Background: Two-pore channels (TPCs) are part of the NAADP-receptor complex, but how and whether they dimerize are unclear.
Results: Human TPCs form homo- and heteromeric complexes.
Conclusion: Multimerization can regulate function and localization of TPCs.
Significance: Multimerization of TPCs is likely to affect fusion events in the endolysosomal system, disturbances of which can lead to the development of lysosomal storage diseases.
Keywords: Calcium Channels, Calcium Signaling, Lysosomes, NAADP, Receptors, Receptor Heteromerization, Two-pore Channel
Abstract
Two-pore channels (TPCs) have been recently identified as NAADP-regulated Ca2+ release channels, which are localized on the endolysosomal system. TPCs have a 12-transmembrane domain (TMD) structure and are evolutionary intermediates between the 24-TMD α-subunits of Na+ or Ca2+ channels and the transient receptor potential channel superfamily, which have six TMDs in a single subunit and form tetramers with 24 TMDs as active channels. Based on this relationship, it is predicted that TPCs dimerize to form functional channels, but the dimerization of human TPCs has so far not been studied. Using co-immunoprecipitation studies and a mass spectroscopic analysis of the immunocomplex, we show the presence of homo- and heteromeric complexes for human TPC1 and TPC2. Despite their largely distinct localization, we identified a discrete number of endosomes that coexpressed TPC1 and TPC2. Homo- and heteromerization were confirmed by a FRET study, showing that both proteins interacted in a rotational (N- to C-terminal/head-to-tail) symmetry. This is the first report describing the presence of homomultimeric TPC1 channels and the first study showing that TPCs are capable of forming heteromers.
Introduction
Two-pore channels (TPCs)3 have been recently described as part of the NAADP-receptor complex and comprise NAADP-gated ion-conducting channel components (1–3). Many organisms express three isoforms (TPC1, TPC2, and TPC3), although humans, rats, and mice express only TPC1 and TPC2 (4–6). All isoforms are localized on organelles of the endolysosomal system: TPC1 is localized on a wide range of vesicles (early, recycling, and late endosomes, as well as lysosomes), whereas the vast majority of TPC2 is restricted to late endosomes and lysosomes. The distribution of TPC3 is less well characterized. It has been found on recycling endosomes, late endosomes, and lysosomes and on other unidentified endosomes (1, 6–9). Thus, the three isoforms of TPCs show distinct localization patterns, although the possibility of coexpression on some late endosomes and lysosomes remains.
It has been shown that TPCs have homologies to voltage-gated calcium channels (VGCCs) and to both the transient receptor potential (TRP) channel superfamily and cation channel of sperm (CatSper) (8–11). Single VGCC α-subunits have 24 transmembrane domains (TMDs) and form functional channels (12). In contrast, TRP and CatSper channels have six TMDs and combine as tetramers containing 24 TMDs to form a functional channel (13, 14). TPCs are predicted to have 12 TMDs, and assuming that the functional TPC again needs 24 TMDs, the dimerization of the channel is predicted (15).
Many ion channels form heteromers, which show altered characteristics compared with the homomeric counterparts, thus allowing a cell to expand its repertoire of channels in signaling pathways (13, 14, 16). Mutations that inhibit the heteromerization can result in mislocalization and lack of channel function (17–20). Heteromerization of human TPCs has so far not been studied. Here, we present results from co-immunoprecipitation (co-IP) and FRET studies that indicate that human TPCs form both homo- and heteromers. This heteromerization was confirmed by MS analysis.
EXPERIMENTAL PROCEDURES
Vector Construction
IMAGE cDNA clones were obtained (Source BioScience) for both human TPC1 (IMAGE clone 40148827, accession number BC150203) and TPC2 (IMAGE clone 5214862, accession number NP620714.2) and subcloned into pcDNA5/TO expression vectors (Invitrogen) with C-terminal HA or mCherry tags. For the FRET constructs, TPC1 and TPC2 were subcloned into pECFP and pEYFP expression vectors (C- and N-terminal forms; Clontech). Insertion of TPC cDNA was done by the In-Fusion protocol (Clontech), and HA and mCherry tags were inserted by conventional subcloning. All primers and restriction enzymes used are listed in supplemental Table S1. Constructs were verified by sequencing.
Cell Culture and Transfection
HEK293T cells were maintained in DMEM supplemented with 10% (v/v) FBS (Biosera), 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). In addition, the medium for the HA-TPC2 stable HEK293 cell line contained 200 μg/ml Geneticin (Invitrogen). Cells were kept at 37 °C in a humidified incubator supplied with 5% (v/v) CO2. Cells were plated on poly-d-lysine-coated coverslips 24 h before transfection. Plasmids were transfected for 24–48 h at a concentration of 20 μg/ml using jetPEI (Qbiogene) according to the manufacturer's instructions.
Immunoprecipitation
Membranes were prepared as described (7). Membrane pellets were resuspended to 1 mg/ml in immunoprecipitation (IP) buffer (150 mm NaCl, 20 mm HEPES (pH 7.2), and 1% CHAPS). After incubation for 1 h at 4 °C with agitation, the mixture was ultracentrifuged (50,000 × g, 30 min, 4 °C) to pellet all insoluble contaminants. Specific antibodies (or non-immune serum/purified non-immune IgGs as a control) were covalently coupled to either protein A or G beads (Sepharose (GE Healthcare) or Dynabeads (Invitrogen)) using dimethyl pimelimidate as a coupling agent and added to the precleared solubilized membranes. The mixture continually agitated for 1 h at room temperature or overnight at 4 °C. Following removal of the supernatant, the beads were washed with IP buffer.
SDS-PAGE/Immunoblot Analysis
This was performed as described (7). Rat anti-HA mAB (clone 3F10; Roche Applied Science), HRP-conjugated rabbit anti-GFP polyclonal antibody (Santa Cruz Biotechnology sc-8334), rat multi-red 5F8 mAB (21), and mouse anti-DsRed mAB (Clontech 632392) were used for IP and detection. Antibodies against TPC1 and TPC2 were custom-made by immunizing rabbits with peptides corresponding to residues 776–792 and 798–816 of human TPC1 and residues 240–254 and 408–422 of human TPC2.
Mass Spectroscopic Analysis
The immunocomplexes purified from HA-TPC2 cell membranes using the anti-HA antibody and rat IgG as a control were trypsin-digested and analyzed via LC-MS/MS. The threshold for inclusion was p < 0.01 and a Mascot score of >60.
Immunofluorescence Staining
This was performed as described (7).
FRET Analysis
Cells were transfected with a combination of the following FRET constructs: 1) N-terminally tagged YFP.TPC1, cyan fluorescent protein (CFP).TPC2, and YFP.TPC2 and 2) C-terminally tagged TPC1.CFP, TPC1.YFP, TPC2.CFP, and TPC2.YFP. 48 h post-transfection, cells were fixed with 2% paraformaldehyde, mounted with ProLong Gold antifade reagent (Invitrogen), and observed on the same microscope as for the immunofluorescence with the following excitation/emission parameters: CFP (457 nm)/Meta-Head (473–612 nm); YFP (514 nm)/band pass (530–600 nm); and FRET channel (457 nm)/long pass (>505 nm). Bleed-through into the FRET channel was 0.25 ± 0.11± from the CFP and 0.18 ± 0.04± from the YFP. Transfected cells were selected, and fluorescence was measured. After subtracting the bleed-through of the respective channel, the normalized FRET ratio was calculated as described (22).
RESULTS
Can Human TPCs Form Homomeric Complexes?
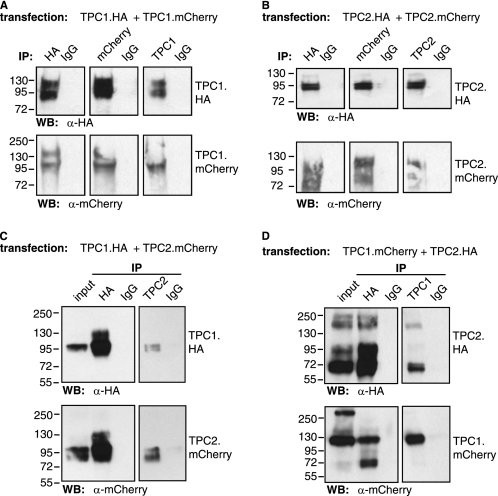
To test whether TPCs form homomeric complexes, we generated HA- and mCherry-tagged constructs of both TPC1 and TPC2. We coexpressed TPC1.mCherry and TPC1.HA and performed IPs with antibodies against HA, mCherry, and TPC1. We probed the immunoblots with antibodies against either HA or mCherry (Fig. 1A). Using the anti-HA antibody, we immunoprecipitated TPC1.HA (Fig. 1A, upper left panel). In addition, we also detected a band when probing an immunoblot from the anti-HA IP with the anti-mCherry antibody, indicating a co-IP of TPC1.mCherry with TPC1.HA (Fig. 1A, lower left panel). Similarly, when using an antibody against mCherry for the IP, we detected a band probing with the anti-HA and anti-mCherry antibodies, again indicating a positive co-IP of the differently tagged TPC1 constructs (Fig. 1A, middle panels). In addition, when using an antibody against human TPC1, we could immunoprecipitate TPC1 carrying both tags (Fig. 1A, right panels). We performed the same IPs in cells coexpressing TPC2.mCherry and TPC2.HA (Fig. 1B) and again found that all antibodies immunoprecipitated both tagged TPC2 proteins, showing that they form homomers. The specificity of the IPs was confirmed by using control IgGs or sera; these did not immunoprecipitate any TPCs. We also tested that the constructs were expressed at their expected full-length by immunoblotting (data not shown). This shows for the first time that human TPC1 and TPC2 both form homomers.
FIGURE 1.
Human TPCs form homo- and heteromeric complexes. HA- and mCherry-tagged TPC1 and TPC2 were coexpressed in HEK293T cells and immunopurified using antibodies against HA, mCherry, or the TPC1 or TPC2 sequences. A, co-IP of HA- and mCherry-tagged TPC1. B, co-IP of HA- and mCherry-tagged TPC2. C, co-IP of TPC1.mCherry and TPC2.HA. D, co-IP of TPC1.HA and TPC2.mCherry. Independent of which antibody was used for the IP, TPCs with both tags were always pulled down, showing the existence of homo- and heteromeric complexes. Each lane corresponds to the eluate from each IP. The input lanes correspond to 5% of the material used in each IP. WB, Western blot.
Can Human TPCs Form Heteromeric Complexes?
We next tested whether human TPCs are also capable of forming heteromers by coexpressing TPC1.HA and TPC2.mCherry. Using the anti-HA antibody for the IP, we detected immunoprecipitated TPC1.HA (Fig. 1C, upper left panel) and TPC2.mCherry (lower left panel) on the immunoblot, showing that both co-immunoprecipitate and form heteromers. This result was confirmed by repeating the IPs in cells coexpressing the alternatively tagged TPCs: mCherry-tagged TPC1 and HA-tagged TPC2 (Fig. 1D). Interestingly, not only antibodies against the tags were able to immunoprecipitate the TPC heteromers but also antibodies against TPC1 and TPC2 sequences. When using an anti-TPC2 antibody for the IP, both TPC1.HA and TPC2.mCherry could be detected in the immunoprecipitated eluate (Fig. 1C, right panels), whereas when using an anti-TPC1 antibody for the IP, TPC2.HA and TPC1.mCherry were present on the immunoblot (Fig. 1D, right panels).
To confirm the formation of TPC heteromers, we performed IPs in a cell line stably expressing HA.TPC2, which we used previously to study NAADP responses in Ca2+ imaging and lipid bilayer experiments (1, 23). By IP via the HA tag, we found both TPC2 and endogenous TPC1 in the immunocomplex, whereas they were not present in the IgG control (supplemental Fig. S1, A and B). This interaction between the two human TPC isoforms was further confirmed by performing analysis of the immunopurified complex, which showed the presence of both TPC1 (two unique peptides and Mascot score of 127) and TPC2 (two unique peptides and Mascot score of 72) (Coomassie Blue-stained gel in supplemental Fig. S1C). This is the first evidence that TPCs can form heteromers.
Do Human TPC1 and TPC2 Co-localize on the Same Vesicles in a Living Cell?
To test whether the two isoforms of human TPCs co-localize in vesicles of the endolysosomal system, we used the same constructs as described above. First, we confirmed the expected subcellular localization of TPC1.mCherry and TPC2.mCherry by cotransfecting them with various GFP-tagged organelle markers (supplemental Fig. S2, A and B). Using Pearson's co-localization coefficient, we found a co-localization of TPC1.mCherry with recycling endosomes, as well as with late endosomes and lysosomes. In contrast, there was no good co-localization of TPC1.mCherry with early endosomes or with the endoplasmic reticulum (ER) (supplemental Fig. S2A). In contrast, TPC2.mCherry showed a very good co-localization with late endosomes and lysosomes. It also overlapped to a certain degree with recycling endosomes, whereas it showed no overlap with either early endosomes or the ER (supplemental Fig. S2B).
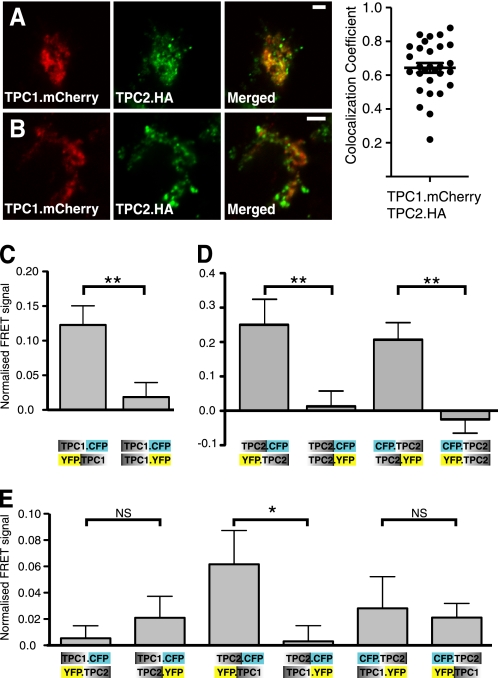
Having confirmed that the overexpressed proteins localize as reported in previous studies, we cotransfected TPC1.mCherry and TPC2.HA and performed immunofluorescence staining for HA-tagged TPC2 (Fig. 2) to determine whether both TPCs co-localize. The degree of co-localization between TPC1.mCherry and TPC2.HA varied between cells, but we always found some overlap of the two TPCs. To indicate this variability, we show examples of cells with a high and low degree of co-localization in Fig. 2 (A and B, respectively). The TPC localization was vesicular and not reticular, indicating that the co-localization occurs in vesicles rather than by retention of proteins in the ER. Repeating the experiments with the alternatively tagged proteins (TPC1.HA and TPC2.mCherry) gave similar results (supplemental Fig. S3). Having found evidence of TPC1 and TPC2 expression on the same vesicles, we decided to investigate whether they are in close enough proximity to form a heteromeric channel.
FIGURE 2.
Overexpressed TPC1 and TPC2 co-localize by immunofluorescence and show positive FRET signals. A, an example of a cell with a good overlap on some vesicles within a cell. B, a cell with a low degree of co-localization between TPC1.mCherry and TPC2.HA. Scale bars = 5 μm. The scatter plot shows Pearson's co-localization coefficient for the different cells. C–E, summary of the FRET study results. C and D, FRET occurs in a rotational symmetry for TPC1 and TPC2 homomers, respectively. E, TPC1-TPC2 heteromers have a larger variation in the FRET signal between different constructs, but positive FRET can be seen for the rotational symmetry between the TPC2.CFP and YFP.TPC1 constructs. n = 12–15 cells per condition. *, p < 0.05; **, p < 0.01; NS, not statistically significant using Student's t test.
Do TPC1 and TPC2 Form Complexes, as Detected by FRET?
FRET signals reveal proximal proteins (<10 nm apart). Using FRET constructs carrying the CFP and YFP tags either at the same end or on opposite ends, we investigated whether TPCs form multimers in a rotational/head-to-tail symmetry (interaction of the N terminus of one channel with the C terminus of the other channel) or whether the assembly is non-rotational (exclusive interaction of either the N or C terminus).
For this, we used TPC1 and TPC2 FRET constructs with a fluorescent tag either at the N terminus (YFP.TPC1, CFP.TPC2, and YFP.TPC2) or at the C terminus (TPC1.CFP, TPC1.YFP, TPC2.CFP, and TPC2.YFP). Control experiments showed that all FRET constructs were expressed at their expected molecular weight (supplemental Fig. S4A) and showed the expected co-localization with LysoTracker Red, a marker for acidic organelles (supplemental Fig. S4, B and C).
To confirm the finding that TPC1 forms homomers, we coexpressed different combinations of the TPC1 FRET constructs and found a positive FRET ratio when the fluorescent proteins were expressed at opposite ends (YFP.TPC1 and TPC1.CFP) (Fig. 2C). In contrast, with the fluorescent tag at the C termini of both proteins (TPC1.CFP and TPC1.YFP) (Fig. 2C), we found a significantly smaller (negligible) FRET ratio. These findings indicate that TPC1 can form homomers with a rotational symmetry. Similar results were obtained with TPC2 (Fig. 2D), which also showed substantially larger FRET ratios when the fluorescent tags were expressed at the opposite ends of the proteins, again indicating a rotational symmetry of the TPC2 homomers.
To confirm the formation of TPC1 and TPC2 heteromers, as determined by the co-IP analysis, we coexpressed the TPC1 and TPC2 FRET constructs in all possible combinations (Fig. 2E). As only a small fraction of TPC1 and TPC2 actually coexpressed on the same vesicles, we did not expect high FRET values. Fig. 2E shows the considerable variability in the FRET signals observed. The highest FRET ratio was once more observed when the constructs were tagged at opposite ends (TPC2.CFP and YFP.TPC1), supporting the theory that TPC heteromers are also formed with a rotational symmetry. One explanation for this particular combination of constructs giving the highest FRET value is that this TPC1 construct (YFP.TPC1) showed the best co-localization with LysoTracker Red (supplemental Fig. S4), thus being more capable of forming heteromers with TPC2 than the other two TPC1 constructs.
The FRET experiments not only confirmed the presence of TPC homo- and heteromers but also suggested that this interaction occurs in a rotational/head-to-tail symmetry for both the homo- and heteromers.
DISCUSSION
Heteromultimerization of ion channels yields modulated channel functions, subcellular localization, and biophysical properties, resulting in a larger repertoire of differentially regulated and differently behaving channels compared with homomeric channels (13, 16). For example, ryanodine receptor heterotetramers show an altered conductance state, with a behavior intermediate to those of the homomeric channels, and an altered Ca2+ selectivity (24). Within heteromeric inositol 1,4,5-trisphosphate receptor complexes, each subunit individually contributes to the overall receptor ligand affinity (25–28). Predictions based on the sequence homology between TPCs and VGCCs suggested dimerization of TPCs when TPC1 was first cloned, and their relationships with TRP and CatSper channels further support this prediction (4, 8, 15).
In this study, we used different approaches to investigate the multimerization of human TPCs and have shown for the first time that they are capable of forming both homo- and heteromers. The only previous study to investigate multimerization of TPCs found homomers of mouse TPC2 when they were overexpressed in HEK cells, but there was no evidence for the presence of heteromers (3). Here, we present the first evidence of TPC1 being able to form homomers, and we report for the first time the existence of TPC1 and TPC2 heteromers. Formation of heteromeric TPC complexes was previously considered unlikely, as TPC1 and TPC2 show distinct localization patterns (1, 2). However, using immunofluorescence, we found evidence of both proteins on the same vesicle.
Both the co-IP and FRET experiments showed the presence of TPC heteromers. The FRET values for the heteromers were lower than those for the homomers (Fig. 2); this is consistent with the smaller number of vesicles on which FRET between TPC1 and TPC2 can potentially occur.
The endolysosomal-based TRP family members, the mucolipins (TRPML1–3) (29–31), show distinct organellar localization patterns, yet heteromeric as well as homomeric assemblies have been reported (17, 33–37). Functionally this is important since heteromers of TRPML1/TRPML3 are important for the induction of autophagy (18). This shows that channels localized on different compartments of the endolysosomal system are capable of forming dynamic interactions in vivo (17, 18, 36) and that heterodimerization affects channel function.
We propose a similar dynamic interaction for TPC1 and TPC2, with TPC1 localized on a wider range of vesicles. Fusion of vesicles carrying TPC1 and TPC2 could lead to the formation of a hybrid organelle. The probable altered channel characteristics of the resulting TPC heteromers might be important for further fusion events in the endocytic pathway, which has been shown to be tightly regulated by Ca2+ (39, 40).
Overexpression of human TPC2, as well as of sea urchin TPC1 and TPC2, results in the formation of enlarged lysosomes (7), typical of many lysosomal storage disorders. The NAADP receptor antagonist Ned-19 (41) not only can reverse that phenotype but can cause defects in basal endolysosomal trafficking events, suggesting that NAADP and TPCs are important for the endolysosomal function (7). A link between NAADP and the induction of autophagy has also been proposed in cultured astrocytes (42), raising the possibility that, like TRPML channels, the heteromerization of TPCs is important for autophagy. Whether stimulation of cells with agonists causing NAADP synthesis might affect the heteromerization of the channels remains to be determined.
Heteromerization of TPCs could also explain the dominant-negative effects found in studies using sea urchin TPC3 (7) or mutant forms of TPCs (2, 43, 44). Assuming the inhibitory channel forms a complex with the endogenous TPCs, the presence of one nonfunctional TPC seems to inhibit NAADP responses of the channel complex. This could be a way of turning off the channel activity or modulating the channel function.
We also studied whether multimerization of the channels occurs in a rotational (head-to-tail) symmetry or whether the interaction involves exclusively the interaction of either N- or C-terminal tails in a non-rotational symmetry. As both the N and C termini of TPCs face the cytosol (5, 45), FRET studies with N- and C-terminal fluorescent tags are possible. In contrast to other ion channels, TPCs have relatively short N- and C-terminal tails (TPC1 N terminus, 105 amino acids; TPC1 C terminus, 126 amino acids; TPC2 N terminus, 80 amino acids; TPC2 C terminus, 54 amino acids), and by comparison with VGCCs, we assume the footprint area of TPCs to be in the range of 10 × 20 nm (46, 47). Therefore, it is unlikely that these tails can interact randomly (which could lead to positive FRET signals regardless of orientation). This assumption was validated by the absence of a FRET signal for homomers in non-rotational symmetry. Preferential interaction in rotational symmetry has been shown for various channels (32, 38), including many of the closely related TRP channels (48–50). The finding that a large number of channels interact in a rotational symmetry is consistent with the results of our FRET study, in which TPCs gave FRET signals when the fluorescent tags were on opposite ends of the two subunits.
Our results show that human TPC1 and TPC2 are able to form both homo- and heteromers. Because of the highly mobile nature of the endolysosomal system and its dependence on Ca2+ for fusion events, we propose that these channel complexes have different Ca2+-signaling properties and different functions for fusion events in the endolysosomal system.
Acknowledgments
We thank E. C. Dell'Angelica (UCLA), P. Woodman (Manchester University), S. Corvera (University of Massachusetts Medical School), and S. Grinstein (University of Toronto) for the GFP-tagged organelle markers; J. McIlhinney (Oxford University) for the enhanced CFP and enhanced YFP constructs; A. Rottach (Ludwig-Maximilians-Universität München) for the multi-red 5F8 antibody; and M. X. Zhu (University of Texas) for the HA.TPC2 (human)-overexpressing cell line. We also thank Jeff McIlhinney and Anthony Morgan for critical discussions.
The work was supported by a Wellcome Trust project programme grant (to A. G. and J. P.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- TPC
- two-pore channel
- VGCC
- voltage-gated calcium channel
- TRP
- transient receptor potential
- TMD
- transmembrane domain
- co-IP
- co-immunoprecipitation
- IP
- immunoprecipitation
- CFP
- cyan fluorescent protein
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Calcraft P. J., Ruas M., Pan Z., Cheng X., Arredouani A., Hao X., Tang J., Rietdorf K., Teboul L., Chuang K. T., Lin P., Xiao R., Wang C., Zhu Y., Lin Y., Wyatt C. N., Parrington J., Ma J., Evans A. M., Galione A., Zhu M. X. (2009) Nature 459, 596–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brailoiu E., Churamani D., Cai X., Schrlau M. G., Brailoiu G. C., Gao X., Hooper R., Boulware M. J., Dun N. J., Marchant J. S., Patel S. (2009) J. Cell Biol. 186, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zong X., Schieder M., Cuny H., Fenske S., Gruner C., Rötzer K., Griesbeck O., Harz H., Biel M., Wahl-Schott C. (2009) Pflugers Arch. 458, 891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galione A., Morgan A. J., Arredouani A., Davis L. C., Rietdorf K., Ruas M., Parrington J. (2010) Biochem. Soc. Trans. 38, 1424–1431 [DOI] [PubMed] [Google Scholar]
- 5. Zhu M. X., Ma J., Parrington J., Calcraft P. J., Galione A., Evans A. M. (2010) Am. J. Physiol. Cell Physiol. 298, C430–C441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brailoiu E., Hooper R., Cai X., Brailoiu G. C., Keebler M. V., Dun N. J., Marchant J. S., Patel S. (2010) J. Biol. Chem. 285, 2897–2901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruas M., Rietdorf K., Arredouani A., Davis L. C., Lloyd-Evans E., Koegel H., Funnell T. M., Morgan A. J., Ward J. A., Watanabe K., Cheng X., Churchill G. C., Zhu M. X., Platt F. M., Wessel G. M., Parrington J., Galione A. (2010) Curr. Biol. 20, 703–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu M. X., Ma J., Parrington J., Galione A., Evans A. M. (2010) FEBS Lett. 584, 1966–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Galione A. (2011) Cold Spring Harb. Perspect. Biol. 3, a004036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patel S., Marchant J. S., Brailoiu E. (2010) Cell Calcium 47, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clapham D. E., Garbers D. L. (2005) Pharmacol. Rev. 57, 451–454 [DOI] [PubMed] [Google Scholar]
- 12. Catterall W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521–555 [DOI] [PubMed] [Google Scholar]
- 13. Schaefer M. (2005) Pflugers Arch. 451, 35–42 [DOI] [PubMed] [Google Scholar]
- 14. Chung J. J., Navarro B., Krapivinsky G., Krapivinsky L., Clapham D. E. (2011) Nat. Commun. 2, 153–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ishibashi K., Suzuki M., Imai M. (2000) Biochem. Biophys. Res. Comm. 270, 370–376 [DOI] [PubMed] [Google Scholar]
- 16. Venkatachalam K., Montell C. (2007) Annu. Rev. Biochem. 76, 387–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeevi D. A., Frumkin A., Offen-Glasner V., Kogot-Levin A., Bach G. (2009) J. Pathol. 219, 153–162 [DOI] [PubMed] [Google Scholar]
- 18. Zeevi D. A., Lev S., Frumkin A., Minke B., Bach G. (2010) J. Cell Sci. 123, 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chubanov V., Waldegger S., Mederos y Schnitzler M., Vitzthum H., Sassen M. C., Seyberth H. W., Konrad M., Gudermann T. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2894–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Abel M., Hoenderop J. G., Bindels R. J. (2005) Naunyn-Schmiedebergs Arch. Pharmacol. 371, 295–306 [DOI] [PubMed] [Google Scholar]
- 21. Rottach A., Kremmer E., Nowak D., Leonhardt H., Cardoso M. C. (2008) Hybridoma 27, 337–343 [DOI] [PubMed] [Google Scholar]
- 22. Gordon G. W., Berry G., Liang X. H., Levine B., Herman B. (1998) Biophys. J. 74, 2702–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pitt S. J., Funnell T. M., Sitsapesan M., Venturi E., Rietdorf K., Ruas M., Ganesan A., Gosain R., Churchill G. C., Zhu M. X., Parrington J., Galione A., Sitsapesan R. (2010) J. Biol. Chem. 285, 35039–35046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu L., Wang Y., Yamaguchi N., Pasek D. A., Meissner G. (2008) J. Biol. Chem. 283, 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joseph S. K., Lin C., Pierson S., Thomas A. P., Maranto A. R. (1995) J. Biol. Chem. 270, 23310–23316 [DOI] [PubMed] [Google Scholar]
- 26. Monkawa T., Miyawaki A., Sugiyama T., Yoneshima H., Yamamoto-Hino M., Furuichi T., Saruta T., Hasegawa M., Mikoshiba K. (1995) J. Biol. Chem. 270, 14700–14704 [DOI] [PubMed] [Google Scholar]
- 27. Nucifora F. C., Jr., Sharp A. H., Milgram S. L., Ross C. A. (1996) Mol. Biol. Cell 7, 949–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwai M., Michikawa T., Bosanac I., Ikura M., Mikoshiba K. (2007) J. Biol. Chem. 282, 12755–12764 [DOI] [PubMed] [Google Scholar]
- 29. Puertollano R., Kiselyov K. (2009) Am. J. Physiol. Renal Physiol. 296, F1245–E1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeevi D. A., Frumkin A., Bach G. (2007) Biochim. Biophys. Acta 1772, 851–858 [DOI] [PubMed] [Google Scholar]
- 31. Yamaguchi S., Jha A., Li Q., Soyombo A. A., Dickinson G. D., Churamani D., Brailoiu E., Patel S., Muallem S. (2011) J. Biol. Chem. 286, 22934–22942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Varnum M. D., Zagotta W. N. (1997) Science 278, 110–113 [DOI] [PubMed] [Google Scholar]
- 33. Kim H. J., Soyombo A. A., Tjon-Kon-Sang S., So I., Muallem S. (2009) Traffic 10, 1157–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Curcio-Morelli C., Zhang P., Venugopal B., Charles F. A., Browning M. F., Cantiello H. F., Slaugenhaupt S. A. (2010) J. Cell. Physiol. 222, 328–335 [DOI] [PubMed] [Google Scholar]
- 35. Karacsonyi C., Miguel A. S., Puertollano R. (2007) Traffic 8, 1404–1414 [DOI] [PubMed] [Google Scholar]
- 36. Martina J. A., Lelouvier B., Puertollano R. (2009) Traffic 10, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grimm C., Jörs S., Saldanha S. A., Obukhov A. G., Pan B., Oshima K., Cuajungco M. P., Chase P., Hodder P., Heller S. (2010) Chem. Biol. 17, 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rosenbaum T., Gordon S. E. (2002) Neuron 33, 703–713 [DOI] [PubMed] [Google Scholar]
- 39. Luzio J. P., Bright N. A., Pryor P. R. (2007) Biochem. Soc. Trans. 35, 1088–1091 [DOI] [PubMed] [Google Scholar]
- 40. Pryor P. R., Mullock B. M., Bright N. A., Gray S. R., Luzio J. P. (2000) J. Cell Biol. 149, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naylor E., Arredouani A., Vasudevan S. R., Lewis A. M., Parkesh R., Mizote A., Rosen D., Thomas J. M., Izumi M., Ganesan A., Galione A., Churchill G. C. (2009) Nat. Chem. Biol. 5, 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pereira G. J., Hirata H., Fimia G. M., do Carmo L. G., Bincoletto C., Han S. W., Stilhano R. S., Ureshino R. P., Bloor-Young D., Churchill G., Piacentini M., Patel S., Smaili S. S. (2011) J. Biol. Chem. 286, 27875–27881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schieder M., Rötzer K., Brüggemann A., Biel M., Wahl-Schott C. A. (2010) J. Biol. Chem. 285, 21219–21222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brailoiu E., Rahman T., Churamani D., Prole D. L., Brailoiu G. C., Hooper R., Taylor C. W., Patel S. (2010) J. Biol. Chem. 285, 38511–38516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hooper R., Churamani D., Brailoiu E., Taylor C. W., Patel S. (2011) J. Biol. Chem. 286, 9141–9149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang M. C., Collins R. F., Ford R. C., Berrow N. S., Dolphin A. C., Kitmitto A. (2004) J. Biol. Chem. 279, 7159–7168 [DOI] [PubMed] [Google Scholar]
- 47. Murata K., Odahara N., Kuniyasu A., Sato Y., Nakayama H., Nagayama K. (2001) Biochem. Biophys. Res. Comm. 282, 284–291 [DOI] [PubMed] [Google Scholar]
- 48. Lepage P. K., Boulay G. (2007) Biochem. Soc. Trans. 35, 81–83 [DOI] [PubMed] [Google Scholar]
- 49. Schilling W. P., Goel M. (2004) Novartis Found. Symp. 258, 18–30; discussion 30–43, 98–102, 263–6 [PubMed] [Google Scholar]
- 50. Fujiwara Y., Minor D. L., Jr. (2008) J. Mol. Biol. 383, 854–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Erler I., Hirnet D., Wissenbach U., Flockerzi V., Niemeyer B. A. (2004) J. Biol. Chem. 279, 34456–34463 [DOI] [PubMed] [Google Scholar]
- 52. García-Sanz N., Fernández-Carvajal A., Morenilla-Palao C., Planells-Cases R., Fajardo-Sánchez E., Fernández-Ballester G., Ferrer-Montiel A. (2004) J. Neurosci. 24, 5307–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hofmann T., Schaefer M., Schultz G., Gudermann T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7461–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang Q., Gyftogianni E., van de Graaf S. F. J., Hoefs S., Weidema F. A., Bindels R. J. M., Hoenderop J. G. J. (2004) J. Biol. Chem. 279, 54304–54311 [DOI] [PubMed] [Google Scholar]