Background: The mechanisms used by chemokine CXCL12 to regulate the functions of mature dendritic cells are unknown.
Results: CXCL12 controls chemotaxis and survival in these cells using CXCR4 and bifunctional signaling molecules.
Conclusion: CXCR4 uses a redundant signaling pathway to control chemotaxis and survival in mature dendritic cells.
Significance: CXCR4 may use a specific signaling signature to regulate the functions of dendritic cells.
Keywords: Apoptosis, Chemokines, Chemotaxis, Dendritic Cells, mTOR Complex (mTORC)
Abstract
Chemokines control several cell functions in addition to chemotaxis. Although much information is available on the involvement of specific signaling molecules in the control of single functions controlled by chemokines, especially chemotaxis, the mechanisms used by these ligands to regulate several cell functions simultaneously are completely unknown. Mature dendritic cells (maDCs) migrate through the afferent lymphatic vessels to the lymph nodes, where they regulate the initiation of the immune response. As maDCs are exposed to chemokine CXCL12 (receptors CXCR4 and CXCR7) during their migration, its functions are amenable to be regulated by this ligand. We have used maDCs as a model system to analyze the mechanisms whereby CXCL12 simultaneously controls chemotaxis and survival in maDCs. We show that CXCL12 uses CXCR4, but not CXCR7, and the components of a signaling core that includes Gi/Gβγ, PI3K-α/-δ/-γ, Akt, ERK1/2 and mammalian target of rapamycin complex 1 (mTORC1), which organize hierarchically to control both functions. Downstream of Akt, Forkhead box class O (FOXO) regulates CXCL12-dependent survival, but not chemotaxis, suggesting that downstream of the aforementioned signaling core, additional signaling molecules may control more selectively CXCL12-dependent chemotaxis or survival. Finally, the data obtained also show that CXCR4 uses a signaling signature that is different from that used by CCR7 to control similar functions.
Introduction
Most studies on chemokines have focused in their chemoattractant properties; however, chemokines may also regulate survival, migratory speed, fugetaxis, endocytosis, cytoarchitecture, and differentiation in leukocytes, suggesting that these ligands may contribute to modulate the response of dendritic cells (DCs)5 in the immune system (1). An important unsolved issue is the mechanisms that allow chemokines regulate several functions in leukocytes simultaneously. Understanding these processes may lead to a more selective modulation of the functions under the control of the chemokines and, consequently, the response of DCs in the immune system.
To regulate the initiation of the immune response, mature dendritic cells (maDCs) have to migrate through the afferent lymphatics, to the lymph nodes (LNs). It is known that the migration of maDCs to the LNs is controlled by chemokines CCL19 and CCL21, both ligands of the chemokine receptor CCR7 (2). Recently, it has been suggested that chemokine CXCL12 (receptors CXCR4 and CXCR7), which is a strong chemoattractant for maDCs in vitro (3, 4), also regulates in vivo the migration of these cells from the skin to regional LNs (5, 6). In this regard, CXCL12 is expressed in tissues through which DCs traffic, including dermal fibroblasts, lymphatic vessels of the skin, and medullar cells in the LNs (6–9). CXCL12 expression may be even further up-regulated in these regions upon exposure to inflammatory stimuli (7, 10). Apart from chemotaxis, it has also been suggested that CXCL12 regulates DC survival (11, 12). However, the relative contribution of chemotaxis and survival to the migration of maDCs to the LNs has not been analyzed.
As CXCL12 recognizes receptors CXCR4 and CXCR7 (13–15), its functional effects in DCs could be mediated by either one of the two or by both receptors. CXCR4 is a well established signaling receptor expressed in maDCs (14), which regulates chemotaxis, fugetaxis, survival, or apoptosis in different cell types (16–19). CXCR7 expression pattern, signaling, and functional capabilities are more controversial (20). Although it was indicated earlier that CXCR7 is expressed in DCs (15), recently it has been reported that it is not expressed in leukocytes (21). The ability of CXCR7 to regulate chemotaxis seems to be cell-dependent because this receptor controls this function only in specific cell types (22, 23).
Studies carried out during the last years have provided a great deal of information on the signaling molecules used by chemokines to regulate its functions. Chemokine receptors use the Gi subfamily of G proteins and Gβγ dimers of these proteins to regulate chemotaxis and other functions (24, 25). Chemokine receptors can also induce activation of the phosphatidylinositol 3-kinase (PI3K) pathway (26–29). These enzymes are classified in class PI3K-IA, which includes PI3K-α, -β and -δ, and class PI3K-IB, which includes PI3K-γ. Although there is sparse information available on the role of specific PI3K isoforms in the regulation chemokine-dependent functions, it is generally accepted that PI3K-γ mediates signaling from G protein-coupled receptors, including chemokine receptors (30), whereas PI3K-α,-β, and -δ mediate signaling from tyrosine kinase receptors (31, 32). Class I PI3Ks catalyze the phosphorylation of phosphatidylinositol 4,5-bisphosphate to generate the lipid second messenger phosphatidylinositol 3,4,5-trisphosphate. This lipid, which is recognized by the pleckstrin homology domain of the serine-threonine kinase Akt, positions this kinase on the membrane so it can be subsequently activated by PDK1 (3-phosphoinositide-dependent protein kinase 1) and mTORC2 (33). Akt is a kinase that regulates survival in some settings and chemotaxis in others (34–36). The effects of Akt can be mediated by several effectors, including the transcription factor FOXO family (FOXO1, FOXO3a, and FOXO4 members) and the mammalian target of rapamycin complex 1 (mTORC1) (33). In maDCs, under chronic stress conditions, FOXO1 translocates to the nucleus, where it regulates the transcription of proapoptotic molecules, including the Bcl-2 family member Bim (28, 29). Upon phosphorylation/inactivation by Akt1, FOXO1 shuttles to the cytoplasm, where it is prevented from up-regulating the expression of Bim. Apart from regulating FOXO, Akt also controls the activation of mTORC1 by modulating the activity of the tuberous sclerosis protein 2 (TSC2) (33). This molecule contains a GTPase-activating protein domain that stimulates the activity of the small GTPase RheB, which is a direct activator of mTORC1. The phosphorylation/inhibition of TSC2 by Akt results in RheB-GTP loading and mTORC1 activation. The latter kinase, in turn, phosphorylates the ribosomal protein S6 kinase (s6K) and the translational regulator eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) (33). Finally, chemokine receptors may also induce activation of the extracellular signal-regulated kinases 1 and 2 (ERK1/2) (34), which regulate chemotaxis, random motility, or survival in different cell types (37).
Although the aforementioned studies have provided a large amount of information on the signaling molecules activated by chemokine receptors, there is almost no information available on the mechanisms whereby a single chemokine receptor can simultaneously control more than one of its functions. This information is important to modulate the function of these receptors and the immune response. Our main goal in this work was to identify the mechanisms used CXCL12 to regulate chemotaxis and survival simultaneously in maDCs. We show that CXCL12 uses CXCR4 but not CXCR7, and a core of bifunctional signaling molecules to control chemotaxis and survival simultaneously in maDCs. Finally, confirming the specificity of the signaling pathways identified, the results also show that CXCR4 uses a signaling mechanism that is different from that used by CCR7 to control similar functions.
EXPERIMENTAL PROCEDURES
Reagents
GM-CSF, IL4, and TNF-α were purchased from Immunotools (Friesoythe, Germany). CMFDA and carboxyfluorescein diacetate were from Molecular Probes. CXCL12, CCL19, and CCL21 were obtained from Peprotech (Rocky Hill, NJ). BSA, poly-l-lysine, LY294002, pertussis toxin (PTX), Hoechst 33342, and the anti-β-actin antibody were from Sigma. Propidium iodide, UO126, PD0325901, the PI3K-α inhibitor PIK-75 (Calbiochem-Merck inhibitor VIII), the PI3K-β TGX-221 (Calbiochem-Merck inhibitor VI), and the PI3K-γ inhibitor AS-605240 were from Calbiochem-Merck. The PI3K-δ inhibitor IC87114 was obtained from Calistoga Pharmaceuticals (Seattle, WA). Rapamycin was obtained from Tocris Bioscience (Ellisville, MO). The polycaspase inhibitor Z-Val-Ala-Asp(OMe)-fluoromethyl ketone (Z-VAD-FMK) was from Biomol (Plymouth Meeting, PA). Annexin V-FITC was from BD Pharmingen. The anti-ERK2 (C14) and the anti-βArk (H222) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-phospho-ERK1/2 (Thr-201/Tyr-204), the anti-phospho-4E-BP1 (Thr-37/Thr-46), the anti-phospho-Akt1 (Ser-473), the anti-phospho-S6K (Thr-389), the anti-phosphotuberin/TSC2 (Thr-1462) (5B12), the anti-phospho-mTOR (Ser-2448), the anti-phospho-mTOR (Ser-2481), and the anti-PI3K p110β were from Cell Signaling Technology (Beverly, MA). The anti-human CXCR4 mAb and the phycoerythrin-conjugated anti-human CCR7 were from R&D Systems. The phycoerythrin-conjugated anti-human CD86, anti-human CD83, and isotype control mAbs were from BD Pharmingen. The anti-CXCR7 mAb clone 9C4 was a gift from Marcus Thelen. The CXCR7 inhibitor CCX733 and the anti-CXCR7 mAb clone 11G8 were gifts from Dr. Thomas Schall.
Mice
C57BL/6 mice (8 weeks old) were maintained in the animal facility at the Centro de Investigaciones Biológicas and treated according to Animal Care Committee guidelines.
Cells and Culture Conditions
Monocyte obtained from human peripheral blood mononuclear cells were isolated from buffy coats from healthy donors over a Lymphoprep (Nycomed) and induced to differentiate to immature DCs by adding GM-CSF and IL4 (both at 1000 units/ml) for 7 days as indicated before (27, 28, 38). To these immature DCs it was added for a further 72-h period GM-CSF, IL4, and 50 ng/ml TNF-α to obtain maDCs (26, 28, 29, 38).
Measurement of Chemotaxis Using Transwell Assays
Chemotaxis in response to chemokines was determined by measuring the number of migrated cells through a polycarbonate filter with 5-μm pore size in 24-well Transwell chambers (COSTAR). The upper chamber included 1 × 105 DCs diluted in 100 μl of RPMI 1640 medium and 0.1% BSA, and the lower chamber contained 600 μl of the same medium with or without CXCL12 (at 100 ng/ml) or, in some experiments, CCL21 (at 200 ng/ml). DCs that migrate to the bottom chamber after 2 h at 37 °C were counted by flow cytometry using the CellQuest software (Becton Dickinson). When pharmacological inhibitors were used, these agents were also added to the lower chambers of the Transwell.
Analysis of Migratory Speed, Endocytosis, and Static Adhesion Assays
Migratory speed (29), endocytosis (39), and static adhesion (40) analysis was performed as indicated before.
Assays of Apoptotic Damage
An equal number of viable DCs (determined by exclusion on trypan blue staining) were incubated in RPMI 1640 medium for 40 h in the presence or absence of CXCL12 (100 ng/ml) and the inhibitors indicated, and then harvested. Apoptotic nuclear morphology was assessed using Hoechst 33342 staining as indicated before (26). Apoptotic (annexin+/propidium iodide−) DCs were stained with FITC-conjugated annexin V and propidium iodide as indicated before (26).
Flow Cytometry
Cytometry analysis was performed as described before (38). Briefly, human monocyte-derived DCs were preincubated with human poly-IgGs (50 μg/ml) and then stained with the anti-CXCR4 mAb (clone 44717, at 2 μg/ml), the anti-CXCR7 mAbs (clones 11G8 and 9C4, both at 10 μg/ml), or the mouse IgG1 isotype control mAbs on ice, for 30 min, and then rinsed with cold buffer (PBS containing 2% BSA). Cells were then incubated on ice for 30 min with Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes), rinsed with cold buffer, and analyzed on a FACScan cytometer using CellQuest software.
RT-PCR
Total RNA, isolated with RNeasy columns (Qiagen), was obtained from maDCs obtained from independent donors and from CXCR7+ HeLa cells, which were used as positive controls. cDNA was synthesized using the High Fidelity cDNA synthesis kit (Roche Applied Science). The cDNA was PCR-amplified with Taq polymerase (Euroclone) using the following oligonucleotide CXCR7 primers (15): 5′TGGTCAGTCTCGTGCAGCAC-3′ and 5′-GCCAGCAGACAAGGAAGACC-3′. Control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was amplified using oligonucleotides 5′-GGCTGAGAACGGGAAGCTTGTCA-3′ and 5′-CGGCCATCACGCCACAGTTTC-3′. The PCR cycle consisted of initial denaturation at 95 °C for 5 min followed by 35 cycles consisting of denaturation at 95 °C for 30 s, annealing at 58 °C for 45 s, and extension at 72 °C for additional 45 s. A final extension step at 72 °C for 10 min was included. PCR products were resolved by gel electrophoresis.
Analysis of the Presence of GFP in the Nucleus of the GFP-transfected DCs
Green fluorescent protein (GFP)-vector or FOXO1-GFP-transfected DCs (50 × 103 cells), suspended in complete medium, were plated onto poly-l-lysine-coated coverslips. The cells were then fixed in 4% paraformadehyde in PBS (10 min at room temperature) and subsequently permeabilized with 0.2% Triton X-100 (10 min at room temperature). DCs were then stained with Hoechst 33342 (2 μm) to assess the position of the nucleus. Coverslips were washed with PBS and distilled water and then mounted in fluorescent mounting medium (DAKO). Cells were analyzed using a Nikon Eclipse E800 microscope. GFP-expressing cells were analyzed using the FITC channel and Hoechst 33342 using UV light.
Cell Lysis and Western Blot Analysis
DCs (100 × 103 cells) were stimulated or not with CXCL12 (100 ng/ml) for the indicated periods of time. Lysis of cells and Western blotting were performed as indicated before (26, 28, 29, 38).
Expression Vectors, siRNAs, and Nucleofections
The pEGFP-C1 (GFP) expression vector was from Clontech. The GFP-FOXO1 and βArk-CT vectors have been described previously (26, 28, 41, 42). Random control and FOXO1 and FOXO3 siRNAs were obtained from Santa Cruz Biotechnology. The plasmidic DNAs and the siRNAs were transfected with the Amaxa nucleoporator system (Amaxa, Koeln, Germany) following the manufacturer's instructions.
Purification of Murine DCs and Labeling of the Cells with Fluorescent Cell Trackers
Murine DCs were purified (97% CD11c) from spleens of donor mice using magnetic beads (Miltenyi Biotec) following the manufacturer's protocol. Murine DCs used in the in vivo studies were labeled for 30 min at 37 °C with 5 μm fluorescent cell tracker probe CMFDA in 0.1% BSA in PBS and then matured with LPS (1 μg/ml).
Analysis of the Migration of CMFDA-DC to the LNs of C57BL/6 Mice
CMFDA-labeled splenic DCs (2 × 106) were dissolved in 25 μl of RPMI 1640 medium, including 1 μg/ml LPS, and then mixed either with PBS (in the control animals) or with a dose of 4 mg/kg AMD3100. Subsequently, the DCs were injected subcutaneously into the hind footpad of recipient C57BL/6 mice. After 18 h, when the DCs had already reached the LNs (43), animals were killed, and the popliteal LNs (PLNs) were extracted. The LNs were dissolved in 5 mm EDTA and then passed through the mesh of a cell strainer. Finally, the cells were centrifuged and pellets dissolved in 10% FCS/RPMI 1640 medium. Flow cytometry was used to assess the percentage of CMFDA-DCs relative to the total number of cells in the LNs.
Two-photon Microscopy Analysis of Apoptotic DCs in LNs
CMFDA-labeled splenic DCs (2 × 106) were dissolved in 25 μl of RPMI 1640 medium including 1 μg/ml LPS and then injected subcutaneously into the hind footpad of recipient C57BL/6 mice. After 18 h treated or control animals were injected intraperitoneally with the CXCR4 inhibitor AMD3100 (4 mg/kg in PBS) or with PBS, respectively. After an additional 22 h, animals were injected intravenously with sulforhodamine B (SR)-FLIVO (λabs 565 nm; λem >600 nm), a reagent that binds irreversibly to caspases up-regulated in apoptotic cells, allowing a selective detection of the latter cells in the LNs by two-photon microscopy. Finally, the mice were killed, and the PLNs were extracted and subsequently subjected to two-photon confocal analysis to visualize among the injected CMFDA-DCs those that present SR-FLIVO staining. Two-photon microscopy analysis was performed as indicated in detail before (28, 29). See additional experimental details in supplemental Experimental Procedures.
Statistics
Data are expressed as mean ± S.E. (n), with n indicating the number of independent experiments performed. Unless otherwise indicated, all data were acquired for at least three independent experiments. Significance of differences between two series of results was assessed using the Student's unpaired t test. Values of p < 0.05 were considered significant.
RESULTS
CXCL12 Uses CXCR4, but Not CXCR7, to Control Chemotaxis and Survival in Human maDCs
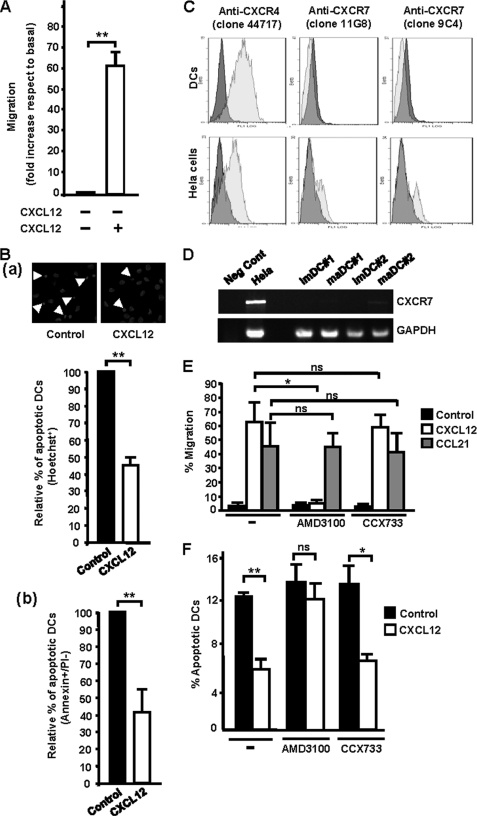
Before analyzing the signaling triggered by CXCL12, we confirmed that this chemokine regulates chemotaxis and survival in TNFα-matured maDCs in vitro and in vivo. Chemotactic assays performed in Transwells confirmed that CXCL12 induced a potent chemotactic response in human monocyte-derived maDCs (Fig. 1A). To determine whether CXCL12 protected the human maDCs from apoptosis, cells were maintained in serum-free medium with or without chemokine. Subsequently, DCs were stained with Hoechst 33342 to assess the percentage of cells that presented condensed or fragmented nucleus, both hallmarks of apoptotic cells. Compared with untreated DCs, stimulation with CXCL12 led to a reduction of almost 55% in the percentage of apoptotic DCs (Fig. 1Ba). Similar protective effects of CXCL12 were observed when apoptosis was assessed by determining the percentage of DCs that display annexin+/propidium iodide− staining, another established apoptotic marker (Fig. 1Bb). Finally, CXCL12, under our conditions, did not modulate migratory speed, fugetaxis, static adhesion, endocytic ability, or maturation of the human maDCs (supplemental Fig. 1).
FIGURE 1.
CXCL12 regulates chemotaxis and survival in human maDCs. A, DCs were allowed to migrate toward CXCL12 in Transwell assays as described under “Experimental Procedures.” Results shown represent the mean ± S.E. (error bars; n = 60). **, p < 0.001. B, DCs were suspended in 0.1% BSA/RPMI 1640 medium and then incubated either with or without CXCL12 for 40 h. a upper, DCs were fixed and stained with Hoechst 33342, and photographs were taken from representative samples. Arrowheads indicate apoptotic DCs. Lower, percentage of apoptotic DCs in RPMI plus CXCL12 is shown with respect to the percentage of apoptotic DCs in RPMI, which was given an arbitrary value of 100. Results represent the mean ± S.E. (n = 12). **, p < 0.0001. At least 200 DCs were analyzed in each experiment. b, DCs were analyzed for annexin V and propidium iodide staining by flow cytometry, and apoptotic DCs (annexin+/propidium iodide−) were quantified. Results shown represent the mean ± S.E. (n = 3). **, p < 0.01. C, flow plots show staining by anti-CXCR4 and anti-CXCR7 mAbs (clear gray histograms) versus isotype-matched control mAbs (dark gray histograms). CXCR7+ HeLa cells were used as positive controls (44). Results are representative of three different experiments performed. D, RNA preparations from human monocyte-derived immature DCs (imDCs) or maDCs were analyzed by RT-PCR and agarose gel electrophoresis. Two donors are shown. CXCR7 mRNA was either not detected or detected at low levels in each sample. Positive controls included CXCR7+ HeLa cells. GAPDH control reactions demonstrated that the substrate RNAs were intact. RNA processed in the absence of reverse transcriptase was used as negative control (Neg Cont). E, DCs suspended in RPMI 1640 medium untreated (−) or treated with AMD3100 (10 μm) or with CCX733 (10 μm) were allowed to migrate toward CXCL12 (100 ng/ml) or CCL21 (100 ng/ml) in chemotactic Transwell assays. Results represent the mean ± S.E. (n = 4). *, p < 0.01. ns denotes no significant differences. F, DCs suspended in RPMI untreated (−), treated with AMD3100 (10 μm) or with CCX733 (10 μm) were incubated for 40 h either in medium without chemokine (Control) or in medium plus CXCL12 (100 ng). Subsequently, the DCs were stained with Hoechst 33342. The number represents the percentage of apoptotic DCs under each condition. At least 200 DCs were analyzed in each experiment. Results represent the mean ± S.E. (n = 3). *, p < 0.01; **, p < 0.001.
Because it has been indicated before that maDCs express both CXCR4 (13, 14) and CXCR7 (15), we analyzed the membrane expression of these receptors in the human maDCs used in our study. Flow cytometry analysis confirmed a high expression of CXCR4 in the human DCs (Fig. 1C). However, using two different anti-CXCR7 mAbs, we detected expression of CXCR7 in HeLa cells, which were used as positive controls (44) (Fig. 1C), but not in human maDCs, which displayed only a marginal expression of CXCR7 (Fig. 1C). Because our results on CXCR7 conflicted with prior published results (15), we analyzed by RT-PCR the expression of CXCR7 in maDCs (15). Consistent with the cytometry data shown above, we found little or no expression of CXCR7 in the maDCs (Fig. 1D). While our work was in progress, another group showed that human or mouse leukocytes do not express CXCR7 (21). Finally, to rule out that the expression of a small amount of receptor could play a functional role in the maDCs, we used CCX733, a selective inhibitor of CXCR7 (44). The results showed that interference with this receptor using CCX733 failed to affect the chemotactic (Fig. 1E) or prosurvival effects (Fig. 1F) of CXCL12 in maDCs. In contrast, treatment of the maDCs with AMD3100, a highly selective inhibitor of CXCR4 (45), blocked the chemotactic (Fig. 1E) and prosurvival effects of CXCL12 (Fig. 1F). The lack of inhibition of the chemotactic response to CCL21 showed the specificity of the inhibitors used (Fig. 1E). In summary, the data obtained indicate that CXCL12 used CXCR4, but not CXCR7, to regulate chemotaxis and survival in human maDCs.
CXCR4 Regulates in Vivo Chemotaxis and Survival
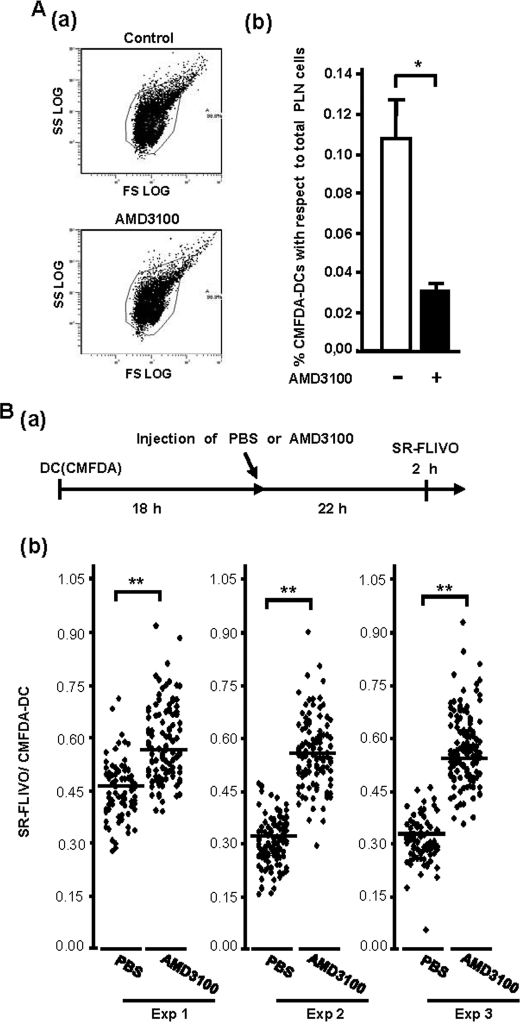
In human and murine DCs the response to CXCL12 is virtually similar when either TNFα or LPS is used to induce maturation of murine or human DCs (3, 4). We used LPS-matured murine splenic DCs to study whether CXCR4 was also important to regulate the migration of murine DCs. Transwell experiments where AMD3100 was used to interfere with CXCR4 showed that this treatment abrogated completely the response to CXCL12 (supplemental Fig. 2), indicating that, similar to the human maDCs, this receptor was also controlling CXCL12-dependent chemotaxis in murine maDCs. To study whether CXCR4 was also able to regulate the migration of maDCs in vivo, we analyzed whether AMD3100 could interfere with the migration of DCs to the LNs. For this purpose, we injected CMFDA-labeled LPS-matured splenic DCs together with either AMD3100 or vehicle subcutaneously into the hind footpad of C57/BL6 mice. 18 h after the injection of the DCs, the mice were killed, the PLNs of control and AMD3100-injected animals were collected, and the CMFDA-DCs in the LNs were analyzed by flow cytometry to assess the percentage of DCs that reached this organ. As shown in Fig. 2A, AMD3100 treatment reduced by almost 70 ± 8% (n = 3) the number of CMFDA-DCs that reached the PLNs.
FIGURE 2.
CXCR4 regulates CXCL12-dependent chemotaxis and survival in murine maDCs in vivo. A, 2 × 106 CMFDA-labeled splenic mature DCs (CMFDA-DCs) injected in the footpads of recipient mice together with AMD3100 or with vehicle. 18 h after the injections, the PLNs of control and AMD3100-treated animals were collected, passed through a mesh, and suspended in PBS, and the number of CMFDA-DCs was analyzed by flow cytometry. a, FL1-FS dot plots showing representative suspension of PLN cells. CMFDA-DCs were analyzed inside the selected gate. b, numbers representing the percentage of CMFDA-labeled DCs with respect to the total number of cells in the LN, obtained from the analysis of the LNs of four mice. Results represent the mean ± S.E. (error bars; n = 4). **, p < 0.001. Ba, experimental protocol. 2 × 106 CMFDA-labeled LPS-matured splenic DCs (CMFDA-DCs) were injected in the footpads of recipient mice. After an additional 18 h, animals were injected intraperitoneally with 4 mg/kg body weight of AMD3100 or vehicle control. After 22 h, the animals were injected intravenously with SR-FLIVO to stain apoptotic DCs in the LNs. After an additional 2 h, the PLNs were extracted, fixed, and subjected to two-photon analysis. b, SR-FLIVO staining presented by CMFDA-DCs obtained from the LNs of animals treated either with AMD3100 or with PBS. The LNs of the mice were extracted and studied by two-photon microscopy. The stacks of optical images of the LNs were examined with the Leica Confocal software, and values of the Maximum Amplitude of the SR-FLIVO and CMFDA channel were obtained (see Ref. 28). Data are presented as maximum intensity of SR-FLIVO over maximum intensity of CMFDA for each individual DC in a LN (28). Three different mice were analyzed. **, p < 0.001.
To study whether CXCL12 also protected the murine maDCs from apoptosis in vivo, we used an experimental approach described before (28, 29). We induced maturation of CMFDA-labeled splenic DCs with LPS, and then these cells were injected subcutaneously in the hind footpad of C57BL/6 mice (see “Experimental Procedures”). After 18 h, when the DCs were largely positioned in the LNs (28), mice were injected intraperitoneally either with 4 mg/kg AMD3100 or with vehicle. After an additional 22 h, the animals were injected intravenously with SR-FLIVO, an agent that specifically binds to caspases and that, consequently, selectively stains apoptotic cells (28, 29) (for more experimental details, see supplemental Experimental Procedures). After additional 2 h, mice were killed, and the PLNs were extracted and analyzed by two-photon microscopy. Inhibiting the signaling in vivo from CXCR4 with AMD3100 in the CMFDA-DCs resulted in an increase in the percentage of DCs that displayed SR-FLIVO staining (34.7 ± 15% increase (n = 3)), compared with the vehicle-treated animals, suggesting that CXCR4 relays prosurvival signals to the DCs also in the LNs.
Stimulation of Human maDCs with CXCL12 Induces Activation of mTORC1 That Is Mediated by Gi, Gβγ, PI3K-α, -δ, -γ, and Akt
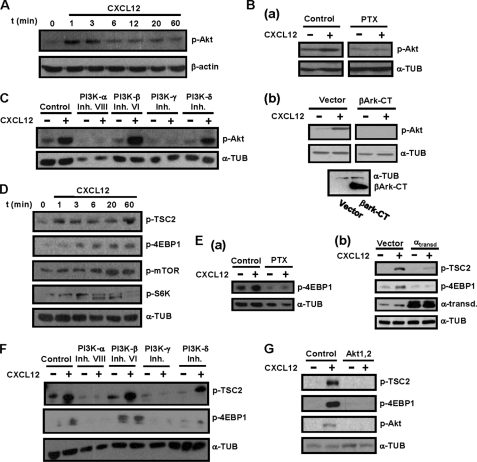
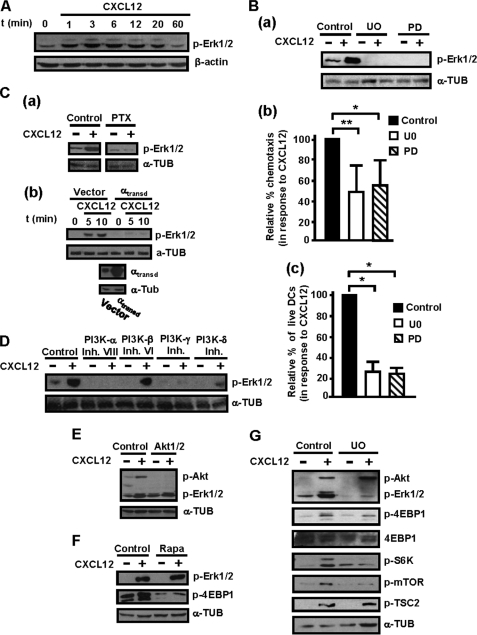
In the next experiments we used human maDCs to analyze the signaling molecules downstream of CXCR4, involved in the control of CXCL12-induced chemotaxis and survival in maDCs. Because Akt is implicated in the regulation of these functions in other cell types (1, 26), we analyzed whether stimulation of maDCs with CXCL12 induced activation of this kinase. Stimulation with CXCL12 induced maximum activation of Akt after 1–3 min, which decayed to near basal levels after 1 h (Fig. 3A; see also Fig. 7). Because chemokine receptors use Gi family members and Gβγ components of the heterotrimeric G proteins to relay intracellular signals (24, 25), we analyzed whether CXCL12-dependent activation of Akt was regulated by Gi and Gβγ. Interference with Gi, using PTX (26, 27, 29) (Fig. 3Ba) or with the Gβγ subunits of G proteins, using βArk-CT, a C-terminal fragment of the kinase βArk that sequesters and inhibit βγ subunits (41) (Fig. 3Bb), blocked CXCL12-dependent phosphorylation of Akt (see also Fig. 7). We used highly selective PI3K family inhibitors to study the role played by specific isoenzymes in mediating the effects of CXCL12 on Akt. The inhibition of PI3K-α, using PIK-75 (46, 47), of PI3K-γ, using AS-605240, (48) and, at lower extent of PI3K-δ, using IC87114 (49), resulted in the inhibition of the phosphorylation of Akt (Fig. 3C). In contrast, inhibition of PI3K-β, using TGX-221, (50) failed to affect CXCL12-dependent Akt phosphorylation (Fig. 3C and see Fig. 7).
FIGURE 3.
Stimulation of human maDCs with CXCL12 induces Gi/Gβγ/PI3K-α/-δ/-γ-mediated activation of Akt and mTORC1. A, DCs stimulated for the indicated times with CXCL12. Subsequently, aliquots were taken to analyze the level of phosphorylated/active Akt (p-Akt) and β-actin by Western blotting. A representative experiment of six performed is shown. Ba, DCs left untreated (Control) or pretreated with PTX (100 ng/ml) for 2 h. Subsequently, the DCs were stimulated or not with CXCL12. After 3 min of stimulation with the chemokine, aliquots were taken to analyze the level of phosphorylated/active Akt1 (p-Akt) and α-tubulin (α-TUB) by Western blotting. A representative experiment of four performed is shown. b upper, DCs transfected either with vector or with βArk-CT. After transfection, aliquots of vector and βArk-CT transfected DCs were stimulated for 3 min with CXCL12, and then levels of phosphorylated/active Akt1, α-tubulin, and βArk-CT (41) were analyzed by Western blotting. Lower, α-tubulin shows equal loading of the gels. A representative experiment of four performed is shown. C, DCs left untreated (Control) or pretreated with PI3Kα (250 nm), PI3Kβ (1 μm), PI3Kγ (10 μm), or PI3Kδ (20 μm) inhibitors for 60 min. Subsequently, DCs were left unstimulated (−) or stimulated (+) with CXCL12. After 3 min of stimulation with chemokines, aliquots were taken to analyze the level of phosphorylated/active Akt1 and α-tubulin. A representative experiment of six performed is shown. D, DCs stimulated for the indicated times with CXCL12 as in A. Aliquots were taken to analyze the level of phosphorylated TSC2 (p-TSC2), mTOR (p-mTOR), 4E-BP1 (p-4EBP1), and S6K (p-S6K) by Western blotting. α-Tubulin levels show equal loading. A representative experiment of four performed is shown. Ea, DCs left untreated (Control) or pretreated with PTX (100 ng/ml) and then stimulated or not with CXCL12 as in B. Aliquots were taken to analyze the level of phosphorylated 4E-BP1 (p-4EBP1) and α-tubulin. A representative experiment of three performed is shown. b, DCs transfected either with vector or with α-transducin (αtransd). After transfection, a similar number of live vector and αtransd-transfected DCs were stimulated for the indicated times with CXCL12, and the levels of phosphorylated TSC2 (p-TSC2), phosphorylated 4E-BP1, transducin, and α-tubulin were analyzed by Western blotting. A representative experiment of three performed is shown. F, experiments performed as described in C except that the following proteins were analyzed by Western blotting: phosphorylated TSC2, phosphorylated 4E-BP1, and α-tubulin. A representative experiment of three performed is shown. G, DCs were left untreated or pretreated with a specific Akt inhibitor (5 μm). Experiments were performed as described in C, except that the levels of phosphorylated/active TSC2, phosphorylated 4E-BP1, and phosphorylated/active Akt were analyzed. α-Tubulin levels show the loading of the gels. A representative experiment of six performed is shown.
FIGURE 7.
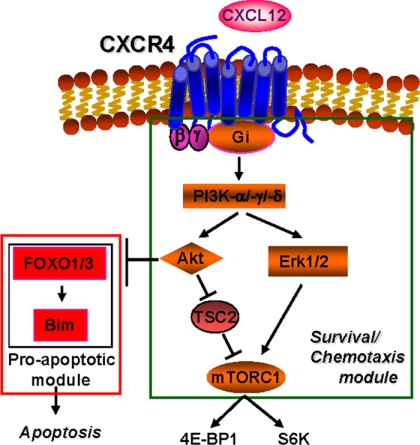
Model indicating the signaling mechanism used by CXCL12 to regulate simultaneously chemotaxis and survival in maDCs. CXCL12 uses CXCR4, Gi, Gβγ, PI3K-α,-γ,-δ, Akt, mTORC1, and ERK1/2 to regulate chemotaxis and apoptosis simultaneously in maDCs. FOXO1/3 regulates, using Bim, apoptosis, but not chemotaxis. This model does not rule out additional unidentified downstream targets of the indicated signaling molecules.
Because Akt regulates mTORC1 (33), we analyzed whether stimulation with CXCL12 also induced activation of the latter kinase complex. Upon phosphorylation by Akt, TSC2 is inhibited, which results in the activation of mTORC1 (see Introduction). Consistent with this model, time course experiments showed that stimulation of DCs with CXCL12 caused a rapid phosphorylation/inhibition of TSC2 (Fig. 3D and see Fig. 7). In line with these results, stimulation with CXCL12 led to an increase in the activity of mTORC1, which was assessed by analyzing the phosphorylation of 4E-BP1 (Thr(P)-37/46–4EBP1) and was further confirmed determining the phosphorylation of S6K (Thr(P)-389-S6K) and mTOR in Ser-2448, a residue that is phosphorylated by active S6K as part of a feedback loop (51) (Fig. 3D and see Fig. 7).
Next, we analyzed the involvement of the axis Gi/Gβγ /PI3K-α/-β/-γ/-δ and Akt in the activation of mTORC1 using the phosphorylation of 4E-BP1 as a readout of the activity of this kinase complex (see also Fig. 7). Inhibition of Gi, using PTX (Fig. 3Ea), of Gβγ, by overexpressing α-transducin (αtransd) (Fig. 3Eb), of PI3K-α, -δ, and -γ, using isoform-selective inhibitors of the respective enzymes (Fig. 3F), and of Akt, using a highly specific inhibitor (52) (Fig. 3G), all blunted the phosphorylation of TSC2 and 4E-BP1. Inhibition of PI3K-β failed to affect the phosphorylation of the latter molecules (Fig. 3F). The results indicate that Gi/Gβγ /PI3K-α/-γ/-δ and Akt mediate CXCL12-dependent activation of mTORC1 (see Fig. 7).
The Signaling Components of the Axis Gi, Gβγ, PI3K-α/-γ/-δ, Akt, and mTORC1 Regulate Both CXCL12-mediated Chemotaxis and Survival in Human maDCs
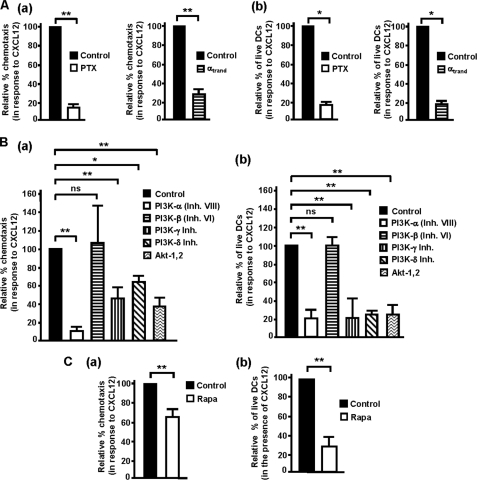
In the next experiments we analyzed whether the molecular components of the signaling axis identified above (i.e. Gi/Gβγ/PI3K-α/-γ/-δ/Akt, and mTORC1) could control CXCL12-regulated chemotaxis and/or survival in maDCs (see Fig. 7). Inhibition of Gi proteins, with PTX, and of Gβγ, by overexpressing αtransd, blocked the chemotactic response (Fig. 4Aa and see Fig. 7) and the prosurvival effects (Fig. 4Ab and see Fig. 7) of CXCL12. Analysis of the role of different PI3K isoforms in the regulation of CXCL12-mediated chemotaxis (Fig. 4Ba) showed that the largest effects were observed upon inhibition of PI3K-α, with lower, but still strong effects, upon inhibition of PI3K-γ and PI3K-δ (Fig. 4Ba). Inhibition of PI3K-α, -γ, and -δ isoforms blocked CXCL12-dependent DC survival to a similar extent (Fig. 4Bb). Inhibition of PI3K-β had no effect on the chemotactic response (Fig. 4Ba) or on the prosurvival effects of CXCL12 (Fig. 4Bb). Inhibition of Akt also reduced both chemotaxis and survival induced by CXCL12 (Fig. 4B, a and b). Interestingly, the effects of blocking PI3K-γ, -δ, and Akt were more potent on CXCL12-dependent survival than on chemotaxis (Fig. 4B).
FIGURE 4.
CXCL12-dependent regulation of chemotaxis and survival in human maDCs is regulated by Gi, Gβγ, PI3K-α, -γ, -δ isoforms, and Akt. Aa left, DCs dissolved in RPMI 1640 medium were kept untreated (Control) or pretreated with PTX (100 ng/ml) for 2 h. Subsequently, DCs were allowed to migrate toward CXCL12 for 2 h. The figure shows the migration of DCs treated with inhibitor relative to the migration of the DCs without inhibitor, which was given an arbitrary number of 100 (control). Results shown represent the mean ± S.E. (error bars; n = 4). **, p < 0.001. Right, aliquots including equal number of live vector pcDNA3 (Control) and α-transducin (αtransd)-transfected maDCs were allowed to migrate toward CXCL12 in Transwell chambers. The figure shows the migration of α-transducin-transfected DCs relative to the migration of vector-transfected DCs that was given an arbitrary number of 100 (control). Results represent the mean ± S.E. (n = 4). **, p < 0.001. b left, DCs were suspended in RPMI 1640 medium and then kept untreated or pretreated with PTX as in A. DCs untreated or pretreated with PTX were maintained in the presence of CXCL12 for an additional 40 h, and apoptotic cells were analyzed after Hoechst 33342 staining. Results represent the percentage of live PTX-treated DCs with respect to the percentage of live untreated DCs, which was given an arbitrary value of 100. Results represent the mean ± S.E. (n = 3). At least 200 DCs were analyzed in each experiment. **, p < 0.01. Right, DCs were transfected with vector pcDNA3 (Control) or α-transducin as in Aa right. Aliquots with similar number of live vectors and α-transducin-transfected DCs were maintained in serum-free medium including CXCL12 for 24 h, and then apoptotic DCs were analyzed by Hoechst 33342 staining. The figure shows the percentage of live α-transducin-transfected DCs with respect to the percentage of live vector-transfected DCs which was given an arbitrary value of 100. Results represent the mean ± S.E. (n = 3). **, p < 0.01. Ba, DCs were suspended in 0.1% BSA/RPMI 1640 medium and then kept untreated or pretreated with PI3Kα (250 nm), PI3Kβ (1 μm), PI3Kγ (10 μm), PI3Kδ (20 μm) or Akt (5 μm) inhibitors for 1 h. Migration was analyzed in a Transwell setting as in Aa. Results represent the mean ± S.E. (n = 6). **, p < 0.001. (b), DCs were suspended in RPMI 1640 medium plus CXCL12 then kept untreated or treated with PI3Kα (250 nm), PI3Kβ (1 μm), PI3Kγ (10 μm), PI3Kδ (20 μm), or Akt (5 μm) inhibitors for 40 h. Apoptotic DCs were determined as in Ab. Results represent the mean ± S.E. (n = 6). **, p < 0.001. Ca, DCs were left untreated or pretreated with the specific mTORC1 inhibitor rapamycin (Rapa, 100 nm) for 1 h. Both untreated DCs or those pretreated with rapamycin were allowed to migrate in Transwells toward CXCL12 for additional 2 h. Migration was analyzed in a Transwell setting as in Aa. Results represent the mean ± S.E. (n = 7). **, p < 0.001. b, DCs untreated or treated with rapamycin as in a were maintained in RPMI 1640 medium in the presence of CXCL12 for 20 h. Subsequently, the DCs were stained with Hoechst 33341 to identify apoptotic DCs as in Ab. Results represent the mean ± S.E. (n = 9). **, p < 0.001.
Finally, to study the involvement of mTORC1 in the regulation of CXCL12-dependent chemotaxis and survival, we treated the maDCs with the highly selective mTORC1 inhibitor rapamycin (52, 53). Importantly, before carrying this analysis, we ensured that the rapamycin concentration used in the experiments (100 nm) inhibited only mTORC1, but not mTORC2. The activity of mTORC2 was assessed by studying the phosphorylation of Akt (Ser-473) and mTOR (Ser(P)-2481) (33, 54) (data not shown). Inhibition of mTORC1 moderately reduced the chemotaxis (Fig. 4Ca) and, more potently, the prosurvival effects (Fig. 4Cb) of CXCL12. The results indicate that Gi/Gβγ/PI3K-α/-γ/-δ, Akt, and mTORC1 control both CXCL12-mediated chemotaxis and survival of maDCs (see also Fig. 7).
CXCL12 Induces Activation of ERK1/2 in Human maDCs, Which Controls Chemotaxis and Survival in These Cells
Because ERK1/2 regulate chemotaxis or survival in different cell types (27, 37, 55), we studied their involvement in the effects elicited by CXCL12. Stimulation of the maDCs with CXCL12 induces activation of ERK1/2 after 1 min, reached the highest level of activity between 3 and 6 min, and decayed to basal levels after 1 h (Fig. 5A; see also Fig. 7). To study the involvement of ERK1/2 in the regulation of CXCL12-dependent survival and chemotaxis, we treated the DCs either with UO126 or with PD0325901, two highly selective compounds that suppress the activation of ERK1/2 by inhibiting MEK (MAPK/ERK kinase) (52, 53). The blocking of ERK1/2 with both agents (Fig. 5Ba) reduced the migration (Fig. 5Bb) and the prosurvival effects (Fig. 5Bc) of CXCL12, indicating that ERK1/2 regulate both CXCL12-mediated chemotaxis and survival in maDCs. The effects of the inhibition of ERK1/2 were, as observed for the other molecules indicated above, more potent on survival than on chemotaxis (compare Fig. 5Bb to Fig. 5Bc; see also Fig. 7).
FIGURE 5.
ERK1/2 regulate CXCL12-mediated chemotaxis and survival downstream of Gi, Gβγ, PI3K-α, -γ, -δ isoforms, but not of PI3Kβ, Akt, and mTORC1 in human maDCs. A, DCs were stimulated for the indicated times with CXCL12 as in Fig. 3A, and aliquots were taken to analyze the level of phosphorylated/active ERK1/2 (p-ERK1/2) by Western blotting. β-Actin levels show equal loading of the gels. A representative experiment of three performed is shown. A and Fig. 3A correspond to the same experiment, and thus similar β-actin control is used. Ba, DCs suspended in 0.1% BSA in RPMI 1640 medium were left untreated (Control) or pretreated with the ERK1/2 inhibitors UO126 (UO) or PD0325901 (PD), both at 5 μm, for 1 h. Subsequently, DCs were stimulated or not with CXCL12. After 3 min of stimulation with CXCL12, aliquots were taken to analyze the level of phosphorylated/active ERK1/2 or α-tubulin (α-TUB). A representative experiment of four performed is shown. b, DCs untreated or pretreated with UO126 or with PD0325901, as in a, were allowed to migrate toward CXCL12. Migration was analyzed in Transwell setting as in Fig. 4Aa. Results represent the mean ± S.E. (error bars; n = 6). **, p < 0.001. c, DCs were suspended in 0.1% BSA in RPMI 1640 medium plus CXCL12 and then kept untreated or treated with UO126 or PD0325901, both at 5 μm, for 40 h. Apoptotic DCs were determined as in Fig. 4Ab. Results represent the mean ± S.E. (n = 8). **, p < 0.001. Ca, DCs were left untreated or pretreated with PTX and, subsequently, were stimulated or not with CXCL12 for 3 min (as in Fig. 3B). Aliquots were taken to analyze the level of phosphorylated/active ERK1/2 and α-tubulin. A representative experiment of four performed is shown. b upper, DCs were transfected either with vector or with α-transducin (αtransd). 18 h after transfection, a similar number of live vector- and α-transducin-transfected DCs were taken and then stimulated for the indicated times with CXCL12. The levels of phosphorylated/active ERK1/2 were analyzed by Western blotting. Lower, expression of α-transducin was analyzed by Western blotting. α-Tubulin levels show equal loading of the gels. A representative experiment of four performed is shown. D, DCs were suspended in 0.1% BSA/RPMI 1640 medium and then kept untreated or pretreated with PI3K-α, PI3K-β, PI3K-γ, or PI3K-δ inhibitors for 1 h as in Fig. 3C. Subsequently, DCs were stimulated or not with CXCL12 for 3 min. Subsequently, aliquots were taken to analyze the level of phosphorylated/active ERK1/2 (p-ERK1/2). α-Tubulin levels show equal loading of the gels. D and 3C correspond to the same experiment, and thus a similar β-actin control is used. A representative experiment of six performed is shown. E, DCs were suspended in 0.1% BSA/RPMI 1640 medium and then kept untreated or pretreated with Akt inhibitor for 1 h as in Fig. 3G. Subsequently, DCs were stimulated or not with CXCL12. After 3 min of stimulation with the chemokine, aliquots were taken to analyze the level of phosphorylated/active Akt (p-Akt), phosphorylated/active ERK1/2 and α-tubulin. A representative experiment of four performed is shown. F, DCs were left untreated or pretreated with a specific mTORC1 inhibitor (rapamycin, 100 nm) for 1 h. Subsequently, the DCs were stimulated or not with CXCL12. After 3 min of stimulation with the chemokine, aliquots were taken to analyze the level of phosphorylated/active ERK1/2, phosphorylated 4E-BP1, and α-tubulin. A representative experiment of four performed is shown. G, DCs were left untreated or pretreated with a specific ERK1/2 inhibitor (UO126, 5 μm) for 60 min. Subsequently, DCs were stimulated or not with CXCL12. After 3 min of stimulation with CXCL12, aliquots were taken to analyze the level of phosphorylated TSC2, phosphorylated/active Akt, phosphorylated/active ERK1/2, phosphorylated 4E-BP1, phosphorylated S6K, phosphorylated mTOR. α-Tubulin and 4E-BP1 levels show equal loading of the gels. A representative experiment of eight performed is shown.
Gi, Gβγ, PI3K-α/-γ/-δ, but Not Akt and mTORC1, Regulate CXCL12-mediated Activation of ERK1/2 in Human maDCs
We analyzed whether ERK1/2 were under the control of the components of the pathway controlled by CXCL12 identified above (i.e. Gi, Gβγ, PI3K-α/-γ/-δ, but not Akt and mTORC1) (see Fig. 7). Treatment of the DCs with PTX or inhibition of Gβγ by overexpressing α-transducin caused a complete inhibition of ERK1/2 (Fig. 5C, a and b). The inhibition of specific PI3K family isoforms showed that PI3K-α, -γ, and -δ, but not PI3K-β, controlled CXCL12-mediated phosphorylation of ERK1/2 (Fig. 5D). Because PI3K-α, -γ, and -δ isoforms are upstream of Akt (see above Fig. 3C), we analyzed whether the latter kinase could mediate CXCL12-dependent phosphorylation of ERK1/2 (see also Fig. 7). However, as shown in Fig. 5E, despite the strong inhibition of Akt, stimulation with CXCL12 was still able to stimulate ERK1/2, indicating that PI3K isoforms regulate the phosphorylation of ERK1/2 independently of Akt (see also Fig. 7). Finally, we studied whether ERK1/2 was downstream of mTORC1. Inhibition of mTORC1, by treating the DCs with rapamycin, one of the most selective inhibitors of this complex (52, 53), had no effect on the CXCL12-stimulated phosphorylation of ERK1/2 (Fig. 5F). In contrast, inhibition of ERK1/2 partially blocked CXCL12-dependent activation of mTORC1, as assessed by analyzing the phosphorylation of 4E-BP1, S6K, and mTOR (at Ser-2448) (Fig. 5G). Inhibition of ERK1/2 did not affect the phosphorylation of Akt either (Fig. 5G), indicating that the effects of ERK1/2 on mTORC1 were not exerted at the level of Akt. Inhibition of ERK1/2 did not affect the phosphorylation of TSC2 at the Akt phosphorylation site (Fig. 5G), indicating that ERK1/2 does not indirectly or directly regulate the phosphorylation of this site. In sum, the experiments indicate, first, that Gi, Gβγ, and PI3K-α, -γ, -δ, but not PI3K-β, Akt, and mTORC1, regulate CXCL12-dependent phosphorylation of ERK1/2 and, second, that ERK1/2 modulate the CXCL12-mediated activation of mTORC1 (see Fig. 7).
Gi, Gβγ, PI3K-α/-γ/-δ, and Akt, but Not mTORC1 and ERK1/2, Regulate CXCL12-mediated Inhibition of FOXO in Human maDCs
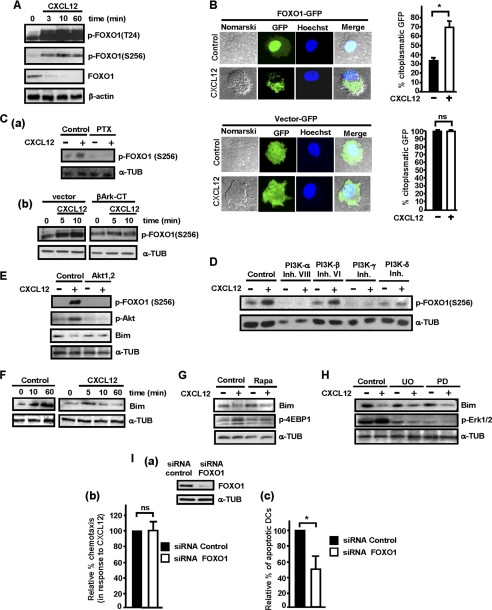
We have shown before (28, 29, 56) that FOXO transcription factors exert proapoptotic roles in maDCs. Because CXCL12 induces activation of Akt, which may potentially phosphorylate/inhibit FOXO1/3, we analyzed whether CXCL12 could induce phosphorylation of FOXO in residues regulated by Akt (Thr-24 and Ser-256 in FOXO1 and Ser-253 in FOXO3). Stimulation of the maDCs with CXCL12 induced a rapid phosphorylation and inhibition of FOXO1 (Fig. 6A) and FOXO3 (data not shown). Phosphorylation of Ser-256 is important because it suppresses transactivation of FOXO1, promotes its exclusion from the nucleus, and is also necessary for the phosphorylation of Thr-24 (42). Because phosphorylation of FOXO by Akt induces its exclusion from the nucleus, we analyzed the percentage of DCs where FOXO remains in the nucleus upon stimulation with CXCL12. The maDCs were nucleofected with a construct that encodes FOXO1-GFP and subsequently were shifted to serum-free medium (Fig. 6B). After 1 h in this medium, FOXO1-GFP was observed in the nucleus in most DCs. However, following stimulation with CXCL12, almost 70% of the transfected DCs showed a clear cytoplasmic FOXO1-GFP location, compared with the 30% of unstimulated DCs that presented cytoplasmic FOXO1 (Fig. 6B). In the DCs transfected with vector-GFP, despite the stimulation with CXCL12, most GFP remained in the cytosol (Fig. 6B).
FIGURE 6.
FOXO regulates CXCL12-mediated survival, but not chemotaxis, downstream of Gi, Gβγ, PI3K-α, -γ, -δ isoforms, and Akt in human maDCs. A, DCs in RPMI were stimulated for the indicated times with CXCL12. Subsequently aliquots were taken to analyze the level of phosphorylated FOXO1 (p-FOXO1 (T24), p-FOXO1 (S256)), FOXO1, and β-actin by Western blotting. A representative experiment of three performed is shown. B, DCs were nucleofected with either pEGFP-C1 vector (Vector-GFP) or with pEGFP-FOXO1 (FOXO1-GFP). A similar number of live nucleofected DCs were washed with RPMI 1640 medium and then either left unstimulated (Control) or stimulated for 1 h with CXCL12. The cells were then plated onto polylysine and the nucleus stained with Hoechst 33342. Left, the figure shows Nomarski optics and GFP protein staining analyzed using the FITC fluorescence channel. A representative experiment of three performed is shown. Right, the bar diagrams present the percentage of DCs, unstimulated (−) or stimulated with CXCL12 (+), that present cytoplasmic vector-GFP (upper) and FOXO1-GFP (lower) after the transfection with these constructions. Results shown represent the mean ± S.E. (error bars; n = 3). *, p < 0.001. Ca, DCs were left untreated or pretreated with PTX as in Fig. 3Ba and then stimulated or not with CXCL12. After 10 min of stimulation, aliquots were taken to analyze the level of phosphorylated FOXO1 (p-FOXO1(S256)) and α-tubulin (α-TUB). A representative experiment of three performed is shown. b, DCs were transfected either with vector or with βArk-CT. 18 h after transfection, aliquots of vector and β-Ark-CT transfected DCs were taken and then stimulated for the indicated times with CXCL12. The levels of phosphorylated FOXO1 were analyzed by Western blotting. α-Tubulin levels show equal loading of the gels. A representative experiment of four performed is shown. D, experiments were performed as indicated in the legend of Fig. 3C. After 3 min of stimulation, aliquots were taken to analyze the level of phosphorylated FOXO1 were analyzed by Western blotting. α-Tubulin levels show equal loading of the gels. D and Fig. 3C correspond to the same experiment, and thus similar β-actin control is used. E, DCs were left untreated or pretreated with a specific Akt inhibitor as in Fig. 3G. Subsequently, DCs were stimulated or not with CXCL12. After 10 min of stimulation with CXCL12, aliquots were taken to analyze the level of phosphorylated/active Akt, phosphorylated FOXO1, Bim, and α-tubulin. A representative experiment of three performed is shown. F, DCs in RPMI 1640 medium were either left unstimulated or were stimulated for the indicated times with CXCL12. Aliquots were taken to analyze the level of Bim and α-tubulin. Results are representative of three experiments performed. G, DCs were left untreated or pretreated with a specific mTORC1 inhibitor as in Fig. 5F. Subsequently, DCs were left unstimulated (−) or stimulated (+) with CXCL12 for 10 min. Aliquots were taken to analyze the level of Bim, phosphorylated 4E-BP1, and α-tubulin. Results are representative of four experiments performed. H, DCs were suspended in RPMI 1640 medium and then kept untreated or pretreated with UO125 (UO) or PD0325901 (PD) as in Fig. 5Ba. Subsequently, DCs were left unstimulated (−) or stimulated (+) with CXCL12 for 10 min. Aliquots were taken to analyze the level of Bim, phosphorylated/active ERK1/2, and α-tubulin. A representative experiment of four performed is shown. Ia, DCs were nucleofected with siRNA control or with siRNA for FOXO1. After 36 h, a similar number of live DCs was taken to analyze the level of FOXO1 and α-tubulin by Western blotting. b, a similar number of live DCs with normal (siRNA control) or reduced levels of FOXO (siRNA FOXO1) was dissolved in RPMI 1640 medium and then allowed to migrate toward CXCL12. The values represent the migration of siRNA FOXO1-nucleofected DCs with respect to the migration of siRNA control-nucleofected DCs that were given an arbitrary number of 100 (control). Results represent the mean ± S.E. (n = 3). ns denotes no significant differences. c, DCs with normal (siRNA control) or with reduced levels of FOXO1 were maintained in RPMI 1640 medium for 24 h, and subsequently, the DCs were stained with Hoechst 33342 to identify apoptotic DCs. The number represents percentage of apoptotic siRNA FOXO1-transfected DCs with respect to the percentage of apoptotic siRNA control DCs, which was given an arbitrary value of 100. Results represent the mean ± S.E. (n = 4). *, p < 0.05.
In the next experiments we tested whether the signaling molecules that control chemotaxis and survival (i.e. Gi/Gβγ /PI3K/Akt/mTORC1 and ERK1/2) were able to regulate CXCL12-dependent phosphorylation of Ser-256 in FOXO1. As expected, inhibition of Gi with PTX (Fig. 6Ca), of Gβγ, by overexpressing βArk-CT (Fig. 6Cb), and inhibition of PI3K-α/-γ/-δ (Fig. 6D) and Akt (Fig. 6E), all abrogated CXCL12-mediated phosphorylation of FOXO1 in maDCs (see Fig. 7). Inhibition of PI3K-β (Fig. 6D), mTORC1 (supplemental Fig. 3A), or ERK1/2 (supplemental Fig. 3B) failed to block the phosphorylation of FOXO1 at Ser-256, indicating that these molecules were not regulating FOXO1 at this residue (Fig. 7).
FOXO1/3 control the transcription of the proapoptotic Bcl2 member Bim (56), which can be used as a readout of the activity of FOXO in maDCs (28, 29). As shown in Fig. 6F, maDCs left in serum-free medium displayed an increase in the expression of Bim, reflecting the enhanced activity of FOXO1 in the nucleus (see Fig. 6B). However, when the maDCs that were maintained in serum-free medium were stimulated with CXCL12, the kinase Akt was activated (Fig. 6E), leading to the phosphorylation/inhibition of FOXO1 (Fig. 6E) and the dampening of Bim expression (Fig. 6E). Because Bim expression reflects the activity of FOXO1, we examined whether mTORC1 and ERK1/2 control the activity of this transcription factor, by studying the effects of the latter kinases on Bim levels. As observed, inhibition of mTORC1 (Fig. 6G) or ERK1/2 (Fig. 6H) failed to prevent the down-regulation of Bim induced by stimulating serum-deprived DCs (that display increased Bim levels) with CXCL12, suggesting that these molecules do not regulate FOXO activity. Together, the results indicate that upon stimulation of maDCs with CXCL12, inhibition of FOXO1 is mediated by Gi, Gβγ, PI3K-α, -γ, -δ, and Akt, but not by mTORC1 and ERK1/2 (see Fig. 7).
FOXO Members Regulate CXCL12-dependent Survival, but Not Chemotaxis, in Human maDCs
As all signaling molecules upstream of FOXO1 that were analyzed were regulating, although at different extent, CXCL12-dependent chemotaxis and survival, we tested whether this transcription factor was also able to regulate both functions. When we compared the chemotactic response to CXCL12 presented by maDCs with normal levels of FOXO, and maDCs where FOXO1 (Fig. 6Ia) or FOXO3 (data not shown) was knocked-down with siRNAs, we did not find any differences between these cells (Fig. 6Ib and data not shown). In contrast, the knocking down of FOXO1 (Fig. 6Ic) and FOXO3 (data not shown) reduced the percentage of apoptotic DCs, indicating that this factor plays proapoptotic roles in maDCs and that its inhibition upon stimulating the maDCs with CXCL12 may contribute to the survival of maDCs (Fig. 6Ic). In summary, the results indicate that FOXO selectively controls CXCL12-dependent survival, but not chemotaxis in maDCs (see Fig. 7).
DISCUSSION
It is emerging that chemokine receptors can regulate a variety of functions in addition to chemotaxis. Although there is much information on how chemokine receptors control chemotaxis and, somewhat less, other functions, surprisingly, there is almost no information on the mechanism whereby chemokine receptors can regulate several functions simultaneously. In the specific case of leukocytes, gaining information on this issue is important to develop strategies that allow the modulation of the functions of chemokines and, eventually, the immune response.
As during the immune response maDCs migrate through afferent lymphatic vessels and inside the LNs (6–8) and CXCL12 is expressed in these regions, this chemokine may potentially control the function/s of these cells in the immune system. Before carrying out a study on how CXCL12 may regulate several functions, we confirmed that CXCL12 regulates both chemotaxis and survival in human maDCs and murine splenic DCs. We found, in line with a recent report (21), that CXCR7 is not expressed in human maDCs, suggesting that in the maDCs that we employ, CXCL12 uses CXCR4 to relays intracellular signals that lead to the regulation of chemotaxis and survival. However, our results largely apply when human DCs are matured with TNFα, and we cannot rule out that CXCR7 could be expressed when the DCs are exposed to other maturation or inflammation signals. Under our conditions, CXCL12 failed to regulate migratory speed, adhesion, fugetaxis, endocytosis, or the maturation of the human DCs. In contrast to our results, other group have shown that interference with CXCR4 using a selective inhibitor blocked the maturation of murine DCs (11, 12), although the discrepancy with our results could be due to the different species of DCs used.
Using splenic DCs we show that CXCR4 also controls the survival and the chemotaxis of the maDCs that migrate in vivo to the LNs. Although previously it was observed that interfering with CXCR4 reduced the number of maDCs that migrate to the LNs (5, 6), so far it has not been assessed how the prosurvival effects of CXCL12 could contribute to the increase in the number of DCs that reach the nodes. Our results indicate that of the observed 70 ± 8% (n = 3) reduction in the percentage of CMFDA-DCs that reach the PLNs when CXCR4 is inhibited, approximately 35%±15% of this percentage could be due to the blocking of the prosurvival effects of CXCR4. The remaining ∼35% inhibition could be explained by interference with other CXCR4-regulated functions, most likely chemoattraction, of the CMFDA-maDCs that migrate toward the PLNs.
To get insight on the mechanisms whereby chemokine receptors, and specifically CXCR4, simultaneously regulate chemotaxis and survival, we first analyzed the signaling molecules controlled by CXCL12 in maDCs. Then, using selective pharmacological agents and siRNA, when this was possible, we studied their role in the regulation of chemotaxis and survival. To reduce the possibility of off-target effects, we have used preferentially inhibitors previously shown to be highly selective in studies with panels of dozens of kinases (52, 53). Using the aforementioned approaches, we observed that a core of signaling molecules, which were hierarchically organized, including Gi, Gβγ, and PI3K-α, -γ, -δ isoforms, Akt, mTORC1, and ERK1/2, were all controlling CXCL12-mediated survival and chemotaxis to different extents in human maDCs (see Fig. 7). Treatment of the maDCs during the time that the chemotaxis assays are in the Transwells with the PI3K-α inhibitor PIK-75, which blocks the prosurvival effects of CXCL12, did not increase the percentage of apoptotic maDCs (supplemental Fig. 4A). Furthermore, when the chemotactic Transwell assays performed with maDCs treated with PIK-75 were carried out in the presence of the polycaspase inhibitor z-VAD-FMK, an agent that blocks apoptotic caspases and, consequently, the apoptosis of maDCs, it was observed that the chemotactic response of the maDCs to the chemokine was not altered (supplemental Fig. 4B). These experiments indicate that the effects that the inhibitors exert on chemotaxis were not caused by an earlier induction of apoptosis that could impair the chemotactic response of the maDCs to CXCL12.
These experiments also indicate that the effect of the inhibition of different molecules takes longer to be evidenced on the prosurvival effects induced by CXCL12 than on the chemotaxis induced by this chemokine. These results indicate a different dynamics in the mechanisms controlling CXCL12-dependent chemotaxis, which have to take place very rapidly in migrating cell, and CXCL12-regulated survival in the maDCs. The delayed effects of the inhibitors on survival may be also related to the increased resistance of the maDCs to apoptosis due to the enhancement in the expression of variety of antiapoptotic molecules, including prosurvival Bcl-2 family members and serine protease inhibitors, that takes place during the maturation of the DCs (1, 57).
Regarding PI3Ks, although it has been indicated that PI3K-γ selectively regulates the signaling from G protein-coupled receptors (31, 32), including chemokine receptors, we observed that PI3K-α and -δ also regulated the signaling and functions of CXCR4 in maDCs. These results are in line with recent reports that indicate G protein-coupled receptors and tyrosine kinase receptors are less selective than previously thought regarding the type of PI3K that they can regulate (58). The activity of PI3K-β was not required to regulate CXCL12-mediated functions or signaling. Of all the molecules tested, only FOXO1/3, which is downstream of Akt, regulated survival, but not chemotaxis, indicating that although most components in the signaling pathway studied are bifunctional, there are signaling molecules, under the control of the components of the main pathway, that control CXCR4 functions more selectively. As most of the signaling molecules identified converge on mTORC1 (see Fig. 7), it is possible that some of the observed effects could be partially mediated by this complex; however, it is most likely that the individual signaling molecules involved may regulate chemotaxis and survival through additional unidentified downstream effectors. In this regard, most interestingly, the inhibition of the different signaling molecules studied did not affect CXCL12-dependent chemotaxis and survival at a similar extent (see for instance Fig. 4, B and C), consistent with the possibility that each of these molecules may use a different number of downstream effectors to control CXCL12-dependent chemotaxis and survival.
Apart from the use of selective inhibitors (52, 53), two types of experimental evidences support the notion that we have identified a bona fide pathway under the control of CXCR4. First is the fact that, the inhibitors affects differently chemotaxis and survival argues against the possibility that the inhibitors may affect unspecifically all of the signaling components and functions studied. Second, the use of similar inhibitors in a prior study on CCR7-dependent function and signaling in maDCs yields a different signaling pathway. In this regard, CCR7 controls chemotaxis and survival in maDCs using independent modules (1, 26, 27). Furthermore, similar signaling molecules seem to play a different regulatory role downstream of CXCR4 and CCR7. For instance, in maDCs inhibition of specific PI3K isoforms failed to block chemotaxis controlled by CCR7 (27, 59).
Although further studies are required to gain more information on the whole gamut of functions regulated by CXCR4 in maDCs, this receptor is apparently less multifunctional than CCR7, which controls endocytosis, differentiation, cytoarchitecture, migratory speed, and survival (1). The effects elicited by CXCL12 in maDCs likely complement the effects of CCL21 and CCL19, the ligands of CCR7, which can also control chemotaxis and survival in maDCs (26, 27, 29). In summary, the results obtained, apart from uncovering an unappreciated signaling mechanism used by CXCR4 to control its functions, may suggest strategies for a more precise modulation of these functions and the immune response.
Acknowledgments
We thank Dr. M. Thelen for the anti-CXCR7 mAb clone 9C4, Dr. T. G. Unterman for the FOXO1-GFP plasmid, Dr. T. Schall for the anti-CXCR7 mAb clone 11G8 and the CXCR7 inhibitor CCX733, and Calistoga Pharmaceuticals for the PI3K-δ inhibitor IC87114. We appreciate the help of L. Gómez Cabañas and Drs. A. Rey-Gallardo and A. Domínguez.
This work was supported by Redes Temáticas de Investigación Cooperativas (RETICS) Program/Instituto de Salud Carlos III Grant RD08/0075 (to J. L. R.-F. and J. L. P.), Ministerio de Educación y Ciencia Grant SAF2005-00801 (to J. L. R.-F.), Proyectos Intramurales de Incorporación, Consejo Superior de Investigaciones Científicas Grant 200620I167, and by Genoma España.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Experimental Procedures.
- DC
- dendritic cell
- CMFDA
- 5-chloromethylfluorescein diacetate
- 4E-BP1
- eukaryotic translation initiation factor 4E-binding protein 1
- FOXO
- Forkhead box class O
- LN
- lymph node
- maDC
- mature DC
- mTORC1 and mTORC2
- mammalian target of rapamycin complex 1 and 2
- PLN
- popliteal LN
- PTX
- pertussis toxin
- S6K
- S6 kinase
- TSC2
- tuberous sclerosis protein 2
- SR
- sulforhodamine B
- Z-VAD-FMK
- Z-Val-Ala-Asp(OMe)-fluoromethyl ketone.
REFERENCES
- 1. Sánchez-Sánchez N., Riol-Blanco L., Rodríguez-Fernández J. L. (2006) J. Immunol. 176, 5153–5159 [DOI] [PubMed] [Google Scholar]
- 2. Randolph G. J., Ochando J., Partida-Sánchez S. (2008) Annu. Rev. Immunol. 26, 293–316 [DOI] [PubMed] [Google Scholar]
- 3. Lin C. L., Suri R. M., Rahdon R. A., Austyn J. M., Roake J. A. (1998) Eur. J. Immunol. 28, 4114–4122 [DOI] [PubMed] [Google Scholar]
- 4. Vecchi A., Massimiliano L., Ramponi S., Luini W., Bernasconi S., Bonecchi R., Allavena P., Parmentier M., Mantovani A., Sozzani S. (1999) J. Leukocyte Biol. 66, 489–494 [DOI] [PubMed] [Google Scholar]
- 5. Ouwehand K., Santegoets S. J., Bruynzeel D. P., Scheper R. J., de Gruijl T. D., Gibbs S. (2008) Eur. J. Immunol. 38, 3050–3059 [DOI] [PubMed] [Google Scholar]
- 6. Kabashima K., Shiraishi N., Sugita K., Mori T., Onoue A., Kobayashi M., Sakabe J., Yoshiki R., Tamamura H., Fujii N., Inaba K., Tokura Y. (2007) Am. J. Pathol. 171, 1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pablos J. L., Amara A., Bouloc A., Santiago B., Caruz A., Galindo M., Delaunay T., Virelizier J. L., Arenzana-Seisdedos F. (1999) Am. J. Pathol. 155, 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hargreaves D. C., Hyman P. L., Lu T. T., Ngo V. N., Bidgol A., Suzuki G., Zhou Y. R., Littman D. R., Cyster J. G. (2001) J. Exp. Med. 194, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. González N., Bermejo M., Calonge E., Jolly C., Arenzana-Seisdedos F., Pablos J. L., Sattentau Q. J., Alcamí J. (2010) J. Virol. 84, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Timmer T. C.., Baltus B., Vondenhoff M., Huizinga T. W., Tak P. P., Verweij C. L., Mebius R. E., van der Pouw Kraan T. C. (2007) Arthritis Rheum. 56, 2492–2502 [DOI] [PubMed] [Google Scholar]
- 11. Kabashima K., Sugita K., Shiraishi N., Tamamura H., Fujii N., Tokura Y. (2007) Biochem. Biophys. Res. Commun. 361, 1012–1016 [DOI] [PubMed] [Google Scholar]
- 12. Hernández-López C., Valencia J., Hidalgo L., Martínez V. G., Zapata A. G., Sacedón R., Varas A., Vicente A. (2008) Immunol. Lett. 120, 72–78 [DOI] [PubMed] [Google Scholar]
- 13. Sozzani S., Allavena P., D'Amico G., Luini W., Bianchi G., Kataura M., Imai T., Yoshie O., Bonecchi R., Mantovani A. (1998) J. Immunol. 161, 1083–1086 [PubMed] [Google Scholar]
- 14. Delgado E., Finkel V., Baggiolini M., Mackay C. R., Steinman R. M., Granelli-Piperno A. (1998) Immunobiology 198, 490–500 [DOI] [PubMed] [Google Scholar]
- 15. Infantino S., Moepps B., Thelen M. (2006) J. Immunol. 176, 2197–2207 [DOI] [PubMed] [Google Scholar]
- 16. Tilton B., Ho L., Oberlin E., Loetscher P., Baleux F., Clark-Lewis I., Thelen M. (2000) J. Exp. Med. 192, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki Y., Rahman M., Mitsuya H. (2001) J. Immunol. 167, 3064–3073 [DOI] [PubMed] [Google Scholar]
- 18. Vlahakis S. R., Villasis-Keever A., Gomez T., Vanegas M., Vlahakis N., Paya C. V. (2002) J. Immunol. 169, 5546–5554 [DOI] [PubMed] [Google Scholar]
- 19. Huttenlocher A., Poznansky M. C. (2008) Trends Cell Biol. 18, 298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thelen M., Thelen S. (2008) J. Neuroimmunol. 198, 9–13 [DOI] [PubMed] [Google Scholar]
- 21. Berahovich R. D., Zabel B. A., Penfold M. E., Lewén S., Wang Y., Miao Z., Gan L., Pereda J., Dias J., Slukvin I. I., McGrath K. E., Jaen J. C., Schall T. J. (2010) J. Immunol. 185, 5130–5139 [DOI] [PubMed] [Google Scholar]
- 22. Balabanian K., Lagane B., Infantino S., Chow K. Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., Bachelerie F. (2005) J. Biol. Chem. 280, 35760–35766 [DOI] [PubMed] [Google Scholar]
- 23. Hartmann T. N., Grabovsky V., Pasvolsky R., Shulman Z., Buss E. C., Spiegel A., Nagler A., Lapidot T., Thelen M., Alon R. (2008) J. Leukoc. Biol. 84, 1130–1140 [DOI] [PubMed] [Google Scholar]
- 24. Thelen M. (2001) Nat. Immunol. 2, 129–134 [DOI] [PubMed] [Google Scholar]
- 25. Neptune E. R., Bourne H. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sánchez-Sánchez N., Riol-Blanco L., de la Rosa G., Puig-Kröger A., García-Bordas J., Martín D., Longo N., Cuadrado A., Cabañas C., Corbí A. L., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2004) Blood 104, 619–625 [DOI] [PubMed] [Google Scholar]
- 27. Riol-Blanco L., Sánchez-Sánchez N., Torres A., Tejedor A., Narumiya S., Corbí A. L., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2005) J. Immunol. 174, 4070–4080 [DOI] [PubMed] [Google Scholar]
- 28. Riol-Blanco L., Delgado-Martín C., Sánchez-Sánchez N., Alonso-C. L. M., Gutiérrez-López M. D., Del Hoyo G. M., Navarro J., Sánchez-Madrid F., Cabañas C., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2009) Nat. Immunol. 10, 753–760 [DOI] [PubMed] [Google Scholar]
- 29. Escribano C., Delgado-Martín C., Rodríguez-Fernández J. L. (2009) J. Immunol. 183, 6282–6295 [DOI] [PubMed] [Google Scholar]
- 30. Del Prete A., Vermi W., Dander E., Otero K., Barberis L., Luini W., Bernasconi S., Sironi M., Santoro A., Garlanda C., Facchetti F., Wymann M. P., Vecchi A., Hirsch E., Mantovani A., Sozzani S. (2004) EMBO J. 23, 3505–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Procko E., McColl S. R. (2005) Bioessays 27, 153–163 [DOI] [PubMed] [Google Scholar]
- 32. Deane J. A., Fruman D. A. (2004) Annu. Rev. Immunol. 22, 563–598 [DOI] [PubMed] [Google Scholar]
- 33. Foster K. G., Fingar D. C. (2010) J. Biol. Chem. 285, 14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ganju R. K., Brubaker S. A., Meyer J., Dutt P., Yang Y., Qin S., Newman W., Groopman J. E. (1998) J. Biol. Chem. 273, 23169–23175 [DOI] [PubMed] [Google Scholar]
- 35. Wang F.., Herzmark P., Weiner O. D., Srinivasan S., Servant G., Bourne H. R. (2002) Nat. Cell Biol. 4, 513–518 [DOI] [PubMed] [Google Scholar]
- 36. Devreotes P., Janetopoulos C. (2003) J. Biol. Chem. 278, 20445–20448 [DOI] [PubMed] [Google Scholar]
- 37. Klemke R. L., Cai S., Giannini A. L., Gallagher P. J., de Lanerolle P., Cheresh D. A. (1997) J. Cell Biol. 137, 481–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Riol-Blanco L., Iglesias T., Sánchez-Sánchez N., de la Rosa G., Sánchez-Ruiloba L., Cabrera-Poch N., Torres A., Longo I., García-Bordas J., Longo N., Tejedor A., Sánchez-Mateos P., Rodríguez-Fernández J. L. (2004) Eur. J. Immunol. 34, 108–118 [DOI] [PubMed] [Google Scholar]
- 39. Yanagawa Y., Onoé K. (2003) Blood 101, 4923–4929 [DOI] [PubMed] [Google Scholar]
- 40. Rodríguez-Fernández J. L., Gómez M., Luque A., Hogg N., Sánchez-Madrid F., Cabañas C. (1999) Mol. Biol. Cell 10, 1891–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Koch W. J., Hawes B. E., Inglese J., Luttrell L. M., Lefkowitz R. J. (1994) J. Biol. Chem. 269, 6193–6197 [PubMed] [Google Scholar]
- 42. Zhang X., Gan L., Pan H., Guo S., He X., Olson S. T., Mesecar A., Adam S., Unterman T. G. (2002) J. Biol. Chem. 277, 45276–45284 [DOI] [PubMed] [Google Scholar]
- 43. Mempel T. R., Henrickson S. E., Von Andrian U. H. (2004) Nature 427, 154–159 [DOI] [PubMed] [Google Scholar]
- 44. Burns J. M., Summers B. C., Wang Y., Melikian A., Berahovich R., Miao Z., Penfold M. E., Sunshine M. J., Littman D. R., Kuo C. J., Wei K., McMaster B. E., Wright K., Howard M. C., Schall T. J. (2006) J. Exp. Med. 203, 2201–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hatse S., Princen K., Bridger G., De Clercq E., Schols D. (2002) FEBS Lett. 527, 255–262 [DOI] [PubMed] [Google Scholar]
- 46. Knight Z. A., Gonzalez B., Feldman M. E., Zunder E. R., Goldenberg D. D., Williams O., Loewith R., Stokoe D., Balla A., Toth B., Balla T., Weiss W. A., Williams R. L., Shokat K. M. (2006) Cell 125, 733–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim S., Garcia A., Jackson S. P., Kunapuli S. P. (2007) Blood 110, 4206–4213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Camps M., Rückle T., Ji H., Ardissone V., Rintelen F., Shaw J., Ferrandi C., Chabert C., Gillieron C., Françon B., Martin T., Gretener D., Perrin D., Leroy D., Vitte P. A., Hirsch E., Wymann M. P., Cirillo R., Schwarz M. K., Rommel C. (2005) Nat. Med. 11, 936–943 [DOI] [PubMed] [Google Scholar]
- 49. Sadhu C., Masinovsky B., Dick K., Sowell C. G., Staunton D. E. (2003) J. Immunol. 170, 2647–2654 [DOI] [PubMed] [Google Scholar]
- 50. Jackson S. P., Schoenwaelder S. M., Goncalves I., Nesbitt W. S., Yap C. L., Wright C. E., Kenche V., Anderson K. E., Dopheide S. M., Yuan Y., Sturgeon S. A., Prabaharan H., Thompson P. E., Smith G. D., Shepherd P. R., Daniele N., Kulkarni S., Abbott B., Saylik D., Jones C., Lu L., Giuliano S., Hughan S. C., Angus J. A., Robertson A. D., Salem H. H. (2005) Nat. Med. 11, 507–514 [DOI] [PubMed] [Google Scholar]
- 51. Holz M. K., Blenis J. (2005) J. Biol. Chem. 280, 26089–26093 [DOI] [PubMed] [Google Scholar]
- 52. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., Cohen P. (2007) Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Copp J., Manning G., Hunter T. (2009) Cancer Res. 69, 1821–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lu Z., Xu S. (2006) IUBMB Life 58, 621–631 [DOI] [PubMed] [Google Scholar]
- 56. Stahl M., Dijkers P. F., Kops G. J., Lens S. M., Coffer P. J., Burgering B. M., Medema R. H. (2002) J. Immunol. 168, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 57. Chen M., Huang L., Shabier Z., Wang J. (2007) Mol. Immunol. 44, 2558–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vanhaesebroeck B., Guillermet-Guibert J., Graupera M., Bilanges B. (2010) Nat. Rev. Mol. Cell Biol. 11, 329–341 [DOI] [PubMed] [Google Scholar]
- 59. Scandella E., Men Y., Legler D. F., Gillessen S., Prikler L., Ludewig B., Groettrup M. (2004) Blood 103, 1595–1601 [DOI] [PubMed] [Google Scholar]