Abstract
Bacterial genome annotations contain a number of coding sequences (CDSs) that, in spite of reading frame disruptions, encode a single continuous polypeptide. Such disruptions have different origins: sequencing errors, frameshift, or stop codon mutations, as well as instances of utilization of nontriplet decoding. We have extracted over 1,000 CDSs with annotated disruptions and found that about 75% of them can be clustered into 64 groups based on sequence similarity. Analysis of the clusters revealed deep phylogenetic conservation of open reading frame organization as well as the presence of conserved sequence patterns that indicate likely utilization of the nonstandard decoding mechanisms: programmed ribosomal frameshifting (PRF) and programmed transcriptional realignment (PTR). Further enrichment of these clusters with additional homologous nucleotide sequences revealed over 6,000 candidate genes utilizing PRF or PTR. Analysis of the patterns of conservation apparently associated with nontriplet decoding revealed the presence of both previously characterized frameshift-prone sequences and a few novel ones. Since the starting point of our analysis was a set of genes with already annotated disruptions, it is highly plausible that in this study, we have identified only a fraction of all bacterial genes that utilize PRF or PTR. In addition to the identification of a large number of recoded genes, a surprising observation is that nearly half of them are expressed via PTR—a mechanism that, in contrast to PRF, has not yet received substantial attention.
Keywords: frameshift mutation, pseudogene, IS element, recoding, programmed ribosomal frameshifting, transcriptional realignment, transcriptional slippage, nonstandard decoding, RNA editing
Introduction
With hundreds of complete bacterial genomes available in current databases, the potential for exploration of genes and their protein products by means of comparative genomics is greater than ever. According to the Genomes OnLine Database (Liolios et al. 2010), 982 completed and 977 draft prokaryotic genomes were available in September 2009. Annotation of prokaryotic genes requires the identification of open reading frames (ORFs) that encode proteins. However, in some cases, a region annotated as protein coding (i.e., a coding sequence or CDS) does not constitute a single ORF. Besides annotation errors, the reasons for such observations are as follows: 1) sequencing and assembly errors; 2) insertions or deletions (indels) of one or more nucleotides due to mutation or recombination; and 3) the presence of a nonstandard, for example, nontriplet, decoding mechanism involved in the gene expression. Figure 1 shows an example of disrupted CDS annotation.
FIG. 1.
An example of a gene with a CDS containing a disrupted ORF (smpB from Buchnera aphidicola). (A) ORF organization. Three translational phases are represented as three boxes. Stop and start codons are shown, respectively, as major and minor vertical dashes within each box. The location of the disruption is shown as a dotted line throughout all three boxes. Regions corresponding to the annotated CDS are highlighted in gray. Three major possible causes of disruptions and their distinct characteristics are summarized in the table below. (B) A fragment of the B. aphidicola completed genome (NC_008513) annotation, corresponding to the smpB gene, in GenBank format.
Nonstandard decoding has been observed in the expression of some genes in virtually all organisms and especially among viruses. Nonstandard (alternative) decoding is known to be involved in regulation of gene expression. The utilization of alternative translational decoding for gene expression (required for synthesis of a product or for regulatory purposes) is termed recoding (Gesteland et al. 1992). For comprehensive reviews on recoding, see Baranov et al. (2002a), Namy et al. (2004), Dinman (2006), and Atkins and Gesteland (2010). The recoding types most relevant to this study are programmed ribosomal frameshifting (PRF) and codon redefinition (also known as stop codon read-through when a stop codon is recognized as encoding one of the 20 standard amino acids; this is not to be confused with codon reassignment; Atkins and Baranov 2010). If PRF is involved in translation of an mRNA, a significant fraction of ribosomes change reading frame at a particular location in response to specific mRNA signal (s) and hence the product of translation is a “fusion” protein encoded by more than one ORF. Stop codon read-through occurs when translation of two ORFs separated by a single stop codon results in the synthesis of a single fusion protein with the stop codon decoded as a codon for an amino acid. The most prominent example of a bacterial gene utilizing PRF is the gene for release factor 2 (RF2), where +1 PRF occurs in nearly 90% of all known bacterial genomes (Bekaert et al. 2006). In the case of RF2, PRF essentially creates a feedback loop for RF2 expression that enables production of RF2 when its concentration in the cell is low. On the other hand, when the RF2 concentration is high, translation termination mediated by RF2 outcompetes PRF and synthesis of RF2 is reduced (Craigen and Caskey 1986; Adamski et al. 1993). Notably, PRF is especially abundant in insertion sequence (IS) elements and in bacteriophages (Baranov et al. 2006). The largest collections of confirmed genes with established recoding (∼1,500) can be found in the Recode database (Bekaert et al. 2010).
Alteration of RNA bases relative to the DNA template is commonly known as RNA editing (for reviews, see Keegan et al. 2001; Bass 2002; Maas et al. 2003; Stuart et al. 2005; Nishikura 2010). The RNA editing community has adapted the term recoding to describe those cases of RNA editing that are responsible for altering DNA templated protein sequences. While posttranscriptional point alterations of RNA sequences are known to be abundant among eukaryotes and their organelles, the only type of RNA editing documented in bacteria is what we refer to here as transcriptional realignment (also known as transcriptional slippage (Wernegreen et al. 2010), stuttering (Iseni et al. 2002), molecular misreading (Ferrer et al. 2008), and reiterative transcription (Turnbough 2011)). In this process, a growing RNA chain realigns to its DNA template in the ternary complex within the RNA polymerase and this realignment results in the insertion or deletion (indel) of a single or multiple nucleotides relative to the template (Chamberlin and Berg 1962; Wagner et al. 1990). The indel usually occurs at a homopolymeric run of nucleotides that form thermodynamically weak RNA–DNA duplexes, allowing for RNA–DNA misalignment, and a shifted realignment. This phenomenon is required for the expression of certain bacterial genes, for example, production of a fusion between the pgk and tim genes in Thermotoga maritima (Schurig et al. 1995), expression of dnaX in Thermus thermophilus (Larsen et al. 2000), expression of genes in IS elements in Deinococcus radiodurans (Baranov et al. 2005), and expression of a number of plasmid-encoded genes in Shigella (Penno et al. 2005; Penno and Parsot 2006). Similarly to ribosomal frameshifting, if the transcriptional realignment is required for the expression of a gene, it can be viewed as a programmed event and we term it programmed transcriptional realignment (PTR). Both PRF and PTR utilize nonstandard decoding that plays a functional role and positively contributes to the organism’s fitness; therefore, both PRF and PTR tend to be evolutionarily conserved.
To achieve the required level of nonstandard decoding, a majority of known examples of PRF require complex signals embedded in the mRNA. However, even relatively short sequence patterns, such as heptanucleotides or homopolymeric runs, could yield significant levels of nonstandard decoding (Weiss et al. 1987, 1990; Wagner et al. 1990; Shah et al. 2002). A logical assumption is that such patterns should be eliminated from most protein-coding regions (i.e., those that do not require recoding) by negative selection, since the nonstandard decoding would result in erroneous expression and yield aberrant protein products. Contrary to this expectation, such patterns seem to be abundant. For instance, a study of ribosomal frameshifting on the sequence A_AA.A_AA.G (here codons in the initial frame are separated by underscores and dots separate codons in the new frame) has shown that this pattern is associated with efficient −1 ribosomal frameshifting in Escherichia coli (Gurvich et al. 2003); for relevant studies in budding yeast, see Shah et al. (2002) and Jacobs et al. (2007). While the underrepresentation of such sequences in protein-coding regions is statistically significant, their overall frequency is still high: ∼70 observed in E. coli K12 versus ∼100 expected (Gurvich et al. 2003). Most of the observed occurrences of A_AA.A_AA.G happened to be located in lowly expressed genes, which agrees with the expectation that negative selection should act primarily on highly expressed genes (Gurvich et al. 2003). When decoding errors occur in lowly expressed genes, only a small number of aberrant molecules are produced. Therefore, the negative effect on the organism’s fitness is less significant. Consequently, these patterns observed in lowly expressed genes may evolve under nearly neutral selection.
An interesting consequence of the presence of PRF or PTR patterns in regular genes is as follows. If a frameshift mutation (an indel) occurs in a gene, it may not necessarily completely inactivate the gene if a compensatory shifty or slippery pattern is present sufficiently close to the frameshift mutation. In such cases, a functional protein product may still be synthesized. To discriminate such genes from those where nonstandard decoding plays a positive role and from those where frameshift mutations fully inactivate a gene (pseudogenes), the term “pseudo pseudogene” has been coined (Baranov et al. 2005). The term signifies genes that resemble pseudogenes by their structure (e.g., presence of frameshifts or premature stop codons) but nevertheless express functional products. Such genes are also known as expressed pseudogenes (Hirotsune et al. 2003). Evidence for the expression of proteins from a few disrupted genes in Sulfolobus solfataricus has been provided recently by mass-spectrometric analysis (Cobucci-Ponzano et al. 2010). In endosymbiotic bacteria with highly AT-rich genomes, long poly-A tracts were observed in nearly every protein-coding gene. Here, transcriptional realignment was experimentally demonstrated for a random selection of poly-A patterns—quite a remarkable finding (Tamas et al. 2008)! It has been suggested that the prevalence of transcriptional realignment in these bacteria provides a mechanism for increased robustness of gene expression that compensates for elevated replication error rates (Tamas et al. 2008; Wernegreen et al. 2010).
We are not aware of a simple computational method available to classify the origin of a disruption in a particular gene as a sequencing error, as a mutation that made an inactive (or expressed) pseudogene or as a programmed nonstandard decoding event such as PRF or PTR (fig. 1). The lack of computational techniques that would correctly annotate genes with disrupted ORFs has resulted in inconsistencies in annotation due to the arbitrary choices made by annotators and/or implemented in annotation pipelines. The same sequences could be treated as genes with sequencing errors, inactive pseudogenes, or active genes that restore the ORF either cotranscriptionally or posttranscriptionally. A comprehensive analysis of genes with disrupted ORFs in a single species suggested that each individual case should be investigated separately (Deshayes et al. 2007). This, however, is an excessively laborious task, whose global application is unfeasible at a time of ongoing diminishing labor, time, and cost needed for DNA sequencing. Therefore, it is necessary to develop universal computational approaches able to characterize CDS disruptions. The present work is the first pilot attempt to systematically analyze the functional status of all currently known disrupted bacterial genes.
Materials and Methods
Collecting Genes with Disrupted ORFs
Nucleotide sequences of protein-coding regions (Fasta files) of 973 bacterial genomes were downloaded from the National Center for Biotechnology Information (NCBI) on 18 September 2009. A custom perl script was used to determine the length of each gene in two ways: 1) the actual length of the sequence present in the Fasta file and 2) the difference between the coordinates of the annotated CDS start and its end, plus one. If these two values did not match, the gene was identified as a gene with a disrupted CDS. The list of 1,033 genes with disrupted CDSs (after excluding genes with large disruptions, poor sequence quality, or truncated N or C termini) is available in supplementary data set 1 (Supplementary Material online).
Identification of Sequencing Errors and Recent Mutations
To identify sequences that contain sequencing errors or recent mutations, a search was performed for homologous nucleotide sequences without such disruptions. All 1,033 sequences were subjected to a BLASTN search against the NCBI nonredundant (nr) database. The resulting alignments (with the exception of the hit to the query sequence itself) with targets that shared identity of >90% and not less than 95% coverage of the query length, were analyzed for the presence of gaps. Sequences for which we found alignments with gaps whose length and position were consistent with a disruption (34 genes), were classified as recent mutations or sequencing errors.
Sequence Clustering and Functional Classification
We clustered the remaining 999 sequences using the BLASTCLUST 2.2.17 program, a part of the standalone BLAST package. Genes with at least 45% identity at the protein level were grouped into one cluster. This identity threshold was found empirically to be the lowest identity level that allowed clustering of all RF2 genes into a single cluster. For functional classification, the protein sequences encoded by the annotated CDSs were subjected to a Pfam search using the “Batch sequence search” option (Finn et al. 2010). Statistically significant hits (with E value < 0.001) were retrieved and used for functional characterization of the genes. To identify GO terms associated with a gene, the Uniprot accession number associated with the protein product of the gene was used to search the GO database (Carbon et al. 2009).
Cluster Enrichment
Annotated CDSs (for 837 genes from 64 clusters) were conceptually translated and subjected to a TBLASTN search against the NCBI nr nucleotide database. Hits with identity of at least 40% in the regions upstream and downstream of the disruption, and whose total length was >75% of that of the query, were considered to be homologous sequences. Nucleotide sequences corresponding to each hit were extracted and added to the clusters.
Generation of Sequence Alignments
Nucleotide sequences in enriched clusters were translated into protein sequences using T-Coffee (Notredame 2010). Multiple protein sequences were aligned using MUSCLE (Edgar 2004). To get a corresponding multiple alignment of nucleotide sequences, the aligned protein sequences were back-translated to the nucleotide sequences using T-Coffee.
Synonymous-Site Conservation Analyses
The degree of conservation at synonymous sites was calculated as described in Firth and Atkins (2009); the procedure was inspired by the SSSV statistic (Simmonds et al. 2008). In order to calculate the conservation statistic for the whole alignment, ORF1 and ORF2 were fused in-frame by artificially inserting one or two “N”s, as appropriate, in each sequence just 5′ of the ORF1-frame stop codon.
Generation of Sequence Logos
Fragments of multiple alignments (30 nt upstream and 40 nt downstream of the first nucleotide in the recoding pattern) were extracted. These alignments were used as input to the Weblogo applet (Crooks et al. 2004) for the generation of sequence logos (Schneider and Stephens 1990).
Test for Purifying Selection
All sequences were split into two regions: the region corresponding to ORF1 (codons separated accordingly) and the region downstream of ORF1 (with the triplet phase defined by ORF2). Highly similar sequences (identity greater than 99%) were removed from the clusters. The remaining sequences were aligned within each cluster, using conceptually translated sequences and back-translated to generate nucleotide sequence alignments (see above). The value of ω was estimated using a maximum likelihood approach under an assumption of uniform ω over the entire phylogenetic tree derived for each cluster. Here, we used the codeml program from the PAML package (Yang 2007).
In addition to estimating ω by maximum likelihood, the dN/dS ratio was calculated directly for each pairwise alignment of each sequence in the cluster and an ancestral sequence inferred by codeml. For each pairwise alignment, the probability P value, of obtaining the observed dN/dS value under a null hypothesis of neutral evolution (ω = 1) was calculated using χ2 statistics. Then the lowest P value from the set of different P values associated with different dN/dS ratios for each sequence pair (extant and inferred ancestral sequence) was chosen for each cluster. Larger clusters tend to produce smaller minimum dN/dS ratios just by chance. Therefore to account for differences between cluster sizes, we adapted the Sidak and the Bonferroni corrections to generate a statistic that could be compared across clusters of different sizes. The Sidak correction is used to correct the statistical significance threshold, α, during multiple testing, where the effective value of α is equal to 1 − (1 − α)1/n, where n is the number of tests. Since our purpose is to correct the P value itself, the effective P value can be calculated as 1 − (1 − P)n, where n is the number of sequences in the cluster. This approximates to Pn for extremely small P values (analogous to the Bonferroni correction). This statistic, for ORF1 and ORF2, is given for each cluster in supplementary data set 2 (Supplementary Material online).
Results
A Model for Discriminating between Errors, Inactivating Mutations, and Programmed Nonstandard Decoding
The first requisite for our study is the formulation of a set of criteria for establishing the nature of disruptions in CDSs. Particularly, a criterion is needed to determine whether a disruption is real and not the result of a sequencing error. We may assume that a truly disrupted gene will have homologous disrupted genes in other genomes, while it is very unlikely that the same sequencing error would occur in the same position in several homologs. Given a sequencing error (an indel) in a particular gene in a given genome, the chance to observe the same error in an homologous gene from another independently sequenced genome is equal to the rate of sequencing indel errors which is estimated as 5.4 × 10−5 per position for sequences in GenBank in 2004 (Wesche et al. 2004). Allowing for a 20 nt vicinity, the probability of error cooccurrence increases to ∼0.001. For a typical bacterial genome size of ∼5 Mb, the total number of sequencing errors in ∼1000 genomes is ∼3 × 105. Only ∼3 × 102 of these will be retained, within a 20 nt vicinity, in their closest homologs; moreover, the expected number of errors retained in the two or three closest homologs drops to ∼3 × 10−1 and ∼3 × 10−4, respectively. Thus, the same gene disruption observed in four or more homologous sequences is highly unlikely to be a result of sequencing errors.
Next, it is important to determine whether a gene containing a disruption is expressed and, if it is expressed, whether its protein product is functional. Computational analysis could provide evidence of expression and functional significance of a disrupted gene product via detection of the presence of purifying selection acting on both parts of the CDS, that is, upstream to, and downstream from, the position of a disruption. These two pieces of evidence—conservation in multiple orthologs and the presence of purifying selection—were chosen as criteria for classifying genes with disrupted CDSs.
Identification of Disrupted Genes and Initial Filtering
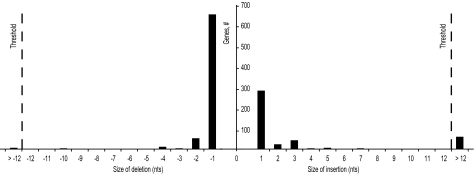
To identify bacterial genes with disrupted ORFs, we searched 973 completed bacterial genomes (see Materials and Methods) for genes with annotated CDSs containing either insertions or deletions (indels) relative to their genomic sequences. This search yielded 1,121 genes with CDS annotations inconsistent with the length of their corresponding genomic sequence (see supplementary data set 1, Supplementary Material online). The absolute values for the difference between CDS and genomic lengths varied from 1 to 11,289 nt. A histogram of the distribution of length differences between CDSs and corresponding genomic regions is shown in figure 2. The majority of disrupted genes differ by an indel of a single nucleotide. Genes with large insertions were very likely inactivated by the insertion of mobile/viral elements. Therefore, genes with disruptions larger than 12 nucleotides were deemed to be inactive and were excluded from further analysis.
FIG. 2.
Distribution of differences between the lengths of CDSs and the lengths of the corresponding genomic sequences for genes with disrupted ORFs. The threshold of 12 nt that was used to select genes for further analysis is indicated.
Furthermore, we removed genes containing ambiguous nucleotide symbols (five instances) as well as genes containing multiple disruptions (seven instances) since multiple disruptions are unlikely to accumulate or evolve in active genes. Genes with CDSs lacking either a start or a stop codon were also removed. The last preliminary filter involved the identification of nearly exact copies of disrupted genes which differed from the disrupted gene only by the disruption itself (see Materials and Methods and supplementary data set 1, Supplementary Material online). Finding such an orthologous gene reveals that the disruption is either a sequencing error or a very recent mutation. Even if the gene is still active despite the mutation, the recent nature of the mutation would not allow us to perform a meaningful phylogenetic analysis as outlined in the previous section. Therefore, we removed 34 such cases from further analysis. A diagram of the various steps involved in the CDS filtering pipeline prior to clustering analysis is shown in figure 3A.
FIG. 3.
Schemes for the analysis of genes with disrupted ORFs: (A) A pipeline for filtering genes with annotated disruptions prior to the initial clustering based on sequence similarity. (B) Scheme for the analysis of the detected clusters.
Clustering Sequences Based on Their Similarity
The genes (999) that passed the initial filtering were clustered based on sequence similarity (see Materials and Methods). As a result, 837 genes were grouped into 64 clusters, whereas 162 genes did not share sufficient sequence similarity and were treated as singletons. Figure 4A shows the distribution of genes among the clusters and among different genomes; a white disk with gray vertical stripes corresponds to singletons. Since we found that functional assignments of the genes in these clusters are sometimes inconsistent, we performed a systematic analysis and reannotation using Pfam (Finn et al. 2010) and Gene Ontology (Carbon et al. 2009), see Materials and Methods. The results can be found in supplementary data set 1 (Supplementary Material online).
FIG. 4.
Representation of genes and genomes in the clusters: axis y indicates the number of genes in each cluster; axis x shows the number of genomes represented in each cluster. The areas of disks are proportional to the number of genes in the clusters. The white disk with vertical gray stripes (also indicated as S) represents singletons–genes that did not cluster. The white disk with horizontal gray stripes (also indicated as RF2) represents cluster 2 (RF2 genes). Clusters containing mobile genetic elements (transposons and IS elements) are shown as gray disks. Clusters of genes with other functions are shown as black disks. (A) Clusters containing genes with annotated disruptions (prior to the enrichment). (B) Clusters after the enrichment (annotated genes and their homologs identified using TBLASTN).
Two functional groups are particularly overrepresented in the initial dataset—RF2 and functional groups corresponding to mobile elements such as transposases and integrases. The RF2 gene cluster has 158 members and represents the largest number of genomes (see fig. 4). However, the number of genes in the RF2 cluster (158) is considerably smaller than what would be expected from our previous estimate (Bekaert et al. 2006) that about 5% of bacterial genomes have lost RF2 genes, whereas close to 90% of species utilize PRF in RF2 expression. This gives ∼800 RF2 genes in the data set of 973 genomes. Such a difference between the expected and observed number of RF2 genes annotated with disruptions indicates that about 80% of RF2 genes are misannotated in bacterial genomes. Notably, the proportion of misannotated RF2 genes is even larger than observed 4 years ago, when we developed a tool (ARFA) specifically for annotating PRF in RF2 (Bekaert et al. 2006). The majority of other clusters are formed by genes located in mobile genetic elements such as IS elements and transposons (fig. 4). Similar to RF2 genes, IS elements with disrupted ORFs are also misannotated in many genomes (e.g., see annotation of IS1, IS2, and IS3 in E. coli K12 [Refseq accession NC_000913]) and therefore escaped our initial selection. The frequent presence of several identical IS element copies within the same genome suggests that these IS elements are currently active in their transposition. An alternative possibility is that these IS element copies are all inactive due to a mutation causing a gene disruption that appeared in one copy and was propagated to the others by gene conversion (Cordaux 2009). However, two pieces of evidence provide support in favor of these genes being active and utilizing nonstandard decoding to express IS-transposase. One is the conservation of a unique gene disruption among phylogenetically related IS elements from different species. The second is the experimental demonstration of functionality through expression by PRF of disrupted ORFs from members of the two most abundant groups revealed by previous studies (for IS1 by Sekine et al. 1992 and for IS3 families by Licznar et al. 2003). The remaining gene clusters contain genes attributed to other functions (neither IS elements nor RF2 genes); these clusters are shown as black disks in figure 4.
Further inspection of ORF organization in the clustered genes also revealed a subset of genes where both ORFs are in the same reading frame, but the CDS annotation misses a triplet of nucleotides relative to the genomic sequence. Genes from six clusters (14, 30, 41, 48, 57, and 64; see supplementary data set 1, Supplementary Material online) are homologs of known selenoproteins. However, instead of annotating the full length ORF containing the in-frame UGA codon (which encodes selenocysteine), each CDS is annotated as a fusion of two ORFs flanking the UGA codon. Consequently, the annotated protein products miss selenocysteine in their sequence despite the fact that many of these genes were correctly annotated as selenoprotein genes. We also identified a cluster (32) that contains genes with another stop codon (UAG) excluded from the CDS. UAG is not known to encode selenocysteine and, not surprisingly, we did not find homologs among known selenoproteins. There were only four UAG-containing sequences in this cluster. Notably, a large number of very close homologs of genes from this particular cluster have CAG in the syntenic location. Since all UAG-containing sequences appeared in the same genome, Streptococcus equi, with the sequence surrounding the UAG identical among them, we concluded that it is likely to be a case of an inactivating mutation that propagated into other gene copies via homologous recombination or gene conversion. Alternatively, it is possible that a C-to-U mutation resulted in a leaky stop codon and that the corresponding variant still represents an active version of the transposon. The lack of other substitutions in the sequences prevents us from discriminating between these two possibilities via analysis of Ka/Ks.
A special case in our study is a class of disruptions that occurs in certain genes due to phase variation. As a result of replicational slippage, indels are inserted in DNA leading to a heterogeneous population of genomes within a population of the same bacterium. This phenomenon contributes to genome plasticity and is particularly common among cell surface and secretory genes of pathogenic bacteria where such plasticity facilitates adaptation to the host and provides resistance to the host immune response (van der Woude and Baumler 2004; Groisman and Casadesus 2005). Since phase variation occurs at the level of replication, the responsible slippery sites should have variable length and hence lead to gaps in the corresponding alignments. Similarly, the reading phase difference between the two ORFs will vary, including some cases where the two ORFs are not in fact disjoint at all. This is in contrast to the cases of PRF, where the sequence patterns at which nonstandard decoding occurs and the phase difference of the relevant ORFs, is expected to be evolutionarily conserved (the shift is either in the −1 or the +1 direction). In the case of PTR, it can be both −1 and +1 since transcriptional slippage could result in insertion or deletion of one or more nucleotides.
For seven clusters, the analysis of identified homologs revealed high variability in the repeat length as well as a large number of orthologs whose coding regions comprise a single undisrupted ORF. Therefore, these clusters were classified as instances of phase variation.
Cluster 50 contains two sequences, which share homology only in the second ORF. These sequences likely correspond to genes with incorrectly annotated initiation codons for the second ORF, which is likely to be expressed independently of the first ORF.
Cluster Enrichment
The larger is the number of genes in a cluster, the stronger is the phylogenetic signal. Therefore, we decided to enrich each cluster by searching for additional homologs in the NCBI nr database. We reasoned that the existence of a bona fide disruption in a gene’s CDS may preclude annotation of the full CDS in a large proportion of homologs. This reasoning is also supported by our observation that only a proportion of RF2 genes appear in the initial data set. Therefore, we performed a TBLASTN search (using protein sequences predicted in our data set) against the NCBI database of nucleotide sequences. Sequences whose proteins were encoded in two ORFs, with at least 40% identity to the query, were extracted and used for cluster enrichment. Overall, we identified 8,046 such sequences (454 sequences containing multiple disruptions were discarded). Importantly, some sequences amongst these 8,046 were repeated since they were found as hits to sequences from more than one cluster. The total number of unique gene sequences in the combined data set was 6,268. Among these genes, 4,001 sequences were derived from the same set of complete genomes that were used for the initial identification of disrupted genes. This large number suggests that genes with disrupted ORFs that are likely to be expressed by nonstandard decoding are frequently missed in annotations of completed genomes.
The distribution of genes in the clusters, and their representation in different genomes after enrichment, is shown in figure 4B. Comparison of gene to genome ratios within clusters prior to the enrichment (fig. 4A) and postenrichment (fig. 4B) shows an interesting transformation. Prior to enrichment, there were many clusters whose genes were derived from either a single or a small number of genomes; therefore, there was no apparent correlation between the sizes of the clusters and the representation of different genomes within them. After enrichment, we observed that the number of genes in clusters positively correlated with the number of genomes represented. Thus, this comparison reveals unsystematic biases in the process of genome annotation. In some genomes, many disrupted genes were annotated, whereas in others, homologous genes have escaped similar annotation. These biases are likely to be caused by different annotation approaches and/or annotation pipelines. As soon as enrichment is performed, which is equivalent to unbiased annotation, the cluster size shows better correlation with the number of genomes in the cluster.
The additional ∼2,260 genes found upon performing the cluster enrichment procedure could come from genomes that were not completed at the time when this study was initiated, from plasmids not included in our original data set, and from other partial genomic sequences or individual gene sequences available in the nr database.
To check that the outlined procedure for cluster enrichment results in the retrieval of true homologs, we analyzed the genes from the RF2 cluster after enrichment (415 genes) using the ARFA program (Bekaert et al. 2006) and confirmed all of them as PRF-containing RF2 genes. Subsequently, we found that 296 of these RF2 genes came from bacterial genomes listed in our original data set. Yet, 296 is less than half of what would be expected based on our previous analysis (Bekaert et al. 2006). This result indicates that the TBLASTN approach has the minimal false positive rate, and thus high specificity, but its sensitivity is lower than that of ARFA. This is not surprising considering the fact that ARFA identifies bacterial class-I release factor homologs using profile-HMMs derived from multiple alignments of release factor sequences (separate models are used for each RF paralog). In contrast, TBLASTN uses similarity scoring matrices that are not position specific and therefore its sensitivity is lower in comparison with ARFA. Therefore, though by using enrichment, we identified a large number of genes, it is likely that we have also missed a substantial number of distant homologs because of the stringent 40% identity threshold we chose. However, for functional characterization of particular gene disruptions it is more important to avoid populating clusters with false positives. Therefore, we found our current TBLASTN-based approach suitable for the purpose of the present work. All the major steps involved in the analysis of clusters are summarized in figure 3B.
Test for Purifying Selection
Although the mere existence of the same disruption in several homologous genes reveals its evolutionary conservation, there are at least two possible scenarios under which a gene-inactivating mutation could be observed in several species. One is simply due to a bias in genome sequencing toward species that are of significant biotechnological and/or medical interest. Several genome projects have been completed for various strains of E. coli, Shewanella, and Staphylococcus aureus. A recent gene-inactivating mutation is likely to be present in all orthologs in a closely related group of organisms. The second possibility is the propagation of inactive genes through gene conversion (Cordaux 2009), as discussed earlier.
A standard test for functional conservation in homologs is the test for purifying selection via calculation of the ratio of nonsynonymous to synonymous substitutions, ω (i.e., dN/dS or Ka/Ks) (Hurst 2002). We attempted to establish whether purifying selection acts on the entire sequence of a disrupted gene or only on one fragment, either the upstream or the downstream ORF, which would indicate that only one ORF is functional. To estimate ω, we used the codeml program from the PAML package (Yang 2007). PAML is a package for phylogenetic analysis of DNA or protein sequences. Codeml allows maximum likelihood estimation of synonymous and nonsynonymous substitution rates and the detection of purifying (or negative) selection in protein-coding DNA sequences. Two multiple sequence alignments were generated for each cluster, one for the upstream ORF and the other for the downstream ORF. The value of ω was estimated for each alignment in each cluster. The results of this analysis are presented in supplementary data set 2 (Supplementary Material online).
Notably, the method is not suitable for the analysis of highly similar sequences where ω estimates may not be accurate due to an insufficient number of substitutions. Therefore, for each cluster, we also considered the P values associated with dN/dS ratios computed for alignments between the inferred ancestral sequence and each sequence in the cluster. For each cluster, we report the lowest P value from the set of P values associated with the different dN/dS ratios between an extant sequence and its ancestral sequence and the respective dN/dS ratio. This P value was also corrected for multiple comparisons, under a null hypothesis of neutral selection (i.e., assuming that synonymous and nonsynonymous substitutions are equally likely, for further details, see Materials and Methods), along with the dN/dS ratio (see supplementary data set 2, Supplementary Material online). The lower is the P value, the less likely it is that the observed substitutions could occur as a result of neutral selection.
For a number of clusters (1, 3, 15, 23, 28, 43, 44, 49, 62), we obtained relatively high P values (>0.01). These clusters may potentially be false positives (i.e., pseudogenes) due to the reasons mentioned above. Still, in the case of large clusters, such as cluster 1 which contains 470 sequences from different genomes, a pseudogene hypothesis is unlikely to be true. A special case is seen in cluster 40 where we cannot reject the neutral selection hypothesis for ORF1 (the sequences are nearly identical); however, ORF2 contains substitutions that are inconsistent with neutral evolution.
Characterization of Individual Enriched Clusters
For all the clusters, where gene sequences involved a shift in the reading frame and which were suspected to utilize either PRF or PTR, we used several complementary approaches for further characterization. First, we translated nucleotide sequences in the reading frame corresponding to the upstream ORF; we generated multiple alignments of these protein sequences; and then, by using reverse translation, we generated alignments of nucleotide sequences. Within each sequence, the region where the true frame transition could occur was determined as the region of overlap between ORF1 (upstream) and ORF2 (downstream). The degree of nucleotide conservation at synonymous sites within the two ORFs was calculated as described previously (Firth and Atkins 2009). Regions of enhanced conservation at synonymous sites are indicative of overlapping functional elements, including overlapping CDSs and frameshift-stimulating elements. To calculate the conservation statistic for the whole alignment, ORF1 and ORF2 were fused in-frame by artificially inserting one or two “N”s, as appropriate, in each sequence just 5′ of the ORF1-frame stop codon. This means that each sequence in the alignment will have a continuous ORF (merged ORF1 and ORF2), while what was the overlap between ORF1 and ORF2 will appear as a nested ORF in a different reading phase. An example of a conservation plot is given in figure 5. In many cases, the region of overlap between ORF1 and ORF2 corresponds to a striking peak in conservation at synonymous sites within the merged ORF. (If a statistically significant peak is not apparent, it is often due to limited sequence data in a particular cluster.) The conservation plots are available online in supplementary data set 3 (Supplementary Material online) and also at the authors’ web site at http://lapti.ucc.ie/dORF/.
FIG. 5.
Alignment statistics for cluster 4: (Panels 1–3) The positions of stop codons in each of the three forward reading frames are shown as blue triangles. ORF1 and ORF2 have been fused in-frame by artificially inserting an “N” in each sequence just 5′ of the first ORF1-frame stop codon. Thus, the region of ORF2 that overlaps ORF1 appears as a short ORF which, in this case, is in the +2 reading frame, while the fusion of ORF1 with the remainder of ORF2 appears as a single long ORF in the +0 frame. (Panel 4) The gray area comprises 106 horizontal bars indicating the region of overlap between ORF1 and ORF2 in each of the 106 distinct sequences in the alignment (the bars are ordered by the location of their 5′ ends). Statistics for the start and end of the overlap region are summarized in the blue and pink boxplots, respectively. (Panels 5–6) Conservation at synonymous sites with respect to the +0 reading frame, for details, see Firth and Atkins (2009). (5) depicts the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, while (6) depicts the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model. There is a striking peak in synonymous site conservation coinciding with the region of frame transition. (Panel 7) Phylogenetically summed sequence divergence for the sequences that contribute to the conservation statistics at each position in the alignment. (In any particular alignment column, some sequences may be omitted from the statistical calculations due to alignment gaps, leading to a reduced statistical signal.) Although we failed to identify the sequence pattern responsible for nonstandard decoding in cluster 4, the plots clearly point to its presence.
The alignments of regions with transitions between frames were further visualized by Sequence Logos and analyzed along with the corresponding multiple alignments for the presence of conserved patterns. The patterns were classified either as PRF or PTR (for details on how such discrimination was carried out, see the following subsection). Notably, for five clusters (4, 28, 44, 45, and 63), we were unable to identify a pattern responsible for nonstandard decoding. We propose that some of these clusters may utilize entirely novel frameshift sites that do not conform to the generalized model of PRF and, thus, have not yet been identified to be the PRF sites.
Nonstandard Decoding: Mechanisms and Sequence Patterns
We used the following considerations to discriminate between PRF and PTR. Since PRF direction should be strictly conserved throughout the cluster, the appearance in the same cluster of sequences of ORF2 situated in both −1 and +1 phases relative to ORF1 was considered to be a strong indication of PTR. Furthermore, we looked for the presence of phased sequence patterns known as typical motifs for nonstandard decoding. For −1 PRF, these are either X_XX.Z_ZZ.N, where XXX and ZZZ are triplets consisting of identical nucleotides or NN.A_AA.R, where A is adenosine and R stands for a purine; the efficiency of −1 frameshifting greatly depends on the identity of the NNA codon (Licznar et al. 2003; Baranov et al. 2004). On the other hand, +1 PRF is known to be associated with a less formalized pattern that requires a codon forming a weak interaction with the anticodon of its cognate tRNA to be followed by either a stop triplet or a rare codon (Baranov et al. 2004; Liao et al. 2008). A PTR-related sequence pattern frequently consists of either a stretch of mononucleotide repeats or a combination of two such adjacent stretches. Interestingly, in a number of clusters (9), we have identified patterns that fit both −1 PRF and PTR models. We believe that both types of nonstandard decoding could work in such clusters to synthesize fusion proteins.
The sequence patterns responsible for PRF and PTR in the gene clusters are listed in table 1 and described in more detail in supplementary data set 2 (Supplementary Material online). Sequence logos for alignments of sequences from particular clusters are shown in figures 6 and 7; sequence logos for all the clusters can be found online in supplementary data set 3 (Supplementary Material online) and at the authors’ web site at http://lapti.ucc.ie/dORF/. The repertoire of patterns identified in this study is limited in comparison with the set of all frameshift-prone patterns described previously (for some of the known bacterial frameshift sites, see Baranov et al. 2006). The most frequent pattern is the −1 frameshift site A_AA.A_AA.C (over 2,000 genes from nine clusters), followed by the best-studied bacterial −1 frameshift site A_AA.A_AA.G (over 900 genes from 13 clusters). In bacteria, ribosomal frameshifting at A_AA.A_AA.C has been previously identified only in IS1 elements (Sekine et al. 1992). The pattern A_AA.A_AA.G is utilized for PRF in a number of bacterial genes, including many IS elements (e.g., IS150, Haas and Rak 2002; IS2, Hu et al. 1996; IS911, Polard et al. 1991) and other bacterial genes (Gurvich et al. 2003), among which dnaX (Tsuchihashi and Brown 1992) is the most prominent. The third most frequent pattern (over 500 genes) is the well-characterized +1 frameshift pattern C.TT_T.GA utilized for PRF in bacterial RF2 genes (Baranov et al. 2002b; Bekaert et al. 2006). All instances of this pattern, along with its minor variant C.TT_T.AA, were found only in cluster 2, which contains RF2 gene sequences.
Table 1.
PRF and PTR Sequence Patterns Identified in This Study.
| Pattern | Number of Clusters/Genes/Genes from 973 Bacterial Genomes/Genomes | Known Genes Utilizing Pattern | Previously Described in |
| PRF patterns | |||
| A_AA.A_AA.C | 9/2133/1538/70 | IS1 | Sekine et al. (1992) |
| A_AA.A_AA.G | 13/971/693/134 | dnaX, IS3 family | Tsuchihashi and Brown (1992), Gurvich et al. (2003), and Fayet and Prere (2010) |
| C.TT_T.GA | 1/564/453/453 | prfB (RF2 gene) | Craigen and Caskey (1986) and Baranov et al. (2002b) |
| G_GG.A_AA.G | 3/413/393/29 | g-t | Xu et al. (2004) |
| A_AA.A_AA.A | 13/343/134/48 | IS3 Family | Weiss et al. (1989), Vogele et al. (1991), Tsuchihashi and Brown (1992), and Fayet and Prere (2010) |
| B_CG.A_AA.G | 3/62/17/8 | cdd, IS1222 | Mejlhede et al. (1999), Licznar et al. (2003), and Mejlhede et al. (2004) |
| TC.A_AA.G | 1/8/8/2 | Licznar et al. (2003) | |
| C.CC_T.GA | 1/10/10/1 | Tsh | Zimmer et al. (2003) |
| G_GT.A_AA.A | 1/47/40/4 | ||
| GC.A_AA.A | Licznar et al. (2003) | ||
| C.TT_T.AA | 1/9/1/1 | prfB (RF2 gene) | Baranov et al. (2002b) |
| G_GG.A_AA.A | 2/8/6/3 | g-t | Xu et al. (2004) |
| G_GG.A_AA.C | 1/13/3/1 | ||
| CA.A_AA.A | 1/27/26/3 | ||
| PTR patterns | |||
| TmAn, m = (2 … 5), n = (5 … 9), n + m = (8 … 11) | 11/686/537/30 | ||
| C5T5 | 1/21/21/1 | ||
| T5C5 | 1/8/8/2 | ||
| AmGn, m = (2 … 7), n = (2 … 7), n + m = (8 … 13) | 10/822/337/60 | ||
| An, n = (7 … 10, 13) | 18/562/327/63 | dnaX, pgk-tim, IS | Schurig et al. (1995), Larsen et al. (2000), and Baranov et al. (2005) |
| Tn, n = (7 … 8) | 3/15/12/3 | MxiE, Spa13 | Penno et al. (2005), Penno et al. (2006), and Penno and Parsot (2006) |
| G5A7G2 | 1/9/9/1 | ||
NOTE.—The frameshift patterns are annotated with underlines to show the separation of codons in the original frame and dots to show the separation of codons in the shifted frame. The notation m = (x . . . y) indicates that the length of the pattern may vary between x and y.
FIG. 6.
Sequence logos representing 70 nucleotides (PRF patterns in the center) of sequence alignments from corresponding clusters. Shading is used for the first and the second positions of codons corresponding to the translational phase of ORF1. Frameshift-prone patterns (with codons in the initial frame separated by vertical dashes) and potential frameshift-facilitating Shine–Dalgarno sequences are indicated below each sequence logo. (A) Cluster 2 (+1 PRF). (B) Cluster 11 (−1 PRF). (C) Cluster 18 (−1 PRF). (D) Cluster 42 (−1 PRF).
FIG. 7.
Sequence logos representing PTR examples. Logos are organized as in figure 6. Sequences of actual PTR patterns occurring in the alignments used for the generation of sequence logos are shown below each logo; only those patterns that have been found in at least five sequences within the alignment are shown. (A) Cluster 6. (B) Cluster 7. (C) Cluster 46. (D) Cluster 60.
The next highly abundant pattern in terms of its presence in different clusters is A_AA.A_AA.A (more than 300 genes from 13 clusters). Causal connection of this pattern with ribosomal frameshifting has been demonstrated in mutagenic studies of dnaX (Tsuchihashi and Brown 1992), IS150 (Vogele et al. 1991), and the MMTV PRF cassette expressed in E. coli (Weiss et al. 1989); also it has been found to occur naturally as a PRF-causing pattern in insertion sequences from the IS3 family (Fayet and Prere 2010). Interestingly, in the present study, we often observed this pattern as a subpattern of long poly-A runs that may be utilized for transcriptional realignment; thus both PTR and PRF could take place at such patterns. Discrimination between these two mechanisms cannot be achieved even on the protein level as their protein products may be indistinguishable. We found a number of cases where a polyA-run overlaps with a known ribosomal frameshift pattern; thus the nonstandard decoding could take place at both transcriptional and translational levels. Yet another relatively frequent pattern G_GG.A_AA.G (over 400 genes from 3 clusters) has been well characterized. This pattern is utilized for PRF in expression of the bacteriophage g-t tail assembly gene fusion (Xu et al. 2004). We also found two variants of this pattern, G_GG.A_AA.C and G_GG.A_AA.A, albeit in a much smaller number of genes (13 and 8, respectively). Less frequent PRF patterns (those occurring in fewer than 100 genes) include CG.A_AA.G, previously characterized as related to −1 PRF in the Bacillus subtilis cdd gene (Mejlhede et al. 1999) and in IS1222 (Mejlhede et al. 2004), as well as the +1 PRF pattern C.CC_T.GA, utilized in the expression of the major tail protein gene of Listeria bacteriophage PSA (Zimmer et al. 2003). We also identified several phased patterns that have not been previously known to be used for natural PRF, such as TC.A_AA.G. Notably, efficient −1 ribosomal frameshifting has been shown to occur at TC.A_AA.G in artificial constructs with variants of N.NA_AA.G (Licznar et al. 2003). The pattern, GC.A_AA.A, differs slightly from the experimentally verified pattern GC.A_AA.G (Licznar et al. 2003); however, this difference is unlikely to affect the activity. For two more newly identified patterns, G_GT.A_AA.A and CA.A_AA.A observed in Cluster 13, ribosomal frameshifting has not been experimentally verified.
The PTR patterns identified in this study were of two types: long runs of As or Ts or a combination of mononucleotide runs, though only two such combinations are particularly abundant, namely a run of Ts followed by a run of As and a run of As followed by a run of Gs. In addition, there were four less abundant patterns (each pattern present in a single cluster only): 1) a combination of five Cs and five Ts; 2) a combination of five Ts and five Cs; and 3) a run of As flanked by Gs on both sides. While PTR has been previously reported for a run of As or Ts in several genes (see Introduction), no examples of PTR with combination patterns have been previously reported with the exception of AnGm runs in paramyxoviruses (Hausmann et al. 1999). The use of combination patterns could facilitate nonstandard decoding by providing a mechanism for specifying directionality of transcriptional realignment. While insertions and deletions of a single or multiple nucleotides have been observed for a run of As (Larsen et al. 2000), realignment of the nascent RNA chain by a specific number of nucleotides has been observed for combination patterns (for mechanism details, see Kolakofsky et al. 2005). Notably, a run of As was observed in the largest number of clusters (18) with a total of 562 genes. At the same time, AmGn patterns occurred in 822 genes from 10 clusters and TmAn patterns occurred in 686 genes from 11 clusters.
Figures 6 and 7 show examples of sequence logos for sequence patterns related to nonstandard decoding. Shading is used for the first and second positions of codons in the upstream ORF (ORF1), while the third position is shown with a white background. This helps to illustrate the differences in the positional pattern of evolutionary selection acting on the nucleotide sequences upstream and downstream of the PRF or PTR patterns. It is well known that more synonymous substitutions are available at the third position of a codon than at the first or second positions of a codon. It can be easily seen that the conservation of nucleotides in the white background (synonymous positions in ORF1, but nonsynonymous in ORF2) is higher downstream from PRF (fig. 6, panels A–D) and PTR patterns (fig. 7, panels A–D). Although we did not systematically analyze the presence of Shine–Dalgarno motifs upstream of PRF patterns, it can be seen that Shine–Dalgarno-like sequences are present upstream of PRF patterns in all cases of PRF in figure 6. Shine–Dalgarno-like sequences have been implicated in the stimulation of both −1 and +1 PRF in bacterial genes (Atkins et al. 2001). In contrast, in the PTR clusters, no Shine–Dalgarno-like sequences were observed upstream of PTR patterns (fig. 7).
Possibility of Independent Initiation of the Second ORF
Some annotated gene disruptions could also arise because of misannotation of two separate adjacent genes as a single fusion gene. To explore this possibility, we searched for potential translation initiation codons for the downstream ORF (ORF2). Notably, the existence of an initiation codon does not necessarily preclude the synthesis of a fusion product, as is clearly evident from consideration of many IS elements where the fusion products are synthesized in addition to separate products of the individual ORFs (reviewed in Baranov et al. 2006). However, the lack of a potential initiation codon for the second ORF is a strong indicator of expression of a fusion product. To find potential initiation codons, we searched for the codons ATG/GTG/TTG/CTG/ATT in the stretch of nucleotides that starts from the 5′ end of the region of overlap between ORF1 and ORF2 and extends downstream to the pattern proposed to be responsible for nonstandard decoding. For clusters where the type of frameshifting mechanism along with the sequence pattern for nonstandard decoding could not be determined, the entire region of overlap was scanned for the presence of start codons in-frame with ORF2. Those clusters where at least 50% of genes contain at least one start codon in the overlapping region are reported in supplementary data set 2 (Supplementary Material online) (21 clusters in total). If a potential start codon is utilized, its position is likely to be conserved. We observed cases when potential start codons did occur in the same position in at least 90% of sequences within a cluster (11 clusters listed in supplementary data set 2, Supplementary Material online). Among them are clusters 28 and 63, where the sequence pattern responsible for nonstandard decoding could not be determined.
Discussion
For a genomics researcher, genes whose sequences are interrupted due to sequencing errors or recent mutations, genes that are subject to phase variations, and those decoded by means of nontriplet decoding all appear to be alike, with the CDS broken into two overlapping ORFs. The problem of gene characterization seems difficult, even with experimental analysis of bacterial transcriptomes, although transcripts corresponding to such genes are easily detectable (Martin et al. 2010). This, in fact, is not surprising. A sequencing error obviously would not interfere with expression of the gene. A recent indel point mutation in the coding region of a gene would certainly affect the ability to produce an active protein product but is unlikely to affect its promoter and hence transcription, even though the same mutation could influence the expression of downstream genes in the same operon by a polar effect. Genes containing recoding sites responsible for the synthesis of fusion proteins by means of programmed nontriplet decoding at the level of either transcription or translation are also clearly expressed. A hope, on the experimental side, is for massive proteomics data which may enable the discrimination between the various types of gene disruption.
In this work, we have explored the power of comparative genomics for the systematic characterization of genes with disrupted CDSs. We focused on the identification of gene disruptions due to programmed nontriplet decoding and used a simple principle: genes where nontriplet decoding plays a functional role should be evolutionarily conserved and occur as orthologous genes with nontriplet decoding in the genomes of different bacteria. Sequence alignments of both parts of such disrupted CDSs are expected to reveal signs of gene evolution under purifying selection.
Application of the comparative genomics approach led to the discovery of over 40 clusters of disrupted genes, where the members of each cluster possess two features: 1) conserved organisation of the overlapping ORF1 and ORF2, 2) common nontriplet decoding patterns involved in the production of functional proteins. The most surprising finding of this work is the identification of a large number of genes containing PTR patterns likely to result in the restoration of full-length ORFs after cotranscriptional insertion or deletion of nucleotides into mRNAs. While a relatively large number of bacterial genes were known to be expressed via PRF (for review, see Baranov et al. 2006), only a limited number of PTR cases had been documented. Our results suggest that, among bacterial genes with annotated disruptions, PTR occurs with a frequency comparable to PRF. The frequency distribution of the different nonstandard decoding mechanisms is shown in figure 8. Here, one can see that there are more clusters with PTR patterns than with PRF patterns. With respect to individual annotated genes, PTR was found in about 50% of all analyzed genes with annotated disruptions; however, upon cluster enrichment, PRF becomes predominant. There might not be a biologically relevant reason for this type of ratio. Most likely, this is simply because PTR patterns are more easily spotted by annotators due to their repetitive character in comparison to PRF patterns whose identification requires prior knowledge of the field of recoding (fig. 8).
FIG. 8.
Frequency distribution of events resulting in disruptions of CDSs. (A and B) Distribution of event frequencies across 64 clusters. (C and D) Distribution of event frequencies among genes with annotated disruptions in the 64 clusters prior to enrichment. (E and F) Distribution of frequencies of nonstandard decoding mechanisms in the gene clusters after enrichment with nonannotated homologs. (A, C, and E) Functional classification of genes in the clusters. The white area represents genes that are expressed via nontriplet decoding for which we found strong evidence of functionality. The gray area (No Evidence of Purifying Selection—NEPS) represents genes for which we have no evidence of purifying selection acting on them. Some of these genes may be pseudogenes. The black area (Dump) contains all genes or pseudogenes that were not considered to be expressed as a result of PRF or PTR, for example, sequencing errors, misannotations, recent mutations, and phase variation. (B, D, and F) Distribution of frequencies of nontriplet decoding mechanisms among the presumably functional disrupted ORFs: The inner disk is divided into four categories. The gray area corresponds to genes in which we were unable to identify the mechanism. The blue area corresponds to PRF. The red area corresponds to PTR. The pink area corresponds to genes where both mechanisms seem to be plausible, that is, both PRF and PTR patterns are present. The areas corresponding to PRF and PRF/PTR are further differentiated on −1 and +1 frameshifting mechanisms within the outer disk.
An important question raised by this study is, how frequent are genes that require nontriplet decoding? Despite the large absolute numbers of discovered cases, the relative number is not high: four genes per genome on average or about 0.1% of all genes. Nonetheless, we believe that the real number of genes with disruptions is higher. Our search for homologous genes incorporated a 40% protein identity threshold which was quite stringent. We chose such a restrictive approach to reduce the false positive rate to a minimum. However, analysis of a control set (i.e., RF2 genes) indicated that we identified only about half of all sequenced RF2 genes. Similarly, for other genes with disruptions, we could as well miss many more homologs than we have identified in this study. Furthermore, our analysis started with a limited set of genes, that is, those where a problem with triplet decoding had already been identified during genome annotation. Many nontriplet decoded genes may simply have escaped annotation in all sequenced genomes where they are present. We also limited our analysis to cases where homologous sequences were identifiable within the initial set of annotated genes with disruptions; thus our analysis discarded the large number of genes that appeared to be singletons at the stage of cluster formation. Therefore, getting an accurate estimate of the fraction of genes that require nontriplet decoding is a rather difficult task. Our study indicates that this fraction is likely to be substantially higher than previously thought. In order to grasp the real picture of gene expression, that is, a picture not oversimplified by the “universal” rules of Genetic Decoding, it is important to develop efficient high-throughput computational and experimental methods for the identification of instances of nontriplet decoding. The first step toward this goal is the development of algorithms that allow the detection of ORF disruptions directly from the sequence, irrespective of its annotation, a problem addressed by the recently developed GeneTack program (Antonov and Borodovsky 2010).
Supplementary Material
Supplementary data sets 1–3 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We would like to acknowledge financial support from Science Foundation Ireland (06/IN.1/B81 to P.V.B. and 08/IN.1/B1889 to J.F.A.); the Wellcome Trust (088789 to A.E.F.); the Agence National de la Recherche (Programme Blanc, NT05-1_44848 to O.F.); the US National Institutes of Health (HG00783 to M.B. and RO1 GM079523 to J.F.A.).
References
- Adamski FM, Donly BC, Tate WP. Competition between frameshifting, termination and suppression at the frameshift site in the Escherichia coli release factor-2 mRNA. Nucleic Acids Res. 1993;21:5074–5078. doi: 10.1093/nar/21.22.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov I, Borodovsky M. Genetack: frameshift identification in protein-coding sequences by the Viterbi algorithm. J Bioinform Comput Biol. 2010;8:535–551. doi: 10.1142/s0219720010004847. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Baranov PV. The distinction between recoding and codon reassignment. Genetics. 2010;185:1535–1536. doi: 10.1534/genetics.110.119016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Baranov PV, Fayet O, et al. (13 co-authors) Overriding standard decoding: implications of recoding for ribosome function and enrichment of gene expression. Cold Spring Harb Symp Quant Biol. 2001;66:217–232. doi: 10.1101/sqb.2001.66.217. [DOI] [PubMed] [Google Scholar]
- Atkins JF, Gesteland RF, editors. Recoding: expansion of decoding rules enriches gene expression. New York: Springer; 2010. [Google Scholar]
- Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. Recoding: translational bifurcations in gene expression. Gene. 2002a;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. Release factor 2 frameshifting sites in different bacteria. EMBO Rep. 2002b;3:373–377. doi: 10.1093/embo-reports/kvf065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov PV, Hammer AW, Zhou J, Gesteland RF, Atkins JF. Transcriptional slippage in bacteria: distribution in sequenced genomes and utilization in IS element gene expression. Genome Biol. 2005;6:R25. doi: 10.1186/gb-2005-6-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M, Atkins JF, Baranov PV. ARFA: a program for annotating bacterial release factor genes, including prediction of programmed ribosomal frameshifting. Bioinformatics. 2006;22:2463–2465. doi: 10.1093/bioinformatics/btl430. [DOI] [PubMed] [Google Scholar]
- Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, Baranov PV. Recode-2: new design, new search tools, and many more genes. Nucleic Acids Res. 2010;38:D69–74. doi: 10.1093/nar/gkp788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M, Berg P. Deoxyribonucleic acid-directed synthesis of ribonucleic acid by an enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1962;48:81–94. doi: 10.1073/pnas.48.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobucci-Ponzano B, Guzzini L, Benelli D, et al. (12 co-authors) Functional characterization and high-throughput proteomic analysis of interrupted genes in the archaeon Sulfolobus solfataricus. J Proteome Res. 2010;9:2496–2507. doi: 10.1021/pr901166q. [DOI] [PubMed] [Google Scholar]
- Cordaux R. Gene conversion maintains nonfunctional transposable elements in an obligate mutualistic endosymbiont. Mol Biol Evol. 2009;26:1679–1682. doi: 10.1093/molbev/msp093. [DOI] [PubMed] [Google Scholar]
- Craigen WJ, Caskey CT. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature. 1986;322:273–275. doi: 10.1038/322273a0. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshayes C, Perrodou E, Gallien S, Euphrasie D, Schaeffer C, Van-Dorsselaer A, Poch O, Lecompte O, Reyrat JM. Interrupted coding sequences in Mycobacterium smegmatis: authentic mutations or sequencing errors? Genome Biol. 2007;8:R20. doi: 10.1186/gb-2007-8-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman JD. Programmed ribosomal frameshifting goes beyond viruses: organisms from all three kingdoms use frameshifting to regulate gene expression, perhaps signaling a paradigm shift. Microbe Wash DC. 2006;1:521–527. doi: 10.1128/microbe.1.521.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O, Prere MF. Programmed ribosomal −1 frameshifting as a tradition: the bacterial transposable elements of the IS3 family. In: Atkins JF, Gesteland RF, editors. Recoding: expansion of decoding rules enriches gene expression. New York: Springer; 2010. pp. 259–280. [Google Scholar]
- Ferrer I, Santpere G, van Leeuwen FW. Argyrophilic grain disease. Brain. 2008;131:1416–1432. doi: 10.1093/brain/awm305. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, et al. (14 co-authors) The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth AE, Atkins JF. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesteland RF, Weiss RB, Atkins JF. Recoding: reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Groisman EA, Casadesus J. The origin and evolution of human pathogens. Mol Microbiol. 2005;56:1–7. doi: 10.1111/j.1365-2958.2005.04564.x. [DOI] [PubMed] [Google Scholar]
- Gurvich OL, Baranov PV, Zhou J, Hammer AW, Gesteland RF, Atkins JF. Sequences that direct significant levels of frameshifting are frequent in coding regions of Escherichia coli. EMBO J. 2003;22:5941–5950. doi: 10.1093/emboj/cdg561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas M, Rak B. Escherichia coli insertion sequence IS150: transposition via circular and linear intermediates. J Bacteriol. 2002;184:5833–5841. doi: 10.1128/JB.184.21.5833-5841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann S, Garcin D, Delenda C, Kolakofsky D. The versatility of paramyxovirus RNA polymerase stuttering. J Virol. 1999;73:5568–5576. doi: 10.1128/jvi.73.7.5568-5576.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- Hu ST, Lee LC, Lei GS. Detection of an IS2-encoded 46-kilodalton protein capable of binding terminal repeats of IS2. J Bacteriol. 1996;178:5652–5659. doi: 10.1128/jb.178.19.5652-5659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002;18:486. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Iseni F, Baudin F, Garcin D, Marq JB, Ruigrok RW, Kolakofsky D. Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids. RNA. 2002;8:1056–1067. doi: 10.1017/s1355838202029977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JL, Belew AT, Rakauskaite R, Dinman JD. Identification of functional, endogenous programmed −1 ribosomal frameshift signals in the genome of Saccharomyces cerevisiae. Nucleic Acids Res. 2007;35:165–174. doi: 10.1093/nar/gkl1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan LP, Gallo A, O'Connell MA. The many roles of an RNA editor. Nat Rev Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Roux L, Garcin D, Ruigrok RW. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J Gen Virol. 2005;86:1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- Larsen B, Wills NM, Nelson C, Atkins JF, Gesteland RF. Nonlinearity in genetic decoding: homologous DNA replicase genes use alternatives of transcriptional slippage or translational frameshifting. Proc Natl Acad Sci U S A. 2000;97:1683–1688. doi: 10.1073/pnas.97.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao PY, Gupta P, Petrov AN, Dinman JD, Lee KH. A new kinetic model reveals the synergistic effect of E-, P- and A-sites on +1 ribosomal frameshifting. Nucleic Acids Res. 2008;36:2619–2629. doi: 10.1093/nar/gkn100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licznar P, Mejlhede N, Prere MF, Wills N, Gesteland RF, Atkins JF, Fayet O. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, Kyrpides NC. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J Biol Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- Martin J, Zhu W, Passalacqua KD, Bergman N, Borodovsky M. Bacillus anthracis genome organization in light of whole transcriptome sequencing. BMC Bioinformatics. 2010;11(Suppl 3):S10. doi: 10.1186/1471-2105-11-S3-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlhede N, Atkins JF, Neuhard J. Ribosomal −1 frameshifting during decoding of Bacillus subtilis cdd occurs at the sequence CGA AAG. J Bacteriol. 1999;181:2930–2937. doi: 10.1128/jb.181.9.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejlhede N, Licznar P, Prere MF, Wills NM, Gesteland RF, Atkins JF, Fayet O. −1 frameshifting at a CGA AAG hexanucleotide site is required for transposition of insertion sequence IS1222. J Bacteriol. 2004;186:3274–3277. doi: 10.1128/JB.186.10.3274-3277.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namy O, Rousset JP, Napthine S, Brierley I. Reprogrammed genetic decoding in cellular gene expression. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C. Computing multiple sequence/structure alignments with the T-coffee package. Curr Protoc Bioinformatics. 2010;8:1–25. doi: 10.1002/0471250953.bi0308s29. [DOI] [PubMed] [Google Scholar]
- Penno C, Hachani A, Biskri L, Sansonetti P, Allaoui A, Parsot C. Transcriptional slippage controls production of type III secretion apparatus components in Shigella flexneri. Mol Microbiol. 2006;62:1460–1468. doi: 10.1111/j.1365-2958.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- Penno C, Parsot C. Transcriptional slippage in mxiE controls transcription and translation of the downstream mxiD gene, which encodes a component of the Shigella flexneri type III secretion apparatus. J Bacteriol. 2006;188:1196–1198. doi: 10.1128/JB.188.3.1196-1198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno C, Sansonetti P, Parsot C. Frameshifting by transcriptional slippage is involved in production of MxiE, the transcription activator regulated by the activity of the type III secretion apparatus in Shigella flexneri. Mol Microbiol. 2005;56:204–214. doi: 10.1111/j.1365-2958.2004.04530.x. [DOI] [PubMed] [Google Scholar]
- Polard P, Prere MF, Chandler M, Fayet O. Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J Mol Biol. 1991;222:465–477. doi: 10.1016/0022-2836(91)90490-w. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurig H, Beaucamp N, Ostendorp R, Jaenicke R, Adler E, Knowles JR. Phosphoglycerate kinase and triosephosphate isomerase from the hyperthermophilic bacterium Thermotoga maritima form a covalent bifunctional enzyme complex. EMBO J. 1995;14:442–451. doi: 10.1002/j.1460-2075.1995.tb07020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Nagasawa H, Ohtsubo E. Identification of the site of translational frameshifting required for production of the transposase encoded by insertion sequence IS 1. Mol Gen Genet. 1992;235:317–324. doi: 10.1007/BF00279376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AA, Giddings MC, Parvaz JB, Gesteland RF, Atkins JF, Ivanov IP. Computational identification of putative programmed translational frameshift sites. Bioinformatics. 2002;18:1046–1053. doi: 10.1093/bioinformatics/18.8.1046. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Karakasiliotis I, Bailey D, Chaudhry Y, Evans DJ, Goodfellow IG. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 2008;36:2530–2546. doi: 10.1093/nar/gkn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Tamas I, Wernegreen JJ, Nystedt B, Kauppinen SN, Darby AC, Gomez-Valero L, Lundin D, Poole AM, Andersson SG. Endosymbiont gene functions impaired and rescued by polymerase infidelity at poly(A) tracts. Proc Natl Acad Sci U S A. 2008;105:14934–14939. doi: 10.1073/pnas.0806554105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi Z, Brown PO. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- Turnbough CL., Jr Regulation of gene expression by reiterative transcription. Curr Opin Microbiol. 2011;14:142–147. doi: 10.1016/j.mib.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogele K, Schwartz E, Welz C, Schiltz E, Rak B. High-level ribosomal frameshifting directs the synthesis of IS150 gene products. Nucleic Acids Res. 1991;19:4377–4385. doi: 10.1093/nar/19.16.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 1990;18:3529–3535. doi: 10.1093/nar/18.12.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Ribosomal frameshifting from −2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol. 1990;39:159–183. doi: 10.1016/s0079-6603(08)60626-1. [DOI] [PubMed] [Google Scholar]
- Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- Wernegreen JJ, Kauppinen SN, Degnan PH. Slip into something more functional: selection maintains ancient frameshifts in homopolymeric sequences. Mol Biol Evol. 2010;27:833–839. doi: 10.1093/molbev/msp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche PL, Gaffney DJ, Keightley PD. DNA sequence error rates in Genbank records estimated using the mouse genome as a reference. DNA Seq. 2004;15:362–364. doi: 10.1080/10425170400008972. [DOI] [PubMed] [Google Scholar]
- Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed +1 translational frameshifting in structural protein synthesis. Mol Microbiol. 2003;50:303–317. doi: 10.1046/j.1365-2958.2003.03684.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.