Abstract
Interactions between single-stranded DNA-binding proteins (SSBs) and the DNA replication machinery are found in all organisms, but the roles of these contacts remain poorly defined. In Escherichia coli, SSB's association with the χ subunit of the DNA polymerase III holoenzyme has been proposed to confer stability to the replisome and to aid delivery of primers to the lagging-strand DNA polymerase. Here, the SSB-binding site on χ is identified crystallographically and biochemical and cellular studies delineate the consequences of destabilizing the χ/SSB interface. An essential role for the χ/SSB interaction in lagging-strand primer utilization is not supported. However, sequence changes in χ that block complex formation with SSB lead to salt-dependent uncoupling of leading- and lagging-strand DNA synthesis and to a surprising obstruction of the leading-strand DNA polymerase in vitro, pointing to roles for the χ/SSB complex in replisome establishment and maintenance. Destabilization of the χ/SSB complex in vivo produces cells with temperature-dependent cell cycle defects that appear to arise from replisome instability.
Keywords: DNA replication, genome maintenance, protein interaction, X-ray crystal structure
Introduction
In Escherichia coli, DNA replication is initiated at a single locus, oriC, at which two divergent replication forks are generated (Johnson and O’Donnell, 2005; van Oijen and Loparo, 2010). Multiprotein complexes called replisomes assemble on each fork to carry out replication (Figure 1A). Within this complex, the genome is unwound to produce single-stranded DNA (ssDNA) templates used by the DNA polymerase III holoenzyme (Pol III HE) for DNA synthesis. Due to the anti-parallel structure of duplex DNA and the unidirectional activity of DNA polymerases, DNA replication is semi-discontinuous—one DNA strand (leading) is replicated continuously whereas the other strand (lagging) is synthesized discontinuously as Okazaki fragments, each of which requires a separate priming and DNA polymerase loading event. An important consequence of this arrangement is that the lagging-strand template is transiently exposed during replication as an extended loop of ssDNA that can be several thousand bases in length (Alberts et al, 1983).
Figure 1.
Structure of the E. coli χ/ψ/SSB-Ct complex. (A) Model of the E. coli replisome. SSB tetramers are depicted as light blue spheres; SSB-Ct elements (thin black lines) are omitted from all but one tetramer for clarity. (Inset) Close-up of the lagging-strand/replisome interface; SSB-Ct elements from one SSB tetramer are illustrated with the sequence of the χ-bound tail provided. (B) Ribbon diagram of the crystal structure of E. coli χ/ψ (green/red) in complex with the SSB-Ct peptide (light blue). (C) Ribbon diagram depicting the SSB-binding site on χ. Key residues from χ are labelled and represented as sticks. Fo−Fc omit electron density for the SSB-Ct is shown. (D, E) Surface representations of the χ/SSB interface depicting χ electrostatics (D, electropositive (blue) and electronegative (red)) or evolutionary conservation (E, invariant (red) and highly conserved (salmon) residues shared among 50 identifiable χ homologues).
To prevent formation of inhibitory secondary structures in the exposed lagging-strand DNA during replication, tetrameric ssDNA-binding proteins (SSBs) coat the ssDNA template. As such, SSB/ssDNA nucleoprotein complexes are the bona fide substrates used in lagging-strand DNA synthesis. To engage this substrate, the E. coli Pol III HE binds directly to SSB forming an interface that has been proposed to be important for stabilization of the replication machinery and for facilitating Okazaki fragment replication initiation (Wu et al, 1992b; Glover and McHenry, 1998; Kelman et al, 1998; Yuzhakov et al, 1999; Downey and McHenry, 2010). Interactions between SSBs and the replication machinery have been found in eukaryotes as well (Oakley and Patrick, 2010), suggesting that such interfaces are broadly utilized in cellular DNA replication pathways.
The E. coli Pol III HE is composed of two DNA polymerases (α, ε and θ subunits), one processivity clamp (β2) for each polymerase and a central complex called the DnaX or γ complex ((τ/γ)3, δ, δ′, χ and ψ subunits). DnaX catalyses loading of β2 onto DNA and integrates replisome function through protein–protein interactions (Johnson and O’Donnell, 2005) (Figure 1A). Each DNA polymerase interacts with β2 to achieve the processivity necessary for extended DNA synthesis during genome replication (Stukenberg et al, 1991). This arrangement necessitates β2 loading during initiation of the leading strand and at the start of each new Okazaki fragment. The ATP-dependent β2 loading reaction is driven by (τ/γ)3/δ/δ′ from the DnaX complex (Bloom, 2006); the χ/ψ subcomplex stimulates this function (Olson et al, 1995; Simonetta et al, 2009). The χ subunit interacts directly with SSB, forming an interface that has multiple proposed functions (Wu et al, 1992b; Glover and McHenry, 1998; Kelman et al, 1998; Yuzhakov et al, 1999; Downey and McHenry, 2010).
One model for the function of the χ/SSB interaction is an architectural/stabilization role in which χ/ψ links the clamp loader machinery to the lagging strand through direct physical interactions (Figure 1A). In this arrangement, ψ binds to the τ/γ subunits in the (τ/γ)3/δ/δ′ subcomplex while χ binds to SSB associated with the lagging-strand template DNA (Glover and McHenry, 1998, 2001; Kelman et al, 1998; Simonetta et al, 2009). Previous studies have mapped the ψ-binding sites on τ/γ, showing that ψ binding augments the stability of the DnaX complex and stimulates its activity (Olson et al, 1995; Simonetta et al, 2009). χ appears to stimulate DnaX complex activities in vitro only when SSB is present, forming a χ/SSB complex that could position the lagging-strand template for clamp loading (Glover and McHenry, 1998; Kelman et al, 1998). In this function, χ binds to SSB's conserved C-terminus (SSB-Ct, –Asp–Asp–Asp–Ile–Pro–Phe in E. coli SSB) (Glover and McHenry, 1998; Kelman et al, 1998; Yuan and McHenry, 2009). An E. coli SSB variant in which the SSB-Ct Pro is changed to Ser (SSB113) has a temperature- and salt-dependent interaction with χ and destabilizes the replisome in vitro (Kelman et al, 1998). In addition, DNA replication in E. coli ssb113 cells is temperature dependent (Greenberg and Donch, 1974; Meyer et al, 1979), suggesting that the χ/SSB interaction is important for replication in vivo. However, since the SSB-Ct acts as a docking site for more than a dozen genome maintenance proteins in E. coli (Shereda et al, 2008), the ssb113 phenotype could be the result of multiple lost interactions rather than unique disruption of the χ/SSB complex. The χ encoding holC gene is not essential in E. coli, but holC mutants have temperature-sensitive viability and have higher rates of deletions in repetitive genomic sequences (Kelman and O’Donnell, 1995; Saveson and Lovett, 1997), consistent with its proposed role in replisome stability. Genetic interactions between χ/ψ and genes involved in DNA replication initiation (Nordman et al, 2007), recombinational repair (Kelman and O’Donnell, 1995; Flores et al, 2001), DNA replication restart and SOS (Flores et al, 2002; Viguera et al, 2003) pathways suggest that the χ/ψ subcomplex may have functions outside of the elongation phase of DNA replication.
A second model for χ/ψ subcomplex function implicates the χ/SSB interaction as a step in the initiation of Okazaki fragment synthesis (Yuzhakov et al, 1999). This model arises from in vitro observations in which lagging-strand replication is blocked when χ is excluded from the Pol III HE or when SSB113 is substituted for SSB. Under either condition, primase binds stably to the primer/template and β2 loading is disallowed. This behaviour is explained by invoking a hand-off mechanism in which χ destabilizes the primase/primer/template/SSB complex by competing with primase for binding to SSB (Yuzhakov et al, 1999). In this model, as primase is released from the primer/template, β2 and DNA polymerase are loaded onto the primed lagging strand. It is unclear, however, whether this model illustrates a required mechanism for lagging-strand polymerase loading, given the viability of holC-mutant E. coli (Kelman and O’Donnell, 1995; Saveson and Lovett, 1997). Moreover, since ψ activity is affected by the absence of χ (Kelman et al, 1998; Yuan and McHenry, 2009), indirect effects stemming from χ-deficient replication are possible. Structural information on the χ/SSB interface would foster directed studies that define the function of the interaction in replication.
In this paper, structural, biochemical and genetic studies are combined to define the roles of the χ/SSB interface in DNA replication. The X-ray crystal structure of the E. coli χ/ψ subcomplex bound to a peptide comprising a portion of the SSB-Ct identifies the SSB-binding site on χ. Based on the structure, χ variants with impaired SSB binding were created and tested for function. In contrast to the proposed essential role of the χ/SSB complex in handing off primers to the lagging-strand DNA polymerase, χ variants with destabilized SSB interactions in vitro support lagging-strand DNA synthesis in reconstituted in vitro replication assays. There are, however, three pronounced replication defects associated with the variants in elevated salt concentrations: leading- and lagging-strand replication is uncoupled, leading-strand polymerase primer engagement is blocked and Okazaki fragments are shorter compared with wild-type (wt) Pol III HE. These findings are consistent with a role of χ/SSB complex formation in replisome stability and point to an unexpected function for the complex in establishing and maintaining coupled leading- and lagging-strand DNA replication. E. coli with SSB-binding-deficient holC mutations display temperature-sensitive growth, cell filamentation, chromosome partition defects and SOS induction, which are consistent with significantly destabilized replication processes. Together, these studies define major roles for the χ/SSB interface in stabilizing the replisome and in coupling leading- and lagging-strand DNA synthesis at replication forks.
Results
X-ray crystal structure of the χ/ψ/SSB-Ct ternary complex
Investigations into the role of the χ/SSB complex in DNA replication have been limited by a lack of structural data defining the molecular basis of the interaction. A crystallographic effort was therefore undertaken to determine the structure of the E. coli χ/ψ/SSB-Ct ternary complex. Crystallization attempts with E. coli χ/ψ bound to a peptide that includes the entire conserved SSB-Ct failed to produce crystals. However, crystals of χ/ψ bound to a shortened SSB-Ct peptide comprising the C-terminal-most four residues of E. coli SSB (with an N-terminal Trp residue added for quantification) formed readily and its structure was determined to 1.85-Å resolution (Figure 1B; Table I). Fo−Fc electron density maps revealed the position of a single SSB-Ct peptide associated with the χ surface (Figure 1C).
Table 1. Diffraction data and crystal structure solution.
| Data collection | |
| Wavelength (Å) | 1.12718 |
| Resolution (last shell) (Å) | 50–1.85 (1.88–1.85) |
| Reflection measured/unique | 289 641/25 843 |
| Multiplicity (last shell) | 11.2 (10.5) |
| Completeness (last shell) (%) | 99.9 (100) |
| Rsym a (last shell) (%) | 7.7 (40.9) |
| I/σ (last shell) | 63.02 (10.9) |
| Refinement | |
| Resolution (Å) | 30–1.85 |
| Rwork/Rfree b (%) | 18.4/20.8 |
| r.m.s. deviation bond lengths (Å) | 0.008 |
| r.m.s. deviation bond angles (deg) | 1.026 |
| Ramachandran statistics (% most favoured/allowed/additionally allowed/disallowed) | 95.5/4.5/0/0 |
| aRsym=ΣΣj∣Ij−〈I〉∣ΣIj, where Ij is the intensity measurement for reflection j and 〈I〉 is the mean intensity for multiply recorded reflections. | |
| bRwork/Rfree=Σ∣∣Fobs∣−∣Fcalc∣∣/∣Fobs∣, where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections (5% of the total) were held aside throughout refinement. | |
The SSB-Ct-binding surface on χ is composed of conserved hydrophobic and charged residues (Figure 1D and E). F177, the C-terminal-most SSB-Ct residue, appears to serve as an anchor in the interaction. Its side chain is bound in a hydrophobic pocket on χ while its α-carboxyl group forms ionic bonds with the R128 side chain from χ (Figure 1C and D). The more N-terminal residues on the SSB-Ct peptide are adjacent to a pair of basic residues in χ (K132 and R135), which could be important for ionic interactions with full-length SSB. A similar binding arrangement involving a hydrophobic pocket and ionic interactions has been observed in a complex formed between E. coli Exonuclease I and the SSB-Ct and has been proposed for E. coli RecQ (Lu and Keck, 2008; Shereda et al, 2009). Residues that form the SSB-binding site on E. coli χ are well conserved among the α-, β- and γ-proteobacteria (Figure 1E), suggesting that the interaction scheme described in the structure is utilized in other bacterial species.
The χ/SSB interface is critical for association in vitro
χ variant proteins in which residues with apparent roles in SSB-Ct binding were substituted with alanine were purified and tested in SSB-binding experiments. Each of the χ variants (R128A, K132A and R135A) retained the ability to bind ψ in size-exclusion chromatographic experiments, indicating that overall folding was not dramatically altered by the sequence changes.
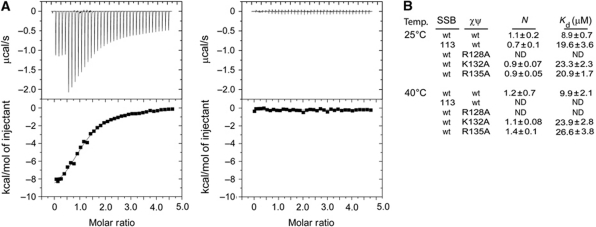
Changes in χ/ψ variant binding to SSB were measured using isothermal titration calorimetry. In these experiments, heat generated by SSB binding to χ/ψ was fitted using a single-site model to determine the stability and stoichiometry of the interaction. At 25°C, SSB binds χ/ψ in an ∼1:1 (SSB monomer:χ/ψ heterodimer) complex with an apparent dissociation constant (Kd) of 8.9±0.7 μM (Figure 2); since SSB is a tetramer, this means that 4 χ/ψ heterodimers can bind to each tetramer. These results are consistent with a previous calorimetry experiment performed under slightly different solution conditions (6.4 μM Kd and 1:1 χ:SSB monomer stoichiometry (Kozlov et al, 2010b)). In contrast, R128A χ/ψ failed to bind SSB (Figure 2). K132A and R135A χ/ψ variants retained the ability to bind SSB but with reduced affinities (23.3±2.3 and 20.9±1.7 μM Kd, respectively). These results match the structural model well; R128 is critical for binding SSB in vitro whereas K132 and R135 play more minor roles. Similar results were also observed in a qualitative co-precipitation experiment (Supplementary Figure S1). These results are consistent with those from a recent study that predicted the SSB-binding site on χ (Naue et al, 2011). The binding affinities of wt and variant χ/ψ complexes to SSB were essentially temperature independent between 25 and 40°C (Figure 2). In contrast, wt χ/ψ binding to SSB113 (an SSB variant with the penultimate proline substituted for serine) was temperature dependent as observed previously (Yuzhakov et al, 1999) (Figure 2).
Figure 2.
Identification of χ variants with compromised SSB binding. (A) Calorimetric analysis of the χ/ψ/SSB interaction for wt χ/ψ (left) and R128A χ/ψ (right). Heats evolved from titration of SSB into χ/ψ solutions are shown in the top panels; derived binding isotherms are shown in the bottom panels. (B) Summary of calorimetry experiments. A single-site model was used to fit the data. ‘N’ is the number of SSB monomer sites per χ/ψ subcomplex; ‘ND’ denotes when no binding was detected. Since SSB is a tetramer, 4 χ/ψ subcomplexes bind to each SSB tetramer when N=1.
χ variants with SSB-binding deficiencies affect DNA replication in vitro
To measure the impact of destabilizing the χ/SSB interface on DNA replication, E. coli Pol III HE complexes were reconstituted by combining Pol III HE lacking β2 (referred to as Pol III*; Wickner et al, 1973) that included either wt χ (wt Pol III*), R128A variant χ (R128A χ Pol III*) or R128E/K132E double charge-reversal variant χ (R128E/K132E χ Pol III*) with β2. R128E/K132E χ Pol III* was included to test a more dramatically destabilized χ variant than the R128A χ protein, which may have some residual affinity for SSB in the context of the actively replicating replisome. Interestingly, SSB co-purified with wt Pol III*, but failed to co-purify with R128A χ Pol III* or R128E/K132E χ Pol III* (Supplementary Figure S2), confirming the identification of the SSB-binding site on χ. The specific primer extension activities of the three Pol III* preparations were nearly identical (Supplementary Figure S2), indicating that each was functional.
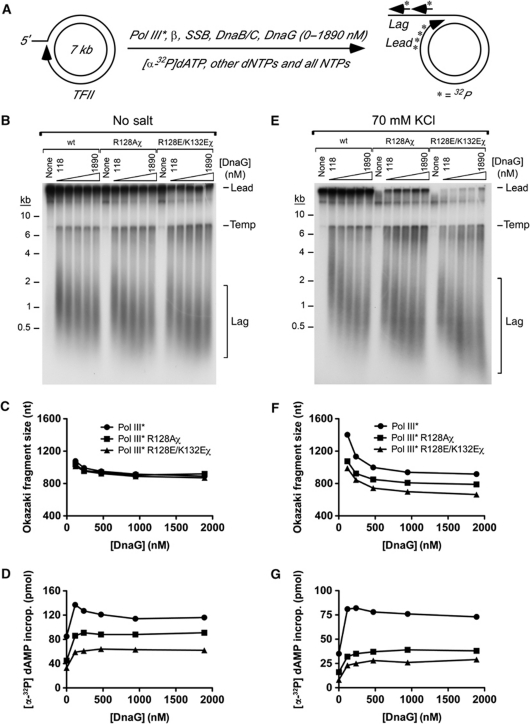
Based on the proposed role of χ in mediating the handoff of primers to the lagging-strand DNA polymerase (Yuzhakov et al, 1999), one prediction is that lagging-strand replication would be severely impaired or, perhaps, non-existent with R128A χ Pol III* or R128E/K132E χ Pol III*. Rolling-circle DNA replication assays using a 7-kbp tailed form II (TFII) DNA template (Xu and Marians, 2000) were used to test this prediction. In these assays, replisomes formed on the TFII DNA produce long leading-strand products and populations of shorter Okazaki fragments (Figure 3A). Okazaki fragment size is set by the frequency of primer synthesis by primase (DnaG) and the efficiency with which these primers are used to initiate lagging-strand synthesis. Therefore, fragment size is inversely related to the concentration of DnaG in the reaction mixture (Zechner et al, 1992). In contrast to the predicted effect of the χ sequence change on lagging-strand DNA synthesis, all three Pol III* preparations supported leading- and lagging-strand DNA replication on these substrates and produced Okazaki fragments of roughly the same sizes (Figure 3B–D). Some reduction in leading-strand DNA synthesis was observed with R128E/K132E χ Pol III* (Figure 3B and D), but the complex was clearly active under the conditions tested.
Figure 3.
Conditional rolling-circle DNA replication by R128A and R128E/K132E χ Pol III HE. Rolling-circle DNA replication reactions (schematically outlined in (A)) were catalysed by wt, R128A χ or R128E/K132E Pol III HE in the absence of added salt (B–D) or with 70 mM KCl added (E–G). Primase (DnaG) was titrated in separate reactions from 0 to 1890 nM. (B, E) Alkaline agarose gel electrophoretic analysis of the replication products. Template (temp) that becomes labelled in the reaction but does not support rolling-circle replication is indicated, as are leading-strand (lead) and lagging-strand (lag) DNA products. (C, F) Average Okazaki fragment size as a function of primase concentration. (D, G) Total DNA synthesis as a function of primase concentration. Equal amounts of radioactivity were applied in each lane of the gels shown in (B, E).
The proposed function of the χ/SSB interaction in stabilizing the replisome (Glover and McHenry, 1998; Kelman et al, 1998) was tested by carrying out rolling-circle DNA replication assays at a moderately elevated salt concentration (70 mM KCl) (Figure 3E–G). Under these conditions, both R128A χ Pol III* and R128E/K132E χ Pol III* were less active than wt Pol III* in DNA synthesis (Figure 3G). Surprisingly, the most dramatic effect was a reduction in the abundance of leading-strand products (Figure 3E). A more subtle effect on lagging-strand synthesis was also observed in which χ-variant-supported DNA replication generated shorter Okazaki fragments than wt Pol III* (Figure 3E and F). The salt sensitivity of R128A χ Pol III* and R128E/K132E χ Pol III* strongly supports a role for the χ/SSB interaction in replisome stability. In general, the observed replication defects are more pronounced in assays using R128E/K132E χ Pol III* than R128A χ Pol III*, consistent with the idea that the R128A χ may have some residual affinity for SSB within the replisome that is further destabilized in the double charge-reversal R128E/K132E χ variant.
The impaired activities of R128A χ Pol III* and R128E/K132E χ Pol III* point to a previously unidentified function for the χ/SSB interaction in stabilizing leading-strand polymerase/primer/template complexes. The novel role was confirmed in a second assay in which oriC-dependent plasmid replication was measured in vitro (Supplementary Figure S3). As with rolling-circle replication, leading-strand synthesis by R128A χ Pol III* and R128E/K132E χ Pol III* is impaired relative to wt Pol III* in a salt-dependent manner. Therefore, in two independent in vitro assays, the χ/SSB interaction appears to be important for replisome stability and the consequence of destabilizing the interface is impaired leading-strand DNA synthesis. When the resplisome is reconstituted in the absence of χ or χ/ψ, the protein complex also exhibits salt-sensitive replication (Glover and McHenry, 1998; Kelman et al, 1998), consistent with the observations reported here.
The activities of R128A χ Pol III* and R128E/K132E χ Pol III* appeared to be most diminished at low primase concentrations (Figure 3; Supplementary Figure S3). Earlier observations of leading-strand priming by primase (Heller and Marians, 2006) led to the postulation that interactions between primase and the replication machinery could help stabilize leading-strand polymerase/primer/template binding. Under primase-deficient conditions with moderate-salt, rolling-circle replication by R128A χ Pol III* and R128E/K132E χ Pol III* is greatly diminished (Figure 3), reinforcing the notion that the χ/SSB interaction is needed for replisome integrity. However, high concentrations of primase appeared to stabilize leading-strand DNA synthesis in moderate salt, possibly because primase/DnaB interactions stabilize binding of the leading-strand DNA polymerase to the replication machinery.
The appearance of Okazaki fragments without concomitant leading-strand synthesis implies that the leading-strand polymerase has either disengaged from the 3′-OH of the nascent leading strand or become non-processive under the modest-salt conditions with the R128A and R128E/K132E χ variants. However, concerted unwinding of the duplex template with priming and synthesis of the lagging strand still occurs. The possibility that uncoupled unwinding of the template could be followed by a general priming-type reaction on the resulting ssDNA is unlikely because SSB, whose presence blocks general priming (Arai and Kornberg, 1979), would coat the ssDNA thereby preventing DNA synthesis. Whereas it is possible that DnaC810 (a DnaC protein variant that loads/reloads DnaB in the absence of DnaA and an origin of replication that are normally required for replication initiation) used in the rolling-circle replication assay could overcome the inhibitory effect of SSB (Figure 3), this cannot happen in the oriC reaction where wt DnaC is present (Supplementary Figure S3). We therefore conclude that within the context of a replisome, the χ/SSB interaction is essential for sustained leading-strand synthesis under conditions of moderate-salt concentrations.
Destabilizing the χ/SSB interface results in temperature-dependent cellular effects
To test the consequences of destabilizing the χ/SSB interaction in vivo, E. coli strains in which the χ structural gene (holC) was mutated to encode R128A, K132A or R135A χ variant proteins (AHM102, AHM103 or AHM104, respectively) were constructed along with an isogenic wt χ strain (AHM101). A second set of charge-reversal mutations was made to holC since the effects of charge reversal on DNA replication in vitro were somewhat more dramatic than charge neutralization (R128E χ (AHM105), K132E χ (AHM106) and R128E/K132E χ (AHM107)). As a first test, strain viability was measured by plating serial dilutions of saturated cultures on rich (Luria Broth) or minimal (MOPS) agar and incubating at several temperatures (Figure 4A; Supplementary Figure S4). Surprisingly, each of the holC E. coli strains encoding an Ala-substituted χ protein, including R128A χ, grew nearly as well as the isogenic wt strain. One possible explanation was the R128A sequence change is insufficient to block χ binding to SSB in vivo. Consistent with this model, holC E. coli strains encoding Glu-substituted χ proteins, which should repulse SSB more strongly than the Ala-substituted variants, manifested measurable defects. AHM107 growth was strongly impaired, with significantly smaller and/or fewer colonies formed at nearly all temperatures tested in either rich or minimal media (Figure 4A; Supplementary Figure S4). Consistent, but more modest, effects were also observed with AHM105. To ensure that the effects observed with AHM107 were not due to misfolding of the R128E/K132E χ variant, the protein was purified and tested for proper folding. The R128E/K132E χ variant maintains the ability to form a complex with ψ and is degraded in limited proteolysis experiments at the same rate as wt χ (Supplementary Figure S5), indicating that it is properly folded. Additionally, the defects in AHM107 could be complemented by transformation with a plasmid encoding wt holC (Supplementary Figure S6), indicating that the phenotype was not due to unanticipated problems in the strain outside of the R128E/K132E χ sequence change.
Figure 4.
Growth and DNA replication in E. coli holC-mutant strains. (A) Saturated cultures of E. coli holC strains spot plated on minimal media were incubated at 30 or 37°C for 48 or 24 h, respectively. (B) Strains were diluted from a saturated culture and grown in minimal media at 30°C (top) or 37°C (bottom) and the number of colony forming units (c.f.u.) per ml of culture were counted over time. Error bars represent 1 s.d. from the mean. (C) Relative incorporation of [3H]-thymidine by strains (each at OD600=0.3) in minimal media at either 30 or 37°C. The cells were grown in MOPS minimal medium to OD600 ∼0.3; individual aliquots of culture were removed at the time indicated and incubated with a labelling solution containing [3H]-thymidine. The [3H]-thymidine incorporation was stopped after 2 min and the counts per minute (c.p.m.) were measured in a scintillation counter. The c.p.m. were normalized based on the amount of incorporation in the wt holC strain at each temperature. Error bars represent 1 s.d. from the mean.
To examine how sequence changes in χ alter cellular function, the growth kinetics of AHM102 (R128A χ), AHM105 (R128E χ), AHM107 (R128E/K132E χ) and AHM101 (isogenic wt χ control), were measured at 30 and 37°C (Figure 4B; Supplementary Figure S7). At 30°C, AHM102 and AHM105 growth curves were indistinguishable from AHM101. AHM107 grew with a 1.4-fold slower generation time relative to the control strain. In contrast, at 37°C, severe differences between AHM107 and the control strain became apparent. Unlike the wt cells, which had a brief lag phase, AHM107 took nearly 15 h to transition to log phase. At either temperature, once AHM107 cultures reached log phase the generation time was 1.4-fold lower than the control strain. Thus, the majority of the temperature dependence of the AHM107 growth characteristics arises from an extended lag phase. One possibility was that the holC mutation in AHM107 entirely blocks growth at 37°C and that a suppressor mutation must be generated to allow entrance into log phase. This possibility was excluded by re-diluting a culture of AHM107 cells that had been grown for 31 h at 37°C into fresh media and examining the culture's growth kinetics. The culture had the same extended lag phase as AHM107 cells, indicating that heritable generic suppressors were not responsible for its eventual transition to log phase.
Destabilization of the χ/SSB interface modestly affects DNA replication rates in E. coli
To test the effects of the χ mutants on DNA replication directly, in vivo DNA replication rates were measured in early log phase E. coli holC-mutant cultures using a [3H]-thymidine incorporation assay. AHM101 incorporation rates were ∼2-fold higher than AHM107 at both 30 and 37°C, consistent with reduced replication activity in the AHM107 χ variant (Figure 4C). This result parallels the 1.4-fold longer doubling time observed for AHM107 in log phase relative to the control strain (Figure 4B). A more modest defect was observed with AHM105 whereas AHM102 was indistinguishable from AHM101.
SOS induction and partitioning defects in holC-mutant strains
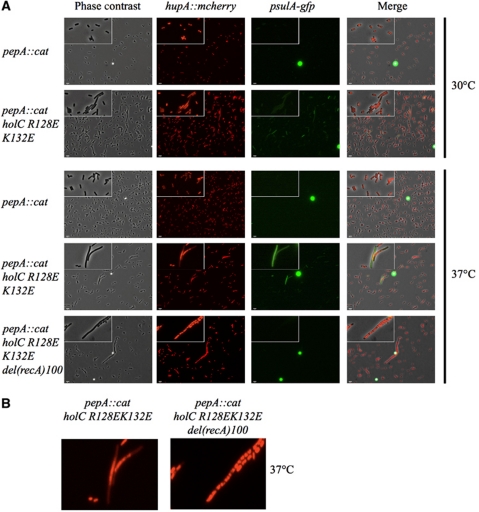
Many temperature-sensitive DNA replication mutants show increased levels of SOS induction and filamentation (Saveson and Lovett, 1997; Viguera et al, 2003). Defects in DNA replication may also lead to aberrant nucleoid structures and/or chromosome partitioning problems. To determine if holC mutations imparted increased SOS induction and defects in nucleoid structure and/or partitioning, their effects were assessed in an E. coli strain containing a psulA-gfp transcriptional fusion at the λ attachment site (McCool and Sandler, 2001; McCool et al, 2004) and a hupA∷mcherry translational fusion. In these strains, psulA-gfp expression is dependent upon SOS induction while hupA∷mcherry expresses a mCherry-tagged version of the histone-like HU-2 protein to label genomic DNA. The strains were grown to mid-log phase in minimal media at 30, 37 or 42°C; cell morphology, psulA-gfp expression (SOS induction) and mCherry staining (nucleoid structure) were then measured. Representative images are shown in Figure 5, with quantitative data from many images provided in Table II. Cells encoding R128E/K132E χ exhibited temperature-dependent SOS induction and cell filamentation: at 30°C, SOS was induced in 2.9% of cells and 1.0% of cells were filamentous, whereas at 37 and 42°C, SOS levels increased (6.5 and 17.3%, respectively) as did filamentation (4.0 and 6.0%, respectively). The same cells also displayed apparent chromosome partitioning defects as evidenced by the diffuse nucleoid structure observed in filamented cells. The observed partition defect was dependent on RecA, as deletion of recA produced cells with more distinct nucleoids (Figure 5). This type of behaviour has been seen previously with the Par− phenotype of priA mutants (McCool and Sandler, 2001). E. coli cells encoding the single-site variant R128E χ also exhibited temperature-dependent induction of SOS and cell filamentation, although the levels were reduced relative to the R128E/K132E χ double mutant (Table II). In contrast, the isogenic wt holC, R128A and K132E strains were all indistinguishable from each other, with no measurable increase in SOS or filamentation over the conditions tested. These results, in conjunction with the growth defects noted earlier, indicate that the interaction between χ and SSB is important for proper and efficient genome replication.
Figure 5.
Filamentation and SOS induction in E. coli holC-mutant strains. (A) Cells were grown at 30 or 37°C as indicated in MOPS minimal medium. Fields of cells were imaged to show cell morphology (phase contrast), nucleoid structure (hupA∷mcherry) or SOS induction (sulAp∷gfp). Bright green spots are beads used for quantification. (Inset) Close-up images of cells. (B) Close-up of hupA∷mcherry of the strains indicated grown at 37°C.
Table 2. Temperature-dependent cellular effects of holC mutations in E. coli.
| Temperature | Strain | holC | pepA | Relative fluorescence intensity | % SOS induced | % Filamented |
|---|---|---|---|---|---|---|
| SS8343 | + | + | 1.0±0.0 | 0.4±0.6 | 0.1 | |
| SS8383 | + | cat | 1.9±0.8 | 1.4±1.0 | 0.4 | |
| 30°C | SS8384 | R128A | cat | 1.7±0.7 | 1.9±1.2 | 0.1 |
| SS8385 | R128E | cat | 1.4±0.5 | 1.0±0.9 | 1.0 | |
| SS8386 | K132E | cat | 1.3±0.9 | 1.0±0.7 | 0.1 | |
| SS8387 | R128E K132E | cat | 2.0±0.7 | 2.9±2.2 | 1.0 | |
| SS8343 | + | + | 1.0±0.0 | 0.5±0.4 | 0.1 | |
| SS8383 | + | cat | 1.0±0.3 | 0.6±0.2 | 0.2 | |
| 37°C | SS8384 | R128A | cat | 1.1±0.7 | 1.0±0.8 | 0.1 |
| SS8385 | R128E | cat | 1.4±0.6 | 2.3±1.6 | 1.4 | |
| SS8386 | K132E | cat | 1.1±0.1 | 1.0±0.7 | 0.0 | |
| SS8387 | R128E K132E | cat | 2.4±0.4 | 6.5±2.3 | 4.0 | |
| SS8343 | + | + | 1.0±0.0 | 1.1±0.9 | 0.2 | |
| SS8383 | + | cat | 1.1±0.6 | 0.3±0.4 | 0.6 | |
| 42°C | SS8384 | R128A | cat | 1.1±0.3 | 0.5±0.1 | 0.4 |
| SS8385 | R128E | cat | 1.9±0.6 | 4.2±1.5 | 2.4 | |
| SS8386 | K132E | cat | 1.1±0.5 | 0.6±0.3 | 0.3 | |
| SS8387 | R128E K132E | cat | 4.1±0.3 | 17.3±3.0 | 6.0 |
Discussion
The interaction between SSB and the cellular DNA replication machinery has been proposed to be important for replisome stability and for delivery of primers to the lagging-strand DNA polymerase throughout replication. These models have been tested using a combination of structural, biochemical and genetic experiments. The crystal structure of an SSB-Ct peptide bound to the χ/ψ replisomal subcomplex from E. coli was determined, allowing χ variant proteins with destabilized SSB-binding sites to be created. In contrast to its proposed role in primer delivery, reconstituted Pol III HE that included χ variants with destabilized SSB-binding sites supported robust lagging-strand DNA synthesis in vitro. However, effects that were consistent with conditional destabilization of the replisome were observed with the variant Pol III HEs in modest-salt conditions. These included uncoupled leading- and lagging-strand replication, obstruction of primer engagement by the leading-strand polymerase, and a reduction in the lengths of Okazaki fragments. In parallel with these observations, destabilization of the χ/SSB complex in vivo produces cells with temperature-dependent SOS induction and cell filamentation along with growth and chromosome partitioning defects that are consistent with replisome instability and aberrant replication. These findings define a role for the χ/SSB complex formation in replisome stability and point to its unexpected importance in establishing and maintaining coupled leading- and lagging-strand DNA replication.
Role of the χ/SSB interaction in DNA replication complexes
Previous experiments examining the role of the χ/SSB interface have been limited to studies in which χ or χ/ψ are omitted, or where SSB variant proteins with altered SSB-Ct sequences were used (Saveson and Lovett, 1997; Yuzhakov et al, 1999; Yuan and McHenry, 2009). Each of these approaches has limitations that arise from the multifunctional nature of the protein complexes involved. For example, omission of χ not only eliminates the interaction between the DnaX complex and SSB but it also has effects on ψ, which has limited solubility in the absence of χ (Xiao et al, 1993). Moreover, alteration of the SSB-Ct has effects on its interactions both with other proteins and with ssDNA (Downey and McHenry, 2010; Kozlov et al, 2010a, 2010b). These complications led us to take a structural approach to identify the SSB-Ct-binding site on χ, which showed that χ relies on an electrostatic surface for interaction with SSB that is similar to sites found in other SSB-interacting proteins (Lu and Keck, 2008; Shereda et al, 2009). This information was used in subsequent biochemical and genetic experiments to test the effects of χ variants that can no longer bind SSB, but that retain the ability to form a complex with ψ.
Several biochemical and cellular defects were associated with sequence changes in χ that destabilized its interaction with SSB. These effects cumulatively point to a role for the χ/SSB interaction in stabilizing the DNA replication machinery in E. coli. One unexpected effect was the particularly strong impact of χ/SSB complex destabilization on leading-strand replication. In both rolling-circle and oriC-dependent replication, extension of the leading strand by DNA polymerase was notably impaired under moderately elevated salt conditions when the χ/SSB interface was weakened (Figure 3; Supplementary Figure S3). This effect may be related to a previous observation in which SSB bound to the lagging-strand template stimulates strand displacement by DNA polymerase on the leading strand; this action is due to a network of interactions important for leading-strand polymerase processivity (Yuan and McHenry, 2009). Our results suggest that under modestly stringent conditions (elevated salt concentrations in vitro or high temperature in vivo), a similar network may be important for leading-strand replication in the context of the full replisome. The protein interactions in this arrangement could stabilize the replication machinery through both favourable thermodynamic interactions in the complex and architectural effects on the replication fork structure. Overall, this scheme would help coordinate the activities of the replisome on the leading strand and lagging strand.
Cellular importance of the χ/SSB interaction
Mutations in E. coli holC that destabilize the χ/SSB interface in vivo had severe temperature-dependent effects on growth rate, induction of SOS and filamentation and chromosomal partitioning that appear to arise from conditionally impaired DNA replication. The growth kinetic defects are largely due to a temperature-dependent extension of the lag phase of cultures—the most impaired holC-mutant strain had a 15-h lag phase (∼7-fold longer than the isogeneic wt strain) but only an ∼2-fold reduction in DNA replication rates once log phase was reached. Several factors could potentially explain the extension of the lag phase. First, the differences could point to a defect in the ability to initiate DNA replication. Replication in vitro under restrictive conditions was characterized by significant uncoupling of the leading- and lagging strands that appeared to be primarily due to blockage of leading-strand primer extension (Figure 3; Supplementary Figure S3). Impacts of the destabilized χ/SSB interaction on leading-strand synthesis were apparent with both pre-primed templates (TFII DNA; Figure 3) and templates that had to be primed de novo (oriC; Supplementary Figure S3). These results highlight the role of the interaction in leading-strand synthesis. A molecular role for the χ/SSB interface in establishment of leading-strand synthesis is consistent with earlier genetic experiments that indicated χ could play an important role in DNA replication initiation (Nordman et al, 2007).
A second effect, which may contribute to the extended lag phase, stems from the observation that the mutant cells had strongly induced SOS, and cell filamentation and partitioning defects that reflect serious genome structural defects. The replisome, weakened by the loss of the χ/SSB interaction, may copy DNA with significantly less efficiency than wt cells, which could produce damaged chromosomal products that require time to be repaired. Similar DNA-damage-dependent delays in entry to log phase in E. coli have been described as a possible bacterial equivalent of a checkpoint that regulates the eukaryotic cell cycle (Murli et al, 2000).
Taken together, our observations have defined the structure of the χ/SSB interface and have shown that this molecular contact is critical for replisomal stability and for productive leading-strand DNA synthesis. These studies help define the roles of interaction between the cellular DNA replication machinery and SSB bound to the lagging-strand template. Similar interactions are likely to be important for replication in all organisms.
Materials and methods
Bacterial strains and primers
Information on bacterial strains and primers used in this study is provided in Supplementary Tables I and II.
Protein overexpression and purification
E. coli χ/ψ
The open-reading frames of holC (encodes χ) and holD (encodes ψ) from E. coli K12 strain MG1655 were PCR amplified and subcloned into either pET15b (creating pET15-χ) or pET28b (creating pET28-ψ) using primers listed in Supplementary Table II. pET15-χ expresses an N-terminal hexahistidine-tagged χ; pET28-ψ encodes untagged ψ with an inserted glycine after the start codon. Site-directed mutations of pET15-χ were generated by the QuikChange scheme (Stratagene). DNA sequencing confirmed the fidelity of the coding region of all plasmids.
BL21 DE3 cells transformed with pLysS (Novagen) and pET15-χ (or a pET15-χ derivative plasmid encoding a χ variant) were grown at 37°C in LB medium supplemented with 100 μg/ml ampicillin (Ap) and 25 μg/ml chloramphenicol (Cm). In all, 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) was added to the culture at mid-log phase (OD600 nm ∼0.6) and growth was continued for 3 h to induce χ overexpression. Cells were centrifuged, suspended in χ-lysis buffer (50 mM Tris–HCl, pH 8.8, 0.5 M NaCl, 10 mM imidazole, 20% glycerol, 2 mM phenylmethanesulfonyl fluoride (PMSF), 2 mM benzamidine) and lysed by sonication on ice. The lysate was centrifuged and the supernatant was incubated with nickel-sepharose resin (GE Healthcare) equilibrated in lysis buffer for 1 h. The resin was then packed in a column, washed with lysis buffer, and χ was eluted with high imidazole buffer (50 mM Tris–HCl, pH 8.8, 0.5 M NaCl, 300 mM imidazole, 20% glycerol). The eluent was dialysed against 20 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 6.0, 150 mM NaCl, 10 mM 2-mercaptoethanol, 20% glycerol, diluted 1:1 with low-salt buffer (20 mM MES, pH 6.0, 50 mM NaCl, 10% glycerol), and loaded onto a S-Fastflow ion-exchange column (GE Healthcare) equilibrated in low-salt buffer. χ-Enriched fractions were eluted with a NaCl gradient, pooled, concentrated and dialysed against 10 mM Tris–HCl, pH 8.0, 150 mM NaCl, 10% glycerol. The histidine tag was cleaved by thrombin and χ was dialysed against storage buffer (50% glycerol, 15 mM dithiothreitol (DTT), 500 mM NaCl, 5 mM ethylenediaminetetraacetic acid (EDTA)).
E. coli ψ was purified as described previously (Xiao et al, 1993), with minor exceptions. BL21 DE3 cells transformed with pLysS and pET28-ψ were grown in LB medium supplemented with 50 μg/ml kanamycin (Km) and 25 μg/ml Cm. IPTG (1 mM) was added to the culture at mid-log phase (OD600 nm ∼0.6) and growth was continued for 3 h to induce ψ overexpression. Cells were centrifuged, suspended in ψ-lysis buffer (50 mM Tris–HCl, pH 7.5, 10% sucrose, 10 mM EDTA, 1 mM PMSF) and lysed by sonication on ice. The lysate was clarified by centrifugation and the insoluble fraction was resuspended in low-salt ψ buffer (20 mM Tris–HCl, pH 7.5, 20% glycerol, 0.5 mM EDTA, 2 mM DTT, 10 mM NaCl), sonicated on ice and centrifuged. The pellet was resuspended in high-salt ψ buffer (20 mM Tris–HCl, pH 7.5, 20% glycerol, 0.5 mM EDTA, 2 mM DTT, 1 M NaCl), centrifuged, then the pellet resuspended with low-salt ψ buffer and centrifuged. ψ was solubilized from the pellet in urea buffer (20 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 6 M urea), clarified by centrifugation and the supernatant was loaded onto Q-Fastflow column (GE Healthcare) equilibrated in QFF buffer A (20 mM Tris–HCl, pH 7.5, 0.5 mM EDTA, 2 mM DTT, 6 M urea). ψ was eluted over by a salt gradient from 0 to 750 mM NaCl.
The χ/ψ complex was made by mixing ψ and χ in a 1:1 molar ratio and resolving the complex on a Sephacryl S-100 column in 5% MPD, 10 mM Tris–HCl pH 8.8, 10 mM imidazole and 100 mM NaCl. χ/ψ-Containing fractions were pooled, concentrated and stored at 4°C.
E. coli SSB
The plasmids for overexpression of E. coli SSB and SSB113 were gifts from Michael Cox. SSB and SSB113 protein was purified as in Lohman et al (1986).
SSB-Ct peptide
SSB-Ct peptide (Trp–Asp–Ile–Pro–Phe) was synthesized and purified by the University of Wisconsin Biotechnology Center.
Pol III*
Approximately 900 ml cell suspension (about 450 g of cells) of BLR(pHOC 2.6.1) (a gift of C McHenry, University of Colorado) or variants in which the holC open-reading frame was mutated to encode R128A or R128E/K132E χ was thawed at 4°C. The density of the suspension was adjusted to OD595 of 200 by the addition of 50 mM Tris–HCl, pH 8.0 at 4°C, 10% sucrose. The suspension was brought to 20 mM EDTA, 150 mM NaCl, 20 mM spermidine and 5 mM DTT, followed by the addition of solid Tris base to adjust the pH to 8.5. Cells were lysed by the addition of lysozyme (200 μg/ml) and incubated for 50 min at 4°C, 4 min at 37°C, then another 10 min at 4°C. After centrifugation for 90 min at 11 000 r.p.m., the supernatant (fraction 1) was made 0.07% in Polymin P by the dropwise addition of a 1% solution. The mixture was cleared by centrifugation for 30 min at 14 500 r.p.m. The protein was precipitated by the addition of 0.226 g/ml of solid ammonium sulphate, and the pellet was collected by centrifugation for 45 min. The protein was back extracted with 1/10 volume buffer A (50 mM Tris–HCl, pH 7.5 at 4°C, 5 mM DTT, 1 mM EDTA, 20% glycerol)+100 mM NaCl containing 0.2 g/ml ammonium sulphate, centrifuged for 30 min at 20 000 r.p.m., then back extracted again with 1/40 volume buffer A+100 mM NaCl containing 0.17 g/ml ammonium sulphate. The pellet was resuspended in 1/80 volume 25 mM Tris–HCl, pH 7.5 at 4°C, 5 mM DTT, 100 mM NaCl, 5% glycerol to give fraction 2. The activity was determined by general priming assay as described (Tougu et al, 1994) (Supplementary Figure S2). A portion of fraction 2 (27.5%) was diluted with buffer A to adjust the conductivity to match buffer A+30 mM NaCl, and applied to a 10-ml Heparin-agarose column that had been equilibrated previously with buffer A+30 mM NaCl. The column was then washed with 25 ml of equilibration buffer, and protein eluted with a 100-ml gradient of 30–400 mM NaCl in buffer A. Active fractions (eluting at ∼200 mM NaCl) were pooled (fraction 3) and precipitated by the addition of an equal volume of 100% saturated ammonium sulphate. The pellet was resuspended in 400 μl buffer A+100 mM NaCl and filtered through a 24-ml Superose 6 column (Amersham) equilibrated and developed with buffer A+100 mM NaCl. Active fractions were pooled (fraction 3), 100% glycerol was added to increase the final concentration to 38% and stored at −80°C. Other replication enzymes (E. coli DnaA, DnaB, DnaC, β2 and gyrase) were purified as described in Wu et al (1992a).
Crystallization and structure determination of χ/ψ/SSB-Ct
E. coli χ/ψ (7.5 mg/ml in 1.5% MPD, 3 mM Tris–HCl, pH 8.8, 3 mM imidazole and 30 mM NaCl) was mixed 1:1 (vol) with well solution (25% PEG 4000, 0.2 M ammonium sulphate, 0.1 M sodium acetate, pH 4.6) in hanging-drop vapour-diffusion crystallization trials to form apo χ/ψ crystals used for seeds in generating χ/ψ/SSB-Ct crystals. χ/ψ (7.5 mg/ml) combined with SSB-Ct peptide (1:2 molar ratio of χ/ψ:SSB-Ct) was mixed with an equal volume of well solution (13–20% PEG 4000, 0.2 M ammonium sulphate, 5% glycerol, 0.1 M sodium acetate pH 4.4) in hanging-drop vapour-diffusion crystallization trails. Apo χ/ψ crystals were streak seeded into the χ/ψ/SSB-Ct drops to generate ternary complex crystals. χ/ψ/SSB-Ct crystals were transferred into cryoprotectant (13–20% PEG 4000, 0.2 M ammonium sulphate, 25% glycerol, 0.1 M sodium acetate, pH 4.4) and flash frozen in liquid nitrogen. Diffraction data were indexed and scaled using HKL2000 (Otwinowski and Minor, 1997). The χ/ψ/SSB-Ct structure was determined by molecular replacement, using Phaser (McCoy et al, 2007) with the previously determined χ/ψ structure (Gulbis et al, 2004) as a search model. Iterative model building and refinement with Coot (Emsley and Cowtan, 2004) and REFMAC (Murshudov et al, 1997) produced the final model. Model coordinates and structure factors have been deposited in the Protein Data Bank (3SXU accession code).
Isothermal titration calorimetry
E. coli χ/ψ (or a χ/ψ subcomplex with a χ variant) (30 μM) and SSB (or SSB113) (600 μM as monomer) were dialysed against 20 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM 2-mercaptoethanol. χ/ψ was thermostatted in a stirred sample cell (2 ml) at either 25 or 40°C and SSB or SSB113 was titrated with the following regime: 1 injection (1 μl for 2 s), 4 injections (4 μl for 8 s each), then 32 injections (8 μl for 16 s each). Data were fitted to a single-site model (1 SSB monomer binding to 1 χ/ψ subcomplex) using a non-linear iterative least squares algorithm (MicroCal ORIGIN). Since SSB is a tetramer, the binding arrangement in solution is 4 χ/ψ subcomplexes per SSB tetramer.
In vitro DNA replication assays
TFII DNA was prepared and purified by sucrose gradient centrifugation as described (Wu et al, 1992a) using the primer TFII annealed to M13mp18 phage DNA. The first 35 nucleotides of the TFII primer forms the tail and the other 35 nucleotides anneal to the DNA. Rolling-circle DNA replication reaction mixtures (10 μl) containing 50 mM Hepes-KOH (pH 8.0), 12 mM Mg(OAc)2, 10 mM DTT, 40 μM [α-32P]dATP (1000–3000 c.p.m./pmol), 40 μM dGTP, dCTP, and TTP, 200 μM GTP, CTP, and UTP, 1 mM ATP, 4 nM TFII DNA, 400 nM DnaC810, 360 nM DnaB, 20 nM Pol III* (or a Pol III* variant), 30 nM β, 250 nM SSB (tetramer), and the indicated concentrations of KCl and DnaG were incubated at 37°C for 5 min as described (Wu et al, 1992a). Reactions were terminated by the addition of EDTA to 20 mM and analysed by electrophoresis through 0.6% denaturing alkaline agarose gels using 30 mM NaOH-2 mM EDTA as the electrophoresis buffer. OriC replication reaction mixtures (10 μl) containing 80 mM Hepes-KOH pH 8.0, 10 mM Mg(OAc)2, 10 mM DTT, 100 μg/ml BSA, 40 μM [α-32P]dATP (1000–3000 c.p.m./pmol), 40 μM dGTP, dCTP, and TTP, 200 μM GTP, CTP, and UTP, 2 mM ATP, 2 nM oriC plasmid DNA, 500 nM DnaA, 200 nM DnaC, 200 nM DnaB, 20 nM Pol III*, 30 nM β2, 250 nM SSB (as tetramer), 20 nM DNA gyrase, and the indicated concentrations of KCl (0 or 70 mM) and DnaG (0–1980 nM) were incubated at 37°C for 10 min as described in (Hiasa et al, 1994). Reactions were terminated and analysed as for the rolling-circle reaction.
Construction of holC-mutant E. coli
All bacterial strains used in this work are derivatives of E. coli K12 and are described in Supplementary Table I. All P1 transductions were selected on 2% agar plates containing minimal media +0.1% glucose. Where appropriate, plates also contained the following antibiotics at these final concentrations: tetracycline 10 μg/ml, Cm 25 μg/ml or Km 50 μg/ml. All transductants were purified on the same type of media on which they were selected, at 30°C for AHM derivatives.
E. coli strain AHM101 was constructed by PCR amplification of holC (including 120 bp 5′ of the open-reading frame) and subcloned into pET15b. The Cm-resistance (CmR) cassette from pKD13 was subcloned 5′ of the holC to create pET15b-AHM101. Site-directed mutagenesis of pET15b-AHM101 was used to create pET15b-AHM102–107. The CmR-holC element of each plasmid was independently electroporated into E. coli strain MG1655 with pKD46 via the Wanner method, and recombinants were selected by plating on Cm-containing media (Datsenko, 2000). Genomic insertions were verified by colony PCR and confirmed by sequencing.
The hupA∷mcherry linear DNA fragment was generated by amplifying genomic DNA by PCR using primers listed in Supplementary Table II for the Km-resistance cassette containing FRT sites from the same strain. Both linear fragments were then purified using the QIAEX II kit (Qiagen). Purified DNA fragments were then combined in a primerless PCR followed by a third PCR using primers extending into a single linear fragment containing mcherry and Km resistance. The linear hupA100∷mcherry∷kan fragment was then transferred to the chromosome by transformation of SS4414 competent cells containing plasmid pKD46 (Datsenko, 2000; Datsenko and Wanner, 2000). Km-resistant colonies were purified and screened for Ap sensitivity, indicating loss of pKD46. Cells were then screened microscopically for red nucleoids and sulAp-gfp expression. To test for wt hupA activity, this mutation was combined del(hupB)∷Tn10. hupA hupB double mutants have diffuse nucleoids, chromosome partitioning defects and are highly filamented where single mutants appear essentially wt (Dri et al, 1991). The hupA100∷mcherry∷kan del(hupB)∷Tn10 double mutants appeared similar to wt (data not shown). Plasmid copies of hupA∷gfp have been shown to give the same pattern of nucleoids as cells stains with DAPI (Wery et al, 2001). The resulting strain with hupA100∷mcherry∷kan was called SS6279.
Phenotypic analysis of holC-mutant E. coli
Saturated cultures of AHM101–AHM107 strains were serially diluted (10-fold) in 145 mM NaCl and 10 μl from each dilution was spot plated onto LB or minimal media Cm agar and incubated at 25, 30, 37 or 42°C to test for changes in colony morphology and viability. For experiments with minimal media agar, the strains were washed once in MOPS media prior to dilution.
Growth curves were performed starting with saturated cultures grown at 37°C in LB Cm from a single colony from freshly streaked cultures. Saturated cultures (5 μl) were added to 50 ml of LB or minimal media (5 μl) with Cm. The cultures were then placed at 30 or 37°C, with shaking. Growth was monitored with colony counts using 10-fold serial dilutions in 145 mM NaCl followed by plating on LB cat and growth at 37°C. Data are the mean of three independent replicates with error bars representing 1 s.d. of the mean. For minimal media growth curves, the saturated cultures were washed with MOPS prior to being resuspended in MOPS. Generation time was calculated using the colony counts during the log phase of cell growth with the equation tgen=1/k; k=(3.32 × log10 [Nt2/Nt1])/(t1−t2), where k is the growth rate constant, t2 is the time when the population is Nt2 and t1 is the time when the population is Nt1.
DNA replication rate determination
Strains were grown at 37°C in minimal media as described above until the OD600 reached 0.3, at which time DNA synthesis was measured by measuring [3H]-thymidine incorporation as described in Courcelle and Courcelle (2006) except that the label solution was composed of MOPS media with 0.1% glucose and 5 μCi/ml of [3H]-thymidine. Data are the mean of three independent replicates of each strain with error bars representing 1 s.d. of the mean.
Microscopy
Cultures were grown in MOPS minimal medium until mid-log phase (OD600=0.3–0.4). Cells were concentrated 10-fold in MOPS minimal and ∼2 μl of this mixture was loaded onto fresh MOPS minimal 1% agarose pads and a cover slip was applied. Agarose pads were prepared using a protocol from P Levin. Microscopy was carried out using an epifluorescent Nikon E600 microscope. An ORCA-ER-cooled charge-coupled device camera (Hammamatsu) and Openlabs software (Improvision) were used for all image acquisition. The exposure time was 300 ms. Approximately nine fields (three on 3 different days) containing calibration beads were photographed. A phase-contrast image and two fluorescent images (green and red) of each field were taken. Openlab 5.0 and Volocity 3.5 (Improvision, Inc.) were used to measure the amount of fluorescence and cell size in individual live cells. Calibration of the fluorescence intensity was set by calibration beads (InSpeck Green (505/515) microscope image intensity calibration kit 2.5 μm I-7219 from Molecular Probes). The relative fluorescence intensity value of an individual cell is calculated by dividing the average calibrated pixel value of a particular cell by the average calibrated pixel value of a strain containing Δattλ∷sulAp Ωgfp-mut2 (SS8343).
Supplementary Material
Acknowledgments
We thank Advanced Photon Source staff (LS-CAT beamline) for assistance with data collection. We thank the Keck laboratory and James Berger for critical reading of this manuscript, and Amber Schuh and Basudeb Bhattacharyya for technical assistance. This work was funded by grants from the NIH (GM068061, JLK, AI059027, SJS and GM34557, KJM). AHM was supported in part by an NIH training grant in Molecular Biosciences (GM07215) and a Shapiro scholarship.
Author contributions: All authors designed and carried out the research. AHM, SB, SCM, SJS, KJM and JLK wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alberts BM, Barry J, Bedinger P, Formosa T, Jongeneel CV, Kreuzer KN (1983) Studies on DNA replication in the bacteriophage T4 in vitro system. Cold Spring Harb Symp Quant Biol 47 (Part 2): 655–668 [DOI] [PubMed] [Google Scholar]
- Arai K, Kornberg A (1979) A general priming system employing only dnaB protein and primase for DNA replication. Proc Natl Acad Sci USA 76: 4308–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom LB (2006) Dynamics of loading theEscherichia coli DNA polymerase processivity clamp. Crit Rev Biochem Mol Biol 41: 179–208 [DOI] [PubMed] [Google Scholar]
- Courcelle CT, Courcelle J (2006) Monitoring DNA replication following UV-induced damage in Escherichia coli. Methods Enzymol 409: 425–441 [DOI] [PubMed] [Google Scholar]
- Datsenko KA (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey CD, McHenry CS (2010) Chaperoning of a replicative polymerase onto a newly assembled DNA-bound sliding clamp by the clamp loader. Mol Cell 37: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dri AM, Rouviere-Yaniv J, Moreau PL (1991) Inhibition of cell division in hupA hupB mutant bacteria lacking HU protein. J Bact 173: 2852–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Flores MJ, Bierne H, Ehrlich SD, Michel B (2001) Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J 20: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Ehrlich SD, Michel B (2002) Primosome assembly requirement for replication restart in the Escherichia coli holDG10 replication mutant. Mol Microbiol 44: 783–792 [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS (1998) The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem 273: 23476–23484 [DOI] [PubMed] [Google Scholar]
- Glover BP, McHenry CS (2001) The DNA polymerase III holoenzyme: an asymmetric dimeric replicative complex with leading and lagging strand polymerases. Cell 105: 925–934 [DOI] [PubMed] [Google Scholar]
- Greenberg J, Donch J (1974) Sensitivity to elevated temperatures in exrB strains of Escherichia coli. Mutat Res 25: 403–405 [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kazmirski SL, Finkelstein J, Kelman Z, O’Donnell M, Kuriyan J (2004) Crystal structure of the chi:psi subassembly of the Escherichia coli DNA polymerase clamp-loader complex. Eur J Biochem 271: 439–449 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2006) Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439: 557–562 [DOI] [PubMed] [Google Scholar]
- Hiasa H, DiGate RJ, Marians KJ (1994) Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J Biol Chem 269: 2093–2099 [PubMed] [Google Scholar]
- Johnson A, O’Donnell M (2005) Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem 74: 283–315 [DOI] [PubMed] [Google Scholar]
- Kelman Z, O’Donnell M (1995) DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem 64: 171–200 [DOI] [PubMed] [Google Scholar]
- Kelman Z, Yuzhakov A, Andjelkovic J, O’Donnell M (1998) Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J 17: 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Cox MM, Lohman TM (2010a) Regulation of single-stranded DNA binding by the C termini of Escherichia coli single-stranded DNA-binding (SSB) protein. J Biol Chem 285: 17246–17252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov AG, Jezewska MJ, Bujalowski W, Lohman TM (2010b) Binding specificity of Escherichia coli single-stranded DNA binding protein for the χ subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry 49: 3555–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman TM, Green JM, Beyer RS (1986) Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry 25: 21–25 [DOI] [PubMed] [Google Scholar]
- Lu D, Keck JL (2008) Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc Natl Acad Sci USA 105: 9169–9174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ (2004) Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol 53: 1343–1357 [DOI] [PubMed] [Google Scholar]
- McCool JD, Sandler SJ (2001) Effects of mutations involving cell division, recombination, and chromosome dimer resolution on a priA2∷kan mutant. Proc Natl Acad Sci USA 98: 8203–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RR, Glassberg J, Kornberg A (1979) An Escherichia coli mutant defective in single-strand binding protein is defective in DNA replication. Proc Natl Acad Sci USA 76: 1702–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murli S, Opperman T, Smith BT, Walker GC (2000) A role for the umuDC gene products of Escherichia coli in increasing resistance to DNA damage in stationary phase by inhibiting the transition to exponential growth. J Bacteriol 182: 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Naue N, Fedorov R, Pich A, Manstein DJ, Curth U (2011) Site-directed mutagenesis of the {chi} subunit of DNA polymerase III and single-stranded DNA-binding protein of E. coli reveals key residues for their interaction. Nucleic Acids Res 39: 1398–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Skovgaard O, Wright A (2007) A novel class of mutations that affect DNA replication in E. coli. Mol Microbiol 64: 125–138 [DOI] [PubMed] [Google Scholar]
- Oakley GG, Patrick SM (2010) Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci 15: 883–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson MW, Dallmann HG, McHenry CS (1995) DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The chi psi complex functions by increasing the affinity of tau and gamma for delta.delta′ to a physiologically relevant range. J Biol Chem 270: 29570–29577 [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Saveson CJ, Lovett ST (1997) Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146: 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol 43: 289–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereda RD, Reiter NJ, Butcher SE, Keck JL (2009) Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J Mol Biol 386: 612–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J (2009) The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell-Vaughan PS, O’Donnell M (1991) Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem 266: 11328–11334 [PubMed] [Google Scholar]
- Tougu K, Peng H, Marians KJ (1994) Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem 269: 4675–4682 [PubMed] [Google Scholar]
- van Oijen AM, Loparo JJ (2010) Single-molecule studies of the replisome. Annu Rev Biophys 39: 429–448 [DOI] [PubMed] [Google Scholar]
- Viguera E, Petranovic M, Zahradka D, Germain K, Ehrlich DS, Michel B (2003) Lethality of bypass polymerases in Escherichia coli cells with a defective clamp loader complex of DNA polymerase III. Mol Microbiol 50: 193–204 [DOI] [PubMed] [Google Scholar]
- Wery M, Woldringh CL, Rouviere-Yaniv J (2001) HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie 83: 193–200 [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R, Geider K, Kornberg A (1973) A new form of DNA polymerase 3 and a copolymerase replicate a long, single-stranded primer-template. Proc Natl Acad Sci USA 70: 1764–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Marians KJ (1992a) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. I. Multiple effectors act to modulate Okazaki fragment size. J Biol Chem 267: 4030–4044 [PubMed] [Google Scholar]
- Wu CA, Zechner EL, Reems JA, McHenry CS, Marians KJ (1992b) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. V. Primase action regulates the cycle of Okazaki fragment synthesis. J Biol Chem 267: 4074–4083 [PubMed] [Google Scholar]
- Xiao H, Crombie R, Dong Z, Onrust R, O’Donnell M (1993) DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J Biol Chem 268: 11773–11778 [PubMed] [Google Scholar]
- Xu L, Marians KJ (2000) Purification and characterization of DnaC810, a primosomal protein capable of bypassing PriA function. J Biol Chem 275: 8196–8205 [DOI] [PubMed] [Google Scholar]
- Yuan Q, McHenry CS (2009) Strand displacement by DNA polymerase III occurs through a tau-psi-chi link to single-stranded DNA-binding protein coating the lagging strand template. J Biol Chem 284: 31672–31679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhakov A, Kelman Z, O’Donnell M (1999) Trading places on DNA—a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell 96: 153–163 [DOI] [PubMed] [Google Scholar]
- Zechner EL, Wu CA, Marians KJ (1992) Coordinated leading- and lagging-strand synthesis at the Escherichia coli DNA replication fork. II. Frequency of primer synthesis and efficiency of primer utilization control Okazaki fragment size. J Biol Chem 267: 4045–4053 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.