Background: γ-Secretase modulators (GSMs) hold great potential as anti-Alzheimer disease drugs, but their molecular target(s) are not established.
Results: The catalytic subunit of γ-secretase, presenilin, was identified as a direct target of novel GSMs.
Conclusion: Enzyme-targeting GSMs establish allosteric modulation as a mechanism of GSM action.
Significance: The identification of presenilin as GSM target may contribute to the development of therapeutically active GSMs.
Keywords: Alzheimer Disease, Amyloid, Drug Action, Presenilin, Secretases, gamma-Secretase, gamma-Secretase Modulator (GSM)
Abstract
γ-Secretase is essential for the generation of the neurotoxic 42-amino acid amyloid β-peptide (Aβ42). The aggregation-prone hydrophobic peptide, which is deposited in Alzheimer disease (AD) patient brain, is generated from a C-terminal fragment of the β-amyloid precursor protein by an intramembrane cleavage of γ-secretase. Because Aβ42 is widely believed to trigger AD pathogenesis, γ-secretase is a key AD drug target. Unlike inhibitors of the enzyme, γ-secretase modulators (GSMs) selectively lower Aβ42 without interfering with the physiological function of γ-secretase. The molecular target(s) of GSMs and hence the mechanism of GSM action are not established. Here we demonstrate by using a biotinylated photocross-linkable derivative of highly potent novel second generation GSMs that γ-secretase is a direct target of GSMs. The GSM photoprobe specifically bound to the N-terminal fragment of presenilin, the catalytic subunit of γ-secretase, but not to other γ-secretase subunits. Binding was differentially competed by GSMs of diverse structural classes, indicating the existence of overlapping/multiple GSM binding sites or allosteric alteration of the photoprobe binding site. The β-amyloid precursor protein C-terminal fragment previously implicated as the GSM binding site was not targeted by the compound. The identification of presenilin as the molecular target of GSMs directly establishes allosteric modulation of enzyme activity as a mechanism of GSM action and may contribute to the development of therapeutically active GSMs for the treatment of AD.
Introduction
Alzheimer disease (AD),5 the major cause of senile dementia, is caused by an accumulation of the aggregation-prone amyloid-β peptide (Aβ) in the brain of affected patients (1). Aβ is a small 37–43-amino acid secreted peptide that is generated from the β-amyloid precursor protein (APP) by the sequential action of two membrane-bound proteases, β- and γ-secretase (1). The latter enzyme is an intramembrane-cleaving protease composed of presenilin (PS) 1 or 2 as the catalytic subunit, APH-1a or -b, nicastrin, and PEN-2 (2–4). γ-Secretase liberates Aβ from the membrane by a stepwise cleavage of the APP C-terminal fragment (CTF) that β-secretase generates via shedding of the APP ectodomain (1). As inhibition and/or modulation of Aβ generation are promising therapeutic strategies to treat AD, β- and γ-secretase are key AD drug targets, and inhibitors of both enzymes are investigated in clinical trial studies (5).
Apart from APP, a number of other γ-secretase substrates have been identified (3). Among these, Notch1 represents a substrate of major importance for the regulation of cell differentiation during embryonic development as well as in adulthood (6). Cleavage of Notch1 by γ-secretase gives rise to the Notch1 intracellular domain (NICD), which acts in the nucleus as an essential transcriptional regulator (6). Unfortunately, direct inhibition of Aβ generation by γ-secretase inhibitors (GSIs) is problematic due to the inhibition of the Notch signaling pathway (5), and a large clinical phase 3 trial has also recently been halted due to the appearance of severe side effects.
In contrast to GSIs, γ-secretase modulators (GSMs) may offer safer alternatives for AD therapy. Such drugs typically shift the cleavage specificity of γ-secretase toward the increased production of the shorter peptide Aβ38, whereas concomitantly reducing the generation of the pathogenic Aβ42 species. A subset of non-steroidal anti-inflammatory drugs (NSAIDs), such as sulindac sulfide and other cyclooxygenase inhibitors, was identified 10 years ago as the first GSMs (7). Although clinical efficacy has yet to be demonstrated, the discovery of Aβ42-lowering GSMs has opened a new opportunity for AD drug discovery, and more potent and brain-penetrant second generation compounds have been identified and developed since the first study (5). These compounds differ structurally from the first generation compounds. Most of the new compounds lack the acidic carboxyl group of NSAIDs and typically consist of bridged aromates (5). Supplemental Table 1 shows representatives of these compounds that were used in this study.
Certain low potency GSMs such as the NSAIDs flurbiprofen and sulindac sulfide have been reported to bind to the APP CTF as well as to Aβ in the N-terminal third of the APP transmembrane domain (8–10), although this interaction was questioned by others (11). Another study identified the γ-secretase subunit PEN-2 as the principal target of non-NSAID-type second generation GSMs (12). However, this study was conducted under conditions in which the γ-secretase complex was dissociated and with isolated PEN-2, i.e. under non-native conditions. Thus, the identity of the binding site(s) of GSMs has been unsettled and remained a major open question. Here we identify the N-terminal fragment of PS, but not PEN-2 or the APP CTF, as the binding site of novel highly potent GSMs with bridged aromatic structural scaffold.
EXPERIMENTAL PROCEDURES
Compounds
Systematic names and synthesis of RO-02, RO-03, RO-57, and RO-57-BpB, as well as synthesis and/or source of all other compounds used, are provided in the supplemental material.
Characterization of GSMs
Modulatory activities of GSMs, including dose-response curves, were analyzed in cell-based and cell-free γ-secretase assays as described previously (13–15). Additional details are described in the supplemental material, including cell lines and antibodies used for analysis.
Photoaffinity Labeling
Membrane fractions of HEK293 cells were incubated in the presence or absence of RO-57-BpB and competitor compounds as indicated and following photoactivation of RO-57-BpB with UV light solubilized in SDS-containing buffer. After a clarifying spin, proteins covalently bound to RO-57-BpB were captured with streptavidin-Sepharose. Following three washes, bound proteins were eluted in biotin- and urea-containing sample buffer and analyzed by SDS-PAGE and immunoblotting. Experimental details and information on the antibodies used for analysis are given in the supplemental material.
RESULTS
We identified a series of a novel class of bridged aromatic aminopyrimidine-derived compounds as highly potent second generation GSMs with IC50 values for inhibition of Aβ42 reaching the low nanomolar range (28). One of these compounds, RO-02 (Fig. 1A), was characterized as a prototype in cell-based and cell-free assays in more detail. RO-02 robustly lowered Aβ42 species in HEK293 cells stably expressing the APPsw mutant (HEK293/sw) (IC50 = 15 nm; log IC50 = −7.82 ± 0.05), increased the short species Aβ38, and consistent with the behavior of GSMs of this structural class (12), also inhibited Aβ40 formation (Fig. 1B). Formation of the NICD and likewise that of the APP intracellular domain (AICD; the counterpart of Aβ) was spared by RO-02 in HEK293 cells stably coexpressing the Notch- and APP-based γ-secretase substrates F-NEXT (16) and C99-6myc (17) up to a concentration of at least 10 μm, i.e. of more than 500 times the IC50 (Aβ42) (Fig. 1C). We next asked whether RO-02 would be effective on distinct γ-secretase complexes using HEK293/sw cell lines stably expressing all possible combinations of PS and APH-1 (Fig. 1D). As shown in Fig. 1E, all six individual complexes generated a similar profile of secreted Aβ species. The compound effectively lowered Aβ42 generation of all six γ-secretase complexes with an apparently increased potency for PS2-containing γ-secretase complexes (Fig. 1F; see also supplemental Fig. 1). Finally, the potent and preferential Aβ42-lowering activity of RO-02 was confirmed in rat primary cortical neurons (IC50 (Aβ42) = 13 nm; log IC50 = −7.90 ± 0.04) (Fig. 1G). To further characterize the mode of action of RO-02 and to address the question whether the compound may target γ-secretase directly, we next investigated whether RO-02 could also modulate γ-secretase activity in a cell-free in vitro assay using purified enzyme complex reconstituted into lipid vesicles (15). Dose-response analysis showed that RO-02 modulated the generation of Aβ species by γ-secretase from the recombinant APP substrate C100-His6 as expected (IC50 (Aβ42) = 0.5 μm under these conditions) and without affecting the levels of total Aβ and total AICD (Fig. 1H). Mass spectrometry analysis showed that RO-02 did not alter the relative levels of the AICD species generated in the cell-free assay when used at a concentration modulatory for the γ-sites (1 μm), suggesting that the compound does not modulate ϵ-cleavage (Fig. 1I).
FIGURE 1.
RO-02, a novel non-acidic GSM, lowers Aβ40 and Aβ42 generation with high potency. A, structure of RO-02. B, dose-response analysis of γ-secretase activity modulation by RO-02 in HEK293/sw cells by Aβ sandwich immunoassay. Data are represented as mean ± S.E. (n = 3). Note that for most data points, error bars are too small to be displayed. C, RO-02 dose-response analysis of NICD and AICD formation in HEK293 cells stably co-expressing F-NEXT and C99-6myc as assessed by immunoblot analysis. Quantitation of NICD and AICD formation is shown in the right panel. Data are represented as mean ± S.E. (RO-02, n = 3–5; DAPT (1 μm), n = 2). veh., vehicle. D–F, RO-02 modulates the activity of all six γ-secretase complexes. D, immunoblot analysis of expression of γ-secretase complex components in HEK293/sw cells stably expressing the individual combinations of PS and APH-1 with calnexin as loading control. NCT, nicastrin; m., mature; im., immature; S, short splice variant; L, long splice variant. Profile of secreted Aβ species (E) and Aβ42 response to RO-02 (F) of the six γ-secretase complexes as determined by Aβ sandwich immunoassay. Data in E and F are represented as mean ± S.E. (n = 3). Asterisks indicate significant differences between corresponding PS1- and PS2-containing complexes for their relative Aβ42 generation in the presence of RO-02 (*, p < 0.05; **, p < 0.01; ***, p < 0.001, two-tailed unpaired Student's t test). G, RO-02 dose-response analysis of Aβ40 and Aβ42 generation in cultured rat primary cortical neurons by Aβ ELISA. Data are represented as mean ± S.E. (n = 2). H, RO-02 dose-response analysis in a cell-free assay using purified γ-secretase by immunoblot analysis of Aβ species separated by Tris-Bicine urea SDS-PAGE and total Aβ and AICD. I, mass spectrometry analysis of AICD species generated in the presence or absence (vehicle) of RO-02 (1 μm).
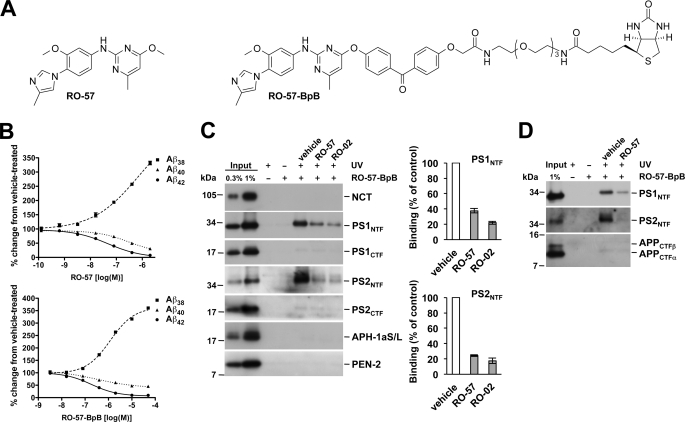
Although the cell-free assay data provide evidence that RO-02 may target γ-secretase directly, they do not prove this as they do not demonstrate direct binding of the drug to its target. Therefore, to unambiguously identify the molecular target(s) of our compounds, a derivative of RO-02 was synthesized (RO-57) that facilitated linkage of a photoactivatable benzophenone group coupled to a biotin moiety (RO-57-BpB) to capture cross-linked target protein(s) by a streptavidin pulldown after photoactivation of the compound (Fig. 2A). The parental compound RO-57 and the photosensitive derivative RO-57-BpB modulated γ-secretase in cultured HEK293/sw cells with an IC50 (Aβ42) of 47 nm (log IC50 = −7.33 ± 0.07) and 185 nm (log IC50 = −6.73 ± 0.07), respectively (Fig. 2B). When total membrane fractions of untransfected HEK293 cells were probed with 1 μm RO-57-BpB, a concentration that inhibits Aβ42 production by ∼30% (supplemental Fig. 2), the PS1 and PS2 N-terminal fragments (NTFs) were identified as molecular targets of RO-57-BpB (Fig. 2C). Capture of the PS NTFs was observed neither in the absence of RO-57-BpB nor in the presence of RO-57-BpB but in the absence of UV irradiation. Labeling of PS1 and PS2 NTFs was strongly inhibited in the presence of a 100-fold molar excess of the parental compound RO-57, demonstrating specific cross-linking. As expected, the close and more potent structural relative of RO-57, RO-02, highly efficiently competed for PS NTF labeling by RO-57-BpB (Fig. 2C). Consistent with our results above (Fig. 1F), we noted that labeling of the PS2 NTF was stronger than that of PS1. These results could be confirmed with RO-03, another potent compound of our GSM series (supplemental Fig. 3). As further shown in Fig. 2C, no capture was observed for any other γ-secretase subunits. Because certain GSM photoprobes, which were based on the low potency compounds flurbiprofen or fenofibrate, have previously been reported to bind to the APP CTF γ-secretase substrates (8), we performed labeling experiments using isolated membranes from HEK293/sw cells. These experiments showed that RO-57-BpB failed to specifically label APP CTFs, suggesting that compounds of this structural class used here are not substrate-targeting GSMs (Fig. 2D).
FIGURE 2.
RO-57-BpB targets the PS N-terminal fragment. A, structures of RO-57 and its photoaffinity labeling derivative RO-57-BpB. B, dose-response analysis of γ-secretase activity modulation by RO-57 and RO-57-BpB in HEK293/sw cells by Aβ sandwich immunoassay. Data are represented as mean ± S.E. (n = 3). Note that for most data points, error bars are too small to be displayed. C, specific labeling by photoactivated RO-57-BpB of the PS1 and PS2 NTFs but not the other γ-secretase subunits as detected by immunoblot analysis. Quantitation of PS NTF binding is shown in the right panel. Data are represented as mean ± S.E. (n = 5). NCT, nicastrin. D, the absence of APP CTF labeling by RO-57-BpB as detected by immunoblot analysis.
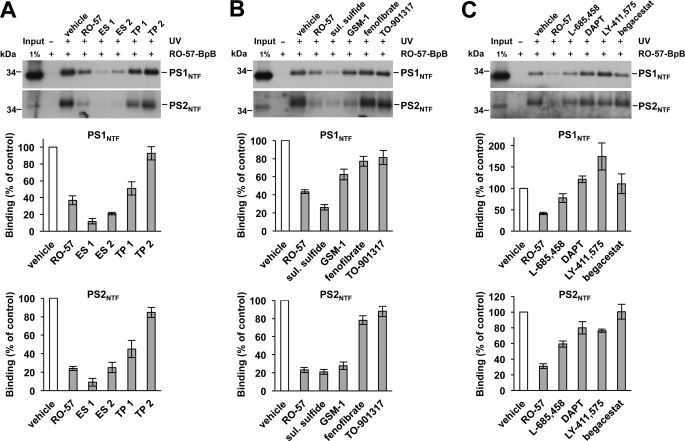
To further investigate whether the binding site of RO-57-BpB may also be targeted by other GSMs, we next tested whether other classes of GSMs could compete for binding of RO-57-BpB. Two structurally similar bridged aromatic second generation GSMs from Eisai Co., Ltd. (ES 1 (= E2012 (18)) and ES 2) (supplemental Table 1) also efficiently competed for labeling of the PS NTFs by RO-57-BpB (Fig. 3A), whereas, interestingly, the bridged aromates from TorreyPines Therapeutics (TP 1 and TP 2) (supplemental Table 1) showed differential effects. While one of the compounds (TP 1) partially prevented PS NTF labeling, the other one (TP 2) did not (Fig. 3A). The acidic GSMs sulindac sulfide, a low potency GSM (7), and GSM-1, a much more potent second generation acidic GSM (13), also competed for PS NTF labeling by RO-57-BpB (Fig. 3B). GSM-1 showed a differential effect and competed for labeling of the PS1 NTF by RO-57-BpB only weakly, whereas PS2 NTF labeling was more strongly reduced. The inverse GSMs fenofibrate (19) and the more potent TO-901317 (20) did weakly reduce the capture of PS NTF by RO-57-BpB. Considering its low potency (IC50 (Aβ42) = 70 μm (14)), the labeling competition of sulindac sulfide was unexpectedly strong. However, although the lack of labeling competition by its inactive analog sulindac sulfone suggested specificity (7, 21) (supplemental Fig. 4), the potential contribution of other factors to the observed labeling competition by sulindac sulfide could not be excluded (see “Discussion”).
FIGURE 3.
GSMs and GSIs differentially compete for PS NTF labeling by RO-57-BpB. A–C, competition assay of PS NTF labeling by RO-57-BpB (1 μm) with other non-acidic GSMs (from Eisai and TorreyPines Therapeutics, each 100 μm) (A), acidic GSMs (sulindac sulfide and GSM-1, each 100 μm) and inverse GSMs (fenofibrate and TO-901317, each 100 μm) (B), and GSIs (L-685,458, DAPT, LY-411,575, and begacestat, each 1 μm) with different mechanisms of action (C). Quantitation of PS NTF binding is shown in the lower panels of A–C. Data are represented as mean ± S.E. (n = 3–6).
Finally, we investigated whether binding of RO-57-BpB to the PS NTF was affected by different classes of highly potent GSIs including the transition-state analog L-685,458, the dipeptidic GSIs DAPT and LY-411,575, and begacestat, a Notch-sparing sulfonamide-type GSI (5). As shown in Fig. 3C, the investigated GSIs did not or only weakly inhibited labeling of the PS NTFs and LY-411,575 rather increased labeling of the PS1 NTF. Taken together, the various GSMs and GSIs differentially competed for PS NTF labeling by RO-57-BpB. With respect to the GSMs, the above results indicate that the binding site of RO-57-BpB might be shared for some GSMs such as the Eisai compounds, whereas for other compounds, the binding site may overlap or be located elsewhere. L-685,458 was the only GSI capable of competing for labeling to some extent, indicating some overlap of the RO-57-BpB binding site with the catalytic site. Alternatively, the competitor compounds used may allosterically alter the RO-57-BpB binding site.
DISCUSSION
The identity of the molecular target(s) of GSMs and thus also the mechanism(s) of GSM action are not yet understood and represent major unresolved questions in the AD drug discovery field (8–12). Here we have provided evidence that γ-secretase is a direct target of new potent second generation GSMs with non-acidic bridged aromatic structure. The compounds target the NTF of its catalytic subunits PS1 and PS2. Binding of PS2 by RO-57-BpB was substantially stronger than that of PS1, indicating that these compounds may preferentially target PS2. This is also supported by the more potent Aβ42 inhibition by RO-02 of γ-secretase containing PS2 as catalytic subunit.
To further characterize the binding site targeted by RO-57-BpB in the PS NTF, we investigated whether other GSMs of similar or different structural scaffold would be able to compete with binding of RO-57-BpB to its target site. Such labeling competition experiments can indicate whether a binding site is shared between different compounds, although they do not discriminate for the alternative possibility that the competitor compound may allosterically alter the binding site of the labeling compound. Each of the two GSMs from Eisai and TorreyPines Therapeutics were tested as representatives of other bridged aromatic GSMs structurally related to our compounds. Both Eisai compounds efficiently competed for labeling of the PS NTF, but only one of the TorreyPines Therapeutics compounds (TP 1) was effective and displayed partial competition. Although it is thus likely that the Eisai compounds target the same site as our compounds, the binding site of the TorreyPines Therapeutics compounds may partially overlap or locate elsewhere (12). The partial labeling competition (TP 1) or its absence (TP 2) could, however, also be due to less favorable physicochemical properties of the TorreyPines Therapeutics compounds, such as high lipophilicity and/or low solubility that may have caused reduced drug availability in the labeling competition experiments.
Consistent with its previously reported direct allosteric interaction with γ-secretase (22), labeling competition was also observed for sulindac sulfide, suggesting that this NSAID-type acidic GSM may target the binding site of our GSMs. However, despite the finding that its inactive analog sulindac sulfone did not compete, it could not be definitively answered whether sulindac sulfide targeted the same site as our GSM photoprobe. For example, the differential effects on labeling competition by these drugs could also have been due to their distinct radical scavenging (23) or membrane-partitioning properties (24).
The second generation acidic compound GSM-1 only weakly competed with RO-57-BpB for PS1 NTF labeling, whereas the drug robustly competed for the labeling of the PS2 NTF. This suggests that the GSM binding sites in the PS1 and PS2 NTFs may have somewhat different conformations and indicates that the binding site of GSM-1 overlaps only partially with that of RO-57-BpB in PS1. The inverse modulators behaved as comparably mild competitors in these assays, suggesting that the PS NTFs are not their principal targets. With the exception of the transition state analog L-685,458, which displayed moderate labeling competition, the GSIs competed for labeling of the PS NTFs very weakly or not at all. The GSM binding site identified here may therefore partially overlap with that of the active site. However, as mentioned before, these interpretations should be taken with some caution as allosteric modulation of the RO-57-BpB binding site by the competitor compounds and/or other factors discussed above influencing labeling competition cannot always be excluded. Vice versa binding experiments using photoactivatable derivatives of the GSMs and GSIs used as competitor compounds in this study against our GSMs would be needed to clarify these issues.
The identification of γ-secretase as a direct target of GSMs also sheds further light on the mechanism of action of GSMs. Although a substrate-targeting mode of GSM action had been suggested (8), the majority of the earlier studies suggested that GSMs target the enzyme (21, 22, 25, 26). By covalent binding of the RO-57-BpB GSM derivative to the PS NTF, but not to APP CTFs, our photoaffinity labeling data now demonstrate this directly for our compounds. Our data thus support previous studies that suggested an allosteric mechanism of GSM action (21, 22, 27), which, however, did not demonstrate direct GSM binding to γ-secretase by GSM photoprobes. Future studies should now aim at a further refinement of the GSM binding site(s) within the PS NTF. Together with this study, such studies will aid in the development of highly effective and hopefully clinically safe GSMs as anti-AD drugs.
Acknowledgments
We thank Edith Winkler and in particular Peer-Hendrik Kuhn for valuable technical advice. We are grateful to Ralph Nixon for antibody PS1N, Manfred Brockhaus for the antibodies BAP15 and BAP24, Masayasu Okochi for the F-NEXT and Alison Goate for the C99-6myc construct, Georg Schmid and Elvira da Silva for HEK293S cell production, Guido Galley for TorreyPines Therapeutics compounds, Helmut Jacobsen for compound analysis in primary neurons, and the Boehringer Ingelheim Pharma KG for DAPT.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB596) (to H. S. and C. H.), the Bundesministerium für Bildung und Forschung (KNDD) (to C. H. and H. S.), and the Center for Integrated Protein Science Munich (CIPSM).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Materials and Methods, Table 1, and Figs. 1–4.
- AD
- Alzheimer disease
- Aβ
- amyloid β-peptide
- AICD
- APP intracellular domain
- APP
- β-amyloid precursor protein
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- CTF
- C-terminal fragment
- GSI
- γ-secretase inhibitor
- GSM
- γ-secretase modulator
- NICD
- Notch1 intracellular domain
- NSAID
- non-steroidal anti-inflammatory drug
- NTF
- N-terminal fragment
- PS
- presenilin
- DAPT
- N-[(3,5-difluorophenyl)acetyl]-l-alanyl-2-phenyl]glycine-1,1-dimethylethyl ester.
REFERENCES
- 1. Haass C., Selkoe D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 101–112 [DOI] [PubMed] [Google Scholar]
- 2. Steiner H., Fluhrer R., Haass C. (2008) J. Biol. Chem. 283, 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wakabayashi T., De Strooper B. (2008) Physiology 23, 194–204 [DOI] [PubMed] [Google Scholar]
- 4. Wolfe M. S. (2009) Semin. Cell Dev. Biol. 20, 219–224 [DOI] [PubMed] [Google Scholar]
- 5. Tomita T. (2009) Expert. Rev. Neurother. 9, 661–679 [DOI] [PubMed] [Google Scholar]
- 6. Bray S. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 7. Weggen S., Eriksen J. L., Das P., Sagi S. A., Wang R., Pietrzik C. U., Findlay K. A., Smith T. E., Murphy M. P., Bulter T., Kang D. E., Marquez-Sterling N., Golde T. E., Koo E. H. (2001) Nature 414, 212–216 [DOI] [PubMed] [Google Scholar]
- 8. Kukar T. L., Ladd T. B., Bann M. A., Fraering P. C., Narlawar R., Maharvi G. M., Healy B., Chapman R., Welzel A. T., Price R. W., Moore B., Rangachari V., Cusack B., Eriksen J., Jansen-West K., Verbeeck C., Yager D., Eckman C., Ye W., Sagi S., Cottrell B. A., Torpey J., Rosenberry T. L., Fauq A., Wolfe M. S., Schmidt B., Walsh D. M., Koo E. H., Golde T. E. (2008) Nature 453, 925–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richter L., Munter L. M., Ness J., Hildebrand P. W., Dasari M., Unterreitmeier S., Bulic B., Beyermann M., Gust R., Reif B., Weggen S., Langosch D., Multhaup G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 14597–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Botev A., Munter L. M., Wenzel R., Richter L., Althoff V., Ismer J., Gerling U., Weise C., Koksch B., Hildebrand P. W., Bittl R., Multhaup G. (2011) Biochemistry 50, 828–835 [DOI] [PubMed] [Google Scholar]
- 11. Beel A. J., Barrett P., Schnier P. D., Hitchcock S. A., Bagal D., Sanders C. R., Jordan J. B. (2009) Biochemistry 48, 11837–11839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kounnas M. Z., Danks A. M., Cheng S., Tyree C., Ackerman E., Zhang X., Ahn K., Nguyen P., Comer D., Mao L., Yu C., Pleynet D., Digregorio P. J., Velicelebi G., Stauderman K. A., Comer W. T., Mobley W. C., Li Y. M., Sisodia S. S., Tanzi R. E., Wagner S. L. (2010) Neuron 67, 769–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Page R. M., Baumann K., Tomioka M., Pérez-Revuelta B. I., Fukumori A., Jacobsen H., Flohr A., Luebbers T., Ozmen L., Steiner H., Haass C. (2008) J. Biol. Chem. 283, 677–683 [DOI] [PubMed] [Google Scholar]
- 14. Kretner B., Fukumori A., Gutsmiedl A., Page R. M., Luebbers T., Galley G., Baumann K., Haass C., Steiner H. (2011) J. Biol. Chem. 286, 15240–15251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Winkler E., Hobson S., Fukumori A., Dümpelfeld B., Luebbers T., Baumann K., Haass C., Hopf C., Steiner H. (2009) Biochemistry 48, 1183–1197 [DOI] [PubMed] [Google Scholar]
- 16. Okochi M., Steiner H., Fukumori A., Tanii H., Tomita T., Tanaka T., Iwatsubo T., Kudo T., Takeda M., Haass C. (2002) EMBO J. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J., Brunkan A. L., Hecimovic S., Walker E., Goate A. (2004) Neurobiol. Dis. 15, 654–666 [DOI] [PubMed] [Google Scholar]
- 18. Portelius E., Van Broeck B., Andreasson U., Gustavsson M. K., Mercken M., Zetterberg H., Borghys H., Blennow K. (2010) J. Alzheimers Dis. 21, 1005–1012 [DOI] [PubMed] [Google Scholar]
- 19. Kukar T., Murphy M. P., Eriksen J. L., Sagi S. A., Weggen S., Smith T. E., Ladd T., Khan M. A., Kache R., Beard J., Dodson M., Merit S., Ozols V. V., Anastasiadis P. Z., Das P., Fauq A., Koo E. H., Golde T. E. (2005) Nat. Med. 11, 545–550 [DOI] [PubMed] [Google Scholar]
- 20. Czech C., Burns M. P., Vardanian L., Augustin A., Jacobsen H., Baumann K., Rebeck G. W. (2007) J. Neurochem. 101, 929–936 [DOI] [PubMed] [Google Scholar]
- 21. Takahashi Y., Hayashi I., Tominari Y., Rikimaru K., Morohashi Y., Kan T., Natsugari H., Fukuyama T., Tomita T., Iwatsubo T. (2003) J. Biol. Chem. 278, 18664–18670 [DOI] [PubMed] [Google Scholar]
- 22. Beher D., Clarke E. E., Wrigley J. D., Martin A. C., Nadin A., Churcher I., Shearman M. S. (2004) J. Biol. Chem. 279, 43419–43426 [DOI] [PubMed] [Google Scholar]
- 23. Santos F., Teixeira L., Lúcio M., Ferreira H., Gaspar D., Lima J. L., Reis S. (2008) Free Radic. Res. 42, 639–650 [DOI] [PubMed] [Google Scholar]
- 24. Fan S. S., Shen T. Y. (1981) J. Med. Chem. 24, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 25. Eriksen J. L., Sagi S. A., Smith T. E., Weggen S., Das P., McLendon D. C., Ozols V. V., Jessing K. W., Zavitz K. H., Koo E. H., Golde T. E. (2003) J. Clin. Invest. 112, 440–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato T., Nyborg A. C., Iwata N., Diehl T. S., Saido T. C., Golde T. E., Wolfe M. S. (2006) Biochemistry 45, 8649–8656 [DOI] [PubMed] [Google Scholar]
- 27. Lleó A., Berezovska O., Herl L., Raju S., Deng A., Bacskai B. J., Frosch M. P., Irizarry M., Hyman B. T. (2004) Nat. Med. 10, 1065–1066 [DOI] [PubMed] [Google Scholar]
- 28. Baumann K., Flohr A., Goetschi E., Jacobsen H., Jolidon S., Luebbers T. (August 27, 2009) World Intellectual Property Organization (WIPO) Patent WO2009103652