Background: Mast cells express cPLA2α and iPLA2β.
Results: Knock-out of cPLA2α, not iPLA2β, hampers arachidonic acid mobilization in mast cells and adjacent fibroblasts.
Conclusion: Mast cell cPLA2α is coupled with stromal synthesis of anti-allergic PGE2, whereas iPLA2β is dispensable for mast cell function.
Significance: The cPLA2α-dependent transcellular PGE2 synthesis opens new insight into the lipid biochemistry and mast cell biology fields.
Keywords: Allergy, Arachidonic Acid, Gene Knock-out, Mass Spectrometry (MS), Mast Cell, Phospholipase A, Phospholipid Metabolism, Prostaglandins
Abstract
Mast cells release a variety of mediators, including arachidonic acid (AA) metabolites, to regulate allergy, inflammation, and host defense, and their differentiation and maturation within extravascular microenvironments depend on the stromal cytokine stem cell factor. Mouse mast cells express two major intracellular phospholipases A2 (PLA2s), namely group IVA cytosolic PLA2 (cPLA2α) and group VIA Ca2+-independent PLA2 (iPLA2β), and the role of cPLA2α in eicosanoid synthesis by mast cells has been well documented. Lipidomic analyses of mouse bone marrow-derived mast cells (BMMCs) lacking cPLA2α (Pla2g4a−/−) or iPLA2β (Pla2g6−/−) revealed that phospholipids with AA were selectively hydrolyzed by cPLA2α, not by iPLA2β, during FcϵRI-mediated activation and even during fibroblast-dependent maturation. Neither FcϵRI-dependent effector functions nor maturation-driven phospholipid remodeling was impaired in Pla2g6−/− BMMCs. Although BMMCs did not produce prostaglandin E2 (PGE2), the AA released by cPLA2α from BMMCs during maturation was converted to PGE2 by microsomal PGE synthase-1 (mPGES-1) in cocultured fibroblasts, and accordingly, Pla2g4a−/− BMMCs promoted microenvironmental PGE2 synthesis less efficiently than wild-type BMMCs both in vitro and in vivo. Mice deficient in mPGES-1 (Ptges−/−) had an augmented local anaphylactic response. These results suggest that cPLA2α in mast cells is functionally coupled, through the AA transfer mechanism, with stromal mPGES-1 to provide anti-anaphylactic PGE2. Although iPLA2β is partially responsible for PGE2 production by macrophages and dendritic cells, it is dispensable for mast cell maturation and function.
Introduction
Mast cells are important effector cells for IgE-associated allergic reactions, such as anaphylaxis, rhinitis, and asthma, and represent an important source of various inflammatory mediators, including vasoactive amines, proteases, arachidonic acid (AA)3 metabolites, cytokines, and chemokines. Mast cells orchestrate various aspects of the IgE-dependent and -independent immune responses not only through the release of these factors but also through cell-cell interaction by which they regulate the function of other cells. Mast cell precursors originating from bone marrow circulate in the blood and migrate into connective or mucous tissues, where they differentiate into mature mast cell phenotypes depending on tissue microenvironments (1–4). Stromal fibroblasts support the homing, growth, differentiation, and survival of mast cells by stem cell factor (SCF) and its receptor c-Kit as well as by other factors, including cytokines, chemokines, and adhesion molecules (5–8). Coculture of IL-3-maintained immature mouse bone marrow-derived mast cells (BMMCs) with fibroblasts is a useful in vitro system for analyzing certain aspects of the change to a mature connective tissue mast cell (CTMC)-like phenotype (9–14). Conversely, mast cells, though as yet unknown mechanisms, can affect the proliferation and functions of surrounding fibroblasts leading to collagen deposition and fibrosis, which are central features of chronic inflammation and tissue repair (15–21).
AA-derived eicosanoids (prostaglandins (PGs) and leukotrienes (LTs)) are produced in substantial amounts in tissues harboring TH2-based allergic responses (22, 23). Activated mast cells produce LTB4 and LTC4, which play crucial roles in allergic responses by facilitating bronchoconstriction, vascular permeability, and inflammatory cell recruitment (23, 24). PGD2 is a major prostanoid produced by activated mast cells (25, 26), and studies using mice deficient in the PGD receptor DP1 or DP2 (CRTH2) have revealed the roles of PGD2 in promotion or prevention of allergic responses in distinct cellular contexts (27–30). In contrast, gene targeting of the PGE receptor EP3 (31) or the biosynthetic enzyme microsomal PGE2 synthase (mPGES-1) (32) has demonstrated that PGE2 acts as a negative modulator of asthmatic reactions and that the PGE2-EP3 axis appears to be the long sought mechanism that explains the aspirin intolerance seen in asthma. Because mast cells produce PGE2 only minimally, stromal cells (e.g. fibroblasts) that surround mast cells may supply PGE2 to tissue microenvironments. However, how stromal PGE2 synthesis is regulated in local tissues where mast cells also reside is not fully understood.
Biosynthesis of prostanoids is initiated by the release of AA from membrane phospholipids by phospholipase A2 (PLA2). PLA2 is also considered to be important for the regulation of membrane remodeling. To date, more than 30 different PLA2 enzymes have been identified in mammals and are subdivided into three major classes as follows: cytosolic PLA2 (cPLA2) and Ca2+-independent PLA2 (iPLA2), which are high molecular weight intracellular PLA2s with a catalytic serine, and secreted PLA2 (sPLA2), which represents a group of low molecular weight, Ca2+-requiring enzymes with a His-Asp catalytic dyad (33, 34). Of the intracellular PLA2s, cPLA2α and iPLA2β are the “big two,” whose functions and regulatory mechanisms have been well studied (reviewed in Ref. 34). In mast cells, group IVA cPLA2α plays a critical role in PGD2 and LTC4 production after various stimuli such as FcϵRI cross-linking or cytokines (35, 36). The AA released by cPLA2α at the perinuclear Golgi membrane is converted by the sequential action of cyclooxygenase (COX) and terminal PG synthases to prostanoids or 5-lipoxygenase and terminal LT synthases to LTs. Group V sPLA2, in cooperation with cPLA2α, reportedly augments PGD2 production by BMMCs in response to zymosan (37) and promotes PGE2 production by neighboring fibroblasts through the transcellular pathway (38). Mast cells also express iPLA2β, a prototypic iPLA2 isoform, and the iPLA2 inhibitor bromoenol lactone (BEL) can attenuate granule exocytosis by mast cells (39), leading to the suggestion that iPLA2β participates in the regulation of mast cell activation. Although iPLA2β has long been thought to play a role in phospholipid remodeling (40), recent studies using mice or cells that are devoid of iPLA2β as a result of gene targeting or siRNA knockdown have defined its roles in various signaling events, such as Ca2+ release-activated Ca2+ (CRAC) channel opening (41), lipid mediator generation (42, 43), exocytosis (44), cytokine secretion (45), cell migration (46), vascular contractility (47), apoptosis (48, 49), cancer (50), and neuronal degeneration (51, 52). Because of its roles in Ca2+ gating and lipid mediator generation, the sequential action of iPLA2β and cPLA2α for the full operation of AA release has been proposed in vascular cells (42). However, the roles of iPLA2β in mast cells have not yet been fully elucidated.
In this study, using BMMCs null for cPLA2α, iPLA2β, group V sPLA2, or mPGES-1, in combination with lipidomic mass spectrometry (MS), we provide the following evidence: (i) cPLA2α plays a fundamental role in AA-selective release from BMMCs during FcϵRI-mediated activation and even during fibroblast-dependent maturation; (ii) iPLA2β minimally mobilizes phospholipids in mast cells during these processes; (iii) the AA selectively released by cPLA2α from BMMCs during maturation is transferred to adjacent fibroblasts to be metabolized to PGE2 by mPGES-1, whereas group V sPLA2 in BMMCs fails to participate in this process; and (iv) mPGES-1-driven PGE2 has a negative regulatory role in the allergen-triggered anaphylactic response. Overall, our findings underscore the importance of mast cell cPLA2α not only in the proximal production of mast cell-derived pro-allergic eicosanoids (PGD2 and LTC4) but also in the distal production of an anti-allergic eicosanoid (PGE2) by fibroblasts through the cell-to-cell AA transfer mechanism. Contrary to our expectation, however, iPLA2β was found to play no role in effector functions or phospholipid membrane remodeling in mast cells.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 mice were purchased from Japan SLC, Inc. Mice deficient in cPLA2α (Pla2g4a−/−) (36), iPLA2β (Pla2g6−/−) (52), group V sPLA2 (Pla2g5−/−) (37), and mPGES-1 (Ptges−/−) (53), all on a C57BL/6 background, were described previously. We used 8–12-week-old mice for all experiments. The genotypes of individual knock-out mice and their littermates were confirmed by PCR analysis of genomic DNA obtained by tail biopsy. All procedures involving animals were performed in accordance with protocols approved by the Institutional Animal Care and Use Committees of the Tokyo Metropolitan Institute of Medical Science and Showa University, in accordance with the Standards Relating to the Care and Management of Experimental Animals in Japan.
Culture and Activation of BMMCs, Macrophages, and Dendritic Cells
To prepare BMMCs, bone marrow cells were obtained from femurs and tibias of mice and cultured in IL-3-containing BMMC complete medium consisting of DMEM, 10% FBS, 2 mm l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 100 mm nonessential amino acids, and an optimal amount of recombinant mouse IL-3 that had been produced by the baculovirus/insect cell system, as described previously (11–14). After 4–5 weeks of culture, the majority of floating cells was confirmed to be immature BMMCs as assessed by Alcian blue-positive and safranin O-negative staining of their granules.
Maturation of immature BMMCs toward CTMC-like cells was described previously in detail (11–14). Briefly, 5 × 106 BMMCs were seeded on a subconfluent Swiss 3T3 fibroblast monolayer in 100-mm culture dishes and cocultured for up to 4 days in the presence of the baculovirus/insect cell-produced soluble form of SCF with replacement of the medium every 2 days. The cells were trypsinized and replated, and nonadherent cells (>97% being mast cells) were collected and used for analyses. Maturation of BMMCs into CTMC-like cells was verified by staining of their granules with Alcian blue and counterstaining with safranin O.
BMMCs (107 cells/ml) were incubated for 1 h in culture medium containing 10 μg/ml anti-dinitrophenyl (DNP) IgE (Sigma) and then treated for appropriate periods with various concentrations of DNP-conjugated albumin as an antigen (Ag) (Sigma) in culture medium. The percentage release of β-hexosaminidase (β-HEX), a degranulation marker, was evaluated as described previously (11). Aliquots of the supernatants were subjected to enzyme immunoassays for lipid mediators (PGD2, PGE2, or LTC4) (Cayman Chemicals) and IL-6 (eBioscience). As required for the experiments, the cells were activated in the presence of (S)- and (R)-BEL (Cayman Chemicals), which preferentially inhibit iPLA2β and iPLA2γ, respectively (53, 54).
Bone marrow-derived macrophages (BMMΦs) and dendritic cells (BMDCs) were prepared by culture of mouse bone marrow cells in medium supplemented for 3 days with 50 ng/ml human macrophage colony-stimulating factor (Kyowa Kirin) or for 9 days with 10 ng/mouse GM-CSF (PeproTech), respectively. BMMΦs and BMDCs were stimulated for 3 h with 100 μg/ml zymosan (Sigma) and for 24 h with 100 ng/ml LPS (Sigma). PGE2 levels in culture supernatants were measured by enzyme immunoassay as above.
Measurement of Intracellular Ca2+ Levels
IgE-sensitized BMMCs on coverslips were loaded for 60 min with the fluorescent Ca2+ indicator fura-2/AM (5 μm) (Invitrogen) in Tyrode/HEPES buffer, pH 7.4, containing 2.5 mm probenecid, 0.04% (v/v) pluronic acid, and 1% serum. Then the cells were washed and stimulated with Ag. Fura-2 fluorescence images were obtained using an ARGUS-50 image analyzer (Hamamatsu Photonics, Shizuoka, Japan) with excitation at 340 nm (F340) and 380 nm (F380) at 5-s intervals. The ratio (F340/F380) was calculated using ImageJ software (National Institutes of Health, Bethesda).
RT-PCR
Total RNA was extracted from BMMCs with TRIzol reagent (Invitrogen). First strand cDNA synthesis was carried out using a SuperScript III reverse transcriptase kit (Invitrogen). Total RNA (5 μg) was used in reactions primed with oligo(dT) (12–18-mer) primer (Invitrogen) to obtain cDNA. Then 1 μl of the synthesized cDNA was used as the template for the mRNA amplification reactions. The PCR amplification was performed with exTaq polymerase (Takara Biomedicals) using a GeneAmp PCR System 9600 (Applied Biosystems). The RT-PCR products were analyzed on a 1.5% agarose gel and visualized using ethidium bromide staining. The primer pairs used were as follows: mouse cPLA2α, 5′-ccactttgttctggccaaca-3′ and 5′-agggaaacagagcaacgaga-3′; mouse cPLA2β, 5′-agccccatgaacagaaactc-3′ and 5′-caaagagctcggaggagatg-3′; mouse cPLA2γ, 5′-agaacttggccagttggatg-3′ and 5′-gcactccttcttccacttgc-3′; mouse cPLA2δ, 5′-cggctgatgaagagacttcc-3′ and 5′-ctggtggaatggcctgccagtcag-3′; mouse cPLA2ϵ, 5′-ctgcatgaggatgaggtaccg-3′ and 5′-cctctcgccatttgtagagc-3′; mouse cPLA2ζ, 5′-gaagaacgtcctggagcttg-3′ and 5′-gtgagccaccaggacaccattgg-3′; mouse iPLA2β, 5′-catgagtacaatcaggacctg-3′ and 5′-caatctgaagtactggatgccga-3′; and mouse mast cell protease (mMCP-4)-4, 5′-tgagagagggttcacagctac-3′ and 5′-tcacagagggagtctctttgg-3′. The PCR thermal conditions were as follows: 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 40 s (for cPLA2s); 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 40 s (for iPLA2β), and 94 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s (for mMCP-4). RT-PCR for mouse GAPDH was performed as described previously (14). After 28 cycles of amplification, the PCR products were run on 1% agarose gels with ethidium bromide. Quantitative RT-PCR was carried out using a Power SYBR Green PCR system (Applied Biosystems) or a TaqMan Gene Expression System (Applied Biosystems) on the ABI7700 real time PCR system (Applied Biosystems), with oligonucleotide primers and probes (Roche Applied Science). The relative abundance of transcripts was normalized relative to the constitutive expression of 18 S ribosomal RNA.
Northern Blotting
Equal amounts (∼5 μg) of total RNA obtained from BMMCs using TRIzol reagent were applied to separate lanes of 1.2% formaldehyde-agarose gels, electrophoresed, and transferred to Immobilon-N membranes (Millipore). The resulting blots were then probed with appropriate cDNA probes that had been labeled with [32P]dCTP (PerkinElmer Life Sciences) by random priming (Takara Biomedicals). Hybridization and subsequent membrane washing were carried out as described previously (11).
Western Blotting
105 BMMCs were lysed in SDS-PAGE sample buffer (63 mm Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, and 0.08% bromphenol blue) containing 5% 2-mercaptoethanol, and then subjected to SDS-PAGE. Proteins were subsequently blotted onto nitrocellulose membranes, followed by blocking with 5% milk powder in PBS containing 0.05% Tween 20. The membranes were incubated for 2 h with rabbit polyclonal antibody against cPLA2α (Santa Cruz Biotechnology), iPLA2β (Cayman Chemicals), cyclooxygenase (COX)-2 (Santa Cruz Biotechnology), or mPGES-1 (Cayman Chemicals) (15, 59, 63) at 1:5000 dilution in PBS, 0.05% Tween 20. After washing with PBS, 0.05% Tween 20, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibody (Zymed Laboratories Inc.) at 1:5000 dilution in PBS, 0.05% Tween 20. After 1 h of incubation, the membranes were washed extensively with PBS, 0.05% Tween 20, followed by washing with PBS without detergent. The membranes were developed with the ECL system (PerkinElmer Life Sciences) in accordance with the protocol provided by the manufacturer.
Electrospray Ionization Mass Spectrometry (ESI-MS)
Total lipids were extracted from BMMCs (107 cells) using the method of Bligh and Dyer (55). ESI-MS analysis was performed using a 4000Q TRAP, quadrupole-linear ion trap hybrid mass spectrometer (AB SCIEX) with an UltiMate 3000 nano/cap/micro-liquid chromatography system (Dionex Corp.) combined with an HTS PAL autosampler (CTC Analytics AG). Phospholipids were subjected directly to ESI-MS analysis by flow injection; typically, 3 μl (3 nmol of phosphorus equivalent) of sample was applied. The mobile phase composition was acetonitrile/methanol/water (6:7:2) (plus 0.1% ammonium formate, pH 6.8) at a flow rate of 10 μl/min. The scan range of the instrument was set at m/z 200–1000 at a scan speed of 1000 Da/s. The trap fill-time was set at 3 ms in the positive ion mode and at 5 ms in the negative ion mode. The ion spray voltage was set at 5500 V in the positive ion mode and at −4500 V in the negative ion mode. Nitrogen was used as a curtain gas (setting of 10, arbitrary units) and as a collision gas (set to “high”). Details of the procedure for ESI-MS have been described previously (56, 57).
AA Transfer Experiments
BMMCs (2 × 106 cells) were preincubated with [3H]AA (0.1 μCi/ml) overnight. The 3H-prelabeled BMMCs were washed with medium and then seeded onto Swiss 3T3 cells (2 × 105 cells/0.5 ml/well) in 24-well plates. After appropriate periods, BMMCs and Swiss 3T3 cells were separated by trypsinization (see above), and the radioactivity incorporated into each cell type was measured using a liquid scintillation counter (Aloka).
Passive Cutaneous Anaphylaxis
Mouse anti-DNP IgE (25 ng in 25 μl of saline) was injected intradermally into the ears of 8–12-week-old male mice. One day later, the mice were intravenously challenged with 60 ng of Ag in 200 μl of PBS containing 0.8% (w/v) Evans blue (Sigma). After 30 min, their ears were removed and dissolved, and the dye extravasation was determined colorimetrically at 630 nm, as described previously (14).
Mast Cell Knock-in
To reconstitute mast cells in mast cell-deficient mice (3, 4), 5 × 106 BMMCs were injected subcutaneously into the ears of KitW-sh/W-sh mice (The Jackson Laboratory). After 2 days, the ears were homogenized in PBS containing 10 μm indomethacin (Sigma); the supernatants were adjusted to pH 3.0 with 1 m HCl and passed through a Sep-Pak C18 cartridge (Waters), and the retained PGE2 were eluted with 3 ml of methanol, as described previously (53). A trace amount of [3H]PGE2 (PerkinElmer Life Sciences) was added to the samples before passage through the cartridges to calibrate the recovery of the PGE2. The sample solvents were evaporated, and then the PGE2 was dissolved in an aliquot of buffer and assayed with EIA kit.
Statistical Analysis
Data were evaluated statistically by unpaired Student's t test at a significance level of p < 0.05.
RESULTS
Expression of cPLA2α and iPLA2β in BMMCs
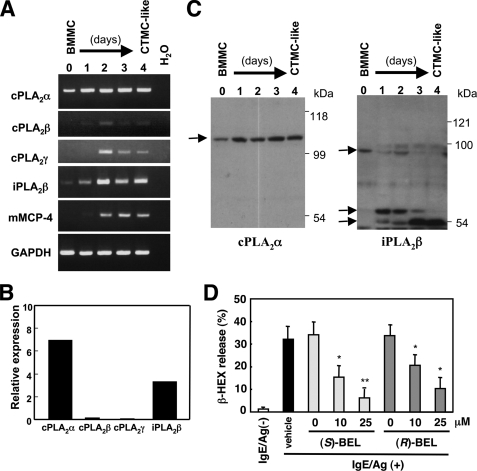
We examined the expression of several intracellular PLA2 enzymes in IL-3-maintained immature BMMCs as well as those placed on Swiss 3T3 fibroblasts in the presence of SCF (designated CTMC-like cells hereafter) by RT-PCR (Fig. 1). As a positive control for the maturation of BMMCs into CTMC-like cells, we monitored the expression of mMCP-4 (Mcpt4) mRNA (58), whose expression was minimal in BMMCs and markedly induced after 2–4 days of coculture with fibroblasts (Fig. 1A). Among six members of the cPLA2 family (α, β, γ, δ, ϵ, and ζ), expression of cPLA2α mRNA was readily detected in BMMCs and increased modestly during the course of maturation toward CTMC-like cells (Fig. 1A). Expression of mRNAs for cPLA2β and cPLA2γ was barely detectable in BMMCs and was elevated (albeit still at low levels) in CTMC-like cells (Fig. 1A), whereas that of mRNAs for cPLA2δ, -ϵ and -ζ was below the detection limit under all conditions tested (data not shown). The prototypic iPLA2 isoform, iPLA2β, was expressed at a significant level in BMMCs and markedly elevated in CTMC-like cells (Fig. 1A). Quantitative RT-PCR confirmed that cPLA2α and iPLA2β are the two major intracellular PLA2s expressed in BMMCs (Fig. 1B).
FIGURE 1.
Expression of various intracellular PLA2 in mast cells. A, RNAs obtained from BMMCs maintained in IL-3 and those cocultured for the indicated periods with Swiss 3T3 cells (CTMC-like cells) were subjected to RT-PCR for various PLA2s (28 cycles). RT-PCR for mMCP-4, a CTMC-specific protease, was performed as a positive control for the differentiation of immature BMMCs into mature CTMCs. Equal loading of each sample was verified by RT-PCR of Gapdh, a housekeeping gene. RT-PCR without RNA (H2O) was done as a negative control. B, quantitative RT-PCR of intracellular PLA2s in BMMCs. C, lysates of replicate cells were subjected to SDS-PAGE (8% gel) followed by immunoblotting with cPLA2α and iPLA2β. Arrows indicate the positions of major bands for individual PLA2s. D, effects of (S)- or (R)-BEL on β-HEX release from IgE/Ag-activated BMMCs (mean ± S.E., n = 5; *, p < 0.05 and **, p < 0.01).
The increase of cPLA2α protein, as assessed by immunoblotting, paralleled that of its mRNA, reaching a plateau level by 1 day after coculture (Fig. 1C, left panel). Interestingly, immunoblotting of iPLA2β revealed that the full-size iPLA2β protein (∼90 kDa), found in BMMCs on day 0, was sequentially converted to an ∼60-kDa form on days 1–2 and then to an ∼50-kDa form on days 3–4, with an incremental expression of the latter (Fig. 1C, right panel). Considering that iPLA2β is known to undergo proteolytic processing leading to its activation during cell activation or apoptosis (48, 49, 59, 60), our results may represent another example of iPLA2β processing during a particular cellular process, i.e. mast cell maturation.
It has been reported that the FcϵRI-dependent and -independent exocytotic responses are attenuated in BMMCs and RBL-2H3 cells (rat mastocytoma) by treatment with BEL, an iPLA2 inhibitor (39). In addition, iPLA2β has been implicated in activation of the CRAC channel (41), which is essential for Ca2+ entry and thereby subsequent effector functions in FcϵRI-activated mast cells (61). Furthermore, contributions of iPLA2β to stimulus-coupled AA release, likely in concert with cPLA2α, have been demonstrated in vascular cells (42). To assess whether iPLA2β indeed participates in mast cell activation or not, we initially examined the effect of (S)-BEL (a preferential inhibitor of iPLA2β) and (R)-BEL (a preferential inhibitor of iPLA2γ) on β-HEX release (a marker of degranulation) by IgE/Ag-stimulated BMMCs. Both agents inhibited β-HEX release in a dose-dependent manner, with slightly better inhibition by (S)-BEL than by (R)-BEL (Fig. 1D).
iPLA2β Plays No Role in Effector Functions of Mast Cells
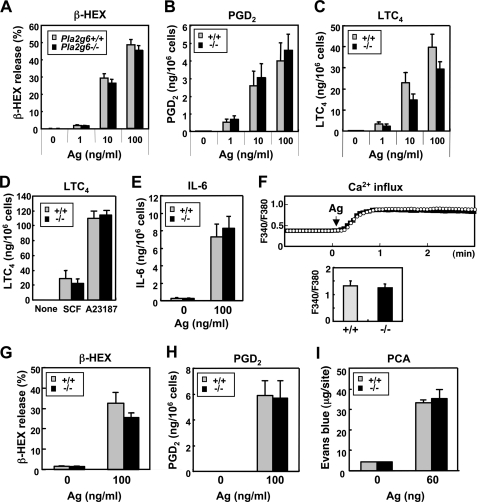
However, because it has become apparent that BEL is not specific for iPLA2β but can also broadly inhibit other iPLA2 isoforms as well as some other signaling molecules and proteases (62, 63), we next attempted to clarify the role of iPLA2β in mast cells using Pla2g6−/− mice, in which iPLA2β is ablated (52). Expression of other PLA2s expressed in BMMCs was unaffected by iPLA2β deficiency (supplemental Fig. S1). When IgE-sensitized BMMCs from Pla2g6+/+ and Pla2g6−/− mice were stimulated with various concentrations of Ag, we observed no difference in β-HEX exocytosis between the genotypes at each dose (Fig. 2A). Likewise, IgE/Ag-dependent generation of the eicosanoids, PGD2 (Fig. 2B) and LTC4 (Fig. 2C), did not differ appreciably between Pla2g6+/+ and Pla2g6−/− mice. Generation of PGD2 (data not shown) and LTC4 (Fig. 2D) in response to SCF or A23187 was also approximately the same between the genotypes. Likewise, IgE/Ag-elicited secretion of IL-6 was unimpaired in Pla2g6−/− mice (Fig. 2E). Consistent with the normal effector functions (exocytosis, eicosanoid synthesis, and cytokine induction), the IgE/Ag-induced increase of intracellular Ca2+ was equivalent between Pla2g6+/+ and Pla2g6−/− mice (Fig. 2F). Even after coculture with fibroblasts for 1 week, a period sufficient to induce functional maturation of mast cells (11–14), IgE/Ag-stimulated release of β-HEX (Fig. 2G) and generation of PGD2 (Fig. 2H) and LTC4 (data not shown) by Pla2g6−/− cells were similar to those by Pla2g6+/+ cells, suggesting that the deficiency of iPLA2β did not affect the functional maturation of BMMCs toward CTMC-like cells. Finally, the Ag-induced PCA reaction, which depends on histamine and LTC4 released by dermal mast cells, occurred normally in IgE-sensitized Pla2g6−/− mice (Fig. 2I), indicating that the differentiation and function of mast cells in vivo are not hampered by the absence of iPLA2β. Thus, iPLA2β plays no role in mast cell activation, and the suppressive effect of BEL on degranulation by BMMCs (Fig. 1D) appears to result from its off-target effects, rather than from inhibition of iPLA2β.
FIGURE 2.
iPLA2β plays no role in the effector functions of mast cells. A–C, IgE-sensitized BMMCs from Pla2g6+/+ and Pla2g6−/− mice were challenged with the indicated concentrations of Ag for 10 min, and the releases of β-HEX (A), PGD2 (B), and LTC4 (C) into the supernatants were evaluated (mean ± S.D., n = 5–6). D, LTC4 generation by Pla2g6+/+ and Pla2g6−/− BMMCs treated for 10 min with 100 ng/ml SCF or 1 μm A23187 (mean ± S.D., n = 3). E, IgE/Ag-stimulated BMMCs from Pla2g6+/+ and Pla2g6−/− mice were cultured for 10 h in the presence of SCF and IL-1β (which amplify cytokine expression in BMMCs (63)) to assess IL-6 secretion (mean ± S.D., n = 3). F, IgE/Ag-triggered Ca2+ influx into BMMCs. A representative monitoring of intracellular Ca2+ levels in Pla2g6+/+ (white circles) and Pla2g6−/− (solid circles) BMMCs after Ag challenge (upper panel) and an average value at 3 min (mean ± S.E.; n = 3) (lower panel) are shown. G and H, IgE-sensitized Pla2g6+/+ and Pla2g6−/− CTMC-like cells, which had been cocultured for 1 week with Swiss 3T3 fibroblasts, were treated with or without Ag for 10 min, and the releases of β-HEX (G) and PGD2 (H) into the supernatants were evaluated (mean ± S.D., n = 3). I, IgE-sensitized Pla2g6+/+ and Pla2g6−/− mice were challenged with or without Ag to assess PCA reaction (mean ± S.D., n = 5).
Role of cPLA2α, but Not iPLA2β, in FcϵRI-activated AA Mobilization from Mast Cell Membranes
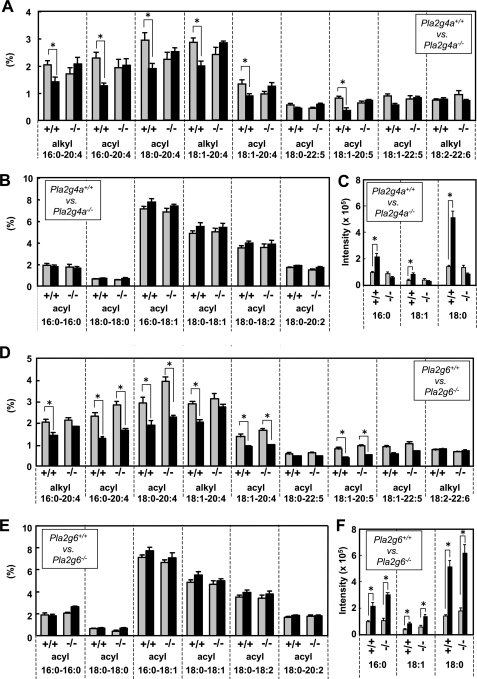
To assess the lipid dynamics during IgE/Ag activation of BMMCs, lipids extracted from BMMCs with or without IgE/Ag stimulation for 2 min (a time point when the rate of AA release was maximal (64)) were subjected to ESI-MS analyses. Quantification of average scores (n = 4) for individual PC molecular peaks revealed that, in wild-type BMMCs, all phosphatidylcholine (PC) species containing sn-2 AA (20:4) or eicosapentaenoic acid (EPA; 20:5) were significantly decreased (Fig. 3A), with a concomitant increase in LPC molecular species (Fig. 3C), following IgE/Ag treatment. In contrast, PC molecular species with sn-2 palmitic acid (16:0), stearic acid (18:0), oleic acid (OA; C18:1), linoleic acid (LA; 18:2), and docosahexaenoic acid (22:6) were unchanged irrespective of the stimulation (Fig. 3, A and B). It is known that cPLA2α shows selectivity for sn-2 AA- and EPA-bearing phospholipids (65), and in fact, the stimulus-dependent selective reductions of AA- and EPA-containing PC species as well as increases in LPC species were entirely absent in BMMCs from Pla2g4a−/− mice (Fig. 3, A–C). Thus, these lipidomic studies confirmed that most of the FcϵRI-dependent AA release by mast cells is mediated by cPLA2α (35, 36).
FIGURE 3.
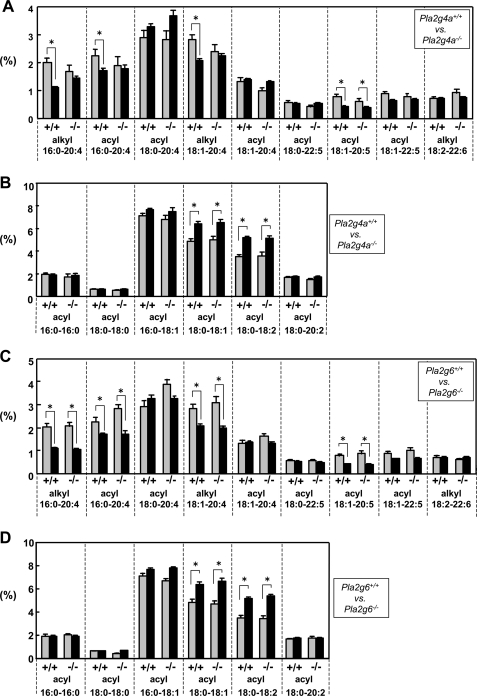
ESI-MS analysis of PC molecular species in BMMCs with or without IgE/Ag stimulation. BMMCs from Pla2g4a+/+ and Pla2g4a−/− mice (A–C) or from Pla2g6+/+ and Pla2g6−/− mice (D–F) were sensitized with IgE and then treated with (filled bars) or without (gray bars) 100 ng/ml Ag for 2 min. Lipids extracted from these cells were subjected to ESI-MS on a positive ion mode. PC species bearing sn-2 fatty acids with a high degree (4) of unsaturation (A and D), those bearing sn-2 fatty acids with no or a low degree (2) of unsaturation (B and E), and LPC species (C and F) were quantified. Values indicate the percentages of individual PC species relative to total PC mass (A, B, D, and E) or signal intensities of individual LPC species on ESI-MS (C and F) (mean ± S.D., n = 3–4; *, p < 0.05).
Although it has been reported that prior activation of iPLA2β and thereby gating of the CRAC channel may be important for full activation of cPLA2α in vascular smooth muscle cells (41, 42), it is unlikely that the iPLA2β-cPLA2α axis is operative in BMMCs because, in agreement with unaltered Ca2+ response and effector functions (Fig. 2), IgE/Ag-evoked decreases in AA- or EPA-containing PC species and reciprocal increases in LPC species occurred almost normally in Pla2g6−/− BMMCs (Fig. 3, D–F). An exception was the propensity for less reduction of some PC species with AA (alkyl 16:0–20:4 and alkyl 18:1–20:4) in IgE/Ag-activated Pla2g6−/− BMMCs in comparison with replicate wild-type BMMCs (Fig. 3D). This subtle change might be related to the modestly decreasing trend of LTC4 synthesis in Pla2g6−/− BMMCs (Fig. 2C), although we did not investigate this issue further because the difference in LTC4 synthesis between the genotypes did not reach statistical significance.
Role of cPLA2α, but Not iPLA2β, in Phospholipid Remodeling during Mast Cell Maturation
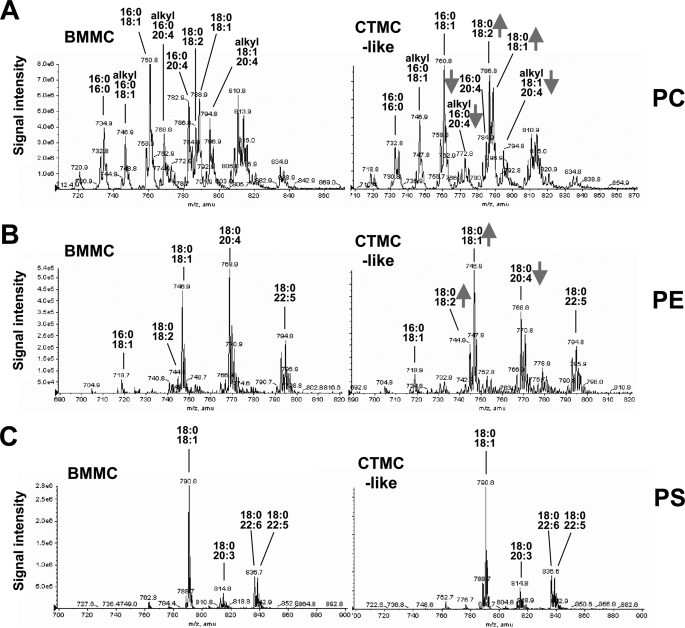
We next examined whether the elevated expression of cPLA2α or iPLA2β in BMMCs during in vitro maturation would be associated with some compositional changes in membrane phospholipids. To this end, total lipids were extracted from BMMCs (i.e. maintained in the presence of IL-3 without coculture) and CTMC-like differentiating cells (i.e. after coculture with fibroblasts plus SCF for 4 days) and then subjected to ESI-MS analyses. Representative ESI-MS profiles of phospholipids (PC, phosphatidylethanolamine (PE), and phosphatidylserine (PS)) in mast cells from wild-type mice are shown in Fig. 4, which indicated that several if not all PC and PE molecular species containing sn-2 AA were decreased, whereas those with sn-2 OA or LA were conversely increased, in CTMC-like cells in comparison with IL-3-maintained BMMCs. In contrast, there were no alterations in PS molecular species between BMMCs and CTMC-like cells (Fig. 4).
FIGURE 4.
ESI-MS profiles of phospholipids in BMMCs and CTMC-like cells from C57BL/6 mice. IL-3-maintained BMMCs (left panel) and CTMC-like differentiated cells after coculture for 4 days with Swiss 3T3 cells (right panel) were subjected to ESI-MS for PC, PE, and PS. Fatty acid compositions of major peaks are indicated. Peaks reduced or increased in CTMC-like cells relative to BMMCs are shown by arrows.
The results shown in Fig. 4 were further validated by quantification of average scores (n = 4) for the proportion of individual PC molecular species (Fig. 5). In CTMC-like cells, several if not all PC molecular species with sn-2 AA and EPA, namely PC with diacyl 16:0–20:4, alkyl 16:0–20:4, alkyl 18:1–20:4, and diacyl 18:1–20:5, were selectively and significantly decreased relative to those in IL-3-maintained BMMCs (Fig. 5A). In contrast, CTMC-like cells contained more PC molecular species with diacyl 18:0–18:1 and 18:0–18:2 than did BMMCs (Fig. 5B). Thus, maturation of BMMCs toward CTMC-like cells was accompanied by decreased unsaturation of sn-2 fatty acids in phospholipids. Notably, the decrease of AA-containing PC species was not seen in Pla2g4a−/− CTMC-like cells (Fig. 5A), suggesting that cPLA2α is mostly responsible for the liberation of AA from phospholipids during mast cell maturation. However, the decrease of PC with EPA and the increase of PC with OA and LA were not influenced by the deficiency of cPLA2α (Fig. 5, A and B), suggesting that these processes are regulated by other PLA2(s) and/or acyltransferase(s). Importantly, these changes in PC molecular species were not impaired at all in Pla2g6−/− cells (Fig. 5, C and D). Thus, although iPLA2β has been believed to be a regulator of phospholipid membrane remodeling (40), it does not contribute significantly to this process in mast cells.
FIGURE 5.
ESI-MS analysis of PC molecular species in BMMCs and CTMC-like cells. IL-3-maintained BMMCs (gray bars) and CTMC-like differentiated cells after coculture for 4 days with Swiss 3T3 cells in the presence of SCF (filled bars) from Pla2g4a+/+ and Pla2g4a−/− mice (A and B) or from Pla2g6+/+ and Pla2g6−/− mice (C and D) were subjected to ESI-MS analyses for PC molecular species on a positive ion mode. PC species bearing sn-2 fatty acids with a high degree (4) of unsaturation (A and C) and those bearing sn-2 fatty acids with no or a low degree (2) of unsaturation (B and D) were quantified. Values indicate the percentages of individual PC species relative to total PC mass (mean ± S.D., n = 3–4; *, p < 0.05).
Mast Cell cPLA2α Is Coupled with Fibroblastic mPGES-1-dependent PGE2 Production
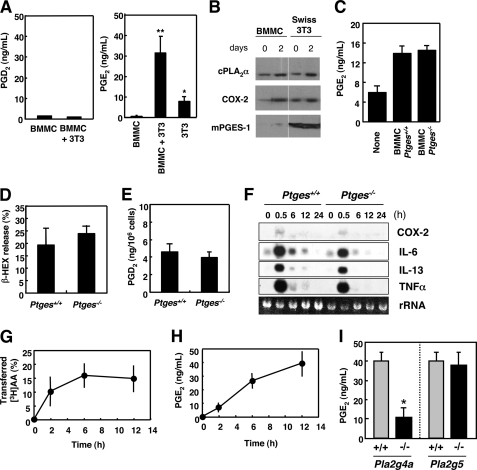
Having established that AA-containing phospholipids are selectively decreased by cPLA2α during coculture of BMMCs with fibroblasts, we next examined whether the free AA thus released would be metabolized to PGD2, a main mast cell-produced prostanoid. However, the level of PGD2 was low in the supernatants of BMMC-fibroblast cocultures (Fig. 6A, left panel). Unexpectedly, the level of PGE2, which was produced to some extent by Swiss 3T3 cells alone and not by BMMCs, was markedly increased in the coculture supernatants (Fig. 6A, right panel). Addition of the coculture supernatants to fresh Swiss 3T3 cells did not increase the PGE2 level further (data not shown), suggesting that the increased PGE2 synthesis requires direct contact between BMMCs and fibroblasts and that it is not mediated by cytokines or other soluble factors secreted from BMMCs.
FIGURE 6.
cPLA2α in mast cell is coupled with mPGES-1 in fibroblasts to provide anti-allergic PGE2. A, BMMCs alone, BMMCs plus Swiss 3T3 cells, and Swiss 3T3 alone were cultured for 2 days, and the resultant supernatants were taken for enzyme immunoassay for PGD2 (left panel) or PGE2 (right panel). Means ± S.D., n = 6; *, p < 0.01 versus BMMCs, or Swiss 3T3 cells alone and **, p < 0.01 versus BMMCs alone. B, expression of PGE2 biosynthetic enzymes in BMMCs and Swiss 3T3 cells before and 2 days after coculture, as assessed by immunoblotting. C, PGE2 production by Swiss 3T3 cells alone and those cocultured with Ptges+/+ or Ptges−/− BMMCs (mean ± S.D., n = 2). D and E, BMMCs from Ptges+/+ and Ptges−/− mice were sensitized with IgE and stimulated with Ag for 15 min to assess β-HEX exocytosis (D) and PGD2 generation (E) (mean ± S.D., n = 8-∼12). F, total RNAs extracted from Ptges+/+ and Ptges−/− BMMCs after stimulation with Ag for the indicated periods were subjected to Northern blotting for COX-2 and various cytokines (IL-6, IL-13, and TNFα). rRNA, 28 S ribosomal RNA visualized in an agarose gel with ethidium bromide. A representative result of three independent experiments is shown. G, [3H]AA-prelabeled BMMCs were cocultured with Swiss 3T3 cells, and percentage transfer of [3H]AA to Swiss 3T3 cells relative to total incorporation of [3H]AA into BMMCs was monitored over time (mean ± S.D., n = 4). H, time course of PGE2 synthesis by BMMC-Swiss 3T3 coculture (mean ± S.D., n = 4). I, BMMCs obtained from Pla2g4a−/− (n = 8) and Pla2g5−/− (n = 4) mice and their littermate wild-type controls were cocultured with Swiss 3T3 cells for 12 h to assess the coculture-dependent increase in PGE2 release (mean ± S.D., *, p < 0.05 versus Pla2g4a+/+ control).
Expression of a panel of enzymes involved in PGE2 biosynthesis in BMMCs and Swiss 3T3 cells before and after coculture, as assessed by immunoblotting, is shown in Fig. 6B. In BMMCs, cPLA2α (also see Fig. 1B) and COX-2 were increased significantly after coculture with fibroblasts, whereas mPGES-1, a terminal enzyme crucial for isomerization of COX-2-produced PGH2 to PGE2, was barely detectable regardless of culture conditions (Fig. 6B, left panel). All three of these enzymes were constitutively expressed in Swiss 3T3 cells, in which only cPLA2α exhibited an obvious increase after coculture (Fig. 6B, right panel). The almost complete absence of mPGES-1 expression in BMMCs suggested that Swiss 3T3 cells, but not BMMCs, were the main source of PGE2 during coculture.
To confirm this finding, we utilized BMMCs obtained from mPGES-1-deficient (Ptges−/−) mice. When BMMCs from Ptges+/+ and Ptges−/− mice were each cocultured with Swiss 3T3 cells, the levels of PGE2 released into the coculture supernatants were comparable (Fig. 6C), implying that the augmented PGE2 during coculture originated from Swiss 3T3 cells and not from BMMCs. Furthermore, although previous studies have shown that PGE2 promotes the differentiation of BMMCs from bone marrow cells (66) and modulates the effector functions of BMMCs (67–69), IL-3- or SCF-dependent growth (data not shown) and FcϵRI-dependent exocytosis of β-HEX (Fig. 6D), generation of PGD2 (Fig. 6E), and transcriptional induction of COX-2 and the cytokines IL-6, IL-13, and TNFα (all of which attained a maximal expression at 0.5 h after stimulation, as assessed by Northern blotting) (Fig. 6F) in Ptges−/− BMMCs were similar to those in replicate Ptges+/+ BMMCs, suggesting that the genetic deletion of mPGES-1 in BMMCs does not affect their proliferation and effector functions.
The results described above led us to hypothesize that the AA released by cPLA2α in BMMCs is transferred to Swiss 3T3 cells and is then metabolized to PGE2. To address whether AA would indeed be transferred from mast cells to fibroblasts, BMMCs were prelabeled with [3H]AA, washed, and then cocultured with Swiss 3T3 cells. A short (hours) rather than long (days) time course was chosen for this experiment to minimize any possibility of metabolic degradation of [3H]AA. As shown in Fig. 6G, [3H]AA was readily transferred from BMMCs to cocultured Swiss 3T3 cells within hours. This [3H]AA transfer was followed by a concomitant increase of PGE2 synthesis in BMMC-3T3 coculture (Fig. 6H).
To acquire clear evidence for the contribution of mast cell-associated cPLA2α to fibroblastic PGE2 synthesis, BMMCs obtained from Pla2g4a−/− and Pla2g4a+/+ mice were cocultured with Swiss 3T3 cells. We also used BMMCs obtained from group V sPLA2-null (Pla2g5−/−) mice, because it has been reported that group V sPLA2, which is expressed in BMMCs, is able to promote transcellular eicosanoid synthesis by adjacent cells (38, 70, 71). As shown in Fig. 6I, the augmented PGE2 production in coculture was partially reduced when Swiss 3T3 cells were incubated with Pla2g4a−/− BMMCs relative to that in coculture with control BMMCs, suggesting that a significant proportion of fibroblastic PGE2 is derived from AA released by cPLA2α in mast cells and that the remaining PGE2 in coculture with Pla2g4a−/− BMMC may be produced by activation of cPLA2α in fibroblasts. In contrast, the PGE2-synthetic responses in Swiss 3T3 cells cocultured with Pla2g5−/− BMMCs and those cocultured with control BMMCs were comparable (Fig. 6I), ruling out the contribution of group V sPLA2 to mast cell-driven fibroblastic (transcellular) PGE2 synthesis in this setting.
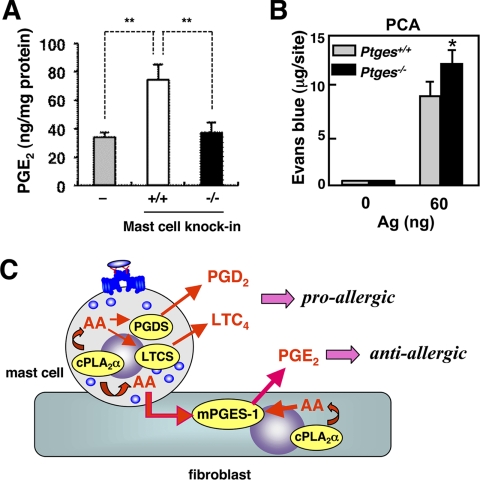
To address whether the mast cell cPLA2α-dependent transcellular PGE2 synthesis could occur in vivo, we carried out the adoptive transfer of Pla2g4a+/+ or Pla2g4a−/− BMMCs into mast cell-deficient mice (mast cell knock-in) (3, 4). Thus, BMMCs from either genotype were transferred subcutaneously into the ears of KitW-sh/W-sh mice, and after 2 days of this mast cell reconstitution, PGE2 levels in the ear homogenates were quantified. As shown in Fig. 7A, a substantial level of PGE2 was already present in the ears of KitW-sh/W-sh mice, indicating that this PGE2 pool is independent of mast cells. Strikingly, the PGE2 level was nearly doubled in the ears reconstituted with wild-type BMMCs, whereas Pla2g4a−/− BMMCs were unable to increase it (Fig. 7A). Proper reconstitution of BMMCs from both genotypes in KitW-sh/W-sh mice was verified by equal histamine levels (data not shown). These results provide evidence that mast cells have the ability to mobilize microenvironmental PGE2 synthesis in vivo and that this process depends on cPLA2α in mast cells.
FIGURE 7.
Mast cell cPLA2α-dependent biosynthesis of anti-anaphylactic PGE2in vivo. A, Pla2g4a+/+ or Pla2g4a−/− BMMCs were subcutaneously transferred into the ears of mast cell-deficient KitW-sh/W-sh mice. After 2 days of reconstitution, PGE2 levels in the ears of KitW-sh/W-sh mice (−), those transferred with Pla2g4a+/+ BMMCs (+/+), and those transferred with Pla2g4a−/− BMMCs (−/−) were quantified (mean ± S.E., n = 12, **, p < 0.01). B, IgE-sensitized Ptges+/+ and Ptges−/− mice were challenged with or without Ag to assess PCA reaction (mean ± S.E., n = 3; *, p < 0.05 versus replicate Ptges+/+ mice). C, schematic diagram of the dual role of cPLA2α in mast cells for the production of pro-allergic and anti-allergic lipid mediators. In mast cells, cPLA2α supplies AA (via COX-1 and -2, which are omitted in the figure) to hematopoietic PGD2 synthase (PGDS) and LTC4 synthase (LTCS) for the biosynthesis of pro-allergic PGD2 and LTC4, respectively (64). In adjacent fibroblasts, AA is supplied by cPLA2α intrinsically expressed in fibroblasts (cell autonomous pathway) and by cPLA2α in mast cells (transcellular pathway) to mPGES-1 for the biosynthesis of anti-allergic PGE2.
To assess the role of mPGES-1-derived PGE2 in the mast cell-dependent allergic response in vivo, we compared IgE/Ag-induced PCA reaction in Ptges+/+ and Ptges−/− mice. As shown in Fig. 7B, extravasation of Evans blue dye was modestly but significantly enhanced in Ag-challenged Ptges−/− mice in comparison with replicate Ptges+/+ mice. Thus, although the mast cell-produced eicosanoids PGD2 and LTC4 exert pro-allergic actions (23, 27, 28), mPGES-1-driven PGE2 produced by stromal fibroblasts, likely through fibroblast-associated intrinsic cPLA2α and through mast cell-resident extrinsic cPLA2α, has a counter-regulatory role in the anaphylactic response (Fig. 7C).
Participation of iPLA2β in PGE2 Production by Dendritic Cells and Macrophages
Finally, to ask whether the apparent absence of iPLA2β requirement is specific for mast cells or could be relevant to other cell types, we investigated the PGE2 biosynthetic response by other bone marrow-derived cells, i.e. dendritic cells and macrophages, from Pla2g6−/− mice as well as from Pla2g4a−/− mice for comparison. To this end, GM-CSF-induced BMDCs or macrophage colony-stimulating factor-induced BMMΦs from Pla2g6−/− mice, Pla2g4a−/− mice, or their littermate controls were stimulated with LPS (a ligand for TLR4) or zymosan (a ligand of TLR2 and dectin-1). As shown in supplemental Fig. S2A, LPS-stimulated PGE2 production by Pla2g6−/− BMDCs or BMMΦs was partially reduced relative to that by replicate control cells, whereas zymosan-stimulated PGE2 production was unaffected by iPLA2β deficiency. In comparison, LPS-stimulated PGE2 release was more profoundly, if not solely, dampened in Pla2g4a−/− BMDCs or BMMΦs, whereas almost no production of PGE2 was evident in these cells in response to zymosan (supplemental Fig. S2B). These results suggest that iPLA2β is partially responsible for LPS-, not zymosan-, stimulated PGE2 generation in dendritic cells and macrophages. Thus, the participation of iPLA2β in eicosanoid synthesis may depend on cell types or stimuli (see “Discussion”).
DISCUSSION
This study has revealed several new lipid-related processes that occur in mast cells during activation after FcϵRI cross-linking and during maturation after mast cell-fibroblast interaction as follows. (i) BMMCs express the two intracellular PLA2s, cPLA2α and iPLA2β, and interaction of BMMCs with fibroblasts results in increased expression of both enzymes (Fig. 1). (ii) Following IgE/Ag stimulation of BMMCs, cPLA2α regulates most of the AA-selective release, whereas iPLA2β plays minimal roles (Figs. 2 and 3). (iii) Maturation of BMMCs toward CTMC-like cells is accompanied by phospholipid remodeling, with reduction of AA-containing phospholipid species and a reciprocal increase of OA- and LA-containing species (Figs. 4 and 5). (iv) A significant proportion (if not all) of these maturation-related changes in membrane phospholipids is regulated by cPLA2α, whereas iPLA2β is again dispensable for this process (Fig. 5). (v) During maturation, the AA released by cPLA2α in CTMC-like differentiating cells is supplied to adjacent fibroblasts to be metabolized to PGE2 via mPGES-1 (Fig. 6). (vi) Finally, the mPGES-1-derived PGE2 can counteract the local anaphylactic reaction (Fig. 7). Although the molecular mechanism underlying the anti-anaphylactic action of PGE2 in vivo has not yet been fully understood, it appears to be mediated by the PGE receptor EP3, because its absence also results in exacerbated allergic reactions in mice (31). Reportedly, the deficiency of mPGES-1 also leads to augmented asthmatic airway inflammation, which could be explained by profound alteration in vascular remodeling (32). Thus, the augmented PCA reaction in Ptges−/− mice observed in our study might also rely on a similar vascular mechanism. Importantly, our present results suggest that not only do mast cells produce pro-allergic PGD2 and LTC4 in response to particular stimuli, such as FcϵRI cross-linking, cytokines, and neurotransmitters, but they also have the capacity to modulate the microenvironmental synthesis of anti-allergic PGE2, thereby controlling physiological balance through cell-autonomous and -extrinsic mechanisms involving lipid mediators.
It has been established that cPLA2α plays a central role in eicosanoid synthesis by activated mast cells, because BMMCs from Pla2g4a−/− mice produce minimal amounts of PGD2 and cysteinyl LTs following IgE-dependent and -independent stimuli (35, 36). In this study, we found that cPLA2α in mast cells can also participate in PGE2 synthesis by adjacent fibroblasts through a cell-to-cell AA transfer mechanism on the basis of the following findings. First, as pointed out above, AA-containing PC and PE were selectively reduced in CTMC-like differentiated cells after coculture with fibroblasts (Figs. 4 and 5). This change was largely abrogated in Pla2g4a−/− CTMC-like cells, in agreement with the view that AA selectivity is one of the most notable characteristics of cPLA2α among the PLA2 enzymes reported so far (33, 65). The PS composition of BMMCs before and after coculture remained unaltered, probably because AA-containing molecular species in PS are present at very low levels in these cells (Fig. 4). Second, [3H]AA pre-incorporated into the membranes of BMMCs was readily transferred to fibroblasts during coculture, accompanied by generation of PGE2 (Fig. 6, G and H). Third, a study using BMMCs from Ptges−/− mice clearly indicated that the augmented PGE2 detected in the coculture supernatants was entirely derived from fibroblasts, and not from mast cells (Fig. 6C). Finally, and most importantly, Pla2g4a−/− BMMCs did not fully support the augmented PGE2 generation upon coculture with fibroblasts (Fig. 6I) or upon adoptive transfer into the skin of mast cell-deficient mice (Fig. 7A). Although mast cells express group V sPLA2, which reportedly has the potential to act on neighboring cells through the transcellular route after secretion (38, 70, 71), our study using Pla2g5−/− BMMCs argues against the contribution of this sPLA2 isoform to coculture augmented fibroblastic PGE2 production. These observations suggest that AA is released mainly by cPLA2α from phospholipids in mast cells, transferred to proximal fibroblasts through the juxtacrine route, and then sequentially metabolized to PGE2 by fibroblastic mPGES-1. As COX-2 was present in both BMMCs and fibroblasts after coculture (Fig. 6B), it is also possible that the cPLA2α-released AA is metabolized by COX-2 in BMMCs to PGH2, which, despite its instability, is then transferred and converted to PGE2 by mPGES-1 in fibroblasts.
The transfer of the intermediate PGH2 or LTA4 from endothelial cells to blood cells, or vice versa, has been demonstrated in cultured cells in vitro and in organ cultures ex vivo (72), and the physiological relevance of this system has recently gained a rationale by sophisticated approaches using adoptive transfer of bone marrow cells from mice lacking enzymes in the 5-lipoxygenase pathway (73, 74). However, even though these bone marrow chimera studies have delineated the transcellular eicosanoid synthesis between hematopoietic cells and parenchyma cells during inflammation, it remains unclear as to which types of hematopoietic cells and which subtypes of PLA2 are actually involved in this process. To our knowledge, this study is the first to demonstrate that cPLA2α in mast cells is capable of regulating prostanoid synthesis by neighboring fibroblasts. Thus, although mast cells can modulate the proliferation and functions of adjacent fibroblasts by several ways (15–20), the transcellular PGE2 synthesis observed in this study represents a novel aspect of mast cell-fibroblast communication.
Despite notable decreases in the proportion of AA-bearing phospholipid species in CTMC-like differentiating cells relative to IL-3-maintained immature BMMCs, there was no accumulation of lysophospholipids in CTMC-like cells (data not shown), suggesting that lysophospholipids are rapidly cleared from the cells by either reacylation or hydrolytic degradation. The occurrence of lysophospholipid reacylation was supported by the fact that PC containing OA and LA, nutrient fatty acids that can be supplied abundantly from the culture medium, are reciprocally increased in CTMC-like cells. Because cPLA2γ, an enzyme that is induced substantially (albeit at a low level) in CTMC-like cells (Fig. 1A), possesses strong lysophospholipase and transacylase activities (75), it might be at least partly responsible for the removal of lysophospholipids in these cells. Although the physiological significance of the enrichment of OA- and LA-bearing phospholipids in CTMC-like cells is unclear, it might be related to the increase in granule membranes in mature mast cells. Nevertheless, despite the decreased proportion of AA-containing phospholipids in CTMC-like cells, they are able to produce PGD2 abundantly after IgE/Ag stimulation (Fig. 2H), implying that the residual AA pool in CTMC-like cells is sufficient for their synthesis of lipid mediators. Unlike FcϵRI-dependent activation in which virtually all AA-containing PC species are promptly decreased (Fig. 3), some AA-containing PC species such as diacyl-PC with 18:0–20:4 or 18:1–20:4 did not decrease or rather showed a tendency to increase modestly in CTMC-like cells compared with BMMCs, suggesting that these phospholipid species are compartmentalized in certain membrane domains to which cPLA2α is hardly accessible or that they are produced from some other phospholipid species by a process involving AA reacylation.
Although iPLA2β was originally proposed to be the main enzyme responsible for phospholipid remodeling, and thereby for the maintenance of cellular homeostasis (40), none of our present results support the remodeling function of iPLA2β in mast cells, because the phospholipid composition in Pla2g6−/− BMMCs was identical to that in wild-type BMMCs before and after coculture with fibroblasts. Dissociation of iPLA2β from phospholipid remodeling has also been demonstrated recently in testis, macrophages, and pancreatic β-cells (44, 76, 77), in which the enzyme appears to play signaling roles in processes such as Ca2+ entry, lipid mediator synthesis, cell migration, exocytosis, and apoptosis (41–49). However, in BMMCs, iPLA2β is apparently nonessential for stimulus-coupled AA mobilization and subsequent eicosanoid generation, granule exocytosis, cytokine secretion, and even upstream Ca2+ entry (Figs. 2 and 3). Indeed, the normal PCA reaction in Pla2g6−/− mice implies that mast cells are numerically and functionally intact in the absence of iPLA2β in vivo. This argues against the proposed idea that iPLA2β-released lysophospholipid is a prerequisite for opening of the Orai1-Stim1 CRAC channel (41), given that perturbation of this Ca2+-sensitive Ca2+ channel profoundly hampers multiple effector functions of FcϵRI-activated mast cells (61). Also, the proposed role of iPLA2β in the inflammasome-dependent IL-1β production, an idea that arose from the pharmacological inhibition exerted by BEL (45), is also unlikely in BMMCs, because our preliminary data show that IL-1β release was unaffected by iPLA2β deficiency (data not shown), and a similar result has recently been obtained by another group (63). Thus far, we have been unable to characterize any functional role of iPLA2β in mast cells, even though it is undeniably present.
Despite the unaltered effector functions of Pla2g6−/− mast cells, BEL potently inhibited β-HEX release by BMMCs (Fig. 1D), indicating that BEL acts on other target(s) that are crucial for mast cell exocytosis. Because nine iPLA2 homologs, known as patatin-like phospholipase A domain-containing lipases (PNPLAs), are encoded in the mammalian genome (31, 78), it remains possible that the genetic deficiency of iPLA2β in mast cells might be compensated by other iPLA2/PNPLA isoform(s) or that other BEL-sensitive iPLA2/PNPLA isoform(s), rather than iPLA2β, might truly contribute to the functions and/or maturation of mast cells. The possibility that iPLA2β deficiency is compensated by iPLA2γ/PNPLA8 is unlikely, because there was no compensatory up-regulation of iPLA2γ in Pla2g6−/− BMMCs (supplemental Fig. S1) and because our preliminary data have shown that Pnpla8−/− BMMCs have normal degranulation capacity.4 It has been recently shown that the lack of PNPLA2 (adipose triglyceride lipase or iPLA2ζ) impairs insulin secretion by pancreatic β-cells (79) and phagocytosis by macrophages (80) due to impaired lipolytic supply of fatty acid fuel. In yeast, Sec14p, a component of the secretory machinery, is functionally coupled with phospholipid deacylation by Nte1p, a yeast homolog of PNPLA6 (neuronal target esterase or iPLA2δ) (81). In analogy, mast cell degranulation might require energy supply by iPLA2ζ/PNPLA2 or vesicular trafficking by iPLA2δ/PNPLA6, a possibility that should await future studies. Note that BEL could also inhibit several enzymes unrelated to the iPLA2/PNPLA family (62, 63). Thus, caution should be exercised when using this drug to study possible roles of iPLA2s in cellular responses.
Several studies have reported the critical role of iPLA2β in lipid mediator synthesis by several cell types. For instance, thapsigargin- or A23187-stimulated AA release is dramatically attenuated in Pla2g6−/− aortic smooth muscle cells, and these cells show decreased migration and proliferation due to reduced PGE2 generation in a model of vascular injury (42). A high glucose-induced iPLA2β activates RhoA/Rho kinase via 12/15-lipoxygenase metabolites, which contributes to vascular smooth muscle hypercontractility in diabetic animals (43). In lung endothelial cells, thrombin- or tryptase-stimulated synthesis of PGI2 and platelet-activating factor is attenuated by iPLA2β deficiency (83). Even in nonvascular cells, scavenger receptor A-dependent adhesion, a process that requires 12/15-lipoxygenase but not COX products, is absent in Pla2g6−/− macrophages (84). Moreover, A23187-induced, but not IL-1β-stimulated, AA release was augmented in iPLA2β-transfected HEK293 cells (70). We have shown here that the PGE2 biosynthetic responses of dendritic cells and macrophages to LPS, but not zymosan, are partially reduced by the deficiency of iPLA2β, confirming the involvement of iPLA2β in the former situation (supplemental Fig. S2). Taken together, the requirement of iPLA2β for cell activation may depend on cell type, stimulus, and/or class of lipid mediators. Although precise molecular mechanisms underlying iPLA2β activation in distinct contexts should need further clarification, mast cells represent a clear example that cell activation is fully operative in the absence of iPLA2β. Presumably, mast cells might be intrinsically devoid of certain unknown factor(s) essential for iPLA2β activation or the lack of iPLA2β might be bypassed by alternative mechanism(s).
It is intriguing, however, that during maturation of BMMCs toward CTMC-like cells, iPLA2β shows an increase of expression accompanied by a molecular mass shift from the full size into smaller forms over time (Fig. 1C). This would suggest that, in addition to transcriptional control of iPLA2β (Fig. 1A), its expression is regulated by proteolytic processing (48, 49, 59, 60) or by alternative splicing (82, 85). It has been reported that proteolytic cleavage of the N-terminal ankyrin repeats of iPLA2β by caspase-3 or unknown proteases occurs in apoptotic cells (48, 49) or even in nonapoptotic migrating cells (60) and agonist-treated islet β-cells (59), leading to activation of this enzyme. Because we did not detect any functional role of iPLA2β in BMMCs, the regulatory mechanisms responsible for the proteolytic processing of iPLA2β and its functional consequences in CTMC-like differentiating cells are still a subject of debate. During apoptosis, the truncated iPLA2β contributes to apoptotic changes in membranes, and the LPC released from apoptotic cells acts as a chemoattractant (“find-me” signal) for phagocytotic cells (48, 49). In this view, iPLA2β might play a role in the phagocytotic clearance of a population of mature mast cells that are destined for apoptosis under certain conditions, a possibility that will need to be examined in a future study.
Acknowledgments
We thank Drs. J. P. Arm (Novartis Institutes for BioMedical Research, Cambridge, MA) and S. Akira (Osaka University, Suita, Tokyo) for providing us Pla2g5- and Ptges-deficient mice, respectively,
This work was supported by grants-in aid for scientific research from the Ministry of Education, Science, Culture, Sports and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Y. Taketomi and M. Murakami, unpublished results.
- AA
- arachidonic acid
- PLA2
- phospholipase A2
- cPLA2
- cytosolic PLA2
- iPLA2
- Ca2+-independent PLA2
- sPLA2
- secreted PLA2
- BMMC
- bone marrow-derived mast cell
- CTMC
- connective tissue mast cell
- DNP
- dinitrophenyl
- ESI-MS
- electrospray ion source-mass spectrometry
- β-HEX
- β-hexosaminidase
- LT
- leukotriene
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PS
- phosphatidylserine
- PG
- prostaglandin
- PNPLA
- patatin-like phospholipase A domain-containing lipase
- SCF
- stem cell factor
- LPC
- lysophosphatidylcholine
- CRAC
- Ca2+ release-activated Ca2+
- EPA
- eicosapentaenoic acid
- OA
- oleic acid
- LA
- linoleic acid
- BEL
- bromoenol lactone
- BMDC
- bone marrow-derived dendritic cell
- Ag
- antigen
- PCA
- passive cutaneous anaphylaxis.
REFERENCES
- 1. Metcalfe D. D., Baram D., Mekori Y. A. (1997) Physiol. Rev. 77, 1033–1079 [DOI] [PubMed] [Google Scholar]
- 2. Wong G. W., Friend D. S., Stevens R. L. (1999) in Signal Transduction in Mast Cells and Basophils (Razin E., Rivera J. eds) pp. 39–53, Springer-Verlag, New York [Google Scholar]
- 3. Kalesnikoff J., Galli S. J. (2008) Nat. Immunol. 9, 1215–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galli S. J., Grimbaldeston M., Tsai M. (2008) Nat. Rev. Immunol. 8, 478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang E., Nocka K., Beier D. R., Chu T. Y., Buck J., Lahm H. W., Wellner D., Leder P., Besmer P. (1990) Cell 63, 225–233 [DOI] [PubMed] [Google Scholar]
- 6. Nocka K., Buck J., Levi E., Besmer P. (1990) EMBO J. 9, 3287–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gurish M. F., Tao H., Abonia J. P., Arya A., Friend D. S., Parker C. M., Austen K. F. (2001) J. Exp. Med. 194, 1243–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abonia J. P., Austen K. F., Rollins B. J., Joshi S. K., Flavell R. A., Kuziel W. A., Koni P. A., Gurish M. F. (2005) Blood 105, 4308–4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Levi-Schaffer F., Austen K. F., Gravallese P. M., Stevens R. L. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6485–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dayton E. T., Pharr P., Ogawa M., Serafin W. E., Austen K. F., Levi-Schaffer F., Stevens R. L. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 569–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogasawara T., Murakami M., Suzuki-Nishimura T., Uchida M. K., Kudo I. (1997) J. Immunol. 158, 393–404 [PubMed] [Google Scholar]
- 12. Takano H., Nakazawa S., Okuno Y., Shirata N., Tsuchiya S., Kainoh T., Takamatsu S., Furuta K., Taketomi Y., Naito Y., Takematsu H., Kozutsumi Y., Tsujimoto G., Murakami M., Kudo I., Ichikawa A., Nakayama K., Sugimoto Y., Tanaka S. (2008) FEBS Lett. 582, 1444–1450 [DOI] [PubMed] [Google Scholar]
- 13. Taketomi Y., Sugiki T., Saito T., Ishii S., Hisada M., Suzuki-Nishimura T., Uchida M. K., Moon T. C., Chang H. W., Natori Y., Miyazawa S., Kikuchi-Yanoshita R., Murakami M., Kudo I. (2003) Biochem. Biophys. Res. Commun. 306, 339–346 [DOI] [PubMed] [Google Scholar]
- 14. Taketomi Y., Sunaga K., Tanaka S., Nakamura M., Arata S., Okuda T., Moon T. C., Chang H. W., Sugimoto Y., Kokame K., Miyata T., Murakami M., Kudo I. (2007) J. Immunol. 178, 7042–7053 [DOI] [PubMed] [Google Scholar]
- 15. Ruoss S. J., Hartmann T., Caughey G. H. (1991) J. Clin. Invest. 88, 493–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levi-Schaffer F., Rubinchik E. (1995) J. Invest. Dermatol. 104, 999–1003 [DOI] [PubMed] [Google Scholar]
- 17. Kendall J. C., Li X. H., Galli S. J., Gordon J. R. (1997) J. Allergy Clin. Immunol. 99, 113–123 [DOI] [PubMed] [Google Scholar]
- 18. Trautmann A., Krohne G., Bröcker E. B., Klein C. E. (1998) J. Immunol. 160, 5053–5057 [PubMed] [Google Scholar]
- 19. Akers I. A., Parsons M., Hill M. R., Hollenberg M. D., Sanjar S., Laurent G. J., McAnulty R. J. (2000) Am. J. Physiol. Lung Cell. Mol. Physiol. 278, L193–L201 [DOI] [PubMed] [Google Scholar]
- 20. Smith T. J., Parikh S. J. (1999) Endocrinology 140, 3518–3525 [DOI] [PubMed] [Google Scholar]
- 21. Smith R. S., Smith T. J., Blieden T. M., Phipps R. P. (1997) Am. J. Pathol. 151, 317–322 [PMC free article] [PubMed] [Google Scholar]
- 22. Murray J. J., Tonnel A. B., Brash A. R., Roberts L. J., 2nd, Gosset P., Workman R., Capron A., Oates J. A. (1986) N. Engl. J. Med. 315, 800–804 [DOI] [PubMed] [Google Scholar]
- 23. Austen K. F. (2008) Nat. Immunol. 9, 113–115 [DOI] [PubMed] [Google Scholar]
- 24. Tager A. M., Bromley S. K., Medoff B. D., Islam S. A., Bercury S. D., Friedrich E. B., Carafone A. D., Gerszten R. E., Luster A. D. (2003) Nat. Immunol. 4, 982–990 [DOI] [PubMed] [Google Scholar]
- 25. Lewis R. A., Soter N. A., Diamond P. T., Austen K. F., Oates J. A., Roberts L. J., 2nd (1982) J. Immunol. 129, 1627–1631 [PubMed] [Google Scholar]
- 26. Murakami M., Matsumoto R., Urade Y., Austen K. F., Arm J. P. (1995) J. Biol. Chem. 270, 3239–3246 [DOI] [PubMed] [Google Scholar]
- 27. Matsuoka T., Hirata M., Tanaka H., Takahashi Y., Murata T., Kabashima K., Sugimoto Y., Kobayashi T., Ushikubi F., Aze Y., Eguchi N., Urade Y., Yoshida N., Kimura K., Mizoguchi A., Honda Y., Nagai H., Narumiya S. (2000) Science 287, 2013–2017 [DOI] [PubMed] [Google Scholar]
- 28. Satoh T., Moroi R., Aritake K., Urade Y., Kanai Y., Sumi K., Yokozeki H, Hirai H., Nagata K., Hara T., Utsuyama M., Hirokawa K., Sugamura K., Nishioka K., Nakamura M. (2006) J. Immunol. 177, 2621–2629 [DOI] [PubMed] [Google Scholar]
- 29. Hammad H., Kool M., Soullié T., Narumiya S., Trottein F., Hoogsteden H. C., Lambrecht B. N. (2007) J. Exp. Med. 204, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shiraishi Y., Asano K., Niimi K., Fukunaga K., Wakaki M., Kagyo J., Takihara T., Ueda S., Nakajima T., Oguma T., Suzuki Y., Shiomi T., Sayama K., Kagawa S., Ikeda E., Hirai H., Nagata K., Nakamura M., Miyasho T., Ishizaka A. (2008) J. Immunol. 180, 541–549 [DOI] [PubMed] [Google Scholar]
- 31. Kunikata T., Yamane H., Segi E., Matsuoka T., Sugimoto Y., Tanaka S., Tanaka H., Nagai H., Ichikawa A., Narumiya S. (2005) Nat. Immunol. 6, 524–531 [DOI] [PubMed] [Google Scholar]
- 32. Lundequist A., Nallamshetty S. N., Xing W., Feng C., Laidlaw T. M., Uematsu S., Akira S., Boyce J. A. (2010) J. Immunol. 184, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kudo I., Murakami M. (2002) Prostaglandins Other Lipid Mediat. 68, 3–58 [DOI] [PubMed] [Google Scholar]
- 34. Murakami M., Taketomi Y., Miki Y., Sato H., Hirabayashi T., Yamamoto K. (2011) Prog. Lipid Res. 50, 152–192 [DOI] [PubMed] [Google Scholar]
- 35. Fujishima H., Sanchez Mejia R. O., Bingham C. O., 3rd, Lam B. K., Sapirstein A., Bonventre J. V., Austen K. F., Arm J. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4803–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakatani N., Uozumi N., Kume K., Murakami M., Kudo I., Shimizu T. (2000) Biochem. J. 352, 311–317 [PMC free article] [PubMed] [Google Scholar]
- 37. Kikawada E., Bonventre J. V., Arm J. P. (2007) Blood 110, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reddy S. T., Herschman H. R. (1996) J. Biol. Chem. 271, 186–191 [DOI] [PubMed] [Google Scholar]
- 39. Fensome-Green A., Stannard N., Li M., Bolsover S., Cockcroft S. (2007) Cell Calcium 41, 145–153 [DOI] [PubMed] [Google Scholar]
- 40. Balsinde J., Balboa M. A., Dennis E. A. (1997) J. Biol. Chem. 272, 29317–29321 [DOI] [PubMed] [Google Scholar]
- 41. Smani T., Zakharov S. I., Csutora P., Leno E., Trepakova E. S., Bolotina V. M. (2004) Nat. Cell Biol. 6, 113–120 [DOI] [PubMed] [Google Scholar]
- 42. Moon S. H., Jenkins C. M., Mancuso D. J., Turk J., Gross R. W. (2008) J. Biol. Chem. 283, 33975–33987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xie Z., Gong M. C., Su W., Xie D., Turk J., Guo Z. (2010) J. Biol. Chem. 285, 8628–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bao S., Song H., Wohltmann M., Ramanadham S., Jin W., Bohrer A., Turk J. (2006) J. Biol. Chem. 281, 20958–20973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Piccini A., Carta S., Tassi S., Lasiglié D., Fossati G., Rubartelli A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 8067–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mishra R. S., Carnevale K. A., Cathcart M. K. (2008) J. Exp. Med. 205, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dietrich H. H., Abendschein D. R., Moon S. H., Nayeb-Hashemi N., Mancuso D. J., Jenkins C. M., Kaltenbronn K. M., Blumer K. J., Turk J., Gross R. W. (2010) Am. J. Physiol. Heart Circ. Physiol. 298, H2208–H2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Atsumi G., Murakami M., Kojima K., Hadano A., Tajima M., Kudo I. (2000) J. Biol. Chem. 275, 18248–18258 [DOI] [PubMed] [Google Scholar]
- 49. Lauber K., Bohn E., Kröber S. M., Xiao Y. J., Blumenthal S. G., Lindemann R. K., Marini P., Wiedig C., Zobywalski A., Baksh S., Xu Y., Autenrieth I. B., Schulze-Osthoff K., Belka C., Stuhler G., Wesselborg S. (2003) Cell 113, 717–730 [DOI] [PubMed] [Google Scholar]
- 50. Li H., Zhao Z., Wei G., Yan L., Wang D., Zhang H., Sandusky G. E., Turk J., Xu Y. (2010) FASEB J. 24, 4103–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morgan N. V., Westaway S. K., Morton J. E., Gregory A., Gissen P., Sonek S., Cangul H., Coryell J., Canham N., Nardocci N., Zorzi G., Pasha S., Rodriguez D., Desguerre I., Mubaidin A., Bertini E., Trembath R. C., Simonati A., Schanen C., Johnson C. A., Levinson B., Woods C. G., Wilmot B., Kramer P., Gitschier J., Maher E. R., Hayflick S. J. (2006) Nat. Genet. 38, 752–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shinzawa K., Sumi H., Ikawa M., Matsuoka Y., Okabe M., Sakoda S., Tsujimoto Y. (2008) J. Neurosci. 28, 2212–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kamei D., Yamakawa K., Takegoshi Y., Mikami-Nakanishi M., Nakatani Y., Oh-Ishi S., Yasui H., Azuma Y., Hirasawa N., Ohuchi K., Kawaguchi H., Ishikawa Y., Ishii T., Uematsu S., Akira S., Murakami M., Kudo I. (2004) J. Biol. Chem. 279, 33684–33695 [DOI] [PubMed] [Google Scholar]
- 54. Jenkins C. M., Han X., Mancuso D. J., Gross R. W. (2002) J. Biol. Chem. 277, 32807–32814 [DOI] [PubMed] [Google Scholar]
- 55. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 56. Houjou T., Yamatani K., Imagawa M., Shimizu T., Taguchi R. (2005) Rapid Commun. Mass Spectrom. 19, 654–666 [DOI] [PubMed] [Google Scholar]
- 57. Taguchi R., Houjou T., Nakanishi H., Yamazaki T., Ishida M., Imagawa M., Shimizu T. (2005) J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 823, 26–36 [DOI] [PubMed] [Google Scholar]
- 58. Gurish M. F., Ghildyal N., McNeil H. P., Austen K. F., Gillis S., Stevens R. L. (1992) J. Exp. Med. 175, 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Song H., Bao S., Lei X., Jin C., Zhang S., Turk J., Ramanadham S. (2010) Biochim. Biophys. Acta 1801, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao X., Wang D., Zhao Z., Xiao Y., Sengupta S., Xiao Y., Zhang R., Lauber K., Wesselborg S., Feng L., Rose T. M., Shen Y., Zhang J., Prestwich G., Xu Y. (2006) J. Biol. Chem. 281, 29357–29368 [DOI] [PubMed] [Google Scholar]
- 61. Vig M., DeHaven W. I., Bird G. S., Billingsley J. M., Wang H., Rao P. E., Hutchings A. B., Jouvin M. H., Putney J. W., Kinet J. P. (2008) Nat. Immunol. 9, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fuentes L., Pérez R., Nieto M. L., Balsinde J., Balboa M. A. (2003) J. Biol. Chem. 278, 44683–44690 [DOI] [PubMed] [Google Scholar]
- 63. Franchi L., Chen G., Marina-Garcia N., Abe A., Qu Y., Bao S., Shayman J. A., Turk J., Dubyak G. R., Núñez G. (2009) J. Innate Immun. 1, 607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Murakami M., Austen K. F., Arm J. P. (1995) J. Exp. Med. 182, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Clark J. D., Lin L. L., Kriz R. W., Ramesha C. S., Sultzman L. A., Lin A. Y., Milona N., Knopf J. L. (1991) Cell 65, 1043–1051 [DOI] [PubMed] [Google Scholar]
- 66. Hu Z. Q., Asano K., Seki H., Shimamura T. (1995) J. Immunol. 155, 2134–2142 [PubMed] [Google Scholar]
- 67. Diaz B. L., Fujishima H., Kanaoka Y., Urade Y., Arm J. P. (2002) J. Immunol. 168, 1397–1404 [DOI] [PubMed] [Google Scholar]
- 68. Gomi K., Zhu F. G., Marshall J. S. (2000) J. Immunol. 165, 6545–6552 [DOI] [PubMed] [Google Scholar]
- 69. Nguyen M., Solle M., Audoly L. P., Tilley S. L., Stock J. L., McNeish J. D., Coffman T. M., Dombrowicz D., Koller B. H. (2002) J. Immunol. 169, 4586–4593 [DOI] [PubMed] [Google Scholar]
- 70. Murakami M., Kambe T., Shimbara S., Kudo I. (1999) J. Biol. Chem. 274, 3103–3115 [DOI] [PubMed] [Google Scholar]
- 71. Wijewickrama G. T., Kim J. H., Kim Y. J., Abraham A., Oh Y., Ananthanarayanan B., Kwatia M., Ackerman S. J., Cho W. (2006) J. Biol. Chem. 281, 10935–10944 [DOI] [PubMed] [Google Scholar]
- 72. Sala A., Folco G., Murphy R. C. (2010) Pharmacol. Rep. 62, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zarini S., Gijón M. A., Ransome A. E., Murphy R. C., Sala A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8296–8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fabre J. E., Goulet J. L., Riche E., Nguyen M., Coggins K., Offenbacher S., Koller B. H. (2002) J. Clin. Invest. 109, 1373–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamashita A., Kamata R., Kawagishi N., Nakanishi H., Suzuki H., Sugiura T., Waku K. (2005) J. Biochem. 137, 557–567 [DOI] [PubMed] [Google Scholar]
- 76. Bao S., Miller D. J., Ma Z., Wohltmann M., Eng G., Ramanadham S., Moley K., Turk J. (2004) J. Biol. Chem. 279, 38194–38200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bao S., Bohrer A., Ramanadham S., Jin W., Zhang S., Turk J. (2006) J. Biol. Chem. 281, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kienesberger P. C., Oberer M., Lass A., Zechner R. (2009) J. Lipid Res. 50, S63–S68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Peyot M. L., Guay C., Latour M. G., Lamontagne J., Lussier R., Pineda M., Ruderman N. B., Haemmerle G., Zechner R., Joly E., Madiraju S. R., Poitout V., Prentki M. (2009) J. Biol. Chem. 284, 16848–16859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chandak P. G., Radovic B., Aflaki E., Kolb D., Buchebner M., Fröhlich E., Magnes C., Sinner F., Haemmerle G., Zechner R., Tabas I., Levak-Frank S., Kratky D. (2010) J. Biol. Chem. 285, 20192–20201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Murray J. P., McMaster C. R. (2005) J. Biol. Chem. 280, 8544–8552 [DOI] [PubMed] [Google Scholar]
- 82. Larsson P. K., Claesson H. E., Kennedy B. P. (1998) J. Biol. Chem. 273, 207–214 [DOI] [PubMed] [Google Scholar]
- 83. Sharma J., Turk J., McHowat J. (2010) Biochemistry 49, 5473–5481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nikolic D. M., Gong M. C., Turk J., Post S. R. (2007) J. Biol. Chem. 282, 33405–33411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Manguikian A. D., Barbour S. E. (2004) J. Biol. Chem. 279, 52881–52892 [DOI] [PubMed] [Google Scholar]