Background: The RNA-binding protein HuR is involved in a range of cellular processes and several diseases.
Results: We reveal the characteristics of HuR binding using genomic methods and explore its network of targets.
Conclusion: Our results reveal the complexity of RBP binding, corroborate the concept of post-transcriptional networks and suggest an interplay between miRNAs and RBPs.
Significance: An understanding of HuR informs our knowledge of RBPs and may lead to effective treatments for related diseases.
Keywords: Alternative Splicing, Cancer Biology, Gene Regulation, RNA Abundance, RNA-binding Protein, ELAVL1, HuR
Abstract
The ubiquitously expressed RNA-binding protein Hu antigen R (HuR) or ELAVL1 is implicated in a variety of biological processes as well as being linked with a number of diseases, including cancer. Despite a great deal of prior investigation into HuR, there is still much to learn about its function. We take an important step in this direction by conducting cross-linking and immunoprecipitation and RNA sequencing experiments followed by an extensive computational analysis to determine the characteristics of the HuR binding site and impact on the transcriptome. We reveal that HuR targets predominantly uracil-rich single-stranded stretches of varying size, with a strong conservation of structure and sequence composition. Despite the fact that HuR sites are observed in intronic regions, our data do not support a role for HuR in regulating splicing. HuR sites in 3′-UTRs overlap extensively with predicted microRNA target sites, suggesting interplay between the functions of HuR and microRNAs. Network analysis showed that identified targets containing HuR binding sites in the 3′ UTR are highly interconnected.
Introduction
RNA-binding proteins (RBPs)3 function as primary regulators of gene expression by controlling processes such as splicing, mRNA transport, localization, decay, and translation (1–3). The human genome has an estimated 1000 RBPs, a large portion of which function in a very specific manner by interacting with particular motifs located in selected groups of precursor mRNAs or mature mRNAs (4). The total number of targets of a given RBP reflects the sequence and structure of its binding motif and can vary from a few dozen to a few thousands (5).
Mapping of RBP target RNAs has been achieved mostly by microarray-based methods. Despite the importance of published studies, it is clear that microarrays have their limitations. Background correction issues, as described in Ref. 6, can affect the number of target RNAs identified, and only limited information can be obtained about the binding motifs. More robust and detailed information can now be obtained using procedures based on deep sequencing, such as iCLIP (7), our method of choice for the analysis described here.
Among human RBPs, Hu antigen R (HuR) or ELAVL1 is probably the most extensively investigated, reflecting its participation in multiple biological processes and diseases (reviewed in 8, 9). HuR is ubiquitously expressed and belongs to the Hu (ELAV) family of RBPs, which also includes the neuro-specific proteins HuB, HuC, and HuD. Although mainly localized in the nucleus, HuR can translocate to the cytoplasm where it prevents decay and modulates translation of target mRNAs (10–12).
It is suggested that HuR targets comprise a large portion of the transcriptome (∼15%). They generally contain AU-rich elements, which are known to mark transcripts for degradation via AU-mediated decay (13). HuR may achieve its stabilization function simply by competing with destabilizing AU-rich-element-binding proteins for the same binding sites or actively protecting the body of the message from degradation (14). HuR contains three RNA recognition motifs, and it is thought to bind single-stranded AU-rich sequences (13), although evidence of an interrelation between AU-mediated decay and miRNA pathways suggests that there may be an added layer of complexity to HuR targeting (15–17). Many HuR targets encode proteins important for cell growth, proliferation, death, and immune response (18), and its link with cancer progression is clearly established (8, 11, 19, 20). Moreover, the gene appears crucial for organism survival, with Elavl1Δ/Δ mice dying within 10 days (18).
A good portion of known HuR targets and binding sites are derived from studies on individual genes (reviewed in Ref. 8), although the broad impact of HuR on gene expression has been illustrated by a few manuscripts describing ribonucleoprotein immunoprecipitation microarray (RIP-Chip) studies (11, 13, 21). To gain a more comprehensive view of HuR function, binding site characteristics, and impact on the transcriptome, we carried out state-of-the-art genomic analyses including RNA-Seq and iCLIP followed by an extensive bioinformatics study.
MATERIALS AND METHODS
Cell Culture and Knockdown Experiments
HeLa cells were grown in RPMI 1640 containing 10% FBS and 1% penicillin/streptomycin. To conduct HuR knockdown, cells were transfected with 100 nm of Dharmacon SMARTpool J-003773-08 using Lipofectamine 2000. RNA levels were quantified using Qiagen QuantiTect primer assay QT00037856 with RPLPO as the endogenous control. RNA was purified using Qiagen RNeasy kit, and quality was accessed with the Agilent Bioanalyzer. RNA (10 μg) was processed for sequencing using Illumina mRNA-Seq per protocol. Clusters were generated on the Illumina cluster station. Samples were run on an Illumina GAIIx 2 × 36 paired end read, and CASAVA 1.6 was used to analyze and generate the raw reads.
iCLIP Experiments
HeLa cells were plated on 100-mm dishes and cross-linked when semi-confluent in 6 ml of PBS with a Spectrolinker XL-1500 two times at 100 mJ/cm2. Cells were scraped and transferred to tubes, pelleted, and snap-frozen. iCLIP was performed according to König et al. (7). Samples were amplified, purified with Beckman Genomics Ampure XP beads, and quantitated with an Agilent 2100 bioanalyzer and DNA 1000 chip. Clusters were generated with the Illumina cBot station. Samples were sequenced on an Illumina GAIIx single read 2 × 36. CASAVA 1.6 was used to analyze and generate the raw reads.
Transcriptomic Analysis
Reads were mapped to hg18 using rmap (22). Differentially expressed transcripts were identified using Fisher's exact test applied to a contingency table of read counts for total RNA and knockdown within and outside each transcript (23). An analogous approach was used on a per-exon and per-junction basis.
HuR Binding Site Mapping
Peaks in the iCLIP read profile were called by fitting a Poisson distribution per-transcript and identifying locations with significant (Bonferroni-corrected p < 0.01) enrichment for reads, similar in principle to previous studies (24); identified locations are hereafter referred to as iCLIP sites. Secondary structure prediction was done using the Vienna RNA package (25). For motif analysis, we used DME (26), and motif matches were identified using Storm from the CREAD package (27). The 44-way multiple alignments from the University of California Santa Cruz (UCSC) Genome Browser are used for analysis of cross-species conservation (28). Targets of miRNAs were taken from TargetScan (29). All p values reported were adjusted for multiple hypothesis testing using the methods of Ref. 30 or the Bonferroni correction where appropriate.
mRNA Decay Analysis
HeLa cells were reverse-transfected with 5 μl of Lipofectamine 2000 and 50 nm ELAVL1 SMARTpool or non-targeting pool (Dharmacon). 24 h later, 10 μg/ml actinomycin D was added. Cells were collected at times 0, 1, 3, and 5 h. RNA was purified using Qiagen RNeasy columns and reverse-transcribed with Applied Biosystems high capacity cDNA reagents, and quantitative PCR was performed using Applied Biosystems TaqMan gene expression assays. Changes in mRNA levels were calculated relative to GAPDH, RPLP0, and time 0. All experiments were done in triplicate.
Gene Ontology and Network Analyses
Functional enrichment analysis was performed using DAVID Bioinformatics Resources 6.7 (31). To eliminate redundant functional categories, we used the Functional Annotation Clustering tool and manually selected 15 clusters with the highest enrichment scores. Within each cluster, we chose the functional category with the most significant p value. The network was constructed using Pathway Studio 8 and drawn with Cytoscape 2.6.3 (32). Full computational analysis details are given in the supplemental material. Read mapping statistics are provided in supplemental Tables 10 and 11.
RESULTS AND DISCUSSION
Identification of HuR Binding Sites and Targets
Concomitant knockdown experiments followed by RNA-Seq analysis and iCLIP experiments in HeLa cells allowed us to get a better picture of HuR impact on gene expression. A reduction in HuR levels produced significant (corrected p < 0.01) changes in gene expression for ∼13% of the RefSeq transcriptome (which covers almost 39% of hg18 when including introns). The large impact on transcriptomic levels is expected for a protein that regulates the expression of a large number of genes as suggested by our iCLIP analysis and previous study (13). However, we expect that most of these variations in gene expression are the result of an indirect effect. Complete RNA-Seq analysis can be found in supplemental Table 1.
HuR binding sites as revealed by the iCLIP analysis are biased toward intronic regions and 3′-UTRs, as shown in Fig. 1 and supplemental Tables 3–6. The presence of HuR binding sites in intronic regions is discussed below. The enrichment of sites in the 3′-UTR of genes is consistent with known HuR patterns of regulation (mRNA decay and translation). Comparison between RNA-Seq and iCLIP data determined that 26% of the genes that show alterations in mRNA expression level upon HuR knockdown have HuR sites in their 3′-UTR, 5′-UTR, or coding sequence (p < 3.9 × 10−23, Fisher's exact test). Supplemental Table 9 lists those genes that show significant expression changes and contain a non-intronic iCLIP site. Bearing in mind the functions of HuR, we suggest that a portion of these genes might have their mRNA decay influenced by HuR. However, some caution is necessary in this interpretation. First, indirect effects are expected as mentioned above; many of the changes in RNA level could be taking place mainly at the transcriptomic level due to the action of other factors affected by HuR. Second, although indicative, changes at the mRNA level by itself cannot be used as a definitive parameter to evaluate mRNA decay, which should be measured over time in the presence of RNA polymerase inhibitors. Even genes not showing significant changes at the mRNA level can have their decay influenced by HuR. In supplemental Fig. 4, we used mRNA decay assays to show, for a group of identified targets, that HuR knockdown influences their stability.
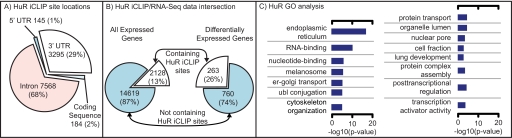
FIGURE 1.
A, breakdown of HuR iCLIP sites based on gene annotations; in line with established patterns of regulation, we see a large proportion of iCLIP sites in 3′-UTRs, although we also, unexpectedly, observe many intronic sites. B, the percentage of expressed genes for which we observe at least one non-intronic iCLIP site (left) and percentage of genes identified as differentially expressed in which we observe at least one non-intronic iCLIP site (right). C, Gene Ontology analysis of HuR target genes containing two or more iCLIP sites in the 3′-UTR. er, endoplasmic reticulum; ubl, ubiquitin-like modifier protein.
To better understand the nature of HuR targets, we conducted Gene Ontology (GO) and network analyses. Because the number of identified iCLIP sites is very high, we restricted the analyses to the group of genes containing two or more iCLIP sites in the 3′-UTR. HuR targets are highly connected, forming a complex network with 300 genes (supplemental Table 8). The core of this network is constituted by a group of 40 cancer-related genes highly associated to each other. Listed genes are those determined to have 15 or more connections to other genes present in the network (ACTB, APP, CALR, CAV1, CCND1, CDC42, CD44, CTNNB1, CYCS, EIF4E, EIF2S1, EGFR, EPAS1, FYN, GSK3B, HSP90AA1, IGF1R, IGF2, IGFBP3, ITGB1, MDM2, MYC, NF2, PTEN, RB1, RHOA, SDC1, SMAD2, SP3, SRBF1, STAT3, TBP, TCF3, TCF4, TFAP2A, TFRC, TGFBR1, XIAP, XPO1, and YY1) (see also supplemental Fig. 1)). Gene Ontology analysis revealed enrichment for endoplasmic reticulum and several processes associated with gene regulation (Fig. 1).
One previous study suggests that HuR might be implicated in alternative splicing regulation (14). Similarly, ELAV, the HuR Drosophila homologue, is a known splicing regulator (33, 34). We decided then to further investigate the subject. Overall, our results suggest that HuR does not play a major role in regulating splicing. Despite the fact that we identified a large number of iCLIP sites in intronic regions, we did not observe a significant enrichment of sites in proximity to known splice sites; more than 80% of identified sites are located 200 nucleotides or more from the closest splice site (supplemental Fig. 3). Further, upon analysis of the RNA-Seq data, we observed no significant changes in junction usage and identified only seven differentially used exons, of which only one has a splice site intersecting a significant iCLIP site for HuR (see supplemental Table 2). Finally, when correcting for region size, the intronic enrichment of reads is diluted to the point where it is comparable with intergenic read levels, indicating that it might be largely attributed to noise (see supplemental Fig. 3).
Characterization of HuR Binding Site
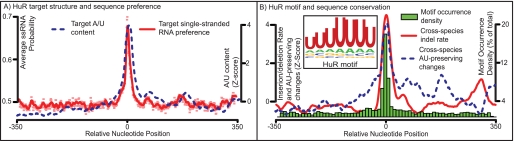
HuR binding site analysis indicated a sizeable increase in AU content at identified iCLIP sites (Fig. 2). HuR iCLIP sites have an overrepresentation of U-rich heptamers but are not as heavily abundant in A/U- or A-rich heptamers (supplemental Fig. 2). We also observed a strong preference for single-stranded RNA at HuR iCLIP sites, and an analysis of multispecies alignments revealed an increased insertion/deletion (indel) rate around sites, coupled with a preference to preserve A/U content (Fig. 2). Taken together, these results suggest that HuR binds to variably sized hairpin loops rich in uracil. These data slightly contradict the concept that HuR binds preferentially A/U rich sequences, but are in line with the findings in Ref. 35.
FIGURE 2.
A, Z-score for single-stranded RNA preference and target A/U content. Previous studies have suggested that HuR binds hairpin loops (13), which the preference for ssRNA supports. B, the histogram shows the occurrence density of the sequence logo (inset), which is built from the top 1% of heptamers enriched at 3′-UTR iCLIP sites. This enrichment suggests that HuR may be preferentially binding uracil-rich sequences, rather than AU-rich sequences in general (see supplemental Fig. 2 for further details). Shown also is cross-species conservation of A/U nucleotides (percentages of non-matching nucleotides to human A/U, which are also A/U or a gap) and cross-species indel rate at 3′-UTR iCLIP sites. Previous studies have suggested that HuR binds hairpin loops (13); our observation of an increased indel rate across species indicates that the size and exact primary sequence of this loop may be highly variable.
HuR Association with miRNAs
The most striking results we obtained concern a significant overlap between iCLIP sites and miRNA target sites predicted by TargetScan (24% of total iCLIP sites present in 3′-UTRs, p < 1 × 10−5; see supplemental Fig. 3 and supplemental Table 7). We also observed a significant overlap with conserved validated miRNA sites from Ref. 36 (p < 0.006). Among the identified sites, we observed a significant enrichment for two tumor suppressor miRNAs, miR-217 (37) and miR-1/206 (38), with ∼3- and ∼2-fold enrichment, respectively (p < 0.008 and p < 0.005). There has been increasing interest in the interplay between RNA-binding proteins and miRNA. In the particular case of HuR, it has been seen that it associates with the 3′-UTR of the CAT1 mRNA after stress, counteracting the effect of miR-122 (reviewed in Ref. 15). Our results suggest that this cross-talk between HuR and miRNAs may be more frequent than expected. However, further detailed analyses are necessary to reveal to what extent HuR antagonizes and/or cooperates with miRNAs.
Acknowledgments
We thank Jernej Ule for advice with iCLIP experiments, Melanie Carless and Roy Garcia for helping with deep sequencing experiments, Devraj Sandhu for help with pathway and GO analyses, Emad Bahrami Samani for ssRNA probability calculations, and Dat Vo for helping with RNA stability assays.
Note Added in Proof
During the preparation of the final version of this manuscript, two articles (39, 40) were published corroborating the findings described herein.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R21HG004664–02 (to L. O. F. P.).
The sequence data reported in this article have been submitted to the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under accession number SRP008069.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Tables 1–11, and Figs. 1–4.
- RBP
- RNA-binding protein
- HuR
- Hu antigen R
- iCLIP
- individual-nucleotide resolution UV cross-linking and immunoprecipitation
- ELAV
- embryonic lethal abnormal vision
- miRNA
- microRNA
- miR
- microRNA.
REFERENCES
- 1. Lunde B. M., Moore C., Varani G. (2007) Nat. Rev. Mol. Cell Biol. 8, 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dreyfuss G., Kim V. N., Kataoka N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195–205 [DOI] [PubMed] [Google Scholar]
- 3. Burd C. G., Dreyfuss G. (1994) Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 4. Anantharaman V., Koonin E. V., Aravind L. (2002) Nucleic Acids Res. 30, 1427–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mittal N., Roy N., Babu M. M., Janga S. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20300–20305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ritchie M. E., Silver J., Oshlack A., Holmes M., Diyagama D., Holloway A., Smyth G. K. (2007) Bioinformatics 23, 2700–2707 [DOI] [PubMed] [Google Scholar]
- 7. König J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D. J., Luscombe N. M., Ule J. (2010) Nat. Struct. Mol. Biol. 17, 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Srikantan S., Gorospe M. (2011) Front. Biomed. Sci., in press [Google Scholar]
- 9. Hinman M. N., Lou H. (2008) Cell. Mol. Life Sci. 65, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fan X. C., Steitz J. A. (1998) EMBO J. 17, 3448–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mazan-Mamczarz K., Hagner P. R., Corl S., Srikantan S., Wood W. H., Becker K. G., Gorospe M., Keene J. D., Levenson A. S., Gartenhaus R. B. (2008) Oncogene 27, 6151–6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yi J., Chang N., Liu X., Guo G., Xue L., Tong T., Gorospe M., Wang W. (2010) Nucleic Acids Res. 38, 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennan C. M., Steitz J. A. (2001) Cell. Mol. Life Sci. 58, 266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kedde M., Agami R. (2008) Cell Cycle 7, 899–903 [DOI] [PubMed] [Google Scholar]
- 16. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. (2006) Cell 125, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 17. von Roretz C., Gallouzi I. E. (2008) J. Cell Biol. 181, 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghosh M., Aguila H. L., Michaud J., Ai Y., Wu M. T., Hemmes A., Ristimaki A., Guo C., Furneaux H., Hla T. (2009) J. Clin. Invest. 119, 3530–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. López de Silanes I., Lal A., Gorospe M. (2005) RNA Biol. 2, 11–13 [DOI] [PubMed] [Google Scholar]
- 20. Filippova N., Yang X., Wang Y., Gillespie G. Y., Langford C, King P. H., Wheeler C., Nabors L. B. (2011) Mol. Cancer Res. 9, 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calaluce R., Gubin M. M., Davis J. W., Magee J. D., Chen J., Kuwano Y., Gorospe M., Atasoy U. (2010) BMC Cancer 10, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Smith A. D., Chung W. Y., Hodges E., Kendall J., Hannon G., Hicks J., Xuan Z., Zhang M. Q. (2009) Bioinformatics 25, 2841–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bloom J. S., Khan Z., Kruglyak L., Singh M, Caudy A. A. (2009) BMC Genomics 10, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yeo G. W., Coufal N. G., Liang T. Y., Peng G. E., Fu X. D., Gage F. H. (2009) Nat. Struct. Mol. Biol. 16, 130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hofacker I.L. (2003) Nucleic Acids Res. 31, 3429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith A. D., Sumazin P., Zhang M. Q. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1560–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith A. D., Sumazin P., Xuan Z., Zhang M. Q. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6275–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rhead B., Karolchik D., Kuhn R. M., Hinrichs A. S., Zweig A. S., Fujita P. A., Diekhans M., Smith K. E., Rosenbloom K. R., Raney B. J., Pohl A., Pheasant M., Meyer L. R., Learned K., Hsu F., Hillman-Jackson J., Harte R. A., Giardine B., Dreszer T. R., Clawson H., Barber G. P., Haussler D., Kent W. J. (2010) Nucleic Acids Res. 38, Suppl. 1, D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benjamini Y., Hochberg Y. (1995) J. R. Stat. Soc. Series B Stat. Methodol. 57, 289–300 [Google Scholar]
- 31. Huang da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koushika S. P., Lisbin M. J., White K. (1996) Curr. Biol. 6, 1634–1641 [DOI] [PubMed] [Google Scholar]
- 34. Lisbin M. J., Qiu J., White K. (2001) Genes Dev. 15, 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ray D., Kazan H., Chan E. T., Peña Castillo L., Chaudhry S., Talukder S., Blencowe B. J., Morris Q., Hughes T. R. (2009) Nat. Biotechnol. 27, 667–670 [DOI] [PubMed] [Google Scholar]
- 36. Chi S. W., Zang J. B., Mele A., Darnell R. B. (2009) Nature 460, 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao W. G., Yu S. N., Lu Z. H., Ma Y. H., Gu Y. M., Chen J. (2010) Carcinogenesis 31, 1726–1733 [DOI] [PubMed] [Google Scholar]
- 38. Nohata N., Sone Y., Hanazawa T., Fuse M., Kikkawa N., Yoshino H., Chiyomaru T., Kawakami K., Enokida H., Nakagawa M., Shozu M., Okamoto Y., Seki N. (2011) Oncotarget 2, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lebedeva S., Jens M., Theil K., Schwanhäusser B., Selbach M., Landthaler M., Rajewsky N. (2011) Mol. Cell 43, 340–352 [DOI] [PubMed] [Google Scholar]
- 40. Mukherjee N., Corcoran D. L., Nusbaum J. D., Reid D. W., Georgiev S., Hafner M., Ascano M., Jr., Tuschl T., Ohler U., Keene J. D. (2011) Mol. Cell 43, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]