Background: BCL10 is an essential molecule for the activation of the transcription factor NF-κB in numerous immune receptor signaling pathways.
Results: Identification of the E3 ubiquitin ligase MIB2 as part of the activated BCL10 complex controlling NF-κB activity.
Conclusion: MIB2 mediates BCL10-dependent NF-κB activation.
Significance: Identification of MIB2 as a regulator of important immune receptor signaling pathways.
Keywords: NF-kappa B (NF-KB), Protein-Protein Interactions, Proteomics, Signal Transduction, T-cell Receptor, Ubiquitin Ligase
Abstract
B-cell CLL/lymphoma 10 (BCL10) is crucial for the activation of NF-κB in numerous immune receptor signaling pathways, including the T-cell receptor (TCR) and B-cell receptor signaling pathways. However, the molecular mechanisms that lead to signal transduction from BCL10 to downstream NF-κB effector kinases, such as TAK1 and components of the IKK complex, are not entirely understood. Here we used a proteomic approach and identified the E3 ligase MIB2 as a novel component of the activated BCL10 complex. In vitro translation and pulldown assays suggest direct interaction between BCL10 and MIB2. Overexpression experiments show that MIB2 controls BCL10-mediated activation of NF-κB by promoting autoubiquitination and ubiquitination of IKKγ/NEMO, as well as recruitment and activation of TAK1. Knockdown of MIB2 inhibited BCL10-dependent NF-κB activation. Together, our results identify MIB2 as a novel component of the activated BCL10 signaling complex and a missing link in the BCL10-dependent NF-κB signaling pathway.
Introduction
BCL10,3 originally identified as a target of a recurrent chromosomal translocation in mucosa-associated lymphoid tissue (MALT) lymphomas (1, 2), has emerged to play essential roles in innate and adaptive immune responses. BCL10-deficient mice are severely immunocompromised because of defects in the induction of the transcription factor NF-κB and MAPK pathways (3, 4), which are important for the regulation of genes associated with cell survival and inflammation (5, 6). BCL10 was shown to be pivotal for NF-κB activation upon engagement of a variety of immune receptors, such as TCR, B-cell receptor, NK-cell receptors, C-type lectin family receptors, Ig receptors, and G protein-coupled receptors (3, 4, 7–16). Furthermore, BCL10 was shown to mediate NF-κB activation triggered by intracellular, nucleotide-sensing RIG-I-like receptors, which control anti-viral immune responses (17).
In unstimulated cells, BCL10 is constitutively associated with the paracaspase MALT1 (18). Receptor engagement, e.g. TCR cross-linking, leads to recruitment of BCL10-MALT1 to the upstream CARD containing adaptor protein CARD-containing MAGUK protein 1 (CARMA1), resulting in the formation of a trimolecular signaling complex consisting of CARMA1, BCL10, and MALT1, referred to as the CBM complex. Oligomerization of these proteins eventually results in the formation of nondegradative polyubiquitin chains and activation of downstream kinases, such as transforming growth factor-β-activated kinase 1 (TAK1) and the TAK1-substrate IκB-kinase β (IKKβ) (19–21). IKKβ is part of the heterotrimeric NF-κB master regulator, the IKK complex, which consists of the catalytic subunits IKKβ and IKKα, and the scaffold protein IKKγ/NEMO. Activated IKKβ phosphorylates the NF-κB inhibitor IkBα, which results in proteosomal degradation of IκBα and subsequent nuclear translocation of NF-κB and activation of NF-κB target genes. Although the constituents of the IKK complex, i.e. IKKβ and IKKγ, are known to be essential for NF-κB activation, their mode of activation is not entirely clear (19, 22). Synthesis of K63-linked polyubiquitin chains appears to be a critical step in pathway activation downstream of BCL10, which is reflected by a defect of TAK1 phosphorylation and IKKγ ubiquitination in T-cells deficient for the ubiquitin-conjugating enzyme Ubc13 (23). The molecular mechanism of NF-κB activation has been investigated in more detail in the Toll-like receptor (TLR)/IL-1R signaling pathway, where TNF receptor-associated factor 6 (TRAF6)-mediated polyubiquitination was shown to control recruitment and activation of TAK1 via the ubiquitin-binding TAB proteins, followed by TAK1-mediated phosphorylation of IKKβ (24). Furthermore, site-specific K63-linked ubiquitination of IKKγ was found to contribute to NF-κB transcriptional activity (25), although the mechanism that governs ubiquitination-dependent IKK activity is still unknown. Even though polyubiquitination and activation of TAK1 and the IKK complex are established events downstream of BCL10, a major question remaining is how the BCL10 complex is linked to these events.
Two proteins, MALT1 and TRAF6 were proposed as candidate ubiquitin ligases (25, 26). However, MALT1 does not contain any identifiable ubiquitin ligase domain, suggesting a more indirect role in the control of polyubiquitin chain synthesis, and TRAF6-deficient T-cells displayed unimpaired NF-κB activation upon TCR triggering (27). As such, it seems very likely that an additional protein ubiquitin ligase exists that transduces signaling from the BCL10 complex toward IKK/NF-κB, either by acting entirely independently of TRAF6 or by compensating for the loss of TRAF6. Here we report the identification of a novel BCL10-associated ubiquitin ligase. Using a proteomic approach, we discovered the E3 ubiquitin ligase MIB2 (mind bomb-2 (Drosophila)) as an essential component of the activated BCL10 signaling complex. MIB2 directly interacts with BCL10 and was found to promote binding and activation of TAK1 and the IKK complex. MIB2-mediated nondegradative ubiquitination and NF-κB activation depended on its C-terminal RING finger domain, which is consistent with the idea that polyubiquitination is critical for NF-κB activation. Knockdown of MIB2 resulted in impaired BCL10-mediated activation of NF-κB. Thus, our findings identify MIB2 as a missing link between the BCL10 complex and the NF-κB activation pathway.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% FCS (Hyclone), penicillin, and streptomycin (Invitrogen). The EL4 mouse T-cell line, human Jurkat T-cell line, or primary mouse thymocytes were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FCS, penicillin, and streptomycin. Stable EL4 or Jurkat cell transfectants were generated by electroporation (250V, 950 microfarad) followed by subcloning in the presence of G418 (1 mg ml−1; Invitrogen).
Reagents and Plasmids
Antibodies against the following proteins were used: M2 FLAG, β-actin (Sigma-Aldrich); MIB2 (Bethyl Laboratories); HA (12CA5) (Roche Applied Science); BCL10, IKKβ, phospho-Tak1 (Thr184/187), and TAK1 (Cell Signaling); His (Santa Cruz); and IKKγ (Pharmingen).
pCMV-Sport6-BCL10, pCMV-SPORT6-MIB2, pCR-BluntII-TOPO-MIB1, and pCR-BluntII-TOPO-MALT1 were obtained from Open Biosystems. Full-length or deletion mutant cDNA was generated by PCR and cloned into an EF1α promoter/FLAG epitope tag containing vector (pEF-SEM-FLAG, corresponding to pcDNA3 (Invitrogen) with an EF1α promoter replacing the CMV promoter), a CMV promoter/HA epitope tag containing vector (pCX-HA, based on pcDNA3, but containing a modified CMV promoter originally obtained from vector RK5 (Pharmingen)), pCMV-Sport6 vector (Open Biosystems) for in vitro translation or pGEX-4T-3 (GE Healthcare) for recombinant protein expression. MyD88 was described previously (28). pCMV-HA-TAK1 was a kind gift from J. Lee (University of California, San Diego) and was cloned into the pEF-SEM-FLAG vector. pRC-HA-IKKγ and pRC-FLAG-IKKγ were kindly provided by M. Karin (University of California, San Diego). pRK5-HA-Ubiquitin, pRK5-HA-Ubiquitin-K33, pRK5-HA-Ubiquitin-K48, and pRK5-HA-Ubiquitin-K63 were generated in T. Dawson's lab (29) and obtained from Addgene. pQCXIP HBT-Ubiquitin was generously provided by C. Tagwerker and P. Kaiser (UCI). pLKO.1-MIB2 shRNA (TRCN0000034114 (hMIB2-shRNA-A)), pGIPZ-MIB2 shRNAmir (RHS4430–98715116 (hMIB2-shRNA-B)) as well as the respective control shRNA vectors were obtained from Open Biosystems. Other reagents used were: phorbol 12-myristate 13-acetate (PMA; 50 ng ml−1; Sigma-Aldrich); ionomycin (50 ng ml−1; Sigma-Aldrich); coumermycin (0.5 μm; Sigma-Aldrich); and TNFα (30 ng ml−1, PeproTech).
Affinity Purification and Mass Spectrometry Analysis
Cells were lysed by one freeze/thaw cycle in buffer A (20 mm Hepes/KOH, pH 7.5, 10 mm KCl, 1.5 mm MgCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm PMSF, 1 mm DTT, 10% glycerol, 1 mm orthovanadate, 10 mm β-glycerophosphate, 5 mm 4-nitrophenyl-phosphate, 10 mm sodium fluoride) supplemented with 'complete proteases inhibitors' (Roche Applied Science) for 20 min. Samples were cleared by centrifugation and loaded onto StrepTactin columns (IBA) equilibrated in Buffer B (20 mm Hepes/KOH, pH 7.5, 150 mm NaCl, 1.5 mm MgCl, 1 mm EDTA, 0.1% Nonidet P-40 supplemented with complete proteases inhibitors (Roche Applied Science)). Proteins were eluted from the column in 1 ml of Buffer B containing desthiobiotin (5 mm; Sigma-Aldrich). Eluted samples were concentrated using 5K NMWL spin columns (Millipore), separated on a 10% Bis-Tris gel (Bio-Rad), and stained with SYPRO-Ruby protein stain (Invitrogen). Entire lanes were cut into several bands, in-gel digested using trypsin, and analyzed by LC/MS using an electrospray ionization coupled to a linear ion trap mass spectrometer (Thermo Scientific LTQ-XL) as previously described (30). The data were analyzed using an automated database search against the SwissProt database with the Mascot software search engine (Matrix Science). Fragment ion tolerance was set to 0.6 Da, and parent ion tolerance was set to 1.5 Da. Identified proteins from the automated search were further verified by manual inspection of the raw data.
Immunoblotting and Immunoprecipitation
The cells were stimulated as indicated in the figure legends. After stimulation, the cells were lysed in lysis buffer containing 20 mm Hepes/KOH (pH 7.5), 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 10% glycerol, 10 mm pyrophosphate supplemented with complete protease inhibitors (Roche Applied Science) for 20 min. For immunoprecipitation (IP) using anti-FLAG M2 resin (Sigma-Aldrich), anti-HA affinity matrix (Roche Applied Science), or IKKγ agarose conjugates (FL-419; Santa Cruz) lysates were precleared with Sepharose beads (Sigma-Aldrich) for 1 h prior to IP overnight. For immunoprecipitation of other endogenous proteins, protein extracts were precleared with protein A anti-rabbit IgG beads (eBioscience) for 45 min, incubated with the indicated antibody for 1 h followed by anti-rabbit IgG beads for 1 h. The immunoprecipitated complexes or cell lysates were resolved by SDS-PAGE (Bio-Rad) and transferred onto nitrocellulose membranes. Membranes were probed with the indicated antibodies and visualized using enhanced chemiluminescence (Pierce) for detection.
Expression of Recombinant Proteins and GST Pulldown Assay
pGEX4T-GST and pGEX4T-GST-BCL10 fusion protein were expressed in Escherichia coli, and the fusion protein was extracted by microfluidization and purified using glutathione-Sepharose 4B affinity matrix (GE Healthcare). In vitro translation and biotinylation of the MIB2 or MALT1 protein was performed using the TNT SP6 high yield wheat germ protein expression system and the Transcend nonradioactive translation detection system (Promega) according to the manufacturers' instructions. GST pulldown assays were performed by incubating GST or GST-BCL10 fusion proteins immobilized on glutathione-Sepharose beads with in vitro translated protein overnight at 4 °C. Proteins complexes were washed five times with lysis buffer, and proteins were eluted with sample buffer (Bio-Rad), resolved on SDS-PAGE, and blotted onto nitrocellulose membranes. The total amounts of proteins on membranes were visualized by MEM Code Stain (Pierce). Biotinylated proteins were visualized by incubation with streptavidin-horseradish peroxidase (GE Healthcare), followed by chemiluminescent detection.
Dual Luciferase Reporter Assays
HEK293T cells were transfected with an NF-κB reporter (firefly luciferase) vector containing a triple NF-κB binding site and minimal promoter, Renilla luciferase vector (pRL-TK; Promega), and indicated expression plasmids using Lipofectamine 2000 (Invitrogen). 24 h after transfection, the cells were lysed in passive lysis buffer (Promega). Luciferase activity was determined using the dual luciferase kit (Promega), and firefly luciferase activity values were normalized to Renilla luciferase activity.
Protein Cross-linking
The cells were lysed in a buffer containing 20 mm Hepes/KOH (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, 10 mm pyrophosphate supplemented with complete protease inhibitors (Roche Applied Science). Lysates were split and either incubated with Me2SO as control or the cross-linker disuccinimidyl suberate (1 mm; Pierce) on ice for 2 h. The reaction was quenched by adding Tris-HCl (pH 7.5; final concentration, 50 mm) at room temperature for 25 min.
Kinase Assay
HEK293T cells were transiently co-transfected with pcDNA3, pCX-MIB2, or pCX-MyD88 and pRC-FLAG-IKKγ using Lipofectamine 2000 (Invitrogen). 24 h after transfection, the cell lysates were prepared and cleared by centrifugation at 10,000 × g for 10 min. IKKγ was immunoprecipitated using anti-FLAG M2 antibody resin (Sigma-Aldrich). Precipitates were washed and incubated with GST-IκBα (1–54) as substrate in kinase buffer (25 mm Hepes, 10 mm MgCl2, 10 mm β-glycerophosphate, 0.5 mm EGTA, 0.5 mm sodium fluoride, 0.5 mm orthovanadate) in the presence of [γ-32P]ATP (1 μCi) and ATP (20 μm) for 20 min at 25 °C. The reactions were stopped by boiling in SDS sample buffer, resolved on a 4–12% SDS gel, and analyzed by PhosphorImager, followed by immunoblot analysis with antibodies against IKKβ.
Lentivirus Production
To generate lentiviruses, 7 × 105 HEK293T cells were seeded in 6-well plates. After 24 h cells were transfected with either pLKO.1-MIB2 shRNA, pGIPZ-MIB2 shRNAmir, or the respective control shRNA vector in combination with packaging vectors that were generously provided by I. Verma (The Salk Institute) and J. Gray (St. Jude Children's Research Hospital) using Lipofectamine 2000. The medium was changed 24 h after transfection. 48 h after transfection the virus containing supernatant was collected and cleared by filtration (0.45 μm). Polybrene (6 μg ml−1) was added to the virus-containing supernatant, and HEK293T cells were transduced by centrifugation at 1500 × g at 37 °C for 1 h. 48 h after transduction, antibiotic was added for stable cell generation. The selected cells were screened for expression of MIB2 by immunoblotting 7–10 days after selection.
RESULTS
Identification of MIB2 as a Novel BCL10-interacting Protein
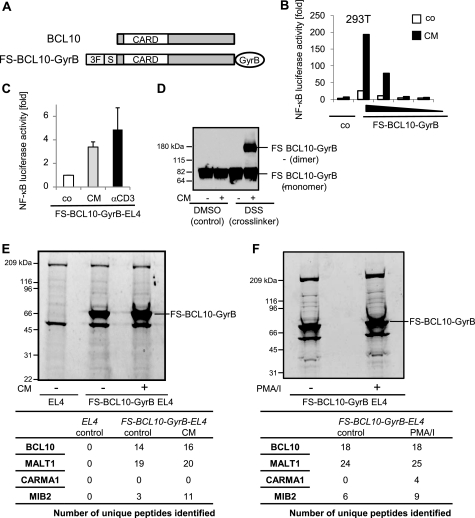
To identify novel BCL10-interacting proteins, we established a biochemical approach that allows us to identify constitutive and activation-dependent components of the BCL10 signaling complex by MS. We took advantage of a technique that is based on the fact that many proteins, including BCL10, are activated through dimerization (28). BCL10 was fused to the subunit B of E. coli DNA gyrase (GyrB; Fig. 1A), which can inducibly be dimerized by adding the bivalent antibiotic coumermycin (CM), thereby mimicking receptor stimulation (Fig. 1B). This construct was further equipped with a tandem epitope tag consisting of a triple FLAG tag and a One-STrEP tag, allowing affinity purification of the BCL10 complex and subsequent analysis of associated proteins by MS. Coumermycin treatment of HEK293T cells transiently transfected with the FS-BCL10-GyrB fusion construct led to transcriptional NF-κB activation, confirming its functional activity (Fig. 1B). Similar results were obtained in EL4 cells stably expressing the FS-BCL10-GyrB protein (Fig. 1C). Dimerization of BCL10 by coumermycin induced transcriptional activity of NF-κB comparable with “physiological” stimulation through TCR cross-linking. Cross-linking experiments confirmed coumermycin-mediated dimerization of a significant amount of the FS-BCL10-GyrB fusion protein (Fig. 1D).
FIGURE 1.
Identification of MIB2 as a novel BCL10-interacting protein. A, schematic representation of wild type BCL10 and the FS-BCL10-GyrB fusion construct. CARD, CARD domain; GyrB, subunit B of E. coli DNA gyrase; 3F, triple FLAG tag; S, One-STrEP tag. B, NF-κB luciferase activity in HEK293T cells transiently transfected with various concentrations of the FS-BCL10-GyrB vector and stimulated with CM (0.5 μm). C, NF-κB luciferase activity in EL4 cells stably expressing FS-BCL10-GyrB stimulated with CM or plate-bound αCD3 (1 μg ml−1). The values represent the means ± S.E., n = 3. D, FS-BCL10-GyrB-expressing EL4 cells were stimulated with CM. The lysates were prepared and either incubated with the cross-linker disuccinimidyl suberate (DSS) or its solvent (dimethyl sulfoxide, DMSO) as control. Dimerization of BCL10 was reflected by its increased molecular weight as revealed by immunoblotting with antibodies against FLAG. E, FS-BCL10-GyrB expressing EL4 cells were left untreated or stimulated with CM for 15 min. EL4 wild type cells were used as control. The cell lysates were prepared, and proteins were purified by one-step affinity purification, followed by SDS-PAGE and SYPRO-Ruby protein staining. Each lane was cut into 24 bands, each of which were processed and analyzed by LC/MS. Peptides were identified by MASCOT analysis and verified manually. The numbers shown represent the numbers of unique peptides of individual proteins identified. F, FS-BCL10-GyrB-expressing EL4 cells were either left untreated or stimulated with PMA/ionomycin (PMA/I; 50 ng ml−1) for 15 min. Cell lysates were prepared as described for E. Each lane was cut into 17 bands, which were processed and analyzed by LC/MS. Peptides were identified by MASCOT analysis and verified manually. The numbers shown represent the numbers of unique peptides of individual proteins identified.
To identify BCL10-interacting proteins, we stimulated EL4-cells stably expressing FS-BCL10-GyrB in two independent experiments with either coumermycin (Fig. 1E) or PMA/ionomycin (Fig. 1F), which mimics physiological, BCL10-dependent TCR activation. EL4-cells were used as a negative control. Protein complexes were isolated by affinity purification, resolved by SDS-PAGE, and stained with SYPRO-Ruby. The identity of proteins was queried by LC/MS. As expected, MALT1 was identified as a constitutive BCL10-interacting protein with a high number of unique peptides in both experimental settings (Fig. 1, E and F, and supplemental Tables S1 and S2). No major difference was observed in the number of peptides identified between nonstimulated and stimulated samples. CARMA1, which acts upstream of BCL10, was not identified upon coumermycin treatment; however, CARMA1 was unequivocally identified as part of the PMA/ionomycin-activated BCL10 complex (Fig. 1, E and F, andsupplemental Tables S1 and S2). These data confirm that GyrB-tagged BCL10 is incorporated into the endogenous CARMA1-marshalled signaling complex. The data also demonstrate that coumermycin-induced dimerization of GyrB-BCL10 acts strictly directionally by mediating downstream signaling but not complex formation with upstream components. This observation is consistent with our former work with GyrB-tagged proteins in the TLR signaling pathway (28). Other proteins suggested to act downstream of BCL10, such as TRAF6, TRAF2, TAB/TAK1, or JNK2 (26, 31), were not identified, possibly because of either the restricted sensitivity of our experimental approach or the absence of these proteins in a stable BCL10 complex. However, one specific BCL10-interacting protein that was identified with a significant number of peptides was the RING finger containing E3 ubiquitin ligase MIB2. MIB2 was constitutively bound to BCL10 at low levels and recruited into the signaling complex upon both coumermycin-induced dimerization of BCL10 and PMA/ionomycin stimulation, indicating that MIB2 acts downstream of BCL10 (Fig. 1, E and F, and supplemental Tables S1 and S2).
MIB2 has previously been associated with Notch and glutamate receptor signaling and was found to be involved in muscle stability in Drosophila (32–34). Its role in BCL10-dependent signaling and NF-κB activation is unclear, even though MIB2 was previously identified as a potential NF-κB-inducing gene in a large scale screen for human NF-κB-activating molecules (35). Overall, the structural similarity of MIB2 to other E3 ligases critically involved in the activation of NF-κB, such as the TRAFs in TNFR and TLR signaling pathways, and the potential function of MIB2 as an activator of NF-κB, led us to focus our studies on MIB2 as a possible new component of the BCL10 signaling complex that links BCL10 to the activation of downstream NF-κB effector kinases.
MIB2 Interacts Directly with BCL10
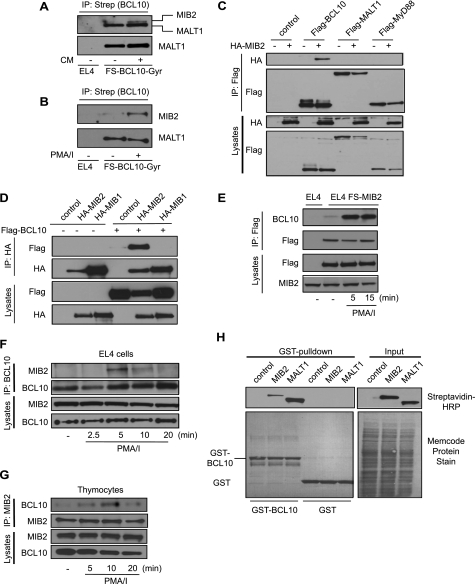
To confirm BCL10-MIB2 interaction, we performed co-IP experiments of BCL10 during stimulation in EL4 cells stably expressing FS-BCL10-GyrB. Endogenous MIB2 was found to be inducibly recruited to BCL10 upon dimerization or PMA/ionomycin treatment (Fig. 2, A and B). Specific interaction between BCL10 and MIB2 was also observed in co-immunoprecipitation experiments in HEK293T cells, using MALT1 and the TLR adaptor protein MyD88 as negative controls (Fig. 2C). Furthermore, no interaction was detectable between BCL10 and MIB1, a paralog of MIB2 with very similar structural domain organization (Fig. 2D). Reverse experiments using EL4 cells stably expressing epitope-tagged MIB2 further confirmed recruitment of MIB2 into the BCL10 complex upon stimulation (Fig. 2E). Moreover, activation of EL4-cells with PMA/ionomycin induced interaction between endogenous BCL10 and MIB2 (Fig. 2F), confirming their interaction under physiological conditions. Comparable results were obtained in thymocytes, where endogenous MIB2 was recruited into the BCL10 complex upon PMA/ionomycin stimulation, confirming MIB2-BCL10 interaction in primary T-cells (Fig. 2G). Using GST-BCL10 as bait protein in in vitro GST pulldown experiments, we found that BCL10 efficiently bound in vitro translated MIB2, strongly suggesting direct protein-protein interaction between BCL10 and MIB2 (Fig. 2G). Taken together, MIB2 is a specific BCL10-interacting protein that is recruited upon BCL10 activation.
FIGURE 2.
MIB2 interacts directly with BCL10. A, FS-BCL10-GyrB-expressing EL4 cells were stimulated with CM (0.5 μm) for 15 min. BCL10 was immunoprecipitated with Strep-Tactin, followed by immunoblot analysis using antibodies against MALT1 (lower panel) and MIB2 (upper panel). Please note that the MALT1-derived signal is also apparent in the MIB2 immunoblot because of incomplete antibody stripping. B, FS-BCL10-GyrB expressing EL4 cells were stimulated with PMA/ionomycin (PMA/I; 50 ng ml−1) for 15 min. BCL10 was immunoprecipitated with Strep-Tactin, followed by immunoblot analysis using antibodies against MIB2 and MALT1, as indicated. C, HEK293T cells were transiently co-transfected with indicated vectors. BCL10, MALT1, and MyD88 were immunoprecipitated using antibodies against FLAG. Immunoprecipitates (IPs) and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. D, FLAG-BCL10 was co-expressed with HA-MIB1 or HA-MIB2 in HEK293T cells. MIB1 and MIB2 were immunoprecipitated with antibodies against HA, and IPs and lysates were analyzed by immunoblotting with antibodies against HA and FLAG. E, FS-MIB2 expressing EL4 cells were stimulated with PMA/ionomycin for the indicated time points, MIB2 was immunoprecipitated with antibodies against FLAG, and IPs and lysates were analyzed by immunoblotting with antibodies against BCL10, FLAG, and MIB2. F, EL4 cells were stimulated with PMA/ionomycin for the indicated time points. Endogenous BCL10 was immunoprecipitated, and IPs and lysates were analyzed by immunoblotting with antibodies against MIB2 and BCL10. G, primary mouse thymocytes were stimulated with PMA/ionomycin for the indicated time points. Endogenous MIB2 was immunoprecipitated, and IPs and lysates were analyzed by immunoblotting with antibodies against MIB2 and BCL10. H, GST pulldown of GST-BCL10 or GST only as control with in vitro translated and biotinylated MIB2 or MALT1. GST-precipitated proteins and input proteins were analyzed by immunoblotting with streptavidin-HRP or by protein staining (Memcode) as indicated.
MIB2 Promotes Activation of TAK1 and IKKγ
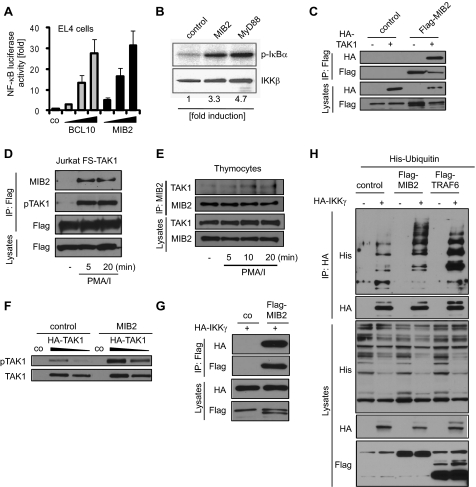
We next investigated whether MIB2 could induce transcriptional NF-κB activity and engage established components of the NF-κB signaling pathway. Similar to BCL10, co-expression of MIB2 with an NF-κB-driven luciferase reporter vector resulted in dose-dependent induction of transcriptional NF-κB activity (Fig. 3A). Furthermore, expression of MIB2 induced catalytic activity of the IKK complex comparable with the known IKK activator MyD88, as determined by in vitro kinase assay (Fig. 3B). Because phosphorylation of TAK1 and ubiquitination of IKKγ are required for activation of the IKK complex in various signaling pathways, we next analyzed MIB2 interaction and modifications of these molecules upon MIB2 expression. MIB2 was indeed found to associate with TAK1 upon co-expression as well as PMA/ionomycin stimulation of Jurkat cells stably expressing epitope-tagged TAK1 (Fig. 3, C and D). Similar results were obtained in primary cells. PMA/ionomycin of thymocytes led to interaction of endogenous MIB2 and TAK1, confirming complex formation of these proteins under physiological conditions (Fig. 3E). Recruitment of TAK1 was accompanied by TAK1 phosphorylation, reflecting increased kinase activity of TAK1 (Fig. 3, D and F) (36). Furthermore, MIB2 was found to bind and ubiquitinate IKKγ upon expression in HEK293T cells (Fig. 3, F and G), comparable with other known E3 ubiquitin ligases critically involved in IKK activation, e.g. TRAF6 (20). Moreover, increased expression of MIB2 led to enhanced ubiquitination of endogenous IKKγ upon PMA/ionomycin stimulation in Jurkat cells (supplemental Fig. S1). Together, the data show that MIB2 interacts with TAK1 and IKKγ, resulting in activation-associated modifications of respective proteins, i.e. TAK1 phosphorylation and IKKγ ubiquitination.
FIGURE 3.
MIB2 induces classical NF-κB signaling. A, NF-κB luciferase activity in EL4 cells transiently transfected with various concentrations of BCL10 or MIB2. The values represent the means ± S.E., n = 3. B, HEK293T cells were co-transfected with control plasmid, MIB2, or MyD88 and FLAG-tagged IKKγ. IKKγ was immunoprecipitated with antibodies against FLAG, and an in vitro kinase assay was performed using GST-IκBα as substrate. C, HEK293T cells were transiently transfected with HA-TAK1 in combination with vector control or with FLAG-MIB2. MIB2 was immunoprecipitated using antibodies against FLAG. IPs and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. D, Jurkat cells stably expressing FS-TAK1 were stimulated with PMA/ionomycin (PMA/I; 50 ng ml−1) for the indicated time points. TAK1 was immunoprecipitated using FLAG antibodies, and IPs and lysates were analyzed by immunoblotting with antibodies against MIB2, pTAK1 (Thr184/187), and FLAG. E, primary mouse thymocytes were stimulated with PMA/ionomycin for the indicated time points. Endogenous MIB2 was immunoprecipitated, and IPs and lysates were analyzed by immunoblotting with antibodies against MIB2 and TAK1. F, HEK293T cells were transiently co-transfected with various concentrations of HA-TAK1 and vector control or MIB2. Levels of phosphorylated TAK1 (Thr184/187) and total TAK1 were determined by immunoblotting. G, HEK293T cells were transiently transfected with HA-IKKγ and vector control or FLAG-MIB2. MIB2 was immunoprecipitated using antibodies against FLAG. IPs and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. H, ubiquitination of IKKγ was determined by co-expressing HA-IKKγ with control vector, FLAG-MIB2 or FLAG-TRAF6 (as control), and His6-Ubiquitin. IKKγ was immunoprecipitated using antibodies against HA. IPs and total lysates were analyzed by immunoblotting using antibodies against His6, HA, or FLAG.
BCL10-MIB2 Interaction Is Mediated via Unconventional MIB/HERC2-CARD Domain Interaction
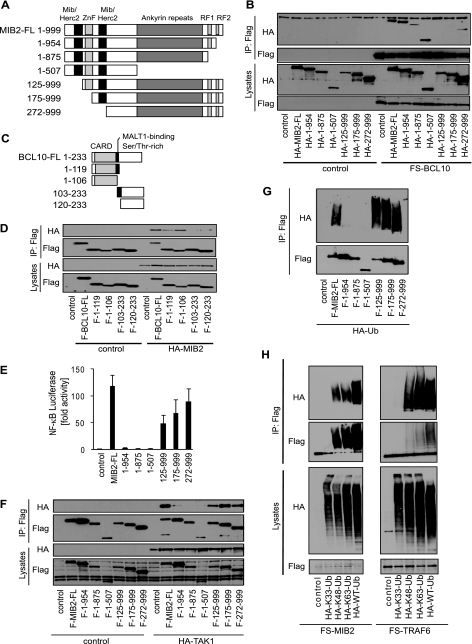
Structurally, several conserved protein domains can be identified in MIB2, such as two MIB/HERC2 domains (37), a zinc finger domain, several ankyrin repeat domains, and two C-terminal RING finger domains (Fig. 4A). Although the function of the MIB/HERC2 domains is not well defined, zinc finger and ankyrin repeat domains are often involved in protein interaction, and RING fingers are signature motifs of E3 ubiquitin ligases. To determine which protein domain mediates MIB2-BCL10 interaction, we tested a series of epitope-tagged MIB2 deletion mutants for BCL10 binding by overexpression and co-IP (Fig. 4B). MIB2 deletion mutants lacking the first 125 amino acids, encompassing the N-terminal MIB/HERC domain failed to interact with BCL10, suggesting that this domain controls BCL10-MIB2 interaction (Fig. 4B). To determine which interacting domain of BCL10 is important for MIB2 binding, we performed similar sets of experiments using a series of BCL10 deletion mutants (Fig. 4C). BCL10 consists of an N-terminal CARD domain, which typically mediates homotypic protein interactions, and a C-terminal serine-threonine-rich region that contains several phosphorylation sites shown to regulate BCL10 function (18). MALT1 was initially found to interact with BCL10 via a region immediately downstream of the CARD domain (38, 39); however, more detailed analysis provided evidence that MALT1 can also interact with the CARD domain in an unconventional non-CARD-CARD interaction, which has also been described for other CARD-containing proteins (39, 40). Co-expression of full-length epitope-tagged MIB2 with a series of epitope-tagged BCL10 deletion mutants demonstrated that CARD domain-containing forms of BCL10 interacted most efficiently with MIB2. Still, residual binding was also observed for the C-terminal truncation mutants of BCL10, which was, however, independent of the described region that contributed to MALT interaction (Fig. 4D). As such, BCL10-MIB2 interaction is chiefly mediated via an unconventional, heterotypic MIB/HERC2-CARD domain interaction.
FIGURE 4.
Functional domains important for MIB2-BCL10 interaction, NF-κB activation, and autoubiquitination of MIB2. A, schematic representation of MIB2 and MIB2 deletion mutants. Mib/Herc2, MIB/HERC2 domain; ZnF, zinc finger domain; Ankyrin repeats, ankyrin repeats domain; RF1 and RF2, ring finger domains 1 and 2. B, HEK293T cells were co-transfected with FS-BCL10 and a vector control, HA-MIB2, or HA-MIB2 deletion mutants. BCL10 was immunoprecipitated using antibodies against FLAG. IPs and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. C, schematic representation of BCL10 and BCL10 deletion mutants. CARD, caspase recruitment domain; MALT1-binding, MALT1-binding region; Ser/Thr-rich, serine/threonine-rich region. D, HEK293T cells were co-transfected with HA-MIB2 and a vector control, FLAG-BCL10, or FLAG-BCL10 deletion mutants. BCL10 was immunoprecipitated using antibodies against FLAG. IPs and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. E, NF-κB luciferase activity in HEK293T cells co-transfected with HA-MIB2 or HA-MIB2 deletion mutants. The data are shown as the means ± S.E., n = 3. F, HEK293T cells were co-transfected with HA-TAK1 and a vector control, FLAG-MIB2, or FLAG-MIB2 deletion mutants. MIB2 was immunoprecipitated using antibodies against FLAG. IPs and total lysates were analyzed by immunoblotting using antibodies against HA or FLAG. G, autoubiquitination of MIB2 was determined by co-expressing HA-Ubiquitin with control vector, FLAG-MIB2 or FLAG-MIB2 deletion mutants. MIB2 was immunoprecipitated using antibodies against FLAG and ubiquitination determined by immunoblotting against HA and FLAG. H, FS-MIB2 or FS-TRAF6 was co-expressed with control vector or various mutants of ubiquitin as well as wild type ubiquitin. MIB2 and TRAF6 were immunoprecipitated using FLAG antibodies. Ubiquitination of MIB2 and TRAF6 was determined by immunoblotting with antibodies against HA and FLAG.
MIB2-induced NF-κB Activation Is Dependent on Its Ligase Activity
To further delineate the mechanisms of MIB2-dependent NF-κB activation, we investigated the various MIB2 deletion mutants in transient NF-κB reporter assays. Consistent with the critical role of protein ubiquitination for NF-κB activation, deletion of the C-terminal RING finger domain of MIB2 led to a complete loss of NF-κB induction (Fig. 4E), accompanied by impaired recruitment and phosphorylation of TAK1 (Fig. 4F and supplemental Fig. S2). A large portion of the N terminus of MIB2, encompassing the described BCL10 interaction domain, was dispensable for NF-κB activation, identifying the RING finger-containing C terminus of MIB2 as effector domain.
MIB2 Supports Synthesis of K48- and K63-linked Polyubiquitin Chains
Polyubiquitination as post-translational modification is well characterized (20, 21). Although K63-linked polyubiquitination was mainly found as nondegradative modification controlling protein function for example during signal transduction, K48-linked ubiquitination typically targets proteins for degradation by the 26 S proteasome (20, 21). Similar to other ubiquitin ligases, we found that MIB2 overexpression mediates autopolyubiquitination, reflecting catalytic activity (Fig. 4G). Similar to TRAF6, MIB2 overexpression supported synthesis of K48-linked, as well as K63-linked, polyubiquitin chains, as determined by co-expression of different ubiquitin lysine mutants (Fig. 4H). This activity depended entirely on the C-terminal RING finger (Fig. 4G) (41). As such, MIB2-mediated autoubiquitination and NF-κB activation depend on the C-terminal RING finger domain, which is consistent with the idea that polyubiquitination is critical for NF-κB activation.
MIB2 Controls BCL10-mediated Activation of NF-κB
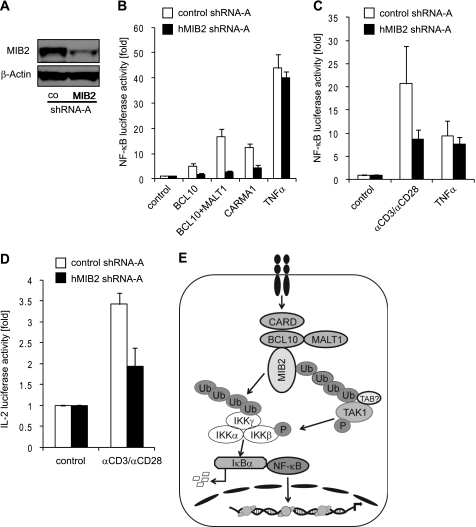
Given our results above, we postulated that MIB2 is required for BCL10-mediated NF-κB activation. Using shRNA-mediated knockdown in HEK293T cells and Jurkat T-cells, we indeed found that MIB2 controls BCL10-mediated NF-κB activation. Knockdown of MIB2 in HEK293T cells led to impaired NF-κB activation upon overexpression of BCL10 alone, BCL10 in combination with MALT1, and CARMA1 (Fig. 5, A and B). In accordance to those results, TCR-dependent NF-κB activity was strongly reduced in Jurkat T-cells upon MIB2 knockdown (Fig. 5C). TNFα-mediated NF-κB activation was largely MIB2-independent, both in HEK293T cells and Jurkat cells. Similar results were obtained for another, independent shRNA construct (supplemental Fig. S3). TCR-mediated activation of the IL-2 promoter, which is known to depend on NF-κB activity (42), was also found to be reduced upon MIB2 knockdown (Fig. 5D), further confirming the critical role of MIB2 in BCL10-mediated activation of NF-κB and NF-κB target genes.
FIGURE 5.
MIB2 is required for BCL10-dependent activation of NF-κB. A, HEK293T cells were stably transduced with control shRNA or MIB2 shRNA (pLKO.1-MIB2). Expression levels of MIB2 were analyzed by immunoblotting using antibodies against MIB2. B, HEK293T cells stably expressing control shRNA or MIB2 shRNA were transiently co-transfected with indicated plasmids and an NF-κB luciferase reporter plasmid. The values represent the means ± S.E., n = 3. C, Jurkat cells were transiently transfected with control shRNA or MIB2 shRNA and an NF-κB luciferase reporter plasmid. 48 h after transfection, the cells were stimulated with plate-bound αCD3 (10 μg ml−1) in combination with αCD28 (10 μg ml−1) or TNFα (30 ng ml−1). The values represent the means ± S.E., n = 4. D, Jurkat cells were transiently transfected with control shRNA or MIB2 shRNA and an IL-2 promoter luciferase reporter plasmid. 48 h after transfection, the cells were stimulated with plate-bound αCD3 in combination with αCD28. The values represent the means ± S.E., n = 3. E, model of proposed function of MIB2 during BCL10 signaling. Receptor engagement leads to the formation of the trimolecular CBM complex containing CARMA1, BCL10, and MALT1, and the recruitment of MIB2 into the activated complex. MIB2 activation leads to assembly of nondegradative, K63-linked polyubiquitin (Ub) chains on MIB2 itself, leading to recruitment of ubiquitin binding TAB proteins along with TAK1 and IKKγ, followed by TAK1 phosphorylation and subsequently IKKβ phosphorylation. MIB2 also promotes ubiquitination of IKKγ, possibly supported by recruitment of IKKγ via MALT1, which together with IKKβ phosphorylation controls IKK activity and ultimately results in NF-κB activation.
DISCUSSION
Polyubiquitination and the subsequent recruitment and activation of TAK1 and the IKK complex are established events in BCL10-mediated activation of NF-κB. Still, a major question remaining was how the BCL10 complex is linked to those downstream events. One reason for this gap in our understanding is that it has been technically difficult to analyze the composition of activated signaling complexes in living cells. We have recently established a proteomic system that allowed us to define the protein composition of signaling complexes in living cells by MS (28). Here, we have extended this biochemical/proteomic approach to the BCL10 signaling pathway and have identified the RING finger domain containing E3 ligase MIB2 as a novel component of the activated BCL10 signaling complex.
So far, MIB2 has been associated with Notch and glutamate receptor signaling and was also shown to play a role in muscle stability in Drosophila (32–34). Although MIB2 was proposed as a potential candidate in a large scale analysis of human NF-κB-inducing genes, its role in physiological signaling pathways remained undefined. We found MIB2 to be important for BCL10-induced activation of NF-κB. MIB2 was constitutively associated with BCL10 at low levels and further recruited into the complex upon activation of the BCL10 signaling pathway. Overexpression of MIB2 induced ubiquitin ligase activity, which promoted autoubiquitination and ubiquitination of IKKγ, as well as recruitment and activation of TAK1 resulting in activation of NF-κB. MIB2-induced autoubiquitination and NF-κB activation were dependent on its ubiquitin ligase activity, which we found to catalyze K63-linked, nondegradative polyubiquitin chains, consistent with the idea that polyubiquitination is critical for NF-κB activation (20, 21, 43). As such, the function of MIB2 closely resembles the function of other E3 ligases known to be critically involved in NF-κB activation in the TNFR and TLR/IL-1 signaling pathways, i.e. the TRAF E3 ligases. A current model of the TRAF-mediated activation of the IKK complex in TNFR and TLR/IL-1R signaling pathways suggests that TRAF activation leads to assembly of nondegradative polyubiquitin chains, in part on the TRAFs themselves, leading to recruitment of ubiquitin binding TAB proteins along with TAK1 and IKKγ, followed by TAK1 and consequently IKKβ phosphorylation, ultimately controlling NF-κB activation (19, 20). IKKγ itself was also found to be ubiquitinated during TCR activation, which contributes to IKK activation (25), although the molecular mechanism that governs ubiquitination-dependent IKK activity is still undefined. Based on these observations and our results described here, it appears that MIB2 acts in BCL10-dependent pathways as the functional equivalent to TRAFs in the TNFR and TLR pathways, mediating nondegradative autopolyubiquitination and polyubiquitination of IKKγ, recruitment and phosphorylation of TAK1, and activation of the IKK complex. Whether any of the three described TAB proteins are required for MIB2-dependent NF-κB activation remains to be determined.
We found MIB2 primarily in the activated BCL10 complex, where BCL10 appeared to interact directly with MIB2. Deletion mutants of BCL10 and MIB2 identified the CARD domain in BCL10 and the MIB/HERC2 domain in MIB2 as major interaction domains. The MIB/HERC2 domain shows high similarity to domains found in other HERC2 ubiquitin ligases (37); however, the function of these domains is not well defined. We found that the first MIB/HERC2 domain in MIB2 was essential for BCL10 binding, which has previously also been implicated to mediate MIB2 interaction with its substrate Delta during Notch signaling (41). As such, the MIB/HERC2 domains appear to control protein interactions, in the case of its described interaction with BCL10 via heterotypic CARD-MIB/HERC2 binding.
Although MIB2 binds BCL10 directly, its relation to other components of the CBM complex, particularly MALT1, remains to be established. The molecular function of MALT1 is still not entirely understood but may entail different activities, such as aiding recruitment of the IKK complex and acting as a protease cleaving the negative regulatory molecule A20 and, possibly, BCL10 (26, 44–47). Although we did not detect any direct interaction between MIB2 and MALT1 (Fig. 1C), it will be interesting to see whether some of the described functions of MALT1 depend on MIB2. Notably, co-expression of MALT1 together with BCL10 led to increased NF-κB activation, which was, however, still dependent on MIB2 as revealed by shRNA experiments (Fig. 5B).
So far, we have characterized the function of MIB2 in general in the BCL10-dependent NF-κB activation pathway. Because BCL10 is critically involved in signaling pathways triggered by various immune receptors, including the TCR, B-cell receptor, and RIG-I-like receptors, it will be important to determine the function of MIB2 in these pathways during physiological cell stimulation. Furthermore, it will also be important to uncover the function of MIB2 for other downstream signaling events, such as MAPK activation. Given the pivotal role of BCL10 in immune defense, it seems likely that MIB2 is also an essential component of adaptive and possibly innate immune responses.
Acknowledgments
We gratefully acknowledge Perdeep Mehta, Vishwajeeth Pagala, Xiang Ding, and Kanisha Shah of the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital for help with the proteomic analysis and bioinformatics support. We also acknowledge the St. Jude Hartwell Center for primer production and DNA sequencing and the Protein Production facility at St. Jude for the generation of recombinant proteins. Furthermore, we thank Rohan Keshwara for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI083443 (to H. H.). This work was also supported by the American Lebanese Syrian Associated Charities.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- BCL10
- B-cell CLL/lymphoma 10
- CARMA1
- CARD-containing MAGUK protein 1
- TAK1
- transforming growth factor-β-activated kinase 1
- IKKβ
- IκB-kinase β
- TLR
- Toll-like receptor
- TRAF6
- TNF receptor-associated factor 6
- TCR
- T-cell receptor
- MALT
- mucosa-associated lymphoid tissue
- CMV
- cytomegalovirus
- PMA
- phorbol 12-myristate 13-acetate
- IP
- immunoprecipitation
- CM
- coumermycin
- MIB2
- mind bomb-2.
REFERENCES
- 1. Willis T. G., Jadayel D. M., Du M. Q., Peng H., Perry A. R., Abdul-Rauf M., Price H., Karran L., Majekodunmi O., Wlodarska I., Pan L., Crook T., Hamoudi R., Isaacson P. G., Dyer M. J. (1999) Cell 96, 35–45 [DOI] [PubMed] [Google Scholar]
- 2. Zhang Q., Siebert R., Yan M., Hinzmann B., Cui X., Xue L., Rakestraw K. M., Naeve C. W., Beckmann G., Weisenburger D. D., Sanger W. G., Nowotny H., Vesely M., Callet-Bauchu E., Salles G., Dixit V. M., Rosenthal A., Schlegelberger B., Morris S. W. (1999) Nat. Genet. 22, 63–68 [DOI] [PubMed] [Google Scholar]
- 3. Ruland J., Duncan G. S., Elia A., del Barco Barrantes I., Nguyen L., Plyte S., Millar D. G., Bouchard D., Wakeham A., Ohashi P. S., Mak T. W. (2001) Cell 104, 33–42 [DOI] [PubMed] [Google Scholar]
- 4. Xue L., Morris S. W., Orihuela C., Tuomanen E., Cui X., Wen R., Wang D. (2003) Nat. Immunol. 4, 857–865 [DOI] [PubMed] [Google Scholar]
- 5. Karin M. (2009) Cold Spring. Harb. Perspect. Biol. 1, a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown K. D., Claudio E., Siebenlist U. (2008) Arthritis Res. Ther. 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gross O., Grupp C., Steinberg C., Zimmermann S., Strasser D., Hannesschläger N., Reindl W., Jonsson H., Huo H., Littman D. R., Peschel C., Yokoyama W. M., Krug A., Ruland J. (2008) Blood 112, 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klemm S., Zimmermann S., Peschel C., Mak T. W., Ruland J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 134–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gross O., Gewies A., Finger K., Schäfer M., Sparwasser T., Peschel C., Förster I., Ruland J. (2006) Nature 442, 651–656 [DOI] [PubMed] [Google Scholar]
- 10. Klemm S., Gutermuth J., Hültner L., Sparwasser T., Behrendt H., Peschel C., Mak T. W., Jakob T., Ruland J. (2006) J. Exp. Med. 203, 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hara H., Ishihara C., Takeuchi A., Xue L., Morris S. W., Penninger J. M., Yoshida H., Saito T. (2008) J. Immunol. 181, 918–930 [DOI] [PubMed] [Google Scholar]
- 12. Hara H., Ishihara C., Takeuchi A., Imanishi T., Xue L., Morris S. W., Inui M., Takai T., Shibuya A., Saijo S., Iwakura Y., Ohno N., Koseki H., Yoshida H., Penninger J. M., Saito T. (2007) Nat. Immunol. 8, 619–629 [DOI] [PubMed] [Google Scholar]
- 13. Malarkannan S., Regunathan J., Chu H., Kutlesa S., Chen Y., Zeng H., Wen R., Wang D. (2007) J. Immunol. 179, 3752–3762 [DOI] [PubMed] [Google Scholar]
- 14. Chen Y., Pappu B. P., Zeng H., Xue L., Morris S. W., Lin X., Wen R., Wang D. (2007) J. Immunol. 178, 49–57 [DOI] [PubMed] [Google Scholar]
- 15. McAllister-Lucas L. M., Ruland J., Siu K., Jin X., Gu S., Kim D. S., Kuffa P., Kohrt D., Mak T. W., Nuñez G., Lucas P. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang D., You Y., Lin P. C., Xue L., Morris S. W., Zeng H., Wen R., Lin X. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Poeck H., Bscheider M., Gross O., Finger K., Roth S., Rebsamen M., Hannesschläger N., Schlee M., Rothenfusser S., Barchet W., Kato H., Akira S., Inoue S., Endres S., Peschel C., Hartmann G., Hornung V., Ruland J. (2010) Nat. Immunol. 11, 63–69 [DOI] [PubMed] [Google Scholar]
- 18. Thome M., Weil R. (2007) Trends Immunol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 19. Häcker H., Karin M. (2006) Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 20. Skaug B., Jiang X., Chen Z. J. (2009) Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 21. Wertz I. E., Dixit V. M. (2010) Cold Spring Harb. Perspect. Biol. 2, a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adhikari A., Xu M., Chen Z. J. (2007) Oncogene 26, 3214–3226 [DOI] [PubMed] [Google Scholar]
- 23. Yamamoto M., Sato S., Saitoh T., Sakurai H., Uematsu S., Kawai T., Ishii K. J., Takeuchi O., Akira S. (2006) J. Immunol. 177, 7520–7524 [DOI] [PubMed] [Google Scholar]
- 24. Wan Y. Y., Chi H., Xie M., Schneider M. D., Flavell R. A. (2006) Nat. Immunol. 7, 851–858 [DOI] [PubMed] [Google Scholar]
- 25. Zhou H., Wertz I., O'Rourke K., Ultsch M., Seshagiri S., Eby M., Xiao W., Dixit V. M. (2004) Nature 427, 167–171 [DOI] [PubMed] [Google Scholar]
- 26. Sun L., Deng L., Ea C. K., Xia Z. P., Chen Z. J. (2004) Mol. Cell 14, 289–301 [DOI] [PubMed] [Google Scholar]
- 27. King C. G., Kobayashi T., Cejas P. J., Kim T., Yoon K., Kim G. K., Chiffoleau E., Hickman S. P., Walsh P. T., Turka L. A., Choi Y. (2006) Nat. Med. 12, 1088–1092 [DOI] [PubMed] [Google Scholar]
- 28. Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
- 29. Lim K. L., Chew K. C., Tan J. M., Wang C., Chung K. K., Zhang Y., Tanaka Y., Smith W., Engelender S., Ross C. A., Dawson V. L., Dawson T. M. (2005) J. Neurosci. 25, 2002–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freibaum B. D., Chitta R. K., High A. A., Taylor J. P. (2010) J. Proteome Res. 9, 1104–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Blonska M., Pappu B. P., Matsumoto R., Li H., Su B., Wang D., Lin X. (2007) Immunity 26, 55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jurd R., Thornton C., Wang J., Luong K., Phamluong K., Kharazia V., Gibb S. L., Ron D. (2008) J. Biol. Chem. 283, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carrasco-Rando M., Ruiz-Gómez M. (2008) Development 135, 849–857 [DOI] [PubMed] [Google Scholar]
- 34. Nguyen H. T., Voza F., Ezzeddine N., Frasch M. (2007) J. Cell Biol. 179, 219–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., Hayashi H., Sugano S. (2003) Oncogene 22, 3307–3318 [DOI] [PubMed] [Google Scholar]
- 36. Kishimoto K., Matsumoto K., Ninomiya-Tsuji J. (2000) J. Biol. Chem. 275, 7359–7364 [DOI] [PubMed] [Google Scholar]
- 37. Itoh M., Kim C. H., Palardy G., Oda T., Jiang Y. J., Maust D., Yeo S. Y., Lorick K., Wright G. J., Ariza-McNaughton L., Weissman A. M., Lewis J., Chandrasekharappa S. C., Chitnis A. B. (2003) Dev. Cell 4, 67–82 [DOI] [PubMed] [Google Scholar]
- 38. Lucas P. C., Yonezumi M., Inohara N., McAllister-Lucas L. M., Abazeed M. E., Chen F. F., Yamaoka S., Seto M., Nunez G. (2001) J. Biol. Chem. 276, 19012–19019 [DOI] [PubMed] [Google Scholar]
- 39. Langel F. D., Jain N. A., Rossman J. S., Kingeter L. M., Kashyap A. K., Schaefer B. C. (2008) J. Biol. Chem. 283, 32419–32431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nam Y. J., Mani K., Ashton A. W., Peng C. F., Krishnamurthy B., Hayakawa Y., Lee P., Korsmeyer S. J., Kitsis R. N. (2004) Mol. Cell 15, 901–912 [DOI] [PubMed] [Google Scholar]
- 41. Koo B. K., Yoon K. J., Yoo K. W., Lim H. S., Song R., So J. H., Kim C. H., Kong Y. Y. (2005) J. Biol. Chem. 280, 22335–22342 [DOI] [PubMed] [Google Scholar]
- 42. Hoyos B., Ballard D. W., Böhnlein E., Siekevitz M., Greene W. C. (1989) Science 244, 457–460 [DOI] [PubMed] [Google Scholar]
- 43. Kanarek N., London N., Schueler-Furman O., Ben-Neriah Y. (2010) Cold Spring Harb. Perspect. Biol. 2, a000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thome M. (2008) Nat. Rev. Immunol. 8, 495–500 [DOI] [PubMed] [Google Scholar]
- 45. Noels H., van Loo G., Hagens S., Broeckx V., Beyaert R., Marynen P., Baens M. (2007) J. Biol. Chem. 282, 10180–10189 [DOI] [PubMed] [Google Scholar]
- 46. Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z. J., Marynen P., Beyaert R. (2008) Nat. Immunol. 9, 263–271 [DOI] [PubMed] [Google Scholar]
- 47. Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E. M., Rufer N., Fasel N., Thome M. (2008) Nat. Immunol. 9, 272–281 [DOI] [PubMed] [Google Scholar]