Abstract
Piwi-interacting RNAs (piRNAs) are a recently discovered class of 24- to 30-nt noncoding RNAs whose best-understood function is to repress transposable elements (TEs) in animal germ lines. In humans, TE-derived sequences comprise ∼45% of the genome and there are several active TE families, including LINE-1 and Alu elements, which are a significant source of de novo mutations and intrapopulation variability. In the “ping-pong model,” piRNAs are thought to alternatively cleave sense and antisense TE transcripts in a positive feedback loop. Because piRNAs are poorly conserved between closely related species, including human and chimpanzee, we took a population genomics approach to study piRNA function and evolution. We found strong statistical evidence that piRNA sequences are under selective constraint in African populations. We then mapped the piRNA sequences to human TE sequences and found strong correlations between the age of each LINE-1 and Alu subfamily and the number of piRNAs mapping to the subfamily. This result supports the idea that piRNAs function as repressors of TEs in humans. Finally, we observed a significant depletion of piRNA matches in the reverse transcriptase region of the consensus human LINE-1 element but not of the consensus mouse LINE-1 element. This result suggests that reverse transcriptase might have an endogenous role specific to humans. Overall, our results elucidate the function and evolution of piRNAs in humans and highlight the utility of population genomics analysis for studying this rapidly evolving genetic system.
Keywords: piRNAs, transposable elements, population genetics, selective constraint, Africans
Introduction
RNA interference (RNAi)–related pathways are characterized by small noncoding RNAs, such as microRNAs or small interfering RNAs, that guide Argonaute proteins to their target RNA transcripts or chromosomal loci. RNAi-related pathways have been the object of a great deal of study in recent years and have been implicated in a myriad of important biological processes (Carthew and Sontheimer 2009). Piwi-interacting RNAs (piRNAs) are a recently discovered class of small RNAs that are conserved in metazoans, including basal metazoans, such as Cnidarians and Poriferans (Grimson et al. 2008). So far they have not been found in plants or fungi. The best-understood function of piRNAs is to repress transposable elements (TEs) in the germ line (Aravin et al. 2007; Malone and Hannon 2009). In addition, a number of other biological functions for piRNAs have been proposed, such as the regulation of non-TE mRNA transcripts (Thomson and Lin 2009). Most human piRNAs map to unique or few (<10) loci in the genome unlike Drosophila piRNAs, which mostly map to repeat regions. Human piRNAs are often clustered in the genome and have largely unknown function. However, a subset of piRNAs map to TEs and are thought to alternately cleave sense and antisense TE transcripts in a positive feedback loop called the “ping-pong model” (Brennecke et al. 2007). There are no known sequence features associated with piRNAs other than a very strong preference for uridine in the first base, although a k-mer scheme to predict new piRNA sequences was recently proposed (Zhang et al. 2011).
TEs have colonized virtually all eukaryotic genomes, and TE-derived sequences comprise ∼45% of the human genome (Lander et al. 2001). There are three known active TE families in humans: Alu, LINE-1, and SVA elements. Alus and LINE-1s are the most active TE families with fixation rates of one Alu insertion for every 21 births and one LINE-1 insertion for every 212 births (Xing et al. 2009). De novo TE insertions are thus a significant source of deleterious mutations and genetic variability in the human population (Cordaux and Batzer 2009).
The evolution of piRNAs has been studied previously by a number of groups. It has been reported that the synteny of piRNA clusters is conserved, but the sequences of the piRNAs have diverged between several pairs of species, including mouse and rat (Assis and Kondrashov 2009), Caenorhabditis elegans and Caenorhabditis briggsae (Ruby et al. 2006), and Drosophila melanogaster and Drosophila simulans (Malone et al. 2009). One exception to these studies is a report that piRNA expression level is positively correlated with conservation across species (Lau et al. 2006). Nonetheless, the overall picture of piRNA sequence evolution between species is consistent with a divergence rate similar to neutrally evolving sequences. Little is known about the evolution of piRNAs within species beyond a recent simulation study in Drosophila that found that piRNA repression of TEs can not only increase host fitness but also allow increased TE copy number (Lu and Clark 2010). In order to study the recent evolution of piRNAs and TEs in humans, we performed a population genomics study using publicly available single nucleotide polymorphism (SNP) genotype data from the HapMap Project phase 3.
Materials and Methods
Data
We downloaded human piRNAs sequenced by Girard et al. (2006) and mouse piRNAs sequenced by Lau et al. (2006) and Girard et al. (2006) from GenBank. We mapped them to the human genome (UCSC Genome Browser version hg18) and mouse genome (UCSC Genome Browser version mm9), respectively. The read mapping was performed with the BWA tool (Li and Durbin 2009) using 0 mismatches unless stated otherwise. We downloaded PhyloP scores (Pollard et al. 2010), RepeatMasker repeat annotations (http://www.repeatmasker.org), and multiZ multiple alignments (Blanchette et al. 2004) of all available primate genomes from the UCSC Genome Browser (Rhead et al. 2010). For the flanking regions of piRNAs, we used 1,000 nt on each side of the piRNA, but we excluded any sequences overlapping RefSeq genes.
To study the fraction of TE bases mapping to at least one piRNA, we obtained consensus sequences for Alus and LINE-1s from GenBank. Our results were unchanged when we used consensus sequences from other sources (Price et al. 2004; Khan et al. 2006).
Results
piRNAs Have Evolved Rapidly between Human and Chimpanzee
We mapped a large data set of previously sequenced human piRNAs (Girard et al. 2006) to the human genome and identified 24,646 piRNAs that mapped uniquely to the genome (Materials and Methods). The uniquely mapping human piRNAs had features consistent with piRNAs from other species. They clustered into 36 broad clusters (<90 kb with >100 uniquely mapping piRNAs). They also showed a strong preference for uridine in position 1 and a weaker but still discernible preference for adenosine in position 10 (supplementary fig. S1, Supplementary Material online). This nucleotide profile is consistent with the ping-pong model (Brennecke et al. 2007) in which primary and secondary piRNAs derived from sense and antisense copies of TEs alternatively cleave each other at the bond between the nucleotides that base pair to nucleotides 10 and 11 of the piRNA.
We measured the sequence conservation of human piRNAs in primates in two ways. First, we used sequence conservation scores computed by the PhyloP method (Pollard et al. 2010). Using this approach, we found a slightly higher rate of conservation in piRNAs than in flanking regions in primates (avg. PhyloP score 0.65 vs. 0.59) that was not statistically significant (P > 0.22). Second, we simply counted nucleotide substitutions between human and chimpanzee. With this method, we found no significant difference in substitution rates between piRNAs and their flanking regions (Material and Methods; Binomial test, P > 0.2). From these two tests, we concluded that human piRNAs have evolved at a similar rate to their flanking regions between human and chimpanzee. This observation is consistent with previous results in rodents, Drosophila, and nematodes and has been previously interpreted to mean that the sequences of the piRNAs might not be functionally important (Girard et al. 2006).
piRNAs Show a Signature of Selective Constraint in African Populations
Although piRNAs are not well conserved between species, we reasoned that they might be under detectable selective constraint at a shorter time scale if they are rapidly evolving genes. For example, such an evolutionary pattern would be expected if piRNAs were involved in transposon repression in primates. This would be consistent with their known role in transposon repression in Drosophila and mouse.
To investigate the strength of selection in humans, we used data from the HapMap Project (phase 3) consisting of SNP genotype data from 1115 individuals in 11 populations (http://www.sanger.ac.uk/humgen/hapmap3). We used the chimpanzee allele to root the SNPs, a procedure which is expected to be accurate in the vast majority of cases. To handle the remaining cases, we corrected for ancestral allele misidentification using a method very similar to a previous method (Hernandez et al. 2007). This correction did not affect our results significantly.
We compared the derived allele frequency distributions of piRNA SNPs with intergenic regions in the genome in each of the 11 HapMap populations separately using a Wilcoxon test (supplementary figs. S1, S4, and S5, Supplementary Material online; table 1). We assumed that intergenic regions are evolving neutrally, but if they are in fact evolving under moderate levels of selective constraint, that would only strengthen our results. For this analysis, we made sure to remove all piRNAs that overlapped with exons to avoid any spurious signatures of selective constraint.
Table 1.
P Values from Wilcoxon Tests for Individual HapMap Phase 3 Populations
| Population | Number of piRNA SNPs | P Value |
| ASW | 248 | 0.000951 |
| CEU | 212 | 0.316 |
| CHB | 202 | 0.219 |
| CHD | 197 | 0.114 |
| GIH | 213 | 0.0651 |
| JPT | 199 | 0.0318 |
| LWK | 246 | 0.00179 |
| MEX | 231 | 0.219 |
| MKK | 230 | 0.000377 |
| TSI | 218 | 0.140 |
| YRI | 245 | 0.000119 |
Note.—The P values shown were not corrected for multiple hypothesis testing. P values significant at the 5% threshold after Bonferroni correction are shown in bold. The population names are from the HapMap Project Phase 3. ASW, African Americans; CEU, Europeans; CHB, Chinese in Beijing; CHD, Chinese in Denver; GIH, Gujarati Indians; JPT, Japanese; LWK, Luhya; MEX, Mexicans; MKK, Masai; TSI, Tuscans; YRI, Yorubans.
The statistical tests showed that piRNAs are evolving under significantly greater selective constraint compared with intergenic regions in all four African populations, namely Yoruba in Ibadan, Nigeria (YRI); individuals of African ancestry in the Southwest USA (ASW); Luhya in Webuye, Kenya (LWK); and Maasai in Kinyawa, Kenya (MKK) (Bonferroni-corrected P < 0.02; table 1). In the seven non-African populations, we observed a trend for piRNAs to be under greater selective constraint than intergenic regions, but the results were not statistically significant after Bonferroni correction for multiple hypothesis testing (Bonferroni-corrected P > 0.35; table 1). Taken together, there is strong statistical support for selective constraint on piRNAs in African populations and only weak evidence for selective constraint in non-African populations.
It has been reported (Lohmueller et al. 2008) that Europeans harbor more deleterious polymorphisms than Africans because in general, non-African groups have smaller population sizes than Africans and therefore are more sensitive to the effects of random drift. At first glance, the higher amounts of selective constraint on piRNA sequences that we observed in Africans compared with non-Africans are consistent with these data. However, we would expect a population-wide effect such as a population size difference to be visible in other classes of functional sites as well, in particular nonsynonymous sites. The fact that we do not observe such an effect (supplementary figs. S1, S4, and S5, Supplementary Material online) suggests that the increased selective constraint we observe in African populations is in fact specific to piRNAs.
Because the biological function of piRNAs in humans is still poorly understood, it is difficult at this point to connect the stronger selective constraint in Africans to a particular biological function. However, if a significant fraction of the uniquely mapping piRNAs are involved in transposon defense, then the patterns we observe are consistent with recent data that show a much higher rate of transposon insertions in African compared with non-African populations (Ewing and Kazazian 2010). Under this scenario, a higher transposition rate in Africans imposes stronger selective pressure on piRNAs to repress the TEs. In principle, piRNA expression could be population dependent, and it might be that the patterns we observed are due to the piRNAs being sequenced from African testis samples. However, the samples were in fact taken from three Caucasian males (Girard et al. 2006) so if anything, the results should be biased toward stronger negative selection in European populations, which we do not observe.
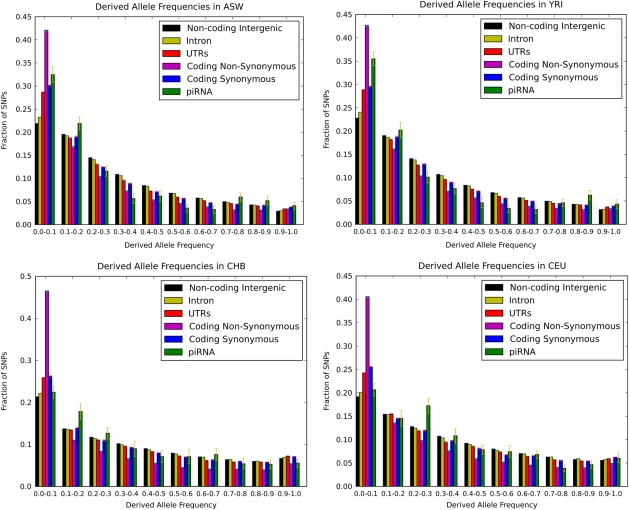
Synonymous SNPs are some times used as a standard for neutral evolution. We note that in our analysis, HapMap synonymous SNPs show an enrichment of low-frequency alleles relative to intergenic regions (fig. 1). We attribute this pattern to the effects of linkage with nonsynonymous alleles as well as ascertainment bias in the HapMap project to oversample SNPs in genes, including synonymous SNPs.
FIG. 1.
Derived allele frequency distributions for different classes of functional sites in the following HapMap phase 3 populations: ASW (African ancestry in Southwest USA), YRI (Yoruba in Ibadan, Nigeria), CHB (Han Chinese in Beijing, China), and CEU (Utah residents with Northern and Western European ancestry from the CEPH collection). An excess of SNPs in piRNAs with low derived allele frequency relative to intergenic SNPs is a signature of selective constraint on piRNA sequences. The error bars were computed by bootstrapping samples of SNPs.
Although the HapMap data are affected by ascertainment bias between different functional classes of sites, ascertainment should be uniform between intergenic regions and thus there should be no ascertainment bias between piRNAs in intergenic regions and other intergenic regions. To further control for ascertainment biases specific to different regions in the genome, we repeated our analysis using only intergenic SNPs in 100-kb flanking regions of piRNA genes as the background set. In this analysis, the P values for all four African-derived populations remained statistically significant after Bonferroni correction (P < 0.014) but not the P values for any of the other populations (P > 0.13). Finally, the Wilcoxon test that we used in our analysis requires that the SNPs be evolving independently and therefore not be in strong linkage disequilibrium with each other. However, this is not expected to be an issue for piRNA genes, which are widely distributed across the genome.
Repeat-Associated piRNAs Directly Repress Active Human LINE-1 and Alu Elements
Thus far, we have studied only the subset of uniquely mapping piRNAs. Although the majority of our piRNAs mapped to nonrepetitive regions of the genome (supplementary fig. S2, Supplementary Material online), in this section, we focused our attention on the subset of piRNAs that map to repetitive elements and might function to repress TEs. We reasoned that if human piRNAs are involved in silencing active copies of TEs in humans, then we should see a signature for more piRNAs to map to the two most active human TE families, namely LINE-1 Ta-1 and AluY elements (Batzer and Deininger 2002; Boissinot et al. 2004) compared with nonactive LINE-1 and Alu subfamilies.
To test this hypothesis, for each subfamily of TEs, we computed the fraction of consensus TE bases for which there is at least one piRNA that maps to that base. We refer to this fraction as the “density” of piRNA matches. For the LINE-1 subfamilies, we found a very strong correlation between the age of the TE family and piRNA mapping density (table 2; fig. 2). The nearly linear relationship between the age of the subfamily and density of piRNA matches suggests that most, if not all, LINE-1 derived piRNAs target or are produced from active LINE-1 elements. The piRNA matches to older LINE-1 subfamilies are likely to result from sequence similarity between the different subfamilies.
Table 2.
Percentage of Bases of LINE-1 Subfamilies That Match piRNAs
| LINE-1 Subfamily | Number of Bases in LINE-1s | Percentage of Bases Matching piRNAs (1 mismatch, 1 indel), % | Percentage of Bases Matching piRNAs (1 mismatch, 0 indel), % | Percentage of Bases Matching piRNAs (0 mismatch, 0 indel), % |
| LINE-1 HS (human specific) | 3,458,046 | 15.96 | 11.87 | 4.23 |
| LINE-1 PA2 (7.6 Ma) | 9,493,804 | 16.56 | 11.35 | 3.83 |
| LINE-1 PA3 (12.5 Ma) | 18,923,178 | 12.71 | 8.91 | 3.23 |
| LINE-1 PA4 (18.0 Ma) | 18,340,583 | 11.18 | 7.17 | 2.23 |
| LINE-1 PA5 (20.4 Ma) | 15,765,637 | 11.55 | 6.92 | 1.64 |
| LINE-1 PA6 (26.8 Ma) | 10,819,404 | 9.26 | 5.90 | 1.48 |
| LINE-1 PA7 (31.4 Ma) | 19,129,677 | 6.14 | 3.55 | 0.96 |
| LINE-1 PA8 (40.9 Ma) | 6,561,140 | 7.06 | 4.23 | 1.00 |
| LINE-1 (all) | 504,651,578 | 3.09 | 1.63 | 0.37 |
Note.—All LINE-1 subfamilies annotated in RepeatMasker (http://www.repeatmasker.org) are listed from youngest to oldest (top to bottom). The age of each LINE-1 subfamily was taken from Khan et al. (2006). The number of bases contained in TEs from each subfamily (column 2) and the percentage of bases that match piRNAs are shown at different matching stringencies (columns 3–5). There is a strong correlation between the age of the subfamily and the percentage of bases that match piRNAs.
FIG. 2.
Correlation between the age of human LINE-1 subfamilies and number of bases in the consensus sequence of that subfamily matching piRNAs.
We repeated the same analysis for subfamilies of Alus and observed a similar trend for the Alus when we grouped the subfamilies into three bins across evolutionary time (table 3). However, the correlation between subfamily age and piRNA density was not apparent over the finer time scale within the bins. The weaker correlation we observed for Alu elements may be due to the greater uncertainty in the dating of the Alu subfamilies because they are much smaller than LINE-1 elements.
Table 3.
Percentage of Bases of Alu Subfamilies That Match piRNAs
| Alu Subfamily | Number of Bases in Alu Subfamily | Percentage of Bases Matching piRNAs (1 mismatch, 1 indel), % | Percentage of Bases Matching piRNAs (1 mismatch, 0 indel), % | Percentage of Bases Matching piRNAs (0 mismatch, 0 indel), % |
| AluYg6 (2 Ma) | 162,316 | 33.99 | 21.86 | 13.86 |
| AluYb9 (5 Ma) | 9,126,467 | 41.17 | 20.77 | 17.29 |
| AluYb8 (5–15 Ma) | 8,802,284 | 50.87 | 31.30 | 20.23 |
| AluYa5 (5–15 Ma) | 1,168,599 | 30.46 | 19.92 | 17.66 |
| AluY (25 Ma) | 39,622,226 | 29.78 | 17.57 | 9.05 |
| AluSg (31 Ma) | 23,605,918 | 28.64 | 15.39 | 4.70 |
| AluSx (37 Ma) | 97,504,435 | 28.07 | 14.04 | 3.69 |
| AluSq (44 Ma) | 26,932,423 | 32.23 | 18.16 | 4.97 |
| Alus (all) | 307,703,885 | 27.01 | 14.35 | 4.20 |
Note.—All Alu subfamilies annotated in RepeatMasker (http://www.repeatmasker.org) are listed from youngest to oldest (top to bottom). The age of the Alu subfamilies was compiled from data in Kapitanov and Jurka (1995), Batzer and Deininger (2002), and Salem et al. (2003). The number of bases contained in TEs from each subfamily (column 2) and the percentage of bases that match piRNAs are shown at different matching stringencies (columns 3–5). The bold horizontal lines demarcate major transitions in Alu evolution (Batzer and Deininger 2002). The correlation between the age of the subfamily and the percentage of bases that match piRNAs is discernable across the major groups of Alus. However, within groups, the correlation is weaker than the correlation for LINE-1 elements, perhaps because of the greater uncertainty in the ages of the Alu subfamilies.
In addition to humans, there are also several large data sets available of mouse piRNAs and LINE-1 elements (Materials and Methods). We were interested if a similar pattern that we observed in humans was also observable in mouse, so we repeated our analysis for mouse piRNAs and LINE-1 subfamilies. In this case, we were only able to divide the mouse LINE-1 subfamilies into active versus inactive subfamilies, and indeed, we found that active mouse LINE-1s have more piRNA matches than inactive mouse LINE-1s (table 4).
Table 4.
Percentage of Bases of Mouse LINE-1 Subfamilies That Match Mouse piRNAs
| Subfamily | 0 Mismatch, % | 1 Mismatch, % | 2 Mismatch, % | |
| Inactive | L1MdF | 1.73 | 6 | 13.2 |
| L1MdF2 | 4.6 | 15.1 | 21.8 | |
| L1MdF3 | 4.85 | 14.2 | 21.6 | |
| Active | L1MdGf | 5.8 | 16.8 | 22.52 |
| L1MdT | 10.3 | 18.9 | 26.5 | |
| L1MdA | 9.45 | 17.5 | 24.0 |
Repeat Masker (http://www.repeatmasker.org) annotates the six youngest LINE-1 subfamilies in the mouse genome (UCSC genome version mm9) as L1MdF, L1MdF2, L1MdF3, L1MdT, L1MdGf, and L1MdA. The different F-subfamilies annotated as L1MdF, L1MdF2 and, L1MdF3 summarize a more complex phylogeny of up to 17 subfamilies of mouse-specific LINE-1s. LINE-1 elements belonging to the subfamilies L1MdT, L1MdGf, and L1MdA have been reported to be currently active (Naas et al. 1998; Hardies et al. 2000; Goodier et al. 2001).
Finally, we wanted to exclude the possibility of putative piRNAs matching TEs due simply to contamination of the sequencing reads by degradation products of highly expressed TE transcripts. To do this, we verified that 89% of the piRNAs matching TEs have a canonical 5′ uridine. Such a pattern would not be expected if we were observing random degradation products. Furthermore, there were roughly equal numbers of sense and antisense piRNAs (data not shown) mapping to human LINE-1 elements, consistent with the ping-pong model. Taken together, our data argue for an active role for human and mouse piRNAs in TE repression.
It would be interesting to study the impact of the loss of piRNA matches on the transposition rate of active LINE-1s. In humans, subfamilies of LINE-1s are characterized by only a small number of diagnostic nucleotides. We found one example of a diagnostic nucleotide that matches to a piRNA. In particular, full-length copies of young subfamilies Ta1-nd, Ta-0, and pre-Ta (Brouha et al. 2003) contain an average of 11 piRNA-binding sites. However, the youngest and most active subfamily, Ta1-d, characterized by just four diagnostic nucleotide differences with respect to the next youngest subfamily Ta1-nd, contains only 10 piRNA matches. This is because one of the diagnostic base substitutions is a mutation at nucleotide 355 of the consensus sequence (Boissinot et al. 2000) from an ancestral A to a G. The A was complementary to the 5′ most base of a piRNA which matches the subsequence between bases 326 and 355 in the Ta1-nd, Ta-0, and pre-Ta subfamilies. Thus, it is tempting to speculate that some cases of increased LINE-1 subfamily transposition are facilitated by small sequence changes that lead to an escape from piRNA repression.
The LINE-1 Reverse Transcriptase Region Is Depleted of piRNA Matches in Human But Not in Mouse
Finally, we examined the pattern of piRNA matches to the consensus LINE-1 element (fig. 3) and Alu element (supplementary fig. S3, Supplementary Material online). We noticed that both families of TEs are depleted of piRNA matches at specific locations in their sequences. For the case of Alu elements, we noted that there is an A-rich element in the region of the Alu element that is depleted of piRNAs. This is consistent with our observation that piRNAs are depleted of long AT-tracts (data not shown). Because low-complexity sequences were not masked in the mapping tool that we used (Materials and Methods), we believe that the depletion of matches that we observed is a real phenomenon and not an artifact of the mapping procedure.
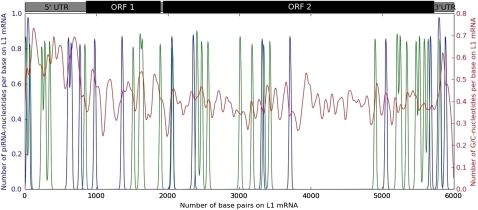
FIG. 3.
Density of piRNA matches to the consensus sequence of human-specific LINE-1 elements. To smooth the density, the plots were made using kernel density estimation with a Gaussian kernel instead of histograms. The blue (green) line shows the density of sense (antisense) piRNA matches to the LINE-1 element. The ∼1-kb region in the coding region of ORF2 that is depleted of piRNA matches is also depleted across all primate-specific LINE-1s in humans. There are 1,134 bases in LINE-1s that match piRNAs.
Intriguingly, there is a clear region of the human LINE-1 transcript that is depleted of piRNAs, and this region contains the domain of LINE-1 ORF2 that functions as reverse transcriptase. We were unable to detect a base composition bias in this region similar to the case of Alu elements that could explain the paucity of piRNA matches (fig. 3). Thus, it is possible that there may be a functional reason for the depletion of piRNA matches in this region.
To further investigate the phenomenon of piRNA depletion in LINE-1s further, we performed a similar analysis in mouse LINE-1 elements. In the mouse case, we did not find a similar depletion of piRNA matches in the reverse transcriptase region of ORF2. It is thus tempting to speculate that at least one copy reverse transcriptase is functional in humans, but not mouse, and therefore is protected from piRNA-mediated repression.
Discussion
In this study, we have examined the evolution of human piRNAs and TEs over a short time scale, namely evolution within the human lineage. We have made three major observations regarding the evolution and function of piRNAs in humans. First, our population genomics study shows that piRNA sequences are under selective constraint within human African populations, even though they have evolved quickly between human and chimpanzee. We also noted a trend for selective constraint in non-African populations that was not statistically significant. The apparent contradiction between the intraspecies analysis and the interspecies analysis can be resolved by one of two nonexclusive interpretations. One explanation is that the strength of selective constraint may simply differ between these two time scales. Such rapid evolution would be expected for genes that mediate defense against parasites such as transposons. The other explanation is that the interspecies substitution rate, but not the derived allele frequency distribution, is affected by mutation rate biases. It is possible that uniquely mapping piRNA loci might be preferentially located in regions of higher mutation rate, leading to a higher substitution rate across species. Such a preference for higher mutation rates would be consistent with a previous result that suggested that genes involved in communication processes such as cell surface receptors and immune response genes tend to be in high mutation rate regions of the human genome (Chuang and Li 2004).
Our second major result is that piRNA mapping density, that is, the fraction of TE bases mapping to at least one piRNA, in Alus and human and mouse LINE-1s correlates with the age of the subfamily. This result is consistent with an active role for piRNAs in transposon repression in these lineages. Although this result was previously known for mouse, it had not been previously shown for humans. We observed that the density of piRNA mapping to Alus is significantly higher (chi-square test, P < 10−15) than the density of piRNAs mapping to LINE-1s. For example, in the youngest subfamilies, we observe a density of 16% for LINE-1s (table 2), which is much lower than 34% for Alus (table 3). One possible explanation for this observation is that the piRNAs were sequenced in the male germ line (Girard et al. 2006) in which there are significantly more hypomethylated Alus, and presumably higher Alu expression, than the female germ line (Rubin et al. 1994). Another possible explanation is that Alus are ∼20 times shorter than LINE-1s so a higher density of piRNAs may be needed to silence them efficiently.
Our finding that a diagnostic nucleotide of the currently most active human LINE-1 subfamily matches to a piRNA motivates the question of how frequent are mutations in piRNA-binding sites and what is the impact of such mutations on the transposition rate of TEs. Currently, the mechanism of piRNA repression is unclear and it is also not known what the relative expression levels of piRNAs and their TE targets are. Therefore, it is not clear if a single mismatch to a piRNA would be enough to alter piRNA repression or the transposition rate of the TE. Nonetheless, our finding is intriguing because of the very small number of nucleotide changes differentiating active from nonactive human LINE-1s. Further research on the piRNA pathway will allow us to elucidate answers to these questions and to further understand the transposition dynamics of LINE-1 elements.
Our third and final result is that the reverse transcriptase region of ORF2 of the human LINE-1 consensus sequence is depleted of piRNA matches, but there is no apparent depletion of piRNA matches anywhere on the mouse LINE-1 consensus sequence. We can only speculate about a possible reason for this depletion, but one possibility is that at least one reverse transcriptase in humans is functional and therefore protected from piRNA repression. One example of a difference between primates and rodents relevant to the reverse transcriptase gene is the presence of Alu elements in primates that rely on the LINE-1 reverse transcriptase to insert themselves into the genome. Nonetheless, more follow-up work is needed before we can be confident of an explanation for the phenomenon we observed.
Supplementary Material
Acknowledgments
We thank David Gould for assistance with the cross-species conservation analysis and Mark Batzer, Abram Gabriel, Jody Hey, Ravi Sachidanandam, Jun Song, Zhiqiang Tan, and Jinchuan Xing for discussions. This work was partially funded by the National Institutes of Health (R00HG004515 to K.C.).
References
- Aravin A, Hannon G, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Assis R, Kondrashov A. Rapid repetitive element-mediated expansion of piRNA clusters in mammalian evolution. Proc Natl Acad Sci U S A. 2009;106:7079–7082. doi: 10.1073/pnas.0900523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer M, Deininger P. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Kent W, Riemer C, Elnitski L, Smit A, Roskin K. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Chevret P, Furano A. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- Boissinot S, Entezam A, Young L. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 2004;14:1221–1231. doi: 10.1101/gr.2326704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin A, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon G. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge R, Lutz-Prigge S, Farley A, Moran J. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R, Sontheimer E. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J, Li H. Functional bias and spatial organization of genes in mutational hot and cold regions in the human genome. PLoS Biol. 2004;2:e29. doi: 10.1371/journal.pbio.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer M. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing A, Kazazian H forthcoming. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2010;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon G, Carmell M. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;7099:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goodier J, Ostertag E, Du K, Kazazian HJ. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Srivastava M, Fahey B, Woodcroft B, Chiang H, King N. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455:1193–1197. doi: 10.1038/nature07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies S, Wang L, Zhou L, Zhao Y, Casavant N, Huang S. LINE-1 (L1) lineages in the mouse. Mol Biol Evol. 2000;17:616–628. doi: 10.1093/oxfordjournals.molbev.a026340. [DOI] [PubMed] [Google Scholar]
- Hernandez R, Williamson S, Bustamante C. Context dependence, ancestral misidentification, and spurious signatures of natural selection. Mol Biol Evol. 2007;24:1792–1800. doi: 10.1093/molbev/msm108. [DOI] [PubMed] [Google Scholar]
- Kapitanov V, Jurka J. The age of Alu subfamilies. J Mol Evol. 1995;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Linton L, Birren B, et al. (12 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lau N, Seto A, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel D. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmueller K, Indap A, Schmidt S, Boyko A, Hernandez R, Hubisz M. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Clark A. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 2010;20:212–227. doi: 10.1101/gr.095406.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C, Brennecke J, Dus M, Stark A, McCombie W, Sachidanandam R. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C, Hannon G. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naas T, DeBerardinis R, Moran J, Ostertag E, Kingsmore S, Seldin M, Hayashikzaki Y, Martin S, Kazazian H. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–597. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K, Hubisz M, Rosenbloom K, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, Eskin E, Pevzner P. Whole-genome analysis of Alu repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn R, Hinrichs A, Zweig A, Fujita P, Diekhans M. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin C, VandeVoort C, Teplitz R, Schmid C. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. 1994;22:5121–5127. doi: 10.1093/nar/22.23.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J, Jan C, Player C, Axtell M, Lee W, Nusbaum C. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Salem A, Kilroy G, Watkins W, Jorde L, Batzer M. Recently integrated Alu elements and human genomic diversity. Mol Biol Evol. 2003;20:1349–1361. doi: 10.1093/molbev/msg150. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, et al. (11 co-authors) Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Kang L forthcoming. A k-mer scheme to predict piRNAs and characterize locust piRNAs. Bioinformatics. 2011;27:771–776. doi: 10.1093/bioinformatics/btr016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.