaired in the ADAR2–PPxY mutant, the cytoplasmic accumulation is obvious. Nuclear and cytoplasmic fractionation was performed with the ADAR2–PPxY mutant in MEF wild-type and Pin1−/− cells and the results were quantified (Figure 7D and E). It is clear that in the absence of Pin1 and if WWP2 cannot bind to ADAR2, there is a cytoplasmic accumulation of ADAR2 that is not observed under normal conditions.
Discussion
These data illustrate the complex regulation of ADAR2 by two proteins with opposing effects; Pin1 is a positive regulator of ADAR2 whereas WWP2 can bind and cause degradation. Pin1 and WWP2 regulate other protein such as the large subunit of RNA pol II and this regulation is conserved from yeast to mammals (Wu et al, 2001).
When ADAR2 is phosphorylated at the amino-terminus, it becomes a substrate for the phosphorylation-dependent prolyl-isomerase Pin1. The enzymatic activity of Pin1 is required for the localization and stability of ADAR2 in the nucleus. In the absence of Pin1, ADAR2 is unstable and is present in the cytoplasm. It can then interact with WWP2, an E3 ligase that results in its poly-ubiquitination and subsequent degradation by the proteasome (Figure 8). One direct consequence of this is a reduction in editing of the Q/R site and R/G sites in GluR2 transcripts. The presence of unedited GluR2Q subunit can have dramatic downstream effects as it can increase the trafficking of GluR2 subunit to the synapse as well as increasing the permeability of AMPA receptors to calcium ions.
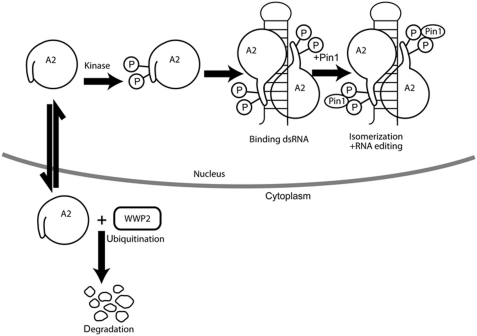
Figure 8.
Schematic representation of the regulation of ADAR2 by Pin1 and WWP2. ADAR2 can exist as a monomer in the nucleus and has sequences at its amino-terminus that inhibit enzymatic activity (Macbeth et al, 2004). However, ADAR2 can be phosphorylated by an unknown kinase either when it is free or bound to dsRNA. ADAR2 is a substrate for Pin1 once it is bound to dsRNA and the active form of ADAR2 is a dimer (Gallo et al, 2003; Poulsen et al, 2006; Valente and Nishikura, 2007). The mechanism of dimer formation is still unclear. After RNA editing has occurred, we do not know the fate of ADAR2. However, in the absence of Pin1, ADAR2 mislocalizes to the cytoplasm where it is poly-ubiquitinated by WWP2 and is degraded by the proteasome.
The function of Pin1 is to isomerize a specific proline from the cis to trans conformation or vice versa (Ranganathan et al, 1997; Yaffe et al, 1997). Most biological processes require proline to be in the trans conformation; however, when the protein is translated the choice in conformation is dependent on the surrounding amino acids. If there is a pool of ADAR2 that is not phosphorylated and therefore not a Pin1 substrate, then this protein may not be fully active. This probably does occur as ADAR2 is expressed in various mammalian cell lines such as HeLa and SH-SY5Y but is not very active and for efficient editing of transcripts, additional ADAR2 must be transfected. There may be sufficient Pin1 present but the kinase required for the phosphorylation of the Pin1-binding site may be limited. This may explain the pool of inactive ADAR2 that has been observed as it may require phosphorylation and subsequent Pin1 activity. For example in the undifferentiated NT2 cell line, ADAR2 is well expressed; however, it requires differentiation of the NT2 to neuronal cells for efficient editing of GluR2 Q/R; this occurs without any major change in ADAR2 expression (Lai et al, 1997). The authors of that study proposed that a post-translational regulatory mechanism is involved.
In the absence of Pin1, ADAR2 mislocalizes to the cytoplasm. It is difficult to detect ADAR2 as it is poly-ubiquitinated by WWP2 in the cytoplasm and degraded. Only when WWP2 is unable to bind to ADAR2 is high level of cytoplasmic accumulation of ADAR2 observed (Figure 7). There are two binding sites for WWP2 on the ADAR2 protein. The site in the amino-terminus of ADAR2 appears to be more important for WWP2 interaction than the site in the deaminase domain despite the deaminase site being more conserved. The crystal structure for the deaminase domain of ADAR2 has been solved (Macbeth et al, 2005) and the PPLY amino acids are on the outside of the protein opposite to the region that has been proposed to interact with the RNA (Supplementary Figure S6). Therefore, these amino acids are easily accessible to WWP2.
The finding that ADAR2 is regulated by Pin1 and WWP2 opens up new avenues of research. Under normal conditions, ADAR2 is present within the nucleolus (Desterro et al, 2003; Sansam et al, 2003). However, once a substrate is transfected into the cell, it relocates to the nucleus and editing can occur. One key factor that is missing is the kinase that phosphorylates ADAR2 and instigates this complex regulation. Also, we would predict from our results both with the ADAR2 triple mutant (Figure 1) and with ADAR2P33A (Figure 4) that Thr32 is phosphorylated and that this is a key phosphorylation event. Pin1 interaction requires the binding of ADAR2 to dsRNA; however, we do not know whether the kinase phosphorylates ADAR2 in the bound or unbound state. Also, we do not know what happens after RNA editing has occurred. Is ADAR2 then a substrate for a phosphatase that results in its relocation to the nucleolus or is it exported and degraded? In the absence of Pin1, ADAR2 mislocalizes to the cytoplasm where it is a substrate for WWP2; however, the molecular mechanism underlying this mislocalization is unknown. It is important to elucidate how these various factors regulate the activity of ADAR2 as this will subsequently impinge on the properties of the AMPA receptor.
This report of post-translational regulation of ADAR2 reveals how ADAR2 is highly coordinated and regulated within the cell as this ultimately controls the calcium permeability and assembly of AMPA receptors. This opposing regulation by Pin1 and WWP2 is very analogous to that of the large subunit of yeast RNA polymerase (pol) II where the yeast orthologue of Pin1; ESS1 binds to the C-terminal domain and positively regulates RNA pol II transcription whereas RSP5; a HECT-type E3 ligase similar to WWP2 mediates its ubiquitination and degradation (Wu et al, 2001). This regulation of RNA pol II large subunit by Pin1 and WWP2 is also conserved in mammals (Li et al, 2007; Xu and Manley, 2007). Therefore, we propose that these two proteins with opposing effects can act coordinately in the regulation and stability of other key cellular proteins.
The interaction of ADAR2 with Pin1 may explain why the Q/R site is edited to 100% in neurons. Pin1 is a key regulator of many proteins and processes within the cell; however, it is itself regulated by phosphorylation, which inhibits its activity (Lu et al, 2002; Lee et al, 2011). We hypothesize that as Pin1 is the hub of a regulatory network and that transient ischaemia or other insults lead to reduction in Pin1 activity. A decrease in ADAR2 activity would then ensue with a subsequent reduction in editing at the Q/R site in GluR2 transcripts. This would result in increased calcium permeability of AMPA receptors that could have major effects depending on the region of the brain and the presence of calcium-binding proteins or calcium pumps within the particular neuron. If calcium-binding proteins are low as in the CA1 pyramidal neurons, then this could lead to neuronal cell death (Liu and Zukin, 2007). Therefore, we propose 100% editing of Q/R in the GluR2 transcript is a ‘quality control’ measure that reflects a healthy neuron. Data from mice support this hypothesis as when the edited GluR2R isoform has been knocked-in, the mice have no apparent phenotype despite a lack of the unedited isoform (Kask et al, 1998). An explanation why rats expressing GluR2R are resistant to forebrain ischaemia in the vulnerable CA1 pyramidal neurons could be that the regulatory network from Pin1 to ADAR2 editing the GluR2 Q/R site has been disrupted (Liu et al, 2004). Other experiments are required to rigorously test this hypothesis; however, if it is correct it will facilitate devising treatments to limit the neuronal damage associated with forebrain ischaemia.
Materials and methods
A more detailed Materials and methods section is provided in Supplementary Data.
Mass spectrometry of ADAR2
A measure of 1 μg of ADAR2 with FLAG and tetra-histidine epitope tags was purified after overexpression in P. pastoris as previously described (Ring et al, 2004). The purified protein was denatured in Novex LDS sample buffer plus 10 mM DTT at 65°C for 30 min and alkylated with 50 mM 4-vinylpyridine for 15 min at room temperature. The protein was separated by SDS–PAGE on a 4–12% MOPS NUPAGE gel, stained with colloidal Coomasie and digested with trypsin (5 μg/ml) in 50 mM ammonium bicarbonate. The phosphopeptides were enriched with PHOS-select resin (Sigma) and analysed on a 4700 TOF–TOF mass spectrometer as described previously (Beullens et al, 2005).
ADAR2 mutagenesis
The pcD3 construct expressing FLAG–ADAR2 has been previously described (Heale et al, 2009). All mutations were generated with the QuickChange mutagenesis strategy (Stratagene, La Jolla, CA) and were sequenced to verify the intended mutations.
S26 to A: 5′-ctggacaacgtggcccccaaggatggc-3′
T32 to A: 5′-tcccccaaggatggcagcgcacctgggcctgg-3′
S26/31 to A: 5′-gtcccccaaggatggcgccacacctgggcctggcga-3′
S26/31/T32 to A: 5′-ggacaacgtggctcccaaggatggcgccgcacctgggcctg-3′
P27 to A: 5′-ctggacaacgtgtccgccaaggatggcagcaca-3′
P33 to A: 5′-cccaaggatggcagcacagctgggcctggcgagggctct-3′
ADAR2 was subcloned into pEGFP-C3 with the oligonucleotides:
EcoRI-ADAR2 5′-ccggaattctgatggatatagaagatgaagaaaacatgagt-3′
ADAR2-SalI (antisense) 5′-ccggtcgacctcagggcgtgagtgagaactggtcctgctc-3′.
WWP2 cloning and mutagenesis
WWP2 was cloned by PCR amplification of cDNA clone IMAGE 100008816 (Gene Service) in the expression vector pENTR221 with the oligonucleotides 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGGACTACAAGGACGACGATGACAAAGCATCTGCCAGCTCTAGCCGGGCA-3′ and antisense 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAATGGTGATGGTGATGGTGCTCCTGTCCAAAGCCTTCGGTCTC-3′ for subsequent gateway cloning (Invitrogen). Similarly, N- and C-terminal truncations of hADAR2 were constructed by PCR amplification of hADAR2 cDNA into the pGEM T-Easy vector with the oligonucleotides. 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGACTACAAGGACGACGATGACAAAGATATAGAAGATGAAGAAAAC-3′ and antisense 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATGGTGATGGTGATGGTGTGGCGTCTGATCCAAGTCCAA-3′ primers were used to produce a construct encoding only the N-terminal portion of hADAR2.
To produce a construct encoding the C-terminal portion of hADAR2 with the oligonucleotides 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATGGACTACAAGGACGACGATGACAAATTGCACTTGGATCAGACGCCA-3′ and antisense 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTAATGGTGATGGTGATGGTGGGGCGTGAGTGAGAACTGGTC-3′ were used. All primers were designed to incorporate 5′ FLAG (bold) and 3′ HIS epitope tags (underlined) at either end of the respective ORF, as well as the att recombination sites required for gateway cloning (italics). The purified PCR products were cloned by site-specific recombination into the donor vector pDONR221 to generate an entry clone. The entry clone was used in a second site-specific recombination reaction with the modified destination vector pcD3 (origin pcDNA3) to generate the expression clone (following the standard protocol, as described by Invitrogen).
To generate FLAG–ADAR2 NH2–PPxY (PPFY to AAFA) and FLAG–ADAR2 COOH2–PPxY (PPLY to AALA) the following oligonucleotides were used:
FLAG–ADAR2NH2–PPxY 5′-gacaaggcggcagcatttgccgtgggctcc-3′ and for FLAG–ADAR2COOH–PPxY 5′-gaggacctggcagctctcgccaccctcaac-3′
Immunoblot analysis, immunoprecipitation and GST pull down
Expression and purification of the GST-tagged proteins were performed as described (Buratti and Baralle, 2001). Immunoprecipitation was performed as described (Rustighi et al, 2009). Immunoblot analysis was performed with primary antibodies: mouse α-FLAG 1:3000 (Sigma), mouse α-HA 1:1000 (Sigma), mouse α-MPM-2 1:1000 (Upstate Cell Signaling) (Davis et al, 1983) mouse α-Pin1 1:500 (G-8) (Santa Cruz Biotechnology, Santa Cruz, CA) rabbit α-ADAR2 1:1000 (Sigma), mouse α-GST 1:5000 dilution (Amersham Pharmacia), overnight at 4°C, followed by an 1-h incubation with the appropriate secondary antibody (Dako).
Ubiquitination assay and proteasome-mediated degradation analysis
Cells were cotransfected with the indicated constructs at the following ratios: FLAG–ADAR2 and FLAG–ADAR2–PPxY 1 μg, FLAG–WWP2 and FLAG–WWP2 (C/A) 4 μg, V5-UBA 1 μg. After 24 h, cells were harvested and lysed under denaturing conditions, and ubiquitinated proteins were purified with Ni2+-NTA agarose beads (QIAGEN) as described previously (Rodriguez et al, 1999). The cells in Figure 7A were incubated for 4.5 h with 20 μM MG132 (Calbiochem), 20 μM MG5 (Sigma) prior to lysis.
Cell lines, transfection conditions and RNA extraction
MEF cells were cultured in a Hypoxic incubator, 10% CO2, 3% O2 (Thermo Scientific HeraCell 150i) (Parrinello et al, 2003). Total RNA was extracted from cells with Trizol reagent (Invitrogen) and treated with Turbo DNA-free DNAseI beads (Ambion). cDNA synthesis was performed with random-hexamer primers. PCR of the GluR2 B13 minigene was performed with primer, 5′-atggaagagaaacacaaagt-3′ that anneals to exon 11 and antisense primer 5′-gaatgataggaaccttctgc-3′ that anneals to intron 11 (Higuchi et al, 1993). PCRs conditions were 94°C for 3 min, followed by 28 cycles of: 94°C for 30 s, 54°C for 30 s, 72°C for 45 s and 72°C for 7 min. For endogenous GluR2 transcript, 1 μg of DNAse-treated total RNA was used for cDNA synthesis and RT–PCR was performed with Superscript™III One step RT–PCR System (Invitrogen), was performed with primer, 5′-atggaagagaaacacaaagt-3′ and the antisense primer 5′-ttccctttggacttccgcac-3′ that anneals to exon 13.
RNAi knockdown
siRNA or pSUPER transfections were performed in HeLa and SH-SY5S cells with Lipofectamine 2000 reagent (Invitrogen). Both siRNA against GAPDH and smart Pool of siRNAs against Pin1 (LPIN1, Dharmacon Thermo Scientific) were added to a final concentration of 100 nM. The pSUPERPin1 and pSUPERLacZ were used as siRNA controls as previously described (Rustighi et al, 2009).
Indirect immunofluorescence
Cells were plated on sterile cover-slips in six-well plates at 2.5 × 105 cells/well and grown overnight before transient transfection of expression constructs with Fugene 6 transfection reagent (Roche). Indirect immunofluorescence was performed as previously (Ayala et al, 2008).
Preparation of cytoplasmic and nuclear extracts
Cytoplasmic and nuclear fractionation was performed with ProteoExtract Subcellular Proteome Extraction Kit (Merck) according to the manufacturer's instructions. Quantification of cytoplasmic and nuclear fractionation was performed with the IMAGEQUANT/TL (GE Heathcare Life Science).
Supplementary Material
Acknowledgments
We thank W Keller for his support over many years, A Leroy for constructs, M Lusic and L Manganaro for scientific discussion, S Thore and C Nicol for figures. M Ditzl, K Nishikura and D Lazinski for reagents. This work was funded by the MRC U.1275.01.005.00001.01 to MO'C, Telethon Foundation Grant GGP07185 and AIRC to GDS.
Author contributions: RM conceived, designed and performed the majority of experiments. JB generated many of the ADAR2 reagents. SP performed some experiments with Pin1. AC performed some experiments with WWP2. SH performed some experiments with WWP2. NM performed mass spectrometry. AB generated some of the Pin1 reagents. LPK was involved in experimental design and contributed to writing the manuscript. GDS was involved in experimental design, provided reagents and contributed to writing the manuscript. MAO'C was involved in experimental design and wrote the manuscript
Footnotes
The authors declare that they have no conflict of interest.
References
- Aizawa H, Sawada J, Hideyama T, Yamashita T, Katayama T, Hasebe N, Kimura T, Yahara O, Kwak S (2010) TDP-43 pathology in sporadic ALS occurs in motor neurons lacking the RNA editing enzyme ADAR2. Acta Neuropathol 120: 75–84 [DOI] [PubMed] [Google Scholar]
- Ayala YM, Misteli T, Baralle FE (2008) TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA 105: 3785–3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beullens M, Vancauwenbergh S, Morrice N, Derua R, Ceulemans H, Waelkens E, Bollen M (2005) Substrate specificity and activity regulation of protein kinase MELK. J Biol Chem 280: 40003–40011 [DOI] [PubMed] [Google Scholar]
- Buratti E, Baralle FE (2001) Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem 276: 36337–36343 [DOI] [PubMed] [Google Scholar]
- Davis FM, Tsao TY, Fowler SK, Rao PN (1983) Monoclonal antibodies to mitotic cells. Proc Natl Acad Sci USA 80: 2926–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP (2008) A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA 105: 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Lafarga M, Berciano MT, O'Connell M, Carmo-Fonseca M (2003) Dynamic association of RNA-editing enzymes with the nucleolus. J Cell Sci 116: 1805–1818 [DOI] [PubMed] [Google Scholar]
- Fujimori F, Takahashi K, Uchida C, Uchida T (1999) Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun 265: 658–663 [DOI] [PubMed] [Google Scholar]
- Gallo A, Keegan LP, Ring GM, O'Connell MA (2003) An ADAR that edits transcripts encoding ion channel subunits functions as a dimer. EMBO J 22: 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greger IH, Khatri L, Kong X, Ziff EB (2003) AMPA receptor tetramerization is mediated by q/r editing. Neuron 40: 763–774 [DOI] [PubMed] [Google Scholar]
- Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA (2009) Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J 28: 3145–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406: 78–81 [DOI] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 75: 1361–1370 [DOI] [PubMed] [Google Scholar]
- Hollmann M, Hartley M, Heinemann S (1991) Ca2+ permeability of KA-AMPA—gated glutamate receptor channels depends on subunit composition. Science 252: 851–853 [DOI] [PubMed] [Google Scholar]
- Kask K, Zamanillo D, Rozov A, Burnashev N, Sprengel R, Seeburg PH (1998) The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function. Proc Natl Acad Sci USA 95: 13777–13782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S (2004) Glutamate receptors: RNA editing and death of motor neurons. Nature 427: 801. [DOI] [PubMed] [Google Scholar]
- Lai F, Chen CX, Lee VM, Nishikura K (1997) Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J Neurochem 69: 43–52 [DOI] [PubMed] [Google Scholar]
- Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, Lu KP (2011) Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell 42: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang Z, Wang B, Zhang J, Zhao Y, Jin Y (2007) Wwp2-mediated ubiquitination of the RNA polymerase II large subunit in mouse embryonic pluripotent stem cells. Mol Cell Biol 27: 5296–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lau L, Wei J, Zhu D, Zou S, Sun HS, Fu Y, Liu F, Lu Y (2004) Expression of Ca(2+)-permeable AMPA receptor channels primes cell death in transient forebrain ischemia. Neuron 43: 43–55 [DOI] [PubMed] [Google Scholar]
- Liu SJ, Zukin RS (2007) Ca2+-permeable AMPA receptors in synaptic plasticity and neuronal death. Trends Neurosci 30: 126–134 [DOI] [PubMed] [Google Scholar]
- Lu KP, Zhou XZ (2007) The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol 8: 904–916 [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP (2002) Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem 277: 2381–2384 [DOI] [PubMed] [Google Scholar]
- Lu PJ, Zhou XZ, Shen M, Lu KP (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283: 1325–1328 [DOI] [PubMed] [Google Scholar]
- Macbeth MR, Lingam AT, Bass BL (2004) Evidence for auto-inhibition by the N terminus of hADAR2 and activation by dsRNA binding. RNA 10: 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell MA, Gerber A, Keller W (1997) Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J Biol Chem 272: 473–478 [DOI] [PubMed] [Google Scholar]
- Parrinello S, Samper E, Krtolica A, Goldstein J, Melov S, Campisi J (2003) Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol 5: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng PL, Zhong X, Tu W, Soundarapandian MM, Molner P, Zhu D, Lau L, Liu S, Liu F, Lu Y (2006) ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron 49: 719–733 [DOI] [PubMed] [Google Scholar]
- Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM (1997) Identification of novel human WW domain-containing proteins by cloning of ligand targets. J Biol Chem 272: 14611–14616 [DOI] [PubMed] [Google Scholar]
- Poulsen H, Jorgensen R, Heding A, Nielsen FC, Bonven B, Egebjerg J (2006) Dimerization of ADAR2 is mediated by the double-stranded RNA binding domain. RNA 12: 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP (1997) Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89: 875–886 [DOI] [PubMed] [Google Scholar]
- Ring GM, O'Connell MA, Keegan LP (2004) Purification and assay of recombinant ADAR proteins expressed in the yeast Pichia pastoris or in Escherichia coli. Methods Mol Biol 265: 219–238 [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT (1999) SUMO-1 modification activates the transcriptional response of p53. EMBO J 18: 6455–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustighi A, Tiberi L, Soldano A, Napoli M, Nuciforo P, Rosato A, Kaplan F, Capobianco A, Pece S, Di Fiore PP, Del Sal G (2009) The prolyl-isomerase Pin1 is a Notch1 target that enhances Notch1 activation in cancer. Nat Cell Biol 11: 133–142 [DOI] [PubMed] [Google Scholar]
- Ryo A, Nakamura M, Wulf G, Liou YC, Lu KP (2001) Pin1 regulates turnover and subcellular localization of beta-catenin by inhibiting its interaction with APC. Nat Cell Biol 3: 793–801 [DOI] [PubMed] [Google Scholar]
- Ryo A, Suizu F, Yoshida Y, Perrem K, Liou YC, Wulf G, Rottapel R, Yamaoka S, Lu KP (2003) Regulation of NF-kappaB signaling by Pin1-dependent prolyl isomerization and ubiquitin-mediated proteolysis of p65/RelA. Mol Cell 12: 1413–1426 [DOI] [PubMed] [Google Scholar]
- Sansam CL, Wells KS, Emeson RB (2003) Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc Natl Acad Sci USA 100: 14018–14023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Kohler M, Sprengel R, Seeburg PH (1991) RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19 [DOI] [PubMed] [Google Scholar]
- Valente L, Nishikura K (2007) RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J Biol Chem 282: 16054–16061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoorn TA, Burnashev N, Monyer H, Seeburg PH, Sakmann B (1991) Structural determinants of ion flow through recombinant glutamate receptor channels. Science 252: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Wong SK, Sato S, Lazinski DW (2003) Elevated activity of the large form of ADAR1 in vivo: very efficient RNA editing occurs in the cytoplasm. RNA 9: 586–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Chang A, Sudol M, Hanes SD (2001) Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr Genet 40: 234–242 [DOI] [PubMed] [Google Scholar]
- Xu YX, Manley JL (2007) Pin1 modulates RNA polymerase II activity during the transcription cycle. Genes Dev 21: 2950–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP (1997) Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 278: 1957–1960 [DOI] [PubMed] [Google Scholar]
- Zacchi P, Gostissa M, Uchida T, Salvagno C, Avolio F, Volinia S, Ronai Z, Blandino G, Schneider C, Del Sal G (2002) The prolyl isomerase Pin1 reveals a mechanism to control p53 functions after genotoxic insults. Nature 419: 853–857 [DOI] [PubMed] [Google Scholar]
- Zheng H, You H, Zhou XZ, Murray SA, Uchida T, Wulf G, Gu L, Tang X, Lu KP, Xiao ZX (2002) The prolyl isomerase Pin1 is a regulator of p53 in genotoxic response. Nature 419: 849–853 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.