Abstract
The hypomethylating agent 5-aza-2′-deoxycytidine (5-aza-CdR) and its derivatives have been successfully used for the treatment of myelodysplastic syndromes, and they frequently improve the anemia that usually accompanies these disorders. However, the molecular mechanisms underlying this action remain poorly understood. In this study, we used two erythroid models, murine erythroid leukemia cells and erythroid burst-forming unit-derived erythroblasts, to show that 5-aza-CdR induced erythroid differentiation and increased the expression of transferrin receptor 1 (TfR1) and ferrochelatase (Fech), thereby increasing iron uptake and heme biosynthesis. We have identified new regulatory E-boxes that lie outside of CpG islands in the TfR1 and Fech promoters, and the methylation status of these sites can be altered by 5-aza-CdR treatment. This in turn altered the binding of the transcription factor c-Myc to these promoter elements. Furthermore, 5-aza-CdR promoted the nuclear translocation of c-Myc and its binding to Max to form functional complexes. The coordinated actions of 5-aza-CdR on the methylation status of the target genes and in stimulating the nuclear translocation of c-Myc provide new molecular insights into the regulation of E-boxes and explain, at least in part, the increased erythroid response to 5-aza-CdR treatment.
Keywords: DNA Methylation, Erythropoiesis, Heme, Protein DNA-Interaction, Transcription Regulation, 5-Aza-CdR, TfR1, c-Myc, Myelodysplastic Syndromes
Introduction
Myelodysplastic syndromes (MDS)4 represent a heterogeneous group of hematopoietic progenitor cell disorders characterized by impaired proliferation and maturation of a range of cell types (1–3). Anemia is the most common feature of the initial diagnosis, and over 80% of patients have a hemoglobin concentration below 10 g/dl and reduced reticulocytes (4). Defects in heme synthesis appear to play an important role in many cases of MDS, and ring sideroblasts are characteristic of some forms of the disease (5).
Hypomethylating agents, such as the azanucleosides, including azacytidine (5-azacytidine) and decitabine (5-aza-2′-deoxycytidine (5-aza-CdR)), have recently been approved by the United States Food and Drug Administration (FDA) for the treatment of MDS (6). Compared with conventional care regimens, azanucleosides have been shown to significantly prolong the survival of MDS patients and reduce the risk of transformation to acute myeloid leukemia (AML) (6). However, only a proportion (37%) of MDS patients are responsive to azanucleoside treatment (1), and their underlying mechanisms of action are still largely unknown (6). One working hypothesis is that the demethylating agents reactivate tumor suppressor genes, such as p15, the inhibitor of cyclin-dependent kinase N2B, through demethylation of CpG islands in the promoter regions of the target genes (7). Bisulfite sequencing analysis indicated that the normally hypermethylated promoter of p15 was demethylated in a high proportion of MDS patients (65%) following 5-aza-CdR treatment (7). Further studies in a large MDS patient population showed that fewer than 20% of the cytosines in CpG islands in the p15 promoter were methylated (8), suggesting that the CpG islands of tumor suppressor gene promoters may not be the only target of 5-aza-CdR. Bioinformatic analysis showed that many genes (approximate 50%) up-regulated by 5-aza-CdR treatment did not contain CpG islands at all in their promoter regions (9). These observations suggest that many genes are regulated by 5-aza-CdR through pathways that do not involve CpG island demethylation, and we hypothesize that there are multiple molecular targets of 5-aza-CdR.
Early studies with 5-aza-CdR reported that the compound could induce erythroid differentiation of murine erythroid leukemia (MEL) cells (10) and leukemic blasts from patients with AML (11). However, the mechanism of differentiation induced by 5-aza-CdR is not clear. Recent studies on the biological functions of 5-aza-CdR have focused on its role in regulating transcription factors involved in the cell cycle (12). Several transcription factors have been shown to play important roles in hematopoiesis, such as GATA1, EKLF, NF-E2, and c-Myc (13–17). In particular, c-Myc has been shown to activate transferrin receptor 1 (TfR1) (18, 19), ferritin, and iron regulatory protein 2 (IRP2) (20). These proteins are all important in cellular iron acquisition (and therefore are critical for erythroid differentiation) and may play pivotal roles in MDS and AML. Furthermore, global gene expression profiling has shown that the expression of target genes of c-Myc differs significantly between MDS patients and normal subjects (21, 22). These observations suggest a potential key role for c-Myc in MDS.
To study the molecular mechanism of 5-aza-CdR-induced erythroid differentiation, we chose MEL cells and mouse erythroid burst-forming units (BFU-Es) as erythroid cell models. We found that several major genes involved in iron uptake and heme biosynthesis were up-regulated in these cells following 5-aza-CdR treatment and that the methylation status of cytosines that are not localized in the CpG islands of these genes is essential for their transcriptional activation by c-Myc. Furthermore, c-Myc transnuclear localization and the formation of functional Myc-Max binding complexes were enhanced by 5-aza-CdR treatment. This coordinated mechanism of c-Myc translocation and target gene demethylation may explain, at least in part, the increased erythroid response to 5-aza-CdR treatment.
EXPERIMENTAL PROCEDURES
Cell Culture
MEL cells were grown in RPMI 1640 medium (Thermo Scientific, Beijing, China) supplemented with 10% fetal calf serum (FCS) (Thermo Scientific, South Logan, UT). MEL cells (5 × 106) in logarithmic growth were treated with either 1.8% (v/v) dimethyl sulfoxide (DMSO) or 0.01–1 μm 5-aza-CdR dissolved in water (Sigma-Aldrich). After a 48-h incubation, cells were collected by centrifugation, washed twice with PBS, and stored at −80 °C for later analysis. For BFU-E culture, the femurs and tibias of BALB/c mice were obtained by surgery, and the ends of these long bones were trimmed to expose the interior marrow shaft. A 3-ml syringe with a 21-gauge needle containing 1 ml of cold RPMI 1640 medium with 2% FCS was used to flush the marrow. Marrow collections were resuspended in fresh, cold RPMI 1640 medium with 2% FCS. Cell number and viability were determined with a Vi-CellTM XR cell viability analyzer (Beckman-Coulter, Fullerton, CA). Bone marrow cells (1 × 106) were separated and cultured in RPMI 1640 medium with different concentrations of 5-aza-CdR for 24 h. Additional 5-aza-CdR was added at 0, 8, 16, and 24 h to keep the concentration of 5-aza-CdR consistent. The cells were further cultured for 8 days in MethoCult® semisolid medium (Stemcell Technologies, Vancouver, Canada) in the absence of 5-aza-CdR prior to the analysis of gene expression.
Total RNA Isolation, RT-PCR, and Quantitative Real-time RT-PCR
Total RNA from MEL cells and BFU-Es was isolated with TRIzol® reagent (Invitrogen) following the manufacturer's instructions. RNA was treated with RQ1 RNase-free DNase to remove contaminating genomic DNA, and 2 μg of DNase-treated RNA was used to synthesize cDNA with oligo(dT) primers and Moloney murine leukemia virus (M-MLV) reverse transcriptase. No genomic DNA contamination was found in the cDNA because the GAPDH transcripts (as a control) cannot be amplified by PCR with the total RNA without reverse transcription reaction as templates. Diluted cDNA (20 ng) was amplified in a 25-μl reaction containing reaction buffer, a 50 μm concentration of each dNTP, 0.01% SYBR Green I (Invitrogen), 1.25 units of GoTaq DNA polymerase, and 50 pmol of each primer. The primers used in this study are listed in Table 1. RT-PCR and real-time PCR were performed using a Mastercycler ep realplex4 thermocycler (Eppendorf, Hamburg, Germany) under the following conditions: 95 °C for 5 min, 30 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. All results were analyzed by the 2−ΔΔCt method (23) and normalized with GAPDH. RQ1 RNase-free DNase, oligo(dT), M-MLV reverse transcriptase, dNTPs, and GoTaq DNA polymerase were obtained from Promega (Madison, WI).
TABLE 1.
Primers used in real-time PCR, bisulfite sequencing, ChIP, and MS-PCR
| Genes | Forward | Reverse |
|---|---|---|
| Real-time PCR | ||
| GAPDH | 5′ -AGGTCGGTGTGAACGGATTTG -3′ | 5′ -TGTAGACCATGTAGTTGAGGTCA -3′ |
| TfR1 | 5′ -GTGGAGTATCACTTCCTGTCGC -3′ ′ | 5′ -CCCCAGAAGATATGTCGGAAAGG -3′ ′ |
| Fech | 5′ -CAGACAGATGAGGCTATCAAAGG -3′ | 5′ -CACAGCTTGTTGGACTGGATG -3′ |
| Alas2 | 5′ -TGGGCTAAGAGCCATTGTCCT -3′ | 5′ -GTAGGTGTGGTCCTGTTTCTTC -3′ |
| Ftl | 5′ -CCATCTGACCAACCTCCGC -3′ | 5′ -CGCTCAAAGAGATACTCGCC -3′ |
| Hmox1 | 5′ -AGCGTCCACAGCCCGACAGCAT -3′ | 5′ -GCCATCACCAGCTTAAAGCCTTCTC -3′ |
| GATA-1 | 5′ -TGGGGACCTCAGAACCCTTG -3′ | 5′ -GGCTGCATTTGGGGAAGTG -3′ |
| Eklf | 5′ -GTACACTCACCACCCTGGGACAG -3′ | 5′ -CGGGAGACTCGGAACCTGGAAAG -3′ |
| Irp2 | 5′ -GGATTCTTGGGTGGGGAGTTGGTGG -3′ | 5′ -CCTACTTGCCTGAGGTGCTTTGTAA -3′ |
| c-Myc | 5′ -TTTCCCTACCCGCTCAACGACAG -3′ | 5′ -TTCCTCATCTTCTTGCTCTTCTTCA -3′ |
| Hba1 | 5′ -AAGCCCTGGAAAGGATGTTTGCTAG -3′ | 5′ -GGCAGTGGCTCAGGAGCTTGAAGTT -3′ |
| Hbb1 | 5′ -CTGAGAACTTCAGGCTCCTGG -3′ | 5′ -GCTAGATGCCCAAAGGTCTTCA -3′ |
| Bisulfite sequencing | ||
| Tfr1 | 5′ -GAGTAAAGGTTGATATTTT -3′ | 5′ -CTAACAAAAAAATCACTCAC -3′ |
| Fech | 5′ -GTAGATTTAGYGGGAATTAG -3′ | 5′ -CAAACACAACCCAACTTACR -3′ |
| Irp2 | 5′ -TCTGGAGAATAATAGAGGT -3′ | 5′ -CCTAAAAACTACAACCAACAAAC -3′ |
| Eklf | 5′ -GGAAGAGTTATATTAAGAGTTYGTATTTTAA -3′ | 5′ -AAATAACCTCRAAATCACTACTATCC -3′ |
| c-Myc | 5′ -TTYGGAGTYGGAGTATTGGGT-3′ | 5′ -CCTARCTCRCAAATTATAAATTCCAATAAA-3′ |
| ChIP | ||
| TfR1 + 285 bp | 5′ -CTTCCTAGTGAGTGACTCCC -3′ | 5′ -AATGTCCGTGACACTAGTAACC -3′ |
| TfR1 −6 kb | 5′ -CAAGCCGTTTATTCTGCTCC -3′ | 5′ -TGTCTCAGCTCTTCCCTACC -3′ |
| Alas2 −300 bp | 5′ -GCCGAGAGTGTATTCTATGTCTG -3′ | 5′ -TTCTTCCTCTACTCCCTCTG -3′ |
| Fech −3 kb | 5′ -TCTTCGTCTTTGTCATAGACAG -3′ | 5′ -AAGGAACTCACAAGTAGCTC -3′ |
| Luciferase assay | ||
| 700 bp | 5′ -GGCGAGCTCGCCGACAGGCCAACTCCAAAGAC-3′ | 5′ -CCGCTCGAGCGGGTCCGTGACACTAGTAACCGA-3′ |
| 1700 bp | 5′ -GGCGAGCTCGCCTGTGCTTCTCTTACAGGGAC-3′ | 5′ -CCGCTCGAGCGGGTCCGTGACACTAGTAACCGA-3′ |
| −6 kb E-box | 5′ -CCCAAGCTTGGGTCATCACAGACAGGTCATACTACC-3′ | 5′ -GGCGAGCTCGCCGGTCACCTCAATCAGTGTTTCTC-3′ |
| −8 kb E-box | 5′ -CCCAAGCTTGGGCAAAGTTGCACTGCAAAGTC-3′ | 5′ -GGCGAGCTCGCCCCAGAATTCAAAGCTAGGCCA-3′ |
Western Blot Analysis
For total protein isolation, cells were lysed with radioimmune precipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, a complete protease inhibitor mixture tablet (Roche Applied Science), and 0.1% SDS). Nuclear fractions were isolated using the NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions. Total protein (50 μg) or nuclear protein (100 μg) samples were electrophoresed on 10% SDS-polyacrylamide gels for 120 min at 70 V. The proteins were then transferred onto nitrocellulose membranes for 120 min at 380 mA in 10% methanol transfer buffer. The nitrocellulose membranes were then probed with the appropriate antibodies. Immunoreactive proteins were visualized using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) and were scanned with a Typhoon TrioTM variable mode imager (Amersham Biosciences). Antibodies against ferrochelatase (Fech), erythroid aminolevulinic acid synthase 2 (Alas2), c-Myc, Max, Gata1, and β-actin were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and an antibody against TfR1 was obtained from Cell Signaling Technology (Danvers, MA).
Iron Uptake
Human apotransferrin (apo-Tf) (Sigma-Aldrich) was labeled with 55FeCl3 (PerkinElmer Life Sciences) as described previously (24). Equal numbers of cells (1 × 107 cells) were incubated with or without 5-aza-CdR (0.01, 0.1, or 1 μm) in RPMI 1640 medium containing 10% FCS at 37 °C for 48 h and then incubated with 1 μm 55Fe-labeled transferrin (55Fe-Tf) at 37 °C in RPMI 1640 medium for up to 3 h. Cells were washed three times with ice-cold PBS, and cell pellets were collected and lysed with radioimmune precipitation assay buffer. Ultima Gold Mixture (1 ml) (PerkinElmer Life Sciences) was added to the cell lysates (25), and the amount of 55Fe in the mixtures was measured using a 1450 MicroBeta TriLux microplate scintillation and luminescence counter (PerkinElmer Life Sciences).
Measurement of Heme Concentration
Cells (5 × 106) were lysed with radioimmune precipitation assay buffer, and the protein concentration was measured with the BCA assay (Applygen, Beijing, China). Each protein sample (10 μg) was added to 0.5 ml of 2.5 m oxalic acid followed by heating at 100 °C for 30 min to remove the iron from the heme. The autofluorescence of protoporphyrin in each sample was quantitatively measured with an Infinite M200 plate reader (TECAN, Grödig, Austria) at an excitation wavelength of 400 nm and an emission wavelength of 620 nm. Samples without heating were used to correct for background autofluorescence of endogenous protoporphyrin (26).
Bisulfite Sequencing
Genomic DNA was isolated using a QIAamp DNA minikit (Qiagen, Hilden, Germany), and 2 μg of DNA was bisulfite-converted and cleaned with an EZ DNA MethylationTM kit (Zymo Research, Orange, CA). Specific sequencing primers for TfR1, Fech, Irp2, and Eklf were used to amplify target sequences (Table 1). The fragments were ligated into pGEM®-T Easy vector (Promega, Madison, WI) and transformed into Escherichia coli. At least 10 individual clones were picked for sequencing.
Co-immunoprecipitation Assay
Nuclear fractions were isolated using a NE-PER nuclear and cytoplasmic extraction kit (Thermo Scientific, Inc.) according to the manufacturer's instructions for the co-immunoprecipitation assay. Anti-c-Myc antibody (0.6 μg) was added to 100 μg of nuclear faction in a volume of 50 μl and incubated for 2 h at 4 °C. Then 10 μl of Protein A-agarose beads (10 μl) (Santa Cruz Biotechnology, Inc.) were used to collect antigen-antibody complexes. The expression of Max was detected following Western blotting as described above using an anti-Max primary antibody (Santa Cruz Biotechnology, Inc.).
Chromatin Immunoprecipitation Assay
For chromatin immunoprecipitation (ChIP) assays, formaldehyde solution was added to cultured cells (1 × 107) at 1% final concentration to cross-link DNA and proteins. After incubation at room temperature for 10 min, 1.25 m glycine was added to quench free formaldehyde. Cells were collected by centrifugation, washed twice with PBS, and lysed with lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1). Sonication was used to shear the genomic DNA into 200–1000-bp fragments, and this was verified by agarose gel electrophoresis. After incubating with anti-c-Myc antibodies (Santa Cruz Biotechnology, Inc.) overnight, Protein A-agarose beads were added to precipitate the DNA-protein complexes. The beads were washed sequentially with wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1) containing first 150 mm NaCl and then 500 mm NaCl. The complexes were eluted from the beads with 1 m NaHCO3. Five molar NaCl was added to separate protein-DNA complexes. The DNA fragments were purified with a ZymocleanTM gel DNA recovery kit (Zymo Research) and used as templates for PCR analysis of the TfR1, Fech, and Alas2 promoters (Table 1).
Methylation-sensitive PCR (MS-PCR)
The restriction enzyme PmlI specifically cleaves DNA at GACGTG sequences, and E-boxes that are methylated are resistant to PmlI cleavage. Thus, genomic DNA was digested with PmlI (New England Biolabs, Beijng, China) following the manufacturer's instructions. Digested genomic DNA (10 ng) was used as a template for the detection of the E-boxes in the promoter regions of genes of interest. If the E-box is unmethylated, PmlI will cleave the E-box and prevent synthesis of the PCR product, whereas when the E-box is methylated, cleavage by PmlI is prevented, and a specific PCR product can be detected (27).
Plasmid Construction and Luciferase Assay
The pGL3-basic, pGL3-promoter, and pR-TK vectors were obtained from Promega (Madison, WI). The pGL3-700, pGL3-1700, pGL3-6kE-box, and pGL3-8kE-box vectors were generated by inserting a 700-bp fragment (−183 to +550), 1700-bp fragment (−1230 to +550), 700-bp fragment plus −6 kb E-box (−6267 to −6061), and 700-bp fragment plus −8 kb E-box (−8689 to −8421) to pGL3-basic vector, respectively. c-Myc and Max expression vectors were obtained by cloning the c-Myc or Max coding regions into the pcDNA3.1(−) vector (Invitrogen). To study the influence of methylation status on the promoter activities, pGL3-700 and pGL3-1700 constructs were first treated with DNA methyltransferase M.SssI (New England BioLabs, Beijng, China) according to the manufacturer's instructions before the report assays. All vectors were transfected into human hepatoma cell line (HepG2) for 48 h of culture. Activities of firefly and Renilla luciferase were measured with the dual luciferase assay kit (Promega, Madison, WI) according to the manufacturer's instructions.
Statistical Analysis
All values are expressed as mean ± S.E. of at least three samples. Statistical comparisons were performed using Student's t test. Results were considered statistically significant when p values were lower than 0.05.
RESULTS
5-Aza-CdR Treatment Alters Iron Homeostasis and Intracellular Heme Levels in MEL Cells
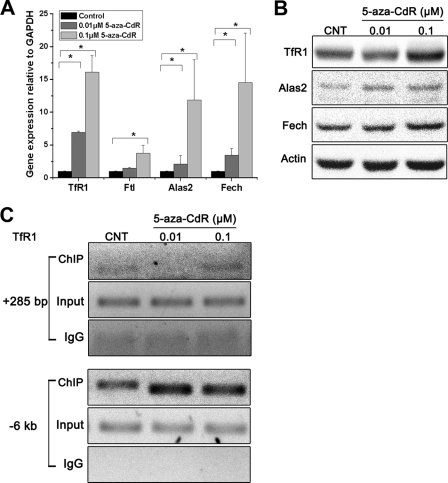
The hypomethylating agent 5-aza-CdR has been reported to induce erythroid differentiation and decrease global methylation status in MEL cells (6). In this study, we investigated the effects of 5-aza-CdR on the expression of a number of genes involved in iron and heme metabolism because iron acquisition and heme synthesis are intimately linked to hematopoiesis. MEL cells were incubated with low (0.1 μm) or high (1 μm) doses of 5-aza-CdR. Consistent with erythroid differentiation, the mRNA expression of Tfr1, which mediates transferrin-dependent iron uptake, and Alas2 and Fech, which are involved in heme biosynthesis, was significantly increased after 5-aza-CdR treatment. As shown in Fig. 1A, TfR1 message levels increased ∼5-fold after 0.1 μm 5-aza-CdR treatment, similar to the effects of DMSO, a known inducer of MEL cell differentiation (28). A higher concentration of 5-aza-CdR further stimulated TfR1 mRNA expression (∼8-fold over the control). We also observed that expression of the mRNA encoding Alas2, the first enzyme in the heme biosynthetic pathway, increased 4- and 32-fold after 0.1 or 1 μm 5-aza-CdR treatment, respectively. The mRNA encoding the terminal enzyme of heme biosynthesis, Fech, showed a smaller increase than the other genes, but it is not normally rate-limiting for heme synthesis (29). We also observed that the mRNA levels of the adult α-globin (Hba1) and β-globin (Hbb1) genes increased significantly after 5-aza-CdR treatment. As a positive control, DMSO treatment also significantly increased the expression of Alas2, Fech, Hba1, and Hbb1. No changes were found in the expression of the genes encoding the ferritin L-subunit (Ftl) or heme oxygenase 1 (Hmox1) (Fig. 1A). The levels of TfR1, Alas2, and Fech protein were analyzed by Western blotting using specific antibodies (Fig. 1B). Densitometry results are shown in Fig. 1C. TfR1 and Fech protein expression increased significantly after 0.1 μm 5-aza-CdR treatment, with a smaller increase observed for Alas2. As a positive control, DMSO treatment also significantly increased the expression of each protein. These results indicate that 5-aza-CdR can stimulate the expression of components of the iron uptake and heme biosynthesis machinery of MEL cells at both the mRNA and protein levels.
FIGURE 1.
5-Aza-CdR stimulated the expression of genes associated with erythroid iron acquisition and heme synthesis. MEL cells were treated with 1.8% DMSO or 5-aza-CdR (0.1 or 1 μm) for 48 h. Cells were collected for total RNA and protein isolation. A, quantitative real-time RT-PCR showed increases in the mRNA levels of TfR1, Hba1, Hbb1, Alas2, and Fech. *, p < 0.01 compared with the control MEL cells. B, a representative Western blot showing the levels of TfR1, Alas2, and Fech protein in 5-aza-CdR-treated MEL cells. C, densitometry analysis of the expression of TfR1, Alas2, and Fech protein levels (the densitometry was normalized to the levels of β-actin). *, p < 0.01 compared with the control MEL cells. D, cellular iron uptake in control and 5-aza-CdR-treated MEL cells. MEL cells treated with or without 5-aza-CdR for 48 h were incubated for 0.5, 1.5, or 3 h with 1.85 μm 55Fe-Tf. After washing three times with cold PBS, the radioactivity was measured using a scintillation counter. *, p < 0.01 compared with 1.5 h control MEL cells; **, p < 0.01 compared with 3 h control MEL cells. E, heme levels in MEL cells treated with or without 5-aza-CdR. Cellular extracts (10 μg of total protein) were analyzed by measurement of fluorescent PPIX levels after removing iron from endogenous heme. *, p < 0.01 compared with control MEL cells. Error bars, S.E.
To investigate whether the increased expression of TfR1 is functional in terms of mediating iron uptake, we measured the iron uptake capacity of MEL cells with or without 5-aza-CdR treatment. After 0.5 h of incubation with 55Fe-Tf, untreated MEL cells took up 0.2 nmol of 55Fe-Tf per million viable cells, whereas those treated with the positive control (DMSO) took up significantly more iron (5.3 nmol of 55Fe-Tf per million viable cells). The stimulation in iron uptake in response to 5-aza-CdR treatment was not as strong (Fig. 1D, left); however, after 1.5 h of incubation, we observed significantly more 55Fe-Tf uptake in cells treated with 0.01 and 0.1 μm 5-aza-CdR compared with the control cells (Fig. 1D, middle). By 3 h, there was a significant increase in iron uptake at all 5-aza-CdR studied (Fig. 1D, right). These observations indicate that 5-aza-CdR treatment results in a notable increase in iron uptake via the TfR1-mediated iron uptake pathway.
To study whether the increased expression of Alas2 and Fech is functional in terms of heme biosynthesis, the levels of heme, another marker of erythroid differentiation in MEL cells, were measured. Heme levels were significantly increased at all concentrations of 5-aza-CdR tested (Fig. 1E). The basal level of heme in MEL cells was 36.7 pmol/106 viable cells. After treatment with various concentrations of 5-aza-CdR, increases in intracellular heme levels of 3.5–8.2-fold relative to the control cells were observed (Fig. 1E). As expected, DMSO was found to be a very strong inducer of heme synthesis (up to 500 pmol/106 viable cells).
CpG Island Demethylation Is Not Involved in the Increased Expression of TfR1 and Fech after 5-Aza-CdR Treatment
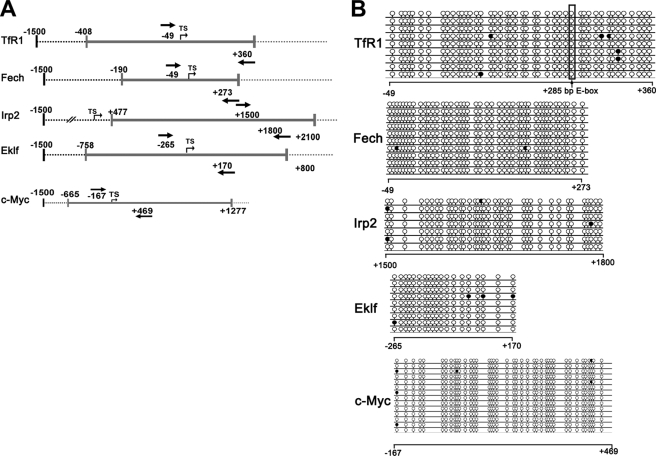
CpG islands are usually close to the transcription start sites (TSS) of specific genes and play important roles in the regulation of gene transcription (30). Methylated cytosine residues may prevent binding of transcription factors to TSS and consequently inhibit transcription of these genes, especially in proto-oncogenes or tumor suppressor genes (31). Demethylation of CpG islands is usually responsible for reactivation of the silenced genes (32). We analyzed 1.5 kb of DNA sequence up- and downstream of the TSS of TfR1, Alas2, and Fech by Methyl Primer Express Software version 1.0 (Applied Biosystems). CpG islands were found in both the TfR1 and Fech promoters (Fig. 2A) but not in the Alas2 promoter. Similar to some known genes with CpG islands (32), bisulfite sequencing showed hypomethylation of cytosine residues in the CpG islands of TfR1 and Fech in control MEL cells (Fig. 2B and supplemental Fig. S1, A and B). Furthermore, 5-aza-CdR and DMSO treatments did not change the methylation status of these CpG islands (data not shown). Thus, the possibility of direct regulation of methylation status in the CpG islands of TfR1 and Fech by 5-aza-CdR treatment was excluded.
FIGURE 2.
Bioinformatic analysis and bisulfite sequencing of CpG islands. A, schematic illustrations of the promoter regions of the genes of interest. CpG islands were predicted and analyzed by Methyl Primer Express Software version 1.0 (Applied Biosystems). The TSS of each gene is shown with an arrow, and gray lines indicate CpG islands. The positions of sequencing primers are shown with boldface arrows (numbers start from the TSS). B, genomic DNA of MEL cells was bisulfite-converted, PCR-amplified, and cloned. Single clones were sequenced. The sequencing results for each clone are shown, with lollipops representing cytosines. Methylated cytosines are shown as black lollipops, and unmethylated cytosines are shown as open lollipops.
If methylation of CpG islands is not critical for TfR1 and Fech promoter activity during erythroid differentiation, one or more of their upstream regulators could be subject to such control. There are several candidates. Gata1 and Eklf are considered key regulators of erythroid differentiation, and genome-wide analysis with ChIP has confirmed that TfR1 and Fech are targets of Eklf and Gata1 (33, 34). The transcription factor c-Myc has also been reported to activate TfR1 during tumorigenesis (18). In addition, the iron regulatory proteins (IRPs) are key factors coordinating intracellular iron homeostasis, and TfR1 mRNA stability is regulated by the IRPs. It is feasible that Eklf, Gata1, c-Myc, and the IRPs are regulated by demethylation of their CpG islands and that the observed effects on TfR1 and Fech are secondary to this. We therefore analyzed 1.5 kb of DNA sequences up- and downstream of the TSS of Eklf, Gata1, c-Myc, and Irp2 by Methyl Primer Express Software version 1.0. CpG islands were found in the Eklf, Irp2, and c-Myc promoters (Fig. 2A), but not in the Gata1 promoter. Similar to TfR1 and Fech, Eklf, Irp2, and c-Myc promoters were hypomethylated in their CpG islands in both control (Fig. 2B and supplemental Fig. S1, C–E) and 5-aza-CdR-treated MEL cells (data not shown). These results also excluded CpG island demethylation in modulating the expression of these key regulators of erythroid differentiation and iron metabolism.
Increased c-Myc Translocation to the Nucleus and Binding to Max after 5-Aza-CdR Treatment
Because Gata1, Eklf, and c-Myc are key regulatory factors in erythroid differentiation, we measured the expression of their mRNAs with quantitative real-time RT-PCR following 5-aza-CdR treatment (Fig. 3A). No significant differences in the expression of any of these transcription factors were observed at the mRNA level. Thus, 5-aza-CdR treatment did not change the expression levels of these transcription factors.
FIGURE 3.
5-Aza-CdR treatment led to c-Myc accumulation and increased binding to Max in nuclei. A, mRNA levels of c-Myc, Gata1, and Eklf were analyzed by quantitative real-time RT-PCR as described in Fig. 1A. B, analysis of the levels of the transcription factor c-Myc in MEL cells treated with or without 5-aza-CdR. MEL cells were collected and separated into two populations for the isolation of nuclei and total protein. c-Myc was detected in both the nuclear and total protein fractions. For co-immunoprecipitation of c-Myc-Max complexes, nuclear fractions were precipitated with anti-Max antibodies and c-Myc levels were detected in the precipitates by Western blotting with anti-c-Myc antibodies. β-Actin and IgG were included as controls. C, densitometry analysis of the levels of c-Myc (normalized to untreated cells). *, p < 0.01 compared with the control MEL cells. Error bars, S.E.
In Western blotting analysis, no significant changes were observed in Gata1 in 5-aza-CdR-treated cells compared with the control cells (data not shown). However, we consistently observed that the nuclear fraction of c-Myc was increased compared with the control cells after 5-aza-CdR treatment, despite no changes in total c-Myc levels (Fig. 3B). Furthermore, c-Myc translocation into the nucleus was positively correlated with the 5-aza-CdR concentration (Fig. 3B). To further investigate whether the accumulation of c-Myc in nuclei is functional, we then performed co-immunoprecipitation experiments to quantitatively measure the levels of c-Myc-Max complexes (Fig. 3, B and C). The Myc-Max complex is believed to be the functional form of c-Myc for specific binding to its targets and initiating transcription (35). Immunoprecipitation of nuclear extracts with anti-Max antibodies followed by Western blotting for c-Myc showed increased c-Myc in cells treated with 5-aza-CdR. This indicates that the enhanced translocation of c-Myc is probably functional in regulating the expression of target genes.
c-Myc Occupies Unmethylated E-box Sites in the Promoters of the TfR1 and Fech Genes
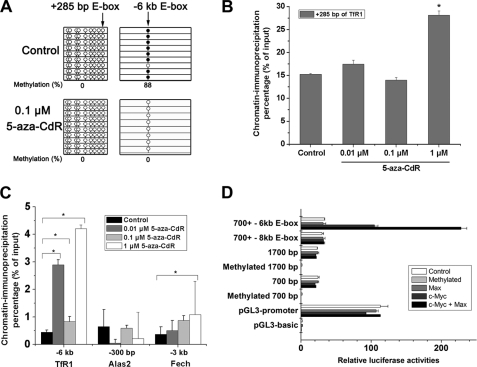
The activation of TfR1 by c-Myc has been reported in tumor cells (18, 19), and the binding sequence CACGTG (E-box) has been located +285 bp downstream of the TSS (18). However, bioinformatic analysis of 7 kb 5′ upstream of the TfR1 TSS indicated that there was one more putative E-box at −6 kb. E-boxes far away from the TSS may also be involved in transcriptional activation (27). DNA methylation is usually found on the cytosine of CG pairs, and thus the CG pairs in E-boxes (CACGTG) are candidates for methylation regulation. To determine whether c-Myc associates with unmethylated E-box sites of TfR1 and whether the E-box methylation status contributes to controlling c-Myc binding, the specific methylation status of E-boxes in the TfR1 gene was studied. Bisulfite sequencing results confirmed that +285 E-box was unmethylated in both normal and 5-aza-CdR-treated MEL cells (Fig. 4A). Interestingly, −6 kb E-box was almost fully methylated (88%) in normal MEL cells, and 5-aza-CdR treatment completely converted this E-box into unmethylated status (Fig. 4A). For comparison, these E-boxes in the TfR1 promoter were also investigated by the MS-PCR method. MEL cell genomic DNA was digested with PmlI and tested for specific methylation status using the MS-PCR assay. If the E-box is unmethylated, the endonuclease PmlI will cleave the E-box and prevent synthesis of the PCR product, whereas when the E-box is methylated, cleavage by PmlI is prevented, and synthesis of a specific PCR product can be measured (27). As shown in supplemental Fig. S6, we observed that after PmlI digestion, there was little amplification of the region containing the +285 bp E-box of TfR1. In contrast, DNA could not be digested by PmlI in either control or 5-aza-CdR (0.1 μm)-treated MEL cells at the −6 kb E-box of TfR1 (supplemental Fig. S6), indicating that this E-box was methylated. However, we found that the −6 kb E-box was partially digested by PmlI when the concentration of 5-aza-CdR rose to 1 μm. This observation further indicates that the E-box was methylated in control MEL cells and that this could be reversed by 5-aza-CdR treatment. Similar results were also observed in the E-boxes in the Alas2 (−300 bp) and Fech (−3 kb) promoters (supplemental Fig. S6).
FIGURE 4.
5-Aza-CdR treatment led to E-box demethylation and occupation by c-Myc in the promoters of the TfR1, Alas2, and Fech genes. A, bisulfite sequencing of the methylation status of the E-boxes in TfR1 promoters (+285 bp and −6 kb E-boxes) before and after 5-aza-CdR treatment. B, ChIP assay of the E-box in the CpG island of the TfR1 promoter (+285 bp). *, p < 0.01 compared with the control MEL cells. C, ChIP assay of E-boxes outside the CpG islands of the TfR1, Alas2, and Fech genes. *, p < 0.01 compared with the control MEL cells. A specific antibody to c-Myc was used for the ChIP assays of the TfR1, Fech, and Alas2 promoters, followed by sequence-specific PCR analysis of specific binding sites of c-Myc. D, transcriptional activities of the constructs containing different E-boxes of TfR1 promoters (+285, −6 kb, or −8 kb E-boxes). Luciferase activities of the constructs containing different fragments of TfR1 promoter, including a 700-bp fragment (−183 to +550), 1700-bp fragment (−1230 to +550), 700-bp fragment plus −6 kb E-box (−6267 to −6061), and 700-bp fragment plus −8 kb E-box (−8689 to −8421) were measured under co-transfection with the expression vectors of c-Myc, Max, or both. All of these E-boxes in these promoter fragments were in demethylated status. Luciferase activities of the constructs containing methylated 700- and 1700-bp fragments were also measured. Firefly luciferase activities were normalized by Renilla luciferase. Error bars, S.E.
To directly measure changes in c-Myc binding at these E-box positions after 5-aza-CdR treatment, ChIP was used. In the +285 bp E-box of the TfR1 promoter, constitutive occupation by c-Myc was observed (Fig. 4B). Only high dose 5-aza-CdR significantly increased the binding capacity of c-Myc at this E-box (Fig. 4B). This observation was also supported by the MS-PCR results (supplemental Fig. S6). The hypomethylation of E-boxes in CpG islands may be necessary for continuous activation of TfR1 in vivo (27). Thus, E-boxes could be occupied by c-Myc or other transcription factors to facilitate the transcription of TfR1. In contrast, the binding of c-Myc at the −6 kb E-box was much lower than binding to the +285 E-box in control MEL cells (Fig. 4C and supplemental Fig. S2). c-Myc binding at this site was increased significantly by 5-aza-CdR treatment (Fig. 4C and supplemental Fig. S2), consistent with demethylation. Elevated occupation of c-Myc was also observed in the promoter of Fech following 5-aza-CdR treatment (Fig. 4C). However, the binding of c-Myc to the Alas2 promoter was not increased by 5-aza-CdR. Importantly, neither the −6 kb E-box of the TfR1 promoter nor the −3 kb E-box of the Fech promoter were in CpG islands (Fig. 2A). Taken together, these results suggest that the increased expression of TfR1 and Fech correlates with enhanced c-Myc binding to demethylated E-boxes after 5-aza-CdR treatment.
To further address the direct connections between c-Myc binding to specific regulatory elements and induction of TfR1 expression, a series of constructs containing the different E-box sequences have been generated, followed by luciferase report assays when co-transfected with c-Myc, Max, or both. As shown in Fig. 4D, enforced expression c-Myc did not increase the transcriptional activities of the constructs containing 700-, 1700-, and 700-bp plus −8 kb E-box fragments but significantly enhanced the expression of the construct containing 700 bp plus −6 kb E-box fragment (more than 3-fold compared with the control). These results clearly demonstrated that increased binding of c-Myc to the specific regulatory element (−6 kb E-box, in demethylated status) is the primary cause of the increased gene expression of TfR1. Moreover, luciferase assays showed that the transcriptional activities of 700-bp, 1700-bp, and 700-bp plus −8 kb E-box constructs were similar when co-transfected with c-Myc and Max. Interestingly, co-transfection of c-Myc and Max further stimulated 700-bp plus −6 kb E-box construct expression, and the relative luciferase activity increased ∼6-fold compared with the control (−6 kb E-box only). This result indicated that the −6 kb E-box of the TfR1 promoter was a strong enhancer that could induce TfR1 expression when c-Myc and Max form functional complexes and bind to it. The methylation status of the −6 kb E-box was also analyzed by bisulfite sequencing (Fig. 4A) before and after 5-aza-CdR treatment. Similar to the MS-PCR results (supplemental Fig. S6), the −6 kb E-box was totally converted to demethylation status after 5-aza-CdR treatment.
5-Aza-CdR Stimulates Gene Expression through Increased Binding of c-Myc to E-boxes of TfR1, Alas2, and Fech in BFU-E-derived Erythroblasts
Although MEL cells are a good model to study erythroid differentiation and heme biosynthesis, they have their limitations (36). Furthermore, our data suggest that the mechanisms of erythroid differentiation induced by DMSO and 5-aza-CdR are different because c-Myc-Max complexes did not increase after DMSO treatment but did after 5-aza-CdR (Fig. 3B). To confirm our observations, BFU-E-derived erythroblasts from mouse bone marrow were used to analyze the effects of 5-aza-CdR. We used lower concentrations of 5-aza-CdR in this study due to the high sensitivity of the primary cells to 5-aza-CdR treatment (37). The levels of TfR1, Alas2, and Fech mRNA increased in 5-aza-CdR-treated BFU-Es (Fig. 5A). The effects of 5-aza-CdR onTfR1, Alas2, and Fech protein levels were less pronounced, but all were increased, particularly at the 0.1 μm concentration (Fig. 5B and Fig. S4). Direct binding of c-Myc to E-boxes in the promoters of the target genes in BFU-Es was measured by ChIP (Fig. 5C and Fig. S5). Binding to the E-boxes at +285 bp and −6 kb of the TfR1 promoter was demonstrated in both control and 5-aza-CdR-treated BFU-Es. However, only at the −6 kb E-box was c-Myc binding significantly increased by 5-aza-CdR treatment. These results indicate that increased c-Myc binding to the E-boxes in response to 5-aza-CdR treatment was directed specifically to the −6 kb E-box of the TfR1 promoter in BFU-Es.
FIGURE 5.
5-Aza-CdR stimulated the expression of specific genes associated with iron uptake and heme synthesis in mouse BFU-Es. Murine bone marrow cells were cultured in MethoCult semisolid medium with different concentrations of 5-aza-CdR for 24 h. The cells were cultured for a further 8 days in the absence of 5-aza-CdR. BFU-Es were collected for quantitative real-time RT-PCR (A), Western blotting analysis (B), and status of c-Myc binding to the TfR1 promoter (C) as described in the legends to Figs. 1 (A and B) and 4B, respectively. Error bars, S.E.
DISCUSSION
Demethylating agents, such as 5-azacytidine and 5-aza-CdR, are FDA-approved drugs for treatment of MDS. These compounds have been shown to significantly prolong the survival of MDS patients and reduce the risk of transformation to AML (6). Anemia is the most common early clinical feature of MDS; however, this anemia resolves in ∼40% of MDS patients in response to 5-aza-CdR treatment. Why this occurs is not known (1). Our current study demonstrated that 5-aza-CdR can specifically stimulate the expression of several genes involved in cellular iron uptake and heme biosynthesis. This direct link between 5-aza-CdR treatment and increased heme synthesis in both MEL cells and BFU-Es may indicate an important action of this MDS drug on erythroid differentiation and particularly on intracellular iron metabolism.
The current working hypothesis for the action of demethylating agents in MDS treatment is that they reactivate tumor suppressor genes, such as p15, through demethylation of the CpG islands in the promoter regions of these genes (7). However, increasingly it is being recognized that mechanisms other than demethylation of CpG islands may play important roles in the biological actions of 5-aza-CdR (38). We have found that c-Myc can specifically bind to demethylated E-boxes in the TfR1 and Fech promoters after 5-aza-CdR treatment, followed by increased expression of these genes. These new molecular targets may explain the differential responses in patients of MDS classifications (39). The binding motifs of c-Myc in the TfR1 and Fech promoters strongly support the notion that demethylation sites that are not in classical CpG islands play important roles in 5-aza-CdR therapy.
The TfR1-mediated uptake of diferric transferrin is the major mechanism for cellular iron acquisition under normal physiological conditions (29). TfR1 is also a well known marker in erythroid differentiation, and several erythroid-related transcription factors have been shown to regulate its expression (33, 34). Although it has been reported that 5-aza-CdR can induce erythroid progenitors to terminal differentiation in AML or anemia patients (11, 37), how it does so is poorly understood. The oncogene c-Myc has been found to be involved in iron metabolism, and the TfR1, ferritin, IRP2 (20), and Nramp1 (40) genes have all been reported to be targets of c-Myc. The +511 bp E-box in the human TfR1 gene and the +285 bp E-box in the mouse TfR1 gene (both relative to the TSS) have been found to mediate the activation of TfR1 by c-Myc (18). In the current study, we observed the occupation of this E-box by c-Myc in both control MEL cells and BFU-Es. The binding of c-Myc to this site did not increase after 5-aza-CdR treatment, indicating its continuous occupation by c-Myc (Figs. 4 and 5). Furthermore, regulatory targets of c-Myc have been found both within and outside CpG islands. In pancreatic cancer cells, downstream targets of N-Myc, whose binding site (CACGTG) is the same as that of c-Myc, were found to be up-regulated by 5-aza-CdR treatment in the absence of CpG island demethylation (38). In addition, CpG island-independent effects of 5-aza-CdR in AML cells support the indications that it is possible to regulate TfR1 at an E-box outside a CpG island (9). Interestingly, in our current study, the −6 kb E-box of the TfR1 promoter showed increased c-Myc occupation after 5-aza-CdR treatment. Bioinformatic analysis showed that its low GC content made this E-box a potential methylation-related but CpG island-independent regulatory element. When the cytosine in the E-box is demethylated by 5-aza-CdR, more c-Myc would occupy the E-box, and the expression of TfR1 would be increased. We also found the conserved −6 kb E-box in the human TfR1 promoter (supplemental Fig. S3). Recent research with MDS patients treated with 5-aza-CdR showed hematologic improvement but no global decreases in DNA methylation (41). In addition, there is no strong correlation between the demethylation of CpG islands and the effectiveness of MDS therapy when hypomethylation agents are used (6). Our data support the hypothesis that CpG island-independent effects of 5-aza-CdR are involved in mediating its therapeutic efficacy.
A second possibility for the enhanced TfR1 gene expression by 5-aza-CdR treatment is via increased c-Myc nuclear localization. No doubt, c-Myc nuclear localization is a necessary step, but it may not be a sufficient step for the enhanced gene expression. Presumably, when c-Myc was translocated into the nuclei, it would form a functional complex with the Max protein, followed by specific binding to the E-boxes within the TfR1 promoter. As shown previously (20, 40), ferritins, IRP2, and Nramp1 are all c-Myc targets; however, we only observed an increase in TfR1 levels after 5-aza-CdR treatment. This may indicate that accessibility (specific gene sequences and their proper conformation) of the E-boxes in these genes plays key roles in enhanced gene expression. There are three putative c-Myc binding E-boxes in the TfR1 promoter, but enforced c-Myc expression did not lead to enhanced reporter gene expression in either +285 bp or −8 kb E-boxes (700-bp, 1700-bp, and 700-bp plus −8 kb vectors in Fig. 4D). Furthermore, when the TfR1 promoter fragments were first treated with CpG DNA methyltransferase M.SssI (recognizing the double-stranded dinucleotide sequence 5′ … CG … 3′ specifically), no transcriptional activities of these TfR1 promoter fragments were observed (Fig. 4D). Therefore, increased c-Myc binding is the determining factor for enhanced gene expression. In turn, c-Myc nuclear translocation and proper DNA methylation status of the E-box elements collectively contribute to the increased binding of c-Myc to the E-boxes.
In the current study, we also observed increased expression of Alas2 and Fech in both MEL cells and BFU-Es after 5-aza-CdR treatment. Aminolevulinic acid synthase 1 (Alas1) has been proposed as the rate-limiting enzyme in heme synthesis in non-erythroid cells. In contrast, in erythroid cells, intracellular iron supply, rather than ALAS activity, limits the rate of heme synthesis (42). Therefore, the contribution of increased Alas2 and Fech expression to enhanced heme and hemoglobin biosynthesis may be less significant than the contribution of enhanced TfR1 expression. Both Fech and Alas2 are predicted targets of c-Myc based on earlier studies (43). Like TfR1, the CpG island in the Fech promoter was demethylated, and the E-box is located outside the CpG island. In the Alas2 promoter region, there is no CpG island. Further research to examine the similarity of TfR1 and these two genes is warranted.
In conclusion, we have found new regulatory E-boxes outside CpG islands in the TfR1, Fech, and Alas2 promoters. These elements were methylated in MEL cells and BFU-Es and can be demethylated by 5-aza-CdR treatment. During erythroid differentiation induced by 5-aza-CdR, c-Myc stimulated the expression of genes involved in intracellular iron metabolism by occupying hypomethylated E-boxes, which are independent of CpG islands. These results provide new molecular insights into 5-aza-CdR therapy in MDS patients. Further investigation of the 5-aza-CdR-induced erythroid response with primary erythroid cells from MDS patients is likely to be informative for 5-aza-CdR therapy for MDS.
This work was supported by National Basic Research Program of China (973 program) Grants 2012CB934000, 2011CB933400, and 2010CB912802 and National Natural Science Foundation of China Grants 30900278 and 10979011.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S4 and S5.
- MDS
- myelodysplastic syndrome(s)
- MEL
- murine erythroleukemia
- 5-aza-CdR
- 5-aza-2′-deoxycytidine
- BFU-E
- burst-forming unit
- AML
- acute myeloid leukemia
- Tf
- transferrin
- MS-PCR
- methylation-sensitive PCR
- IRP
- iron regulatory protein
- TSS
- transcription start site(s)
- TfR1
- transferrin receptor 1
- Eklf
- erythroid kuffer like factor
- Ftl
- ferritin L-subunit
- Alas2
- aminolevulinic acid synthase 2
- Fech
- ferrochelatase
- Hba1
- adult hemoglobin alpha 1
- Hbb1
- adult hemoglobin beta 1
- Hmox1
- heme oxygenase 1
- Irp2
- iron regulatory protein 2.
REFERENCES
- 1. Nimer S. D. (2008) Blood 111, 4841–4851 [DOI] [PubMed] [Google Scholar]
- 2. Tefferi A., Vardiman J. W. (2009) N. Engl. J. Med. 361, 1872–1885 [DOI] [PubMed] [Google Scholar]
- 3. Aul C., Bowen D. T., Yoshida Y. (1998) Haematologica 83, 71–86 [PubMed] [Google Scholar]
- 4. Hofmann W. K., Koeffler H. P. (2005) Annu. Rev. Med. 56, 1–16 [DOI] [PubMed] [Google Scholar]
- 5. Jädersten M., Hellström-Lindberg E. (2010) Exp. Cell Res. 316, 1390–1396 [DOI] [PubMed] [Google Scholar]
- 6. Quintás-Cardama A., Santos F. P., Garcia-Manero G. (2010) Nat. Rev. Clin. Oncol. 7, 433–444 [DOI] [PubMed] [Google Scholar]
- 7. Daskalakis M., Nguyen T. T., Nguyen C., Guldberg P., Köhler G., Wijermans P., Jones P. A., Lübbert M. (2002) Blood 100, 2957–2964 [DOI] [PubMed] [Google Scholar]
- 8. Fandy T. E., Herman J. G., Kerns P., Jiemjit A., Sugar E. A., Choi S. H., Yang A. S., Aucott T., Dauses T., Odchimar-Reissig R., Licht J., McConnell M. J., Nasrallah C., Kim M. K., Zhang W., Sun Y., Murgo A., Espinoza-Delgado I., Oteiza K., Owoeye I., Silverman L. R., Gore S. D., Carraway H. E. (2009) Blood 114, 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmelz K., Sattler N., Wagner M., Lübbert M., Dörken B., Tamm I. (2005) Leukemia 19, 103–111 [DOI] [PubMed] [Google Scholar]
- 10. Creusot F., Acs G., Christman J. K. (1982) J. Biol. Chem. 257, 2041–2048 [PubMed] [Google Scholar]
- 11. Pinto A., Attadia V., Fusco A., Ferrara F., Spada O. A., Di Fiore P. P. (1984) Blood 64, 922–929 [PubMed] [Google Scholar]
- 12. Valdez B. C., Li Y., Murray D., Corn P., Champlin R. E., Andersson B. S. (2010) Leuk. Res. 34, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotkow K. J., Orkin S. H. (1995) Mol. Cell. Biol. 15, 4640–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weiss M. J., Orkin S. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9623–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crossley M., Tsang A. P., Bieker J. J., Orkin S. H. (1994) J. Biol. Chem. 269, 15440–15444 [PubMed] [Google Scholar]
- 16. Perkins A. C., Sharpe A. H., Orkin S. H. (1995) Nature 375, 318–322 [DOI] [PubMed] [Google Scholar]
- 17. Dang C. V. (1999) Mol. Cell. Biol. 19, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Donnell K. A., Yu D., Zeller K. I., Kim J. W., Racke F., Thomas-Tikhonenko A., Dang C. V. (2006) Mol. Cell. Biol. 26, 2373–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Okazaki F., Matsunaga N., Okazaki H., Utoguchi N., Suzuki R., Maruyama K., Koyanagi S., Ohdo S. (2010) Cancer Res. 70, 6238–6246 [DOI] [PubMed] [Google Scholar]
- 20. Wu K. J., Polack A., Dalla-Favera R. (1999) Science 283, 676–679 [DOI] [PubMed] [Google Scholar]
- 21. Sridhar K., Ross D. T., Tibshirani R., Butte A. J., Greenberg P. L. (2009) Blood 114, 4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mills K. I., Kohlmann A., Williams P. M., Wieczorek L., Liu W. M., Li R., Wei W., Bowen D. T., Loeffler H., Hernandez J. M., Hofmann W. K., Haferlach T. (2009) Blood 114, 1063–1072 [DOI] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Ponka P., Schulman H. M. (1985) J. Biol. Chem. 260, 14717–14721 [PubMed] [Google Scholar]
- 25. Oshiro S., Nakamura Y., Ishige R., Hori M., Nakajima H., Gahl W. A. (1994) J. Biochem. 115, 849–852 [DOI] [PubMed] [Google Scholar]
- 26. Sinclair P. R., Gorman N., Jacobs J. M. (2001) in Current Protocols in Toxicology, pp. 8.3.1–8.3.7, John Wiley & Sons, Inc., New York: [DOI] [PubMed] [Google Scholar]
- 27. Perini G., Diolaiti D., Porro A., Della Valle G. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12117–12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christman J. K., Weich N., Schoenbrun B., Schneiderman N., Acs G. (1980) J. Cell Biol. 86, 366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ponka P. (1997) Blood 89, 1–25 [PubMed] [Google Scholar]
- 30. Bird A. P. (1986) Nature 321, 209–213 [DOI] [PubMed] [Google Scholar]
- 31. Jaenisch R., Bird A. (2003) Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 32. Suzuki M. M., Bird A. (2008) Nat. Rev. Genet. 9, 465–476 [DOI] [PubMed] [Google Scholar]
- 33. Welch J. J., Watts J. A., Vakoc C. R., Yao Y., Wang H., Hardison R. C., Blobel G. A., Chodosh L. A., Weiss M. J. (2004) Blood 104, 3136–3147 [DOI] [PubMed] [Google Scholar]
- 34. Tallack M. R., Whitington T., Yuen W. S., Wainwright E. N., Keys J. R., Gardiner B. B., Nourbakhsh E., Cloonan N., Grimmond S. M., Bailey T. L., Perkins A. C. (2010) Genome Res. 20, 1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pelengaris S., Khan M., Evan G. (2002) Nat. Rev. Cancer 2, 764–776 [DOI] [PubMed] [Google Scholar]
- 36. Gusella J. F., Weil S. C., Tsiftsoglou A. S., Volloch V., Neumann J. R., Keys C., Housman D. E. (1980) Blood 56, 481–487 [PubMed] [Google Scholar]
- 37. Humphries R. K., Dover G., Young N. S., Moore J. G., Charache S., Ley T., Nienhuis A. W. (1985) J. Clin. Invest. 75, 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Angst E., Dawson D. W., Nguyen A., Park J., Go V. L., Reber H. A., Hines O. J., Eibl G. (2010) Pancreas 39, 675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fenaux P., Mufti G. J., Hellstrom-Lindberg E., Santini V., Finelli C., Giagounidis A., Schoch R., Gattermann N., Sanz G., List A., Gore S. D., Seymour J. F., Bennett J. M., Byrd J., Backstrom J., Zimmerman L., McKenzie D., Beach C., Silverman L. R. (2009) Lancet Oncol. 10, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bowen H., Biggs T. E., Phillips E., Baker S. T., Perry V. H., Mann D. A., Barton C. H. (2002) J. Biol. Chem. 277, 34997–35006 [DOI] [PubMed] [Google Scholar]
- 41. Soriano A. O., Yang H., Faderl S., Estrov Z., Giles F., Ravandi F., Cortes J., Wierda W. G., Ouzounian S., Quezada A., Pierce S., Estey E. H., Issa J. P., Kantarjian H. M., Garcia-Manero G. (2007) Blood 110, 2302–2308 [DOI] [PubMed] [Google Scholar]
- 42. Laskey J. D., Ponka P., Schulman H. M. (1986) J. Cell. Physiol. 129, 185–192 [DOI] [PubMed] [Google Scholar]
- 43. Fernandez P. C., Frank S. R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., Amati B. (2003) Gene Dev. 17, 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]