Abstract
Cell migration is essential for several important biological outcomes and is involved in various developmental disorders and disease states including cancer cell invasiveness and metastasis. A fundamental step in cell migration is the development of a leading edge. By using HeLa carcinoma cells as an initial model system, we uncovered a surprising role for the heat shock protein 70 (Hsp70) and its ability to bind the protein cross-linking enzyme, tissue transglutaminase (tTG), in cancer cell migration. Treatment of HeLa cells with EGF results in the activation of a plasma membrane-associated pool of tTG and its redistribution to the leading edges of these cells, which are essential events for EGF-stimulated HeLa cell migration. However, we then found that the ability of tTG to be localized to the leading edge is dependent on Hsp70. Similarly, the localization of tTG to the leading edges of MDAMB231 breast carcinoma cells, where it also plays an essential role in their migration, has a strict requirement for Hsp70. Treatment of these different cell lines with inhibitors against the ATP hydrolytic activity of Hsp70 prevented tTG from localizing to their leading edges and thereby blocked EGF-stimulated HeLa cell migration, as well as the constitutive migration normally exhibited by MDAMB231 cells. These findings highlight a new and unconventional role for the chaperonin activity of Hsp70 in the localization of a key regulatory protein (tTG) at the leading edges of cancer cells and the important consequences that this holds for their ability to migrate.
Keywords: Cell Migration, Growth Factors, Heat Shock Protein, Membrane Bilayer, Signal Transduction, EGF, Cancer Cells, Chaperonin, Invasion, Tissue Transglutaminase
Introduction
Cell migration is a highly complex, multistep process that is carried out by virtually all cell types at some point during their lifetime, and it underlies a number of important biological outcomes that range from embryonic development to immune responses and tissue repair/regeneration (1–5). It is also a cellular function that is frequently targeted for deregulation in human cancers, because acquiring the ability to migrate aberrantly is a crucial step in the development of the invasive and metastatic phenotypes exhibited by advanced stage cancers (6, 7). Although many important questions regarding how migration is regulated in different cellular contexts remain unanswered, the general process of cell migration is fairly well established. Common to all forms of directional cell migration is the formation of a polarized cell, where the asymmetric distribution of signaling proteins, phospholipids, and cytoskeletal components gives rise to a cell with both leading and trailing edges (8). The leading edge is characterized by Arp2/3-driven actin polymerization and plasma membrane protrusions that extend from the cell body in the direction of movement. These membrane protrusions then make new contact points between a cell and its underlying substrate, providing the necessary adhesion that a cell needs to move forward. However, in order for a cell to successfully migrate, immediately following the formation of new adhesion sites along the leading edge, changes at the trailing edge of the cell must also occur. In particular, contraction of the actomyosin cytoskeleton and the disassembly of established cell adhesion sites at the trailing edge take place to allow the back end of a migrating cell to detach from its substrate, retract, and then shift the remaining part of the cell body toward the leading edge.
Given the connections between cell migration and numerous biological outcomes including the progression of certain human disease states (i.e. cancer), it is not surprising that a good deal of effort has been dedicated toward understanding the mechanisms that are responsible for regulating the ability of cells to migrate. The induction of cell migration is typically triggered by growth factors and/or signals from the extracellular matrix that surrounds cells (9, 10). The abilities of these extracellular stimuli to induce the activation of a series of signaling events within target cells help coordinate the extensive remodeling of the actin cytoskeleton and changes in the adhesion properties of cells that are necessary for cell migration. For example, stimulating the human cervical carcinoma cell line, HeLa, with EGF causes these cells to acquire a polarized morphology (forming both leading and trailing edges) and increases their ability to migrate and exhibit invasive activity (11).
Among the most extensively investigated group of signaling proteins that have been linked to EGF-induced cell migration are members of the Rho family of small GTPases including Cdc42, Rho, and Rac (12, 13). Each of these highly related GTPases are regulated in a spatially and temporally specific manner in actively migrating cells. Cdc42 is necessary for establishing and maintaining cell polarity by properly positioning the nucleus and orienting microtubules in the direction of cell movement and by helping to recruit signaling and motor/assembly proteins to the leading edges of cells. On the other hand, Rac and Rho act antagonistically toward each other, with Rac being predominantly localized to leading edges where it promotes Arp2/3-dependent actin polymerization and the formation of cellular protrusions, whereas Rho is found along trailing edges where it participates in the contraction and bundling of the actin cytoskeleton.
Although several other proteins have been implicated in EGF-stimulated cell migration, the vast majority of these are traditional signaling proteins (i.e. Ras, PI3K, phospholipase C, ERK, and JNK) (7, 14, 15). However, there are also a few examples of nontraditional signaling proteins that are important for certain types of cells to migrate. One such example is tTG,2 a dual functioning protein that combines an ability to bind and hydrolyze GTP with an enzymatic transamidation activity that generates covalent cross-links between two proteins or between a protein and a polyamine (16, 17). tTG is overexpressed in a significant percentage of advanced stage and high grade human cervical, lung, brain, prostate, and breast tumors, and its transamidation (cross-linking) activity has been shown to be essential for the invasive/metastatic behavior of highly aggressive cancer cells, such as the MDAMB231 human breast cancer cell line (18–21). Recently, we showed that the stimulation of HeLa carcinoma cells with EGF resulted in tTG activation and its accumulation at their leading edges, whereas knock-downs of tTG or exposure of the cells to the tTG inhibitor monodansylcadaverine (MDC) inhibited the EGF-stimulated migration and invasive activity of these cells (11). Although these findings demonstrate a fundamental role for tTG and in particular its enzymatic cross-linking activity, in EGF-stimulated cancer cell migration what remains to be determined is how EGF triggers the accumulation of tTG at the leading edges of cells and whether this event is important for the ability of tTG to promote cell migration.
In this study, we have taken an important step toward answering these questions by uncovering a novel connection between tTG, a component of the chaperonin network, Hsp70, and the ability of human cancer cells to migrate. We identify Hsp70 as a novel tTG-interacting partner and show that the ability of plasma membrane-associated tTG to localize to the leading edges of EGF-stimulated HeLa cells, as well as to the leading edges of constitutively migrating MDAMB231 breast cancer cells, is sensitive to inhibition of Hsp70. Importantly, we further demonstrate that exposure of these different cell lines to inhibitors of the ATP hydrolytic activity of Hsp70, while having no effect on the protein cross-linking activity of tTG, inhibits EGF-induced HeLa cell migration and the constitutive migration normally exhibited by MDAMB231 breast carcinoma cells. To our knowledge, these findings show for the first time that the chaperonin capabilities of heat shock proteins can participate in cell migration by helping to target key regulatory proteins (i.e. tTG) to the leading edges of migrating cells and demonstrate that the proper localization of tTG to leading edges is crucial for its ability to promote cancer cell migration.
EXPERIMENTAL PROCEDURES
Materials
All cell culture reagents, the Colloidal Blue staining kit, EGF, Lipofectamine, Lipofectamine 2000, protein G beads, and V5 antibody, as well as the control, tTG, and Hsp70 siRNAs, were from Invitrogen. MDC, 6-diamidino-2-phenylindole (DAPI), myricetin, methylene blue, and the fibronectin antibody were obtained from Sigma, whereas VER 155008 was from Tocris Biosciences. The tTG and actin antibodies were from Neomarkers; the IκBα, Hsp70, and cortactin antibodies were from Cell Signaling; and the HA and Myc antibodies were from Covance. The biotinylated pentylamine (BPA) was from Pierce, and T101 was from Zedira. The Quick Blue staining kit was from Boston Biologicals.
Cell Culture
HeLa and MDAMB231 cells were grown in RPMI 1640 medium containing 10% FBS. The pcDNA3 constructs encoding V5-tagged Hsc70, the various forms of Myc-tagged tTG, and the HA-tagged forms of activated Rac and Ras were transfected into cells using Lipofectamine, whereas the control, tTG, and Hsp70 siRNAs were introduced into cells using Lipofectamine 2000. As indicated, cell cultures or cell extracts were treated with various combinations of 0.1 μg/ml EGF, 50 μm myricetin, 10 μm methylene blue, 50 μm VER 155008, and 1.0 μm T101. The cells were then either collected for cell fractionation, fixed with 3.7% formaldehyde, or lysed with cell lysis buffer (25 mm Tris, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, 1 mm DTT, 1 mm NaVO4, 1 mm β-glycerol phosphate, 1 μg/ml aprotinin, and 1 μg/ml leupeptin). The Bio-Rad DC protein assay was used to determine protein concentrations.
Immunofluorescence
The fixed cells were permeabilized with PBS containing 0.1% Triton X-100, blocked in PBS containing 10% BSA, and incubated with the indicated primary antibodies for 2 h. The cells were then incubated with either Oregon Green 488- or Rhodamine Red-conjugated secondary antibody (Molecular Probes) for 1 h. Where indicated, Rhodamine-conjugated phalloidin was used to stain actin filaments, whereas DAPI was used to stain nuclei. Following the secondary incubations, the cells were washed extensively with PBS, mounted, and visualized using the 63× objective on a Zeiss Axioskop fluorescent microscope. The images were captured and processed using IPLAB.
Cell Migration (Scratch) Assays
Parental cells or cells expressing the control siRNA or tTG siRNAs were grown to confluence and then put in serum-free medium without or with 0.1 μg/ml EGF and without or with 20 μm MDC, as indicated. Fifteen hours later, a wound was struck using a pipette tip, and the culturing medium on the cells was replenished to remove detached cells. When examining the effects of myricetin, methylene blue, and VER 155008 on cell migration, the cells maintained in serum-free medium for 15 h with or without 0.1 μg/ml EGF were treated without or with one of the inhibitors for 1 h before striking a wound and replenishing the medium. After the indicated incubation period, the cells were fixed and visualized by light microscopy. Each of these experiments was performed at least three times.
Cell Fractionation
To fractionate cells into their cytosolic and membrane components, harvested cells were resuspended in homogenization buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 200 mm sucrose, and 1 mm PMSF) and homogenized using a Dounce homogenizer. The cell extracts were then centrifuged at 1,000 rpm for 10 min to remove the intact nuclei, followed by centrifugation at 47,000 rpm for 1.5 h to pellet cellular membranes. The soluble cytosolic fraction was removed and saved, whereas the membrane fraction was lysed in membrane lysis buffer (10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm PMSF, and 0.5% Triton X-100) for 1 h, followed by centrifugation at 13,000 rpm for 10 min to remove any insoluble material.
Transamidation Assays
Cell lysates (15 μg of each) were incubated in a buffer containing 10 mm dithiothreitol, 10 mm CaCl2, and 50 μm BPA for 10 min. The reactions were stopped with the addition of Laemmli sample buffer, followed by boiling, and the proteins were resolved on a gel, transferred to PDVF membranes, and blocked for at least 1 day in BBST (100 mm boric acid, 20 mm sodium borate, 0.01% SDS, 0.01% Tween 20, and 80 mm NaCl) containing 10% BSA. The membranes were incubated with horseradish-peroxidase-conjugated streptavidin, diluted at 1:2000 in BBST containing 5% BSA for 1 h, and then washed extensively with BBST. The proteins that incorporated BPA were visualized on x-ray film after exposing the membranes to ECL reagent.
In Vitro Liposome Fractionation Assays
Synthetic liposomes were prepared from a lipid mixture containing 35% phosphatidylethanolamine, 25% phosphatidylserine, 5% phosphatidylinositol, and 35% cholesterol resuspended in TBSM buffer (20 mm Tris, pH 7.5, 150 mm NaCl, and 2 mm MgCl2). The lipids were extruded through an 8-micron filter, pelleted by centrifugation at 13,000 rpm for 15 min, and resuspended in TBSM buffer. Equal amounts of the lipid preparation were then incubated with either recombinant wild-type tTG or BSA for 15 min, followed by centrifugation at 13,000 rpm for 10 min at room temperature. The supernatant was concentrated to ∼30 μl using a microcentrifuge concentrator with a 10,000 molecular weight cut-off, whereas the pelleted liposomes were resuspended in 30 μl of TBSM buffer. Each of the samples was resolved on a gel and then stained with Quick Blue to detect proteins.
Immunoprecipitations
Cell lysates (∼1.2 mg) that had been precleared with protein G beads were incubated with nothing, a Myc antibody, or a nonspecific mouse IgG control antibody for 1.5 h as indicated. Protein G beads were then added to the lysates and incubated for an additional 1.5 h, at which time the beads were washed extensively with cell lysis buffer.
Immunoblot Analysis
Whole cell extracts, various isolated subcellular fractions, and immunoprecipitated proteins were resolved by SDS-PAGE, followed by transfer to PVDF membranes. The membranes were incubated with the indicated primary antibodies diluted in TBST (in 20 mm Tris, 135 mm NaCl, and 0.02% Tween 20). The primary antibodies were detected with horseradish-peroxidase-conjugated secondary antibodies followed by exposure to ECL reagent. Some of the resulting blots were quantified using ImageJ software.
RESULTS
tTG Is Recruited to the Leading Edges of Migrating Cancer Cells
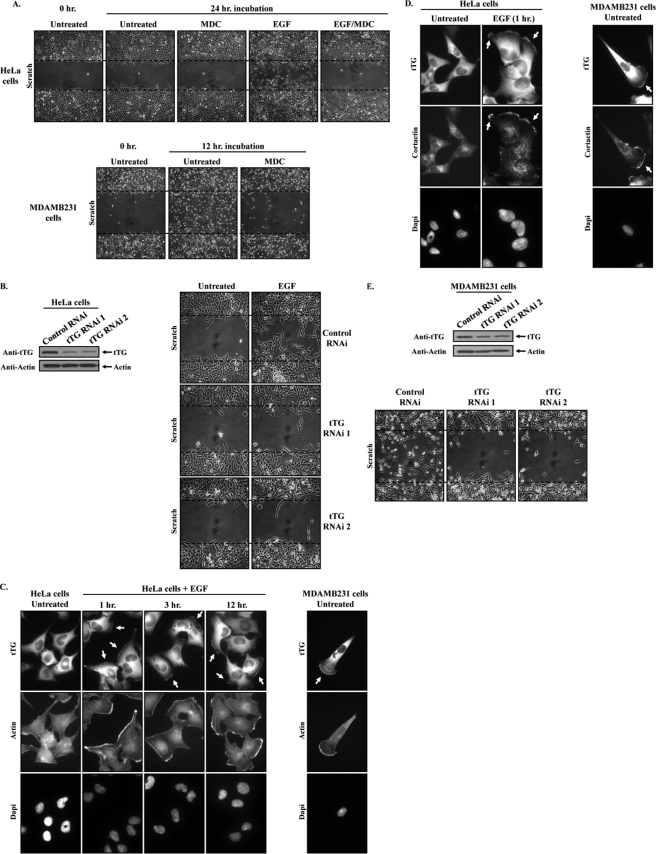
The accumulating evidence implicating tTG as an important contributor to cancer cell migration, invasion, and metastasis (11, 19, 20) prompted us to investigate further the possible regulatory mechanisms that underlie its ability to promote cell migration. We chose the HeLa cervical carcinoma cell line as our initial model system because we had previously established that stimulating HeLa cells with EGF induced tTG activation and showed that knocking down tTG or treatment of these cells with the tTG cross-linking inhibitor MDC blocked their EGF-stimulated migration and invasive activity (11). Using scratch or wound healing assays as a means to determine the rates of cell migration, we confirmed the importance of tTG activity in mediating EGF-stimulated HeLa cell migration by showing that the ability of EGF to promote the migration of HeLa cells, as indicated by their movement into the wound, is severely ablated when the cells are treated with MDC (Fig. 1A, compare the last two images in the top panels). Likewise, knocking down tTG in HeLa cells using two different siRNAs (Fig. 1B, left panels) inhibited the EGF-dependent migration of these cells (Fig. 1B, right panels). However, we also noticed by immunofluorescent analysis that treating serum-starved HeLa cells with EGF for increasing lengths of time caused a detectable change in the subcellular distribution of tTG (Fig. 1C, top row on the left). Although in untreated HeLa cells, tTG was expressed primarily throughout the cytoplasm, EGF treatment of the cells for as little as 1 h caused an accumulation of tTG at their plasma membranes, particularly along leading edges, as indicated by its co-localization with sites of F-actin build-up (Fig. 1C, middle row), as well as with the leading edge marker cortactin (22) (Fig. 1D, left panels). This effect of EGF on the subcellular localization of tTG persisted even after 12 h of EGF stimulation (Fig. 1C, top row), suggesting that tTG could be a leading edge-resident protein. Consistent with this idea, immunofluorescence performed on serum-starved MDAMB231 breast carcinoma cells, a highly invasive/metastatic human cancer cell line whose constitutive migration capabilities are dependent on tTG as determined by MDC treatment (Fig. 1A, compare the last two images in the bottom panels) and by tTG knockdown (Fig. 1E), revealed that tTG accumulated along their leading edges as well (Fig. 1, C, right panels, and D, right panels). Taken together, these findings raise the intriguing possibility that the recruitment of tTG to the leading edges of actively migrating cells may represent an important regulatory step underlying its ability to promote cell migration.
FIGURE 1.
tTG is localized to the leading edges of actively migrating cells, and its cross-linking activity is necessary for cell migration. A, scratch assays were performed on HeLa cells (top panels) and MDAMB231 cells (bottom panels) treated without (Untreated) or with EGF and without or with MDC as indicated. MDAMB231 cells were fixed 12 h after striking the wound; HeLa cells were fixed after 24 h. The cells were then visualized using light microscopy, and the extent of wound closure was determined. One set of untreated cells was fixed immediately after striking the wound (Untreated 0 h.) to indicate the size of the initial wounds. The widths of the initial wounds are indicated by dashed lines. B, the extracts collected from HeLa cells transfected with control-RNAi, tTG-RNAi 1, or tTG-RNAi 2 were immunoblotted with tTG and actin antibodies (left panels). Scratch assays were then performed on cells transfected with the same siRNAs, treated without (Untreated) or with EGF. The cells were processed as outlined in A (right panels). C and D, duplicate sets of serum-starved cultures of HeLa cells and MDAMB231 cells were treated without (Untreated) or with EGF for increasing lengths of time, as indicated, and then were fixed. C, immunofluorescence was performed on one set of the cells using a tTG antibody, rhodamine-conjugated phalloidin (Actin), and DAPI (to stain nuclei). D, immunofluorescence was performed on the second set of cells using tTG and cortactin antibodies and DAPI. Representative fluorescent images of the cells are shown, and the localization of tTG and cortactin at leading edges is indicated with arrows. E, the extracts collected from MDAMB231 cells transfected with control-RNAi, tTG-RNAi 1, or tTG-RNAi 2 were immunoblotted with tTG and actin antibodies (top panels). Scratch assays were then performed on cells transfected with the same siRNAs, and the cells were processed as outlined in A (bottom panels).
A Plasma Membrane-associated Pool of tTG Is Trafficked to Leading Edges
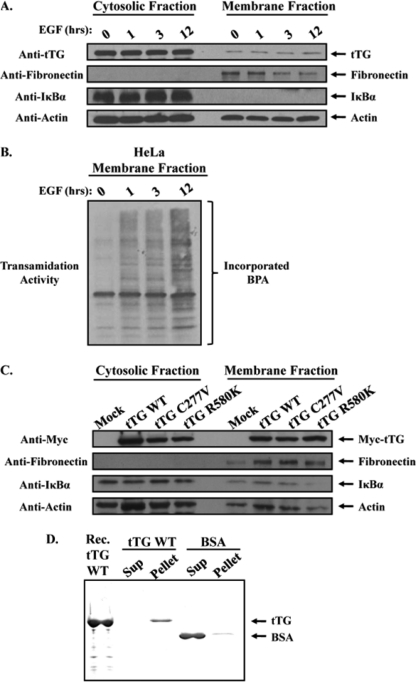
To determine whether the localization of tTG at leading edges plays an essential role in cell migration, we set out to better understand how tTG accumulates at these distinct regions of the plasma membrane. Based on our immunofluorescence experiments (Fig. 1, C and D) it appears that tTG is predominantly expressed in the cytoplasm of untreated HeLa cells, but can then be detected along their leading edges in response to EGF stimulation. Thus, we initially suspected that the appearance of tTG at the leading edges of EGF-stimulated HeLa cells was an outcome of its recruitment from the cytoplasm to the plasma membrane. To determine whether this was indeed the case, we examined whether the changes in the distribution of tTG in HeLa cells treated with EGF, as observed by immunofluorescence, could also be detected using a cell fractionation approach (i.e. as monitored by increases in the amount of tTG present in the membrane fractions collected from EGF-stimulated HeLa cells). Serum-starved HeLa cells that had been left untreated or were stimulated with EGF for 1, 3, or 12 h were collected and divided into their cytosolic and membrane fractions, and the resulting extracts were immunoblotted. Fig. 2A shows that the fractionation procedure was successful, as indicated by the enrichment of the plasma membrane-associated protein, fibronectin, in the membrane fractions, and the cytosolic protein, IκBα, in the cytoplasmic fractions (Fig. 2A, second and third panels from the top, respectively). However, when the blot was probed for tTG, we obtained a surprising result. Rather than detecting increased amounts of tTG in the membrane fractions from the EGF-stimulated HeLa cells compared with those collected from untreated HeLa cells, we found that the levels of tTG were essentially equivalent (determined by densitometry to be 10 ± 2.5% of the tTG expressed in the cells) in these different membrane fractions (Fig. 2A, top panel). This indicated that EGF does not direct the translocation of tTG from the cytoplasm to the surface of HeLa cells. Instead, it appears that a small but discrete pool of tTG associates with the plasma membrane in serum-starved HeLa cells (that cannot be detected by immunofluorescence) and is redistributed in an EGF-dependent manner such that it accumulates at the leading edges of cells.
FIGURE 2.
A pool of tTG is constitutively associated with the plasma membrane. Serum-starved HeLa cells that had been treated without (lane 0) or with EGF for increasing lengths of time, as indicated, were homogenized and then subjected to differential centrifugation to separate cytosolic and membrane fractions. A, the cellular fractions were immunoblotted with tTG, fibronectin, IκBα, and actin antibodies. B, the same membrane fractions were also assayed for tTG transamidation activity by determining the incorporation of BPA into lysate proteins. C, actively growing HeLa cells that had been either mock transfected without DNA (Mock) or transfected with various Myc-tagged forms of tTG including wild-type (tTG WT), a transamidation-defective mutant (tTG C277V), or a GTP-binding-defective mutant (tTG R580K) were homogenized and then subjected to differential centrifugation to isolate cytosolic and membrane fractions. The fractions were immunoblotted with Myc, fibronectin, IκBα, and actin antibodies. D, synthetic liposomes were prepared by extrusion, and then equal amounts of this preparation were combined with either 5 μg of recombinant tTG (tTG WT) or 5 μg of BSA. After a 15-min incubation, the liposomes were pelleted by centrifugation, and the resulting supernatant (Sup) and liposome (Pellet) fractions were resolved by SDS-PAGE. The gel was then stained with Quick Blue to detect the proteins. A lane containing recombinant tTG (Rec. tTG WT) was included as a standard.
We next examined whether EGF treatment would affect the activity of the plasma membrane-associated pool of tTG. The same membrane fractions analyzed in Fig. 2A were assayed for tTG-catalyzed transamidation activity, as read-out by the incorporation of BPA into lysate proteins. Fig. 2B shows that little tTG activity was detectable in the membrane fraction collected from serum-starved HeLa cells. In contrast, EGF stimulation caused a marked increase in the cross-linking activity of the plasma membrane-associated tTG. The increase in tTG activity was detectable within 1 h of EGF treatment and was maintained, if not slightly enhanced, through 12 h of continuous EGF stimulation. It is also worth noting that the abilities of EGF to stimulate the enzymatic cross-linking activity of the membrane-associated tTG and to cause tTG to accumulate at the leading edges of HeLa cells occur on similar time scales (compare Fig. 1C, left panel, and Fig. 2B), highlighting the fact that both the activation and the localization of tTG to the leading edges of cells are tightly coupled EGF-dependent signaling events.
To gain a better understanding for how tTG associates with the plasma membrane, two additional experiments were performed. In the first experiment, we examined whether the GTP-binding capability or the transamidation activity of tTG was required for it to associate with plasma membranes. Actively growing HeLa cells ectopically expressing Myc-tagged forms of wild-type tTG or mutant forms of tTG that were defective in their abilities to either catalyze the formation of protein cross-links (tTG C277V) or to bind GTP (tTG R580K) (23, 24) were collected and fractionated into their cytosolic and membrane components. The resulting lysates were then immunoblotted with a Myc antibody to detect the different ectopically expressed forms of tTG. The top panel in Fig. 2C shows that the ectopically expressed transamidation-defective form of tTG (tTG C277V) and the GTP-binding-defective mutant (tTG R580K) were localized to the membrane fractions from HeLa cells just as efficiently as the ectopically expressed wild-type tTG, indicating that the ability of tTG to associate with plasma membranes does not require its GTP binding or transamidation activity.
In the second experiment, we examined whether tTG could directly associate with plasma membranes or whether it requires additional proteins to do so. To address this question, we used an in vitro approach similar to one that was developed to examine the ability of the small GTPase Cdc42 to interact with synthetically derived liposomes (25). Purified recombinant wild-type tTG or BSA serving as a control was combined together with synthetically derived liposomes whose lipid composition was similar to that of the inner leaflet of the plasma membrane in mammalian cells. Following a brief incubation, the vesicles were pelleted by centrifugation, and then the resulting supernatants and pellets were resolved by SDS-PAGE and stained with Quick Blue to detect the proteins. Fig. 2D shows that tTG has a relatively high affinity for lipids, because nearly all of the recombinant tTG (tTG WT) was found to have pelleted with the synthetic vesicles. On the other hand, BSA only weakly pelleted with the liposomes, with most of the control protein remaining in the supernatant.
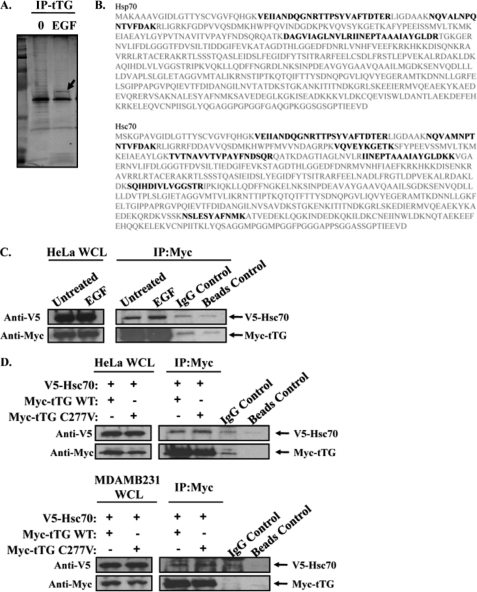
Hsp70 Is a Novel tTG-interacting Protein
The findings above describe the identification and initial characterization of a plasma membrane-associated pool of tTG that may have important consequences for cell migration. However, they did not shed light on how tTG accumulates at the leading edges of migrating cells. In particular, we wondered whether tTG may need to interact with another protein(s) to achieve this localization. Thus, we set out to identify proteins that interacted with tTG in the plasma membranes from EGF-stimulated HeLa cells by mass spectrometry. Serum-starved cultures of HeLa cells that had been left untreated or were stimulated with EGF for 12 h (i.e. a time period that we knew to be sufficient for the accumulation of tTG at the leading edges of cells) were collected and their corresponding membrane fractions isolated and then subjected to immunoprecipitation using an anti-tTG antibody. The resulting immunocomplexes were analyzed by SDS-PAGE followed by Colloidal Blue staining to detect proteins that co-immunoprecipitated with tTG. Whereas several proteins co-immunoprecipitated with tTG from the membrane fractions prepared from untreated HeLa cells, the majority of these proteins were not detected when tTG was immunoprecipitated from the membrane fractions derived from EGF-stimulated cells (Fig. 3A). A notable exception was a protein band with an apparent molecular mass of ∼70 kDa (Fig. 3A, arrow). Because we were searching for proteins that would interact with tTG in EGF-stimulated cells, as candidates for helping to mediate the translocation of tTG to leading edges, we determined the identity of the ∼70-kDa protein by mass spectrometry. In fact, we found that this protein band consisted of two highly homologous and functionally redundant members of the heat shock protein family, namely Hsp70 and Hsc70 (26) (Fig. 3B). What makes the identification of Hsc70 and Hsp70 as novel interacting partners for plasma membrane-associated tTG particularly intriguing is that they were recently shown to be uniquely capable of integrating into artificial lipid bilayers, as well as into plasma membranes derived from cancer cell lines (27, 28). We first confirmed this interaction by performing immunoprecipitations with a Myc antibody on the cellular lysates from HeLa cells transiently co-expressing a V5-tagged form of Hsc70 and a Myc-tagged form of tTG, treated without or with EGF. Fig. 3C shows that V5-tagged Hsc70 can be co-immunoprecipitated with Myc-tagged tTG, independent of EGF treatment. Next we compared the ability of Hsc70 to interact with wild-type tTG and a transamidation-defective form of tTG (C277V) by performing immunoprecipitations with a Myc antibody on the cellular extracts from HeLa cells (Fig. 3D, top panels), as well as MDAMB231 breast cancer cells (Fig. 3D, bottom panels), that were transiently co-expressing a V5-tagged form of Hsc70 and a Myc-tagged form of tTG. Fig. 3D shows that V5-tagged Hsc70 can be co-immunoprecipitated with Myc-tagged tTG (Myc-tTG WT) in each of these cell types. Moreover, the transamidation-defective form of tTG (Myc-tTG C277V), when ectopically expressed in HeLa cells or MDAMB231 cells, was able to bind Hsc70 as efficiently as wild-type tTG, suggesting that the ability of tTG to interact with heat shock protein family members does not require its enzymatic transamidation activity.
FIGURE 3.
Hsp70 interacts with plasma membrane-associated tTG. A, serum-starved HeLa cells that had been treated without (lane 0) or with EGF for 12 h were homogenized and then subjected to differential centrifugation to isolate the membrane components of the cells. Immunoprecipitations with a tTG antibody (IP:tTG) were performed on the membrane extracts, and the resulting immunocomplexes were resolved by SDS-PAGE. The gel was then stained with Colloidal Blue to detect the proteins that co-immunoprecipitated with tTG. One protein band (Mr = ∼70 kDa), denoted with an arrow, was determined to contain two isoforms of the heat shock protein 70 family, Hsp70 and Hsc70, by mass spectrometry. B, the protein sequences of human Hsp70 and Hsc70 are shown in gray. The peptide fragments identified by mass spectrometry are shaded in black. C, immunoprecipitations with a Myc antibody were performed on the extracts of HeLa cells ectopically expressing V5-tagged Hsc70 and Myc-tagged tTG, treated without (Untreated) or with EGF. The whole cells lysates (WCL) and the resulting immunocomplexes (IP:Myc) were immunoblotted with V5 and Myc antibodies. Nonspecific mouse IgG control antibody and beads-only control immunoprecipitations were also performed on the extracts to show that the Hsc70-tTG interaction was specific. D, immunoprecipitations with a Myc antibody were performed on the extracts of actively growing HeLa (top panels) and MDAMB231 cells (bottom panels) that were ectopically expressing V5-tagged Hsc70 and a Myc-tagged form of either wild-type tTG (tTG WT) or a transamidation-defective form of tTG (tTG C277V). The whole cell lysates (WCL) and the resulting immunocomplexes (IP:Myc) were immunoblotted with V5 and Myc antibodies. Nonspecific mouse IgG control antibody and beads-only control immunoprecipitations were performed on the extracts collected from HeLa and MDAMB231 cells expressing V5-Hsc70 and Myc-tTG WT to show that the Hsc70-tTG interaction was specific.
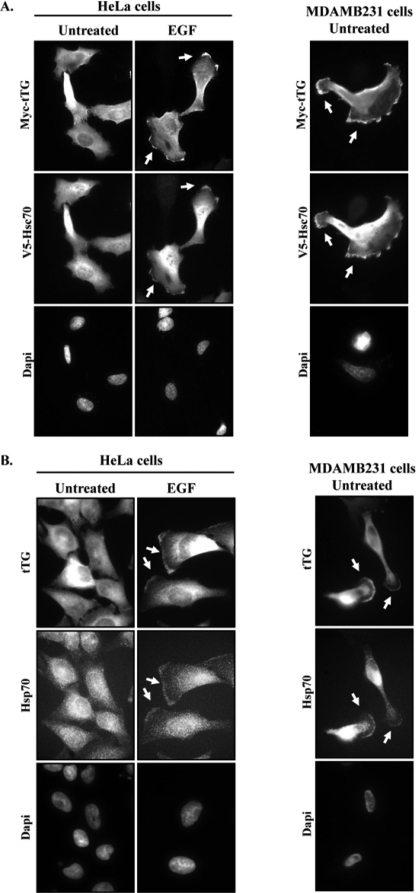
We next used immunofluorescence approaches to determine whether Hsp70, like tTG, was expressed along the leading edges of migrating cells. Our reasoning was that if the binding of tTG to Hsp70 family members is necessary for the recruitment and accumulation of tTG at leading edges, then Hsp70 and tTG should be present together at these cellular sites. Cultures of serum-starved HeLa cells co-expressing V5-tagged Hsc70 and Myc-tagged tTG that were either untreated or stimulated with EGF were fixed and then stained with Myc and V5 antibodies. The resulting fluorescent images showed that the V5-tagged Hsc70 was predominantly cytoplasmic in the serum-starved HeLa cells (Fig. 4A, left panels). However, following EGF stimulation, the V5-tagged form of Hsc70 was readily detectable at the leading edges of these cells, where it co-localized with the ectopically expressed form of tTG (Myc-tTG WT).
FIGURE 4.
tTG and Hsp70 co-localize to the leading edges of cells. A, HeLa and MDAMB231 cells ectopically expressing V5-Hsc70 and Myc-tTG were fixed and then subjected to immunofluorescence using V5 and Myc antibodies and DAPI (to stain nuclei). Representative images of the transfectants are shown, with the co-localization of the ectopically expressed forms of tTG and Hsc70 at leading edges being highlighted with arrows. B, serum-starved HeLa cells and MDAMB231 cells were treated without (Untreated) or with EGF for 12 h, as indicated, and fixed. Immunofluorescence was performed on the cells using tTG and Hsp70 antibodies and DAPI. Representative images of the cells are shown, with the co-localization of tTG and Hsp70 at leading edges being indicated with arrows.
An analogous experiment was then performed on the constitutively migrating MDAMB231 breast cancer cell line. Consistent with the findings from HeLa cells, immunofluorescent analysis performed on MDAMB231 cells co-expressing V5-tagged Hsc70 and Myc-tagged tTG showed that both of these ectopically expressed proteins co-localized along the leading edges of these cells (Fig. 4A, right panels). Endogenous Hsp70 was also shown to consistently localize to the developing leading edges in EGF-treated HeLa cells and in MDAMB231 cells (Fig. 4B, left and right panels, respectively).
Plasma Membrane-associated tTG Is Recruited to Leading Edges by Hsp70
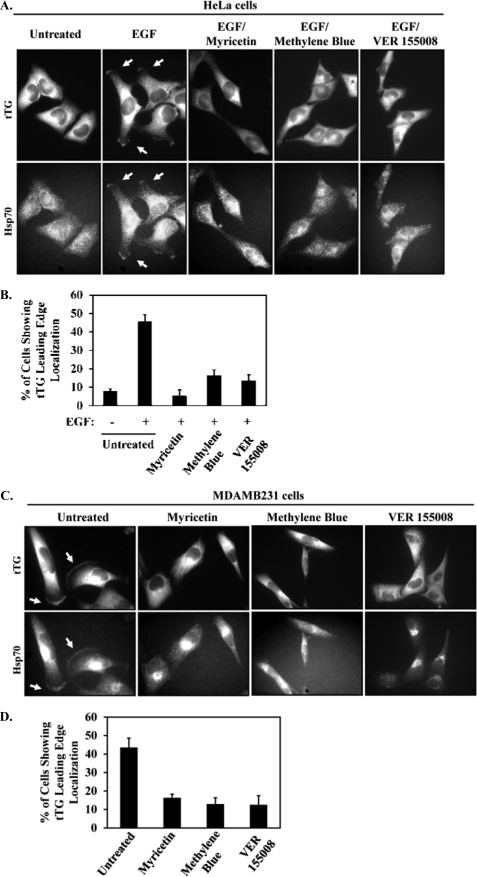
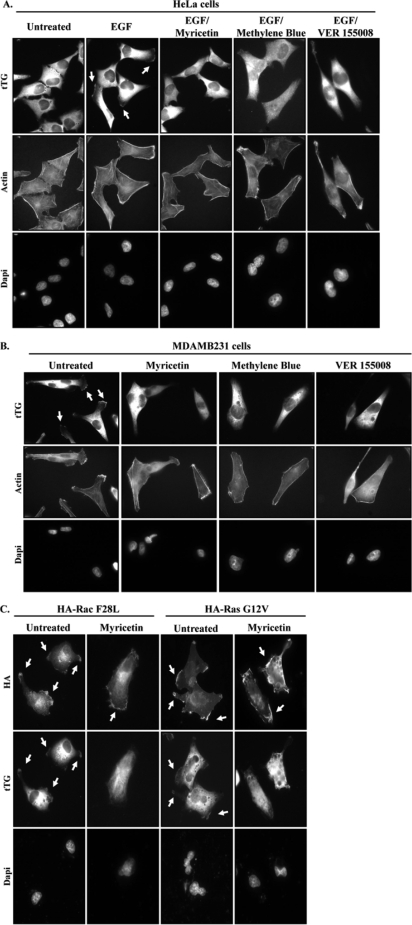
We next set out to determine whether Hsp70 was responsible for the accumulation of tTG at the leading edge. Initially we tried to examine this by knocking down Hsp70 family members from cells and seeing whether the ability of tTG to be localized to leading edges was disrupted. However, despite a number of attempts to knock down Hsp70 and Hsc70, either individually or in combination, in HeLa cells or MDAMB231 cells (including our use of six different siRNAs targeting Hsp70 and/or Hsc70 in varying amounts for transfection), we have thus far been unable to significantly reduce their expression levels (data not shown). Consequently, we turned to three different inhibitors of the ATP hydrolytic activity of Hsp70, namely, myricetin, methylene blue, and VER 155008, to investigate the involvement of Hsp70 in targeting tTG to leading edges. Fig. 5A shows that the exposure of EGF-stimulated HeLa cells to any of these inhibitors was sufficient to block the ability of tTG, as well as Hsp70, to accumulate along leading edges. The results of these experiments are quantified in Fig. 5B and show that the Hsp70 inhibitors reduced the percentage of HeLa cells with tTG along their leading edges from ∼40% to between ∼5 and 15%, depending on which inhibitor was used. Hsp70 inhibition also blocked the leading edge accumulation of tTG in MDAMB231 breast cancer cells (Fig. 5C), with the results of these experiments being quantified in Fig. 5D, indicating that Hsp70 activity is essential for redistributing the plasma membrane-associated pool of tTG to leading edges in at least two different cell types. Moreover, it is worth emphasizing that the effect of the three Hsp70 inhibitors at preventing the recruitment of tTG to leading edges appears to be specific and not simply a consequence of disrupting leading edge formation in cells, based on two additional pieces of data. First, the characteristic build-up of actin that occurs at the leading edges of EGF-stimulated HeLa cells or in constitutively migrating MDAMB231 cells that is necessary to promote membrane protrusions and the generation of new cell-to-substrate contacts was not inhibited when the cells were exposed to either myricetin, methylene blue, or VER 155008 (Fig. 6, A and B, respectively). Second, we also found that myricetin treatment did not disrupt the ability of two ectopically expressed, activated forms of the leading edge-resident proteins, Rac (HA-tagged Rac F28L) and Ras (HA-tagged Ras G12V), to be localized to these sites in MDAMB231 cells (Fig. 6C).
FIGURE 5.
Inhibiting Hsp70 activity blocks the ability of tTG and Hsp70 to localize at leading edges. A, serum-deprived HeLa cells were treated without (Untreated) or with EGF, with or without myricetin, methylene blue, or VER 155008, and then immunofluorescence was performed on the cells using tTG and Hsp70 antibodies. The resulting fluorescent images are shown with the tTG and Hsp70 at leading edges being indicated with arrows. B, quantification of the cells shown in A with tTG at their leading edges. C, serum-starved MDAMB231 cells were treated without (Untreated) or with myricetin, methylene blue, or VER 155008, and then immunofluorescence was performed on the cells using tTG and Hsp70 antibodies. The resulting fluorescent images are shown with the tTG and Hsp70 at leading edges being indicated with arrows. D, quantification of the cells shown in C with tTG at their leading edges. Three independent experiments were performed, with at least 250 cells being scored for each condition. The results from the experiments were then averaged together and graphed. The error bars indicate standard deviation.
FIGURE 6.
Inhibition of Hsp70 does not have a global effect on leading edge proteins. A, serum-deprived HeLa cells were treated without (Untreated) or with EGF, with or without myricetin, methylene blue, or VER 155008, and then immunofluorescence was performed on the cells using a tTG antibody, rhodamine-conjugated phalloidin (Actin), and DAPI. The resulting fluorescent images are shown with the tTG at leading edges being indicated with arrows. B, serum-starved MDAMB231 cells were treated without (Untreated) or with myricetin, methylene blue, or VER 155008, and then immunofluorescence was performed on the cells using a tTG antibody, rhodamine-conjugated phalloidin (Actin), and DAPI. The resulting fluorescent images are shown with the tTG at leading edges being indicated with arrows. C, MDAMB231 cells were transiently transfected with either of two HA-tagged constructs: constitutively active Rac (F28L) or GTP hydrolysis-defective Ras (G12V), and immunofluorescence was performed using HA and tTG antibodies and DAPI. The resulting fluorescent images are shown with the Rac, Ras, and tTG at leading edges being indicated with arrows.
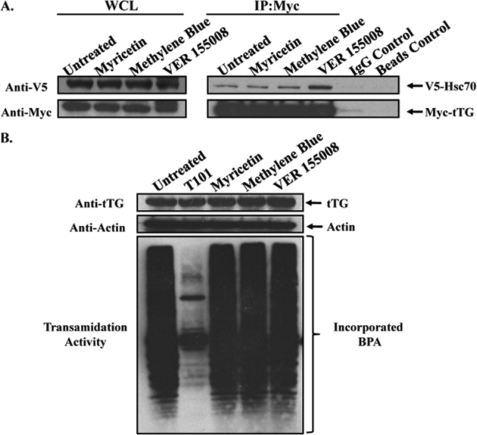
The ATP hydrolytic activity of heat shock proteins is essential for their ability to associate with client proteins (29). Although traditionally the binding of heat shock proteins to client proteins occurs in response to cellular stresses, such as elevated temperatures, as a means to help ensure proper protein folding under stressful conditions, more recently, heat shock proteins have been implicated in promoting human cancer progression (30, 31). Not only are the expression and activation levels of several heat shock proteins, including Hsp70, frequently up-regulated in a variety of primary tumors and tumor-derived cell lines, but the ability of heat shock proteins to bind to and potentiate the signaling capabilities of key mitogenic and pro-survival signaling proteins has been shown to be necessary for inducing/maintaining the transformed state (30, 32). Given our novel finding that the recruitment of tTG to the leading edges of HeLa cells and MDAMB231 cells is dependent on Hsp70 activity, it was logical to consider the possibility that tTG might be a client of Hsp70. If this were the case, then we would predict that the interaction between Hsp70 and tTG in cells would be sensitive to treatment with myricetin, methylene blue, or VER 155008, because these inhibitors function by blocking the ATP hydrolytic activity of Hsp70 and thereby disrupt its ability to bind client proteins through the conserved substrate (client) binding domain. HeLa cells transiently co-expressing V5-tagged Hsc70 and Myc-tagged tTG were either left untreated or were incubated with one of the inhibitors and lysed. The cell extracts were then subjected to immunoprecipitation using a Myc antibody, with the resulting immunocomplexes then analyzed by Western blot. Fig. 7A shows that V5-tagged Hsc70 co-immunoprecipitated with Myc-tagged tTG from each of the inhibitor-treated cells to a similar extent as it did from the untreated (control) HeLa cells. This suggests that tTG is not a client of Hsp70 in the conventional sense and does not directly bind to Hsp70 through its substrate (client) binding site.
FIGURE 7.
Inhibiting the ATP hydrolytic activity of Hsp70 has no effect on the interaction between tTG and Hsp70, or on the protein cross-linking activity of tTG. A, HeLa cells ectopically expressing V5-Hsc70 and Myc-tTG were treated without (Untreated) or with myricetin, methylene blue, or VER 155008, as indicated, and lysed. Immunoprecipitations (IP) with a Myc antibody were performed on the cell extracts, followed by SDS-PAGE and Western blot analysis using V5 and Myc antibodies. Nonspecific mouse IgG control antibody and beads-only control immunoprecipitations were performed on the extracts from the untreated cells to show that the Hsc70-tTG interaction was specific. B, serum-starved cultures of MDAMB231 cells were treated without (Untreated) or with T101, myricetin, methylene blue, or VER 155008 and then lysed. The extracts were immunoblotted with tTG and actin antibodies (top panels) and assayed for transamidation activity as read-out by the incorporation of BPA into lysate proteins (bottom panel).
Next, we asked how inhibiting Hsp70 would affect tTG activity. Serum-starved MDAMB231 cells that had been left untreated or were incubated with an irreversible tTG cross-linking inhibitor T101 or with myricetin, methylene blue, or VER 155008 were collected, and then the enzymatic transamidation activities in each sample were assayed by reading out the incorporation of BPA into lysate proteins. Consistent with previous results, tTG expressed in serum-starved MDAMB231 cells was constitutively active, although as expected, treatment of the cells with the tTG inhibitor T101 reduced tTG activity (Fig. 7B). However, the amount of tTG-catalyzed transamidation activity detected in the inhibitor-treated MDAMB231 cell samples was similar to that detected in the control sample.
The Recruitment of tTG to Leading Edges Is Important for Cell Migration
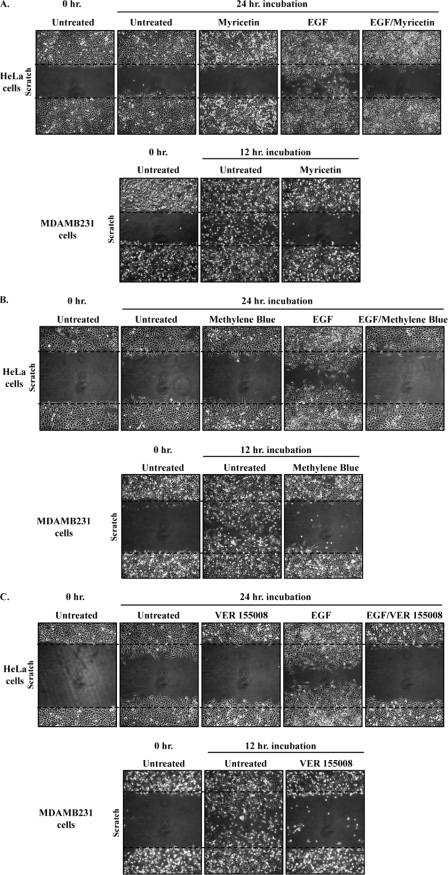
Our findings showing that inhibiting Hsp70 activity prevented the accumulation of tTG at the leading edges of cells, without affecting its transamidation activity, afforded us with a unique opportunity to examine the importance of tTG localization to leading edges on the ability of cells to migrate. Multiple plates of confluent HeLa cells or MDAMB231 cells that either were untreated or incubated with different combinations of EGF and myricetin were subjected to a scratch (or wound healing) assay, and their rates of cell migration were compared. The top panels in Fig. 8A show that the EGF-stimulated migration of HeLa cells, as indicated by the ability of these cells to close the wound, was blocked when the cells were also cultured in the presence of myricetin. Likewise, the constitutive migration activity normally exhibited by the MDAMB231 cells was also ablated by myricetin treatment (Fig. 8A, bottom panels). Similar results were obtained using the two additional Hsp70 inhibitors, methylene blue (Fig. 8B) and VER 155008 (Fig. 8C). Taken together, these findings demonstrate that the proper localization of tTG to leading edges by Hsp70 family members is essential for the migration of certain human cancer cell lines.
FIGURE 8.
Inhibition of Hsp70 blocks the migration of HeLa and MDAMB231 cells. A, scratch assays were performed on serum-deprived HeLa cells treated without (Untreated) or with EGF, with or without myricetin, and on serum-starved MDAMB231 cells with or without myricetin. The MDAMB231 cells were fixed 12 h after striking the wound; HeLa cells were fixed after 24 h. The cells were then visualized using light microscopy, and the extent of wound closure was determined. One set of untreated cells was fixed immediately after striking the wound (Untreated 0 h.) to indicate the size of the initial wounds. The widths of the initial wounds are indicated by dashed lines. B, scratch assays were performed on the cell lines using methylene blue as outlined in A. C, scratch assays were performed on the cell lines using VER 155008 as outlined in A.
DISCUSSION
Cell migration is a fundamental process in biology that underlies key stages of development and tissue regeneration; however, it is frequently deregulated in human diseases such as cancer, where the aberrant migration of cells serves as a precursor to their metastatic and invasive capabilities (1, 3, 5, 7). For this reason, we have chosen to focus on the mechanisms used by cancer cells to enhance migration. We have previously identified tTG as a nontraditional signaling protein that is essential for the EGF-dependent migration of HeLa cells and the constitutive migration of MDAMB231 cells (11). Here we show that Hsp70 plays a critical regulatory role in the localization of tTG to the leading edges of actively migrating cells.
Given that EGF stimulation results in the accumulation of tTG at the leading edges of cells, our initial assumption was that a growth factor-induced trafficking event triggered the movement of a pool of cytosolic tTG to the plasma membrane. However, membrane fractionation studies revealed that a discrete population of tTG, which comprises ∼10% of the tTG expressed in cells, constitutively associates with the plasma membrane and is unaffected by EGF treatment. These findings supported the possibility that the membrane-associated pool of tTG is redistributed to leading edges following exposure to EGF. What is particularly interesting is that EGF can activate this membrane-bound pool of tTG on a similar time scale as the localization of tTG to leading edges. This correlation leads us to suspect that the activation of the membrane-associated tTG and its localization to the leading edge are coupled and need to occur in a coordinated fashion to promote cell migration. Support for this idea comes from previous studies using HeLa cells where we found that ectopically expressed tTG showed protein cross-linking (transamidation) activity, but its ectopic expression alone was not sufficient for enhancing cell migration (11). We believe the reason for this is that the ectopically expressed tTG cannot localize to leading edges without EGF treatment and that such localization is essential for promoting cell migration.
Certainly an important question concerned how tTG is capable of associating with the plasma membrane so that it can ultimately localize to the leading edges of cells. Although tTG appears to have some intrinsic capability to associate with lipid bilayers, as indicated by liposome experiments, sequence and structure analyses do not provide obvious clues as to how this might occur. We also wondered how tTG is recruited in an EGF-dependent manner to the leading edge, and so we searched for interacting proteins that might help tTG to localize to these membrane sites. Immunoprecipitation of tTG from membrane extracts and subsequent mass spectrometry analysis of the immunocomplexes led to the identification of two heat shock protein 70 family members, Hsp70 and Hsc70, as novel binding partners for tTG. We further showed that Hsp70 and tTG co-localize to leading edges. This suggested that Hsp70 could indeed play a role in the ability of tTG to accumulate at these sites. Using several different inhibitors against the ATP hydrolytic activity of Hsp70, including myricetin, methylene blue, and VER 155008, we showed that blocking the chaperonin activity of Hsp70 prevented tTG and Hsp70 from co-localizing to leading edges. Importantly, we further demonstrated that the exclusion of tTG and Hsp70 from leading edges caused by inhibiting Hsp70 activity was specific for these two proteins. In the presence of these inhibitors, actin structures still formed at the leading edges of cells. Moreover, other leading edge-resident proteins including activated forms of the small GTPases Rac and Ras were still able to properly localize to these membrane sites even when the cells were treated with myricetin, thus indicating that this inhibitor is not having a global effect. Therefore, the chaperonin ability of Hsp70 appears to be critical for the localization of tTG to leading edges.
Surprisingly, treatment of cells with the Hsp70 inhibitors does not block the interaction of tTG and Hsp70, suggesting that tTG is not a client of Hsp70. However, given the effect of myricetin, methylene blue, or VER 155008 on the localization of tTG, these findings imply that the interaction of Hsp70 with a certain client protein(s) is necessary for regulating the localization of tTG. It has been shown that a number of signaling proteins may be clients of heat shock proteins including Src, Raf, and Akt and that these chaperones may play an important role in extending their signaling lifetimes (30). This then raises some interesting questions for the future, such as whether specific clients of Hsp70 are required to help tTG properly localize to leading edges.
Previous work by us has shown that the transamidation activity of tTG is important for the process of cell migration, because inhibiting this activity with MDC blocks the EGF-dependent migration of HeLa cells, as well as the constitutive migration of MDAMB231 cells (11). However, this activity is not sufficient for enhancing cell migration in either case, thereby suggesting that in addition to becoming activated, tTG needs to localize to leading edges. Initially, we used a biochemical approach to define the region of tTG that was required for its association with membranes in hopes of designing a tTG mutant that retained protein cross-linking activity but was defective for binding to the plasma membrane. Thus far, we have not been able to identify a point mutant that uncouples these two functions of tTG. However, our identification of Hsp70 as a novel binding partner of tTG and the fact that these two proteins co-localize to leading edges with the localization being blocked by myricetin, as well as methylene blue and VER 155008, provided us with a nice system for specifically investigating the importance of the leading edge localization of tTG. In particular, because these Hsp70 inhibitors do not affect the protein cross-linking activity of tTG, it is possible to uncouple this function from its leading edge localization and determine what effect this has on cell migration. Indeed, we have shown that by blocking either tTG protein cross-linking activity (using MDC) or its leading edge localization (using myricetin, methylene blue, or VER 155008), we are able to inhibit cell migration, demonstrating that these two events are coupled.
These findings now raise a number of intriguing questions regarding the role of heat shock proteins in cancer progression. Recently, it was shown that Hsp70 and Hsc70 are uniquely capable of interacting with lipids, because they can insert into artificial lipid bilayers and selectively bind to phosphatidylserine (28). Others have demonstrated that Hsp70 can bind to cholesterol-rich microdomains in tumor cells (27). Still, the importance of the membrane association of Hsp70 and the consequences this holds for cancer progression are not yet understood. Here we offer a possible insight into why these chaperones localize to the plasma membrane, namely to regulate a key signaling molecule, tTG, and facilitate its redistribution to leading edges. We believe that the identification of Hsp70 client proteins will provide additional insights into the roles of Hsp70 and tTG in cell migration. All of this then leads to the question of why tTG needs to localize to leading edges. We believe that the coupling of tTG localization with the activation of its protein cross-linking activity should shed light on the identity of a key transamidation substrate for tTG. The recent development of “clickable” inhibitors against tTG, which bind to only the transamidation-active form of the enzyme, offer the possibility of visualizing tTG-catalyzed protein cross-linking activity in cells (33). These inhibitors may prove very useful for determining whether tTG is active specifically at leading edges. If this is the case, we will want to identify the substrate(s) of tTG located at these leading edges and determine how cross-linking contributes to enhanced cell migration.
Acknowledgment
We thank Cindy Westmiller for excellent secretarial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM061762.
- tTG
- tissue transglutaminase
- Hsp
- heat shock protein
- MDC
- monodansylcadaverine
- BPA
- biotinylated pentylamine
- DAPI
- 6-diamidino-2-phenylindole.
REFERENCES
- 1. Raja, Sivamani K., Garcia M. S., Isseroff R. R. (2007) Front. Biosci. 12, 2849–2868 [DOI] [PubMed] [Google Scholar]
- 2. Cyster J. G. (2005) Annu. Rev. Immunol. 23, 127–159 [DOI] [PubMed] [Google Scholar]
- 3. Aman A., Piotrowski T. (2010) Dev. Biol. 341, 20–33 [DOI] [PubMed] [Google Scholar]
- 4. Luster A. D., Alon R., von Andrian U. H. (2005) Nat. Immunol. 6, 1182–1190 [DOI] [PubMed] [Google Scholar]
- 5. Keller R. (2005) Curr. Opin. Cell Biol. 17, 533–541 [DOI] [PubMed] [Google Scholar]
- 6. Friedl P., Wolf K. (2003) Nat. Rev. Cancer 3, 362–374 [DOI] [PubMed] [Google Scholar]
- 7. Sahai E. (2007) Nat. Rev. Cancer 7, 737–749 [DOI] [PubMed] [Google Scholar]
- 8. Le Clainche C., Carlier M. F. (2008) Physiol. Rev. 88, 489–513 [DOI] [PubMed] [Google Scholar]
- 9. Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., Ben-Chetrit N., Tarcic G., Lindzen M., Avraham R., Liao Y. C., Trusk P., Lyass A., Rechavi G., Spector N. L., Lo S. H., Schmitt F., Bacus S. S., Yarden Y. (2007) Nat. Cell Biol. 9, 961–969 [DOI] [PubMed] [Google Scholar]
- 10. Hood J. D., Cheresh D. A. (2002) Nat. Rev. Cancer 2, 91–100 [DOI] [PubMed] [Google Scholar]
- 11. Antonyak M. A., Li B., Regan A. D., Feng Q., Dusaban S. S., Cerione R. A. (2009) J. Biol. Chem. 284, 17914–17925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raftopoulou M., Hall A. (2004) Dev. Biol. 265, 23–32 [DOI] [PubMed] [Google Scholar]
- 13. Parsons J. T., Horwitz A. R., Schwartz M. A. (2010) Nat. Rev. Mol. Cell Biol. 11, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang C., Rajfur Z., Borchers C., Schaller M. D., Jacobson K. (2003) Nature 424, 219–223 [DOI] [PubMed] [Google Scholar]
- 15. Yip S. C., El-Sibai M., Coniglio S. J., Mouneimne G., Eddy R. J., Drees B. E., Neilsen P. O., Goswami S., Symons M., Condeelis J. S., Backer J. M. (2007) J. Cell Sci. 120, 3138–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greenberg C. S., Birckbichler P. J., Rice R. H. (1991) FASEB J. 5, 3071–3077 [DOI] [PubMed] [Google Scholar]
- 17. Folk J. E. (1980) Annu. Rev. Biochem. 49, 517–531 [DOI] [PubMed] [Google Scholar]
- 18. Jiang D., Ying W., Lu Y., Wan J., Zhai Y., Liu W., Zhu Y., Qiu Z., Qian X., He F. (2003) Proteomics 3, 724–737 [DOI] [PubMed] [Google Scholar]
- 19. Shao M., Cao L., Shen C., Satpathy M., Chelladurai B., Bigsby R. M., Nakshatri H., Matei D. (2009) Cancer Res. 69, 9192–9201 [DOI] [PubMed] [Google Scholar]
- 20. Mangala L. S., Fok J. Y., Zorrilla-Calancha I. R., Verma A., Mehta K. (2007) Oncogene 26, 2459–2470 [DOI] [PubMed] [Google Scholar]
- 21. Singer C. F., Hudelist G., Walter I., Rueckliniger E., Czerwenka K., Kubista E., Huber A. V. (2006) Clin. Exp. Metastasis 23, 33–39 [DOI] [PubMed] [Google Scholar]
- 22. Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Datta S., Antonyak M. A., Cerione R. A. (2006) Biochemistry 45, 13163–13174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Datta S., Antonyak M. A., Cerione R. A. (2007) Biochemistry 46, 14819–14829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Johnson J. L., Erickson J. W., Cerione R. A. (2009) J. Biol. Chem. 284, 23860–23871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daugaard M., Rohde M., Jäättelä M. (2007) FEBS Lett. 581, 3702–3710 [DOI] [PubMed] [Google Scholar]
- 27. Gehrmann M., Liebisch G., Schmitz G., Anderson R., Steinem C., De Maio A., Pockley G., Multhoff G. (2008) PLoS One 3, e1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arispe N., Doh M., Simakova O., Kurganov B., De Maio A. (2004) FASEB J. 18, 1636–1645 [DOI] [PubMed] [Google Scholar]
- 29. Mayer M. P., Bukau B. (2005) Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jego G., Hazoume A., Seigneuric R., Garrido C. (2010) Cancer Lett., in press [DOI] [PubMed] [Google Scholar]
- 31. Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aghdassi A., Phillips P., Dudeja V., Dhaulakhandi D., Sharif R., Dawra R., Lerch M. M., Saluja A. (2007) Cancer Res. 67, 616–625 [DOI] [PubMed] [Google Scholar]
- 33. Dafik L., Khosla C. (2011) Chem. Biol. 18, 58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]