Background: HCV causes ER stress in the infected cells.
Results: HCV-induced ER stress leads to increased expression of certain proteins that in turn enhance the degradation of HCV glycoproteins and decrease production of virus particles.
Conclusion: HCV infection activates the ERAD pathway, leading to modulation of virus production.
Significance: ERAD plays a crucial role in the viral life cycle.
Keywords: ER Stress, Hepatitis Virus, RNA Viruses, Ubiquitin, Virus Assembly
Abstract
Viral infections frequently cause endoplasmic reticulum (ER) stress in host cells leading to stimulation of the ER-associated degradation (ERAD) pathway, which subsequently targets unassembled glycoproteins for ubiquitylation and proteasomal degradation. However, the role of the ERAD pathway in the viral life cycle is poorly defined. In this paper, we demonstrate that hepatitis C virus (HCV) infection activates the ERAD pathway, which in turn controls the fate of viral glycoproteins and modulates virus production. ERAD proteins, such as EDEM1 and EDEM3, were found to increase ubiquitylation of HCV envelope proteins via direct physical interaction. Knocking down of EDEM1 and EDEM3 increased the half-life of HCV E2, as well as virus production, whereas exogenous expression of these proteins reduced the production of infectious virus particles. Further investigation revealed that only EDEM1 and EDEM3 bind with SEL1L, an ER membrane adaptor protein involved in translocation of ERAD substrates from the ER to the cytoplasm. When HCV-infected cells were treated with kifunensine, a potent inhibitor of the ERAD pathway, the half-life of HCV E2 increased and so did virus production. Kifunensine inhibited the binding of EDEM1 and EDEM3 with SEL1L, thus blocking the ubiquitylation of HCV E2 protein. Chemical inhibition of the ERAD pathway neither affected production of the Japanese encephalitis virus (JEV) nor stability of the JEV envelope protein. A co-immunoprecipitation assay showed that EDEM orthologs do not bind with JEV envelope protein. These findings highlight the crucial role of the ERAD pathway in the life cycle of specific viruses.
Introduction
Quality control of proteins, such as the elimination of misfolded proteins, is largely connected with the endoplasmic reticulum (ER),2 which is an organelle responsible for the folding and distribution of secretory proteins to their sites of action. This pathway is termed ER-associated degradation (ERAD) and is triggered by ER stress. It results in retrotranslocation of misfolded proteins into the cytosol, followed by polyubiquitylation and proteasomal degradation (1). Several viral infections have been reported to trigger the ERAD pathway (2–4); however, the role of this pathway in the life cycle of viruses remains poorly defined.
Initiation of the ERAD pathway occurs from the oligomerization and autophosphorylation of IRE1, an ER stress sensor. The activated IRE1 removes an intron from X-box-binding protein 1 (XBP1) mRNA, which then encodes a potent transcription factor for activation of genes, for example, ER degradation-enhancing α-mannosidase-like protein (EDEM). EDEM1 (5), along with its two homologs EDEM2 (6) and EDEM3 (7), as well as ER mannosidase I (ER ManI), belong to the glycoside hydrolase 47 family. EDEMs are thought to function as lectins that deliver misfolded glycoproteins to the ERAD pathway. However, the precise mechanism by which they assist in glycoprotein quality control remains unclear.
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease. The RNA genome of HCV, a member of the Flaviviridae family, encodes the viral structural proteins Core, E1, E2, and p7, as well as six nonstructural proteins (8, 9). Two N-glycosylated envelope proteins E1 and E2 are exposed on the surface of the virus and are necessary for viral entry.
The aim of this study was to investigate whether the ERAD pathway is activated upon HCV infection and whether this affects the quality control of virus glycoproteins and virion production. We show that HCV infection triggers the ERAD pathway, possibly through IRE1-mediated splicing of XBP1. Moreover, EDEM1 and EDEM3, but not EDEM2, interact with HCV glycoproteins, resulting in increased ubiquitylation. EDEM1 knockdown and chemical inhibition of the ERAD pathway increases glycoprotein stability, as well as production of infectious virus particles, whereas overexpression of EDEM1 decreases virion production. These results provide insight into the mechanism by which HCV triggers the ERAD pathway and subsequently affects the quality control of virus glycoproteins and virus particle production.
EXPERIMENTAL PROCEDURES
Cell Culture and Chemicals
Human hepatoma cells HuH-7 and HuH-7.5.1 (a gift from Dr. F. V. Chisari (The Scripps Research Institute) (10) and human embryonic kidney cells 293T were cultured at 37 °C and 5% CO2 in DMEM containing 10% FBS, 10 mm HEPES, 1 mm sodium pyruvate, nonessential minimum amino acids, 100 units/ml penicillin, and 100 μg/ml streptomycin. Tunicamycin (TM) was purchased from Sigma-Aldrich, and kifunensine (KIF) was purchased from Toronto Research Chemicals (Ontario, Canada).
Preparation of Virus Stock
HCV JFH-1 was generated by introducing in vitro transcribed RNA into HuH-7.5.1 cells by electroporation, and virus stocks were prepared by infecting at a multiplicity of infection (m.o.i.) of 0.01, as described previously (10). Infected cells were grown in culture medium containing 2% FBS, and supernatants were collected after multiple passages to get high titer virus. The supernatants were concentrated using a 500-kDa hollow fiber module (GE Healthcare) resulting in ∼90% recovery of the virus. Focus-forming units were measured with an anti-HCV core antibody to determine virus titration (2H9, described below). Virus stocks containing 1 × 107 focus-forming units/ml were divided into small aliquots and stored at −80 °C until use. rAT strain of Japanese encephalitis virus (JEV) (11) was used to generate virus stock.
Plasmids
cDNAs of mouse EDEM1-HA, EDEM2, and EDEM3-HA, having 92, 93, and 91% amino acid homology with their human orthologs, respectively, were a kind gift from Drs. N. Hosokawa (Kyoto University) and K. Nagata (Kyoto Sangyo University). A HA tag was attached to the C terminus of EDEM2 by PCR, and sequencing analysis was performed to confirm the sequence. To generate pJFH/E1dTM-myc and pJFH/E2dTM-myc, HCV E1 encoding amino acids 170–352 and HCV E2 encoding amino acids 340–714 of JFH-1 polyprotein were amplified by PCR with forward primer and reverse primer containing NotI and XbaI restriction sites, respectively, and cloned into a NotI/XbaI site of the pEF1/Myc-His plasmid (Invitrogen). The pCAGC105E plasmid carrying PrM and E proteins of the rAT strain of JEV has been described (12). Plasmids carrying the firefly luciferase reporter gene under control of the intact promoter of GRP78 and GRP94 or the defective promoter lacking ERSE elements have been described (13) and were a kind gift from Dr. K. Mori (Kyoto University).
Antibodies
Rabbit polyclonal antibodies included anti-HA (Sigma-Aldrich), anti-HCV NS5A (14), anti-SEL1L (Sigma-Aldrich), anti-ubiquitin (MBL, Nagoya, Japan), and anti-JEV E antibodies. The mouse monoclonal antibodies were anti-HA (clone 16B12; Covance, Emeryville, CA), anti-HCV E2 (clone 8D10-3),3 anti-β-actin (clone AC15; Sigma-Aldrich), anti-HCV core (clone 2H9) (15), and anti-Myc (clone 9E10; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies. Anti-JEV antibodies have been described (16) and were a kind gift from Drs. C. K. Lim and T. Takasaki (National Institute of Infectious Diseases).
Analysis of XBP1 Splicing
Total RNA was extracted from cells using Isogen (Nippon Gene, Tokyo, Japan) following the manufacturer's protocol, and 2 μg of RNA was subjected to cDNA synthesis using oligo(dT) and Superscript III (Invitrogen). PCR was carried out using specific primers 5′-AAACAGAGTAGCAGCTCAGACTGC-3′ and 5′-GTATCTCTAAGACTAGGGGCTTGGTA-3′ for XBP1 and 5′-TCCTGTGGCATCCACGAAACT-3′ and 5′-GAAGCATTTGCGGTGGACGAT-3′ for β-actin to generate PCR fragments of 598 bp for unspliced XBP1, 572 bp for spliced XBP1, and 315 bp for β-actin. The following cycling conditions were used to amplify the genes: 1 cycle of 98 °C for 3 min, followed by 30 cycles of 98 °C for 20 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension of 72 °C for 10 min. The PCR product of XBP1 was further digested with PstI enzyme (New England Biolabs) and resolved on a 2% agarose gel prepared in TAE buffer. Unspliced XBP1 yielded two smaller fragments of 291 and 307 bp whereas spliced XBP1 stayed intact due to loss of the restriction site after splicing.
Gene Microarray Analysis
For microarray analysis, RNA was extracted from HuH-7.5.1 cells at 48 and 72 h after JFH-1 infection. Cells treated for 12 h with 5 μg/ml TM served as a positive control. Hybridization was performed on a 3D-Gene (see 3D-Gene web site) Human Oligonucleotide chip 25k (Toray Industries Inc., Tokyo, Japan). For efficient hybridization, this microarray chip has three dimensions and is constructed with a well between the probes and cylinder stems with 70-mer oligonucleotide probes on the top. Total RNA was labeled with Cy3 or Cy5 using the Amino Allyl MessageAMP II aRNA Amplification kit (Applied Biosystems). The Cy3- or Cy5-labeled aRNA pools were subjected to hybridization for 16 h using the supplier's protocol. Hybridization signals were scanned using a ScanArray Express Scanner (PerkinElmer Life Sciences) and processed by GenePixPro version 5.0 (Molecular Devices, Sunnyvale, CA). Detected signals for each gene were normalized using a global normalization method (Cy3/Cy5 ratio median = 1). Genes with Cy3/Cy5 normalized ratios >log2 1.0 or <log2 −1.0 were defined, respectively, as significantly up- or down-regulated genes.
Quantification of Cellular Gene Expression
Gene expression levels were measured using predesigned assay-on-demand (Applied Biosystems). RT-PCR amplification was performed under the following conditions: 48 °C for 30 min, 95 °C for 10 min, 50 cycles of 95 °C for 15 s, and 60 °C for 1 min. Standard curves were constructed on a 1:5 serial dilution of the RNA template. The results were normalized to GAPDH mRNA levels.
Determination of Protein Stability
HuH-7 cells were infected with HCV JFH-1 at a m.o.i. of 2. Six hours after infection, the cells were either treated with KIF or transfected with EDEM1 siRNA. Forty hours later, culture medium was replaced with 100 μg/ml cycloheximide (CHX). Cells, including floating cells, were harvested at different time points after CHX addition, and immunoblotting was performed to determine the amount of HCV E2.
Plasmid Transfection and Immunoprecipitation
HuH-7 or 293T cells were seeded in 6-well cell culture plates at 3 × 105 cells/well and cultured overnight. Plasmid DNA was transfected into cells using TranIT-LT1 transfection reagent (Mirus, Madison, WI). Cells were harvested at 48 h after transfection, washed once with 1 ml of PBS, and lysed in 200 μl of lysis buffer (20 mm Tris-HCl, pH 7.4, 135 mm NaCl, 1% Triton X-100, and 10% glycerol supplemented with 50 mm NaF, 5 mm Na3VO4, and protease inhibitor mixture tablets (Roche Diagnostics). Cell lysates were sonicated at 4 °C for 10 min, incubated for 30 min at 4 °C, and centrifuged at 14,000 × g for 5 min at 4 °C. After preclearing for 2 h, the supernatants were immunoprecipitated overnight by rotating with 1.5 μl of anti-HA monoclonal antibody (16B12) or anti-HCV E2 monoclonal antibody (clone 8D10-3) at 4 °C. The immunocomplexes were then captured on protein G-agarose beads (Invitrogen) by rotation-incubation at 4 °C for 3 h. Beads were subsequently precipitated by centrifugation at 800 × g for 1 min and washed five times with lysis buffer. Finally, proteins bound to the beads were boiled in 40 μl of SDS sample buffer and subjected to SDS-PAGE.
Western Blotting
Proteins resolved by SDS-PAGE were transferred onto PVDF membranes (Immobilon; Millipore). After blocking in 2% skim milk, the membranes were probed with primary antibodies followed by exposure to peroxidase-conjugated secondary antibodies and visualization with an ECL Plus Western blotting detection system (GE Healthcare). The intensity of the bands was measured using a computerized imaging system (ImageJ software; National Institutes of Health).
Small Interfering RNA (siRNA) Transfection
HuH-7 cells were transfected with duplex siRNAs at a final concentration of 10 nm using Lipofectamine RNAiMAX (Invitrogen). Three siRNAs for each gene were examined for knock-down efficiency and cytotoxic effects. The siRNA with best performance was selected for further experiments. Target sequences of the siRNAs which exhibited the best knock-down efficiencies were as follows: EDEM1 (sense) 5′-CAUAUCCUCGGGUGAAUCUtt-3′, EDEM2 (sense) 5′-GAAUGUCUCAGAAUUCCAAtt-3′, EDEM3 (sense) 5′-CAUGAGACUACAAAUCUUAtt-3′, IRE1 (sense) 5′-GGACGUGAGCGACAGAAUAtt-3′. 5′-GGUGUCCUUACCAUACUAAtt-3′ served as a negative control. The lowercase letters denote overhanging deoxyribonucleotides.
Quantification of HCV Core and RNA
HCV core was quantified using an enzyme immunoassay (Ortho HCV antigen ELISA kit; Ortho Clinical Diagnostics, Tokyo, Japan). HCV RNA was quantified as described (17).
Statistical Analysis
Student's t test was employed to calculate the statistical significance of the results. p < 0.05 was considered significant.
RESULTS
HCV Infection Induces XBP1 mRNA Splicing and EDEM Expression
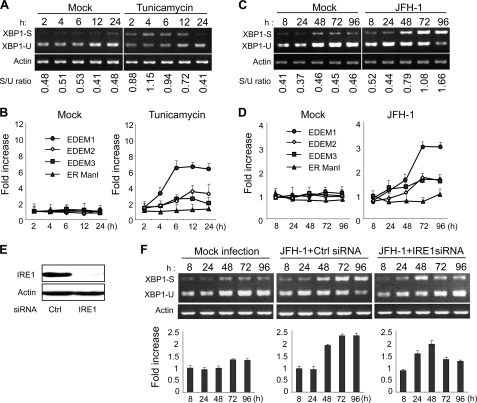
XBP1 plays a key role in activating the ERAD pathway, which mediates unfolded protein response in the ER. Under conditions of ER stress, XBP1 mRNA is processed by unconventional splicing and translated into functional XBP1, which in turn mediates transcriptional up-regulation of a variety of ER stress-dependent genes. The resultant activation of downstream pathways boosts the efficiency of ERAD, which coincides with elevated transcription of EDEMs. To validate our method for detecting activation of the ERAD pathway, we exposed HuH-7.5.1 cells to TM, which is a typical ER stress inducer, and performed an assay to quantify spliced XBP1 mRNA, as described under “Experimental Procedures,” at different time points after treatment. The spliced form of XBP1 mRNA started accumulating within these cells as early as 2 h after exposure to TM (Fig. 1A), and levels remained elevated until at least 12 h after treatment. Quantitative RT-PCR showed that mRNA levels of EDEM1, EDEM2, and EDEM3 were elevated in TM-treated cells whereas ER ManI, which is not an ER stress-responsive gene, did not show any up-regulation (Fig. 1B). To examine involvement of the ERAD pathway in the HCV life cycle, we infected HuH-7.5.1 cells with JFH-1 at m.o.i. of 5 and analyzed XBP1 mRNA splicing and EDEM up-regulation. Upon infection, the fragment corresponding to spliced XBP1 mRNA, was detectable 8 h after infection, and the difference in splicing between mock- and HCV-infected cells became more pronounced at 48 h after infection and then persisted (Fig. 1C). Increased levels of XBP1 mRNA splicing were dependent on the m.o.i. (supplemental Fig. 1A), suggesting that expression of active XBP1 was induced by HCV infection. A small amount of spliced XBP1 was detected in mock-infected cells, presumably because of some intrinsic stress. A 3.1-fold increase in the level of EDEM1 mRNA was observed at 3–4 days after infection (p < 0.05). Increases in EDEM2 and EDEM3 mRNA levels were moderate and reached ∼1.5-fold, whereas ER ManI mRNA exhibited no change after infection (Fig. 1D). Expression of EDEMs, particularly EDEM1, was up-regulated in accordance with HCV infection titers (supplemental Fig. 1B). Knocking down the IRE1 gene (Fig. 1E) effectively reversed the accumulation of spliced XBP1, as well as the transcriptional up-regulation of EDEM1 (Fig. 1F), thus confirming that HCV infection induces ERAD through the IRE1-XBP1 pathway.
FIGURE 1.
Splicing of XBP1 mRNA and induction of ERAD gene expression in HCV JFH-1-infected cells. A, splicing of XBP1 mRNA analyzed in mock- and TM (5 μg/ml)-treated HuH-7.5.1 cells at different time points after treatment. The upper and lower bands represent spliced and unspliced RNA, respectively. The numbers at the bottom of the panel indicate the density ratios of bands corresponding to spliced and unspliced XBP1. B, graphs showing the -fold induction of EDEM1, EDEM2, EDEM3, and ER Man1 mRNA in HuH-7.5.1 cells treated or untreated with TM. Data are normalized to GAPDH expression levels. The mean ± S.D. (error bars) of three independent experiments are shown. C, splicing of XBP1 mRNA analyzed in mock- and HCV JFH-1-infected HuH-7.5.1 cells (m.o.i. 5) at different time points after infection. Numbers at the bottom of the panel indicate the density ratios of bands corresponding to spliced and unspliced XBP1. D, real-time PCR analysis of EDEM1, EDEM2, EDEM3, and ER ManI mRNA induction in mock- and HCV-infected cells. Data are normalized to GAPDH expression. The mean ± S.D. of three independent experiments are shown. Note that a reduction in the level of GAPDH mRNA within infected cells was not observed until 96 h after infection when a slight decrease was observed. This led us to use GAPDH as a housekeeping gene in our experiments. E, Western blotting of IRE1 in cells transfected with mock or gene-specific siRNA of IRE1. F, splicing of XBP1 mRNA and induction of EDEM1 in HCV-infected cells after knocking down of the IRE1 gene. HuH-7.5.1 cells infected with JFH-1 at a m.o.i. of 5 were transfected with mock (center) or IRE1-specific siRNA (right) 48 h after infection, after which splicing of XBP1 (upper) and transcriptional up-regulation of EDEM1 (lower) were examined at the indicated time points after infection. The mean ± S.D. of two independent experiments are shown.
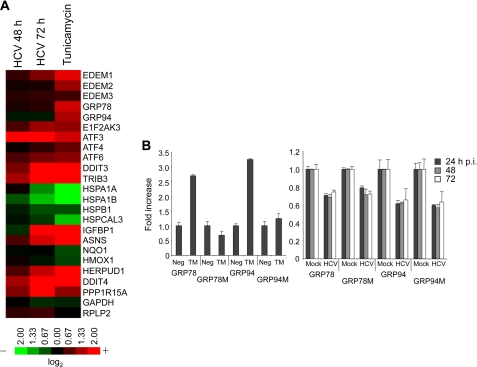
To enable a comprehensive investigation of the transcriptional changes that occur, up- and down-regulation of the transcriptome was examined in HCV-infected cells and in TM-treated cells. The results were compared with those of mock-transfected cells at each time point. A range of genes involved in ER stress was found to be regulated in HCV-infected and in TM-treated cells (Fig. 2A). EDEM1 was significantly up-regulated upon HCV infection, whereas expression levels of EDEM2 and EDEM3 remained unchanged. Although transcriptional changes caused by HCV infection in many of the genes listed are analogous to those that occur in cells treated with TM, up-regulation of two ER chaperone proteins, GRP78 and GRP94, was induced by TM treatment but not by HCV infection. This differential induction was confirmed by a reporter assay for GRP78 promoter and GRP94 promoter activities (Fig. 2B). These results are in agreement with a previously described finding that GRP78 and GRP94 are not responsive to HCV infection in hepatoma cells (18). It remains likely that HCV infection interferes with transcriptional activation of some ER chaperone proteins; however, the mechanism by which this occurs remains to be elucidated.
FIGURE 2.
Comprehensive analysis of ERAD gene expression in JFH-1-infected HuH-7.5.1 cells. A, HuH-7.5.1 cells treated with TM (5 μg/ml) for 12 h or infected with JFH-1 for 48 and 72 h were subjected to microarray analysis, along with their negative controls. Expression of ER stress genes is shown as a heat map. Red and green indicate up- and down-regulation, respectively. Information on each gene shown is indicated on the 3D-Gene web site. B, GRP78 and GRP94 induction in TM-treated (left) and HCV-infected cells (right). GRP78M and GRP94M represent the defective promoters. The mean ± S.D. (error bars) of three independent experiments are shown.
EDEMs Cause Ubiquitylation of HCV Glycoproteins and Enhance Their Degradation
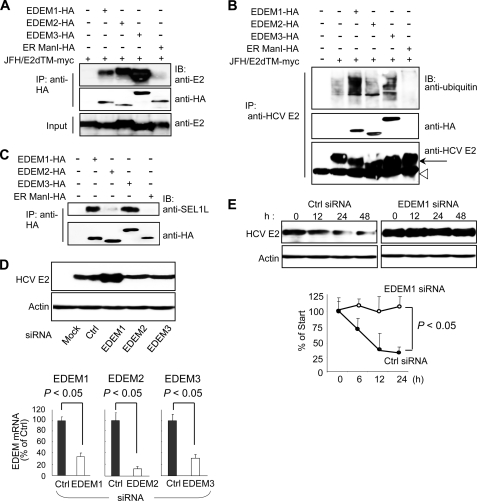
Because EDEMs have been reported to enhance proteasomal degradation of ERAD substrates through direct binding, we investigated the interaction of EDEMs with HCV glycoproteins in 293T cells by co-transfecting the expression plasmids for E1dTM or E2dTM together with plasmids carrying either EDEM or ER ManI genes. Immunoprecipitation and immunoblotting demonstrated that each EDEM, but not ER ManI, was capable of interacting with E2 (Fig. 3A) and E1 (supplemental Fig. S2). HCV glycoproteins displayed enhanced mobility when co-expressed with EDEM1, EDEM3, or ER ManI, which could be due to the mannosidase activity of these proteins, which is lacking in EDEM2 (6). HCV primarily replicates in hepatocytes so we examined the interaction of EDEMs with E2dTM in HuH-7 cells as well, which yielded similar results (data not shown). E2dTM lacks the transmembrane domain, which could affect its folding and ER retention and thus modulate the ability of this protein to interact with EDEMs and ER ManI. Second, E1 and E2 glycoproteins assemble as noncovalent heterodimers to make functional complexes, which may alter the interaction of these proteins with EDEMs. To address these issues, we co-transfected HuH-7 cells with plasmids carrying full-length E1E2 glycoproteins together with plasmids carrying either EDEMs or ER ManI. Similar phenotypes were produced following transfection full-length E1E2 proteins (supplemental Fig. S3A), demonstrating that functional complexes of HCV glycoproteins bind with EDEMs. Recently, we have reported on the development of a packaging system of HCV subgenomic replicon sequences through the provision of viral core NS2 proteins in trans (19). Transcomplementation with core NS2 proteins resulted in successful packaging of the viral sequences; therefore, plasmids carrying these proteins are a valid construct by which to examine the interaction of envelope proteins with ERAD machinery. Thus, we performed an immunoprecipitation assay of HuH-7 cells co-transfected with core NS2 and EDEMs. In agreement with our previous results, EDEMs, but not ER ManI, were observed to bind to HCV E2 protein (supplemental Fig. S3B). To examine the functional importance of this interaction, we analyzed the ubiquitylation of HCV E2 protein in cells co-transfected with HCV E2 and EDEM proteins. An immunoprecipitation assay revealed that overexpression of EDEM1 and EDEM3, but not of EDEM2 and ER ManI, dramatically increased the ubiquitylation of HCV glycoprotein (Fig. 3B). In mammals, the ER membrane ubiquitin-ligase complex involved in the dislocation of ERAD substrates, and their ubiquitylation contains the ER membrane adaptor SEL1L. It has recently been shown that SEL1L interacts with EDEM1 in cells and functions as a cargo receptor for ERAD substrates (20); however, it is unknown whether SEL1L interacts with other EDEMs. We therefore assessed whether SEL1L interacts with EDEM1, EDEM2, EDEM3, and ER ManI in cells (Fig. 3C). Interestingly, endogenous SEL1L co-precipitated with EDEM1 and EDEM3, whereas little to no interaction was observed with EDEM2 and ER ManI. Collectively, it is likely that, although all EDEMs can recognize HCV E1 and E2, EDEM1 and EDEM3 are involved in the ubiquitylation of HCV glycoproteins by delivering them to SEL1L-containing ubiquitin-ligase complexes. To investigate further the role of EDEMs in quality control of HCV glycoproteins, we measured the steady-state level of HCV E2 protein after EDEM knockdown. Transfection of HCV-infected cells with siRNAs against EDEM1, EDEM2, or EDEM3 caused a 60–80% reduction in mRNA levels of the respective genes (Fig. 3D) with no cytotoxic effects observed (data not shown). Immunoblotting showed a considerable increase in the steady-state level of viral E2 in EDEM1 siRNA-treated cells (Fig. 3D). We subsequently examined the turnover of E2 in cells with and without EDEM1 knockdown. In CHX half-life experiments, E2 protein was moderately unstable in control-infected cells, presumably via proteasomal degradation (Fig. 3E). Treatment with MG132, a proteasome inhibitor, blocked its destabilization (data not shown). In contrast, E2 was completely stable in EDEM1-knockdown cells during the chase period of time tested (Fig. 3E). Together, these results strongly suggest that EDEM1 and EDEM3, particularly EDEM1, are involved in the post-translational control of HCV glycoproteins.
FIGURE 3.
EDEMs are involved in the degradation of HCV glycoproteins. A, binding of EDEMs and ER ManI with HCV E2. 293T cells were seeded in 6-well plates at a density of 3 × 105 cells/well. After overnight incubation, cells were co-transfected with plasmids carrying HCV E2-myc (1 μg) and EDEM1-HA, EDEM2-HA, EDEM3-HA, or ER ManI-HA proteins (1 μg each). Forty-eight hours later, cells were harvested, immunoprecipitated (IP) with anti-HA antibodies, and Western blotting (IB) was performed with the indicated antibodies. B, ubiquitylation of HCV E2 protein in cells co-transfected with HCV E2 and EDEM plasmids. 293T cells were seeded in 6-well plates at a density of 3 × 105 cells/well. Twenty-four hours later, the cells were co-transfected with plasmids carrying HCV E2-myc (1 μg) and EDEM1-HA, EDEM2-HA, EDEM3-HA, or ER ManI-HA genes (1 μg each). Forty-eight hours later, the cells were harvested and immunoprecipitated with anti-E2 antibodies, and Western blotting was performed with the indicated antibodies. Arrow, HCV E2; open arrowhead, immunoglobulin heavy chain. C, binding of EDEMs and ER ManI with endogenous SEL1L in cells. D, steady-state level of HCV E2 in HCV-infected HuH-7 cells after EDEM knockdown (upper). The knockdown efficiencies of the respective siRNAs are shown in the lower panel. Values are normalized to GAPDH expression levels, and normalized values in negative control cells have been arbitrarily set at 100%. E, stability of HCV E2 protein in EDEM1 knockdown cells. HCV-infected HuH-7 cells were transfected with control or EDEM1 siRNA. Forty hours later, the cells were exposed to CHX (100 μg/ml) for 0, 12, 24, and 48 h, followed by immunoblotting. Specific signals were quantified by densitometry, and the percent of HCV E2 remaining was compared with initial levels. The mean ± S.D. (error bars) of two independent experiments are shown.
Involvement of EDEM1 in the Production of Infectious HCV
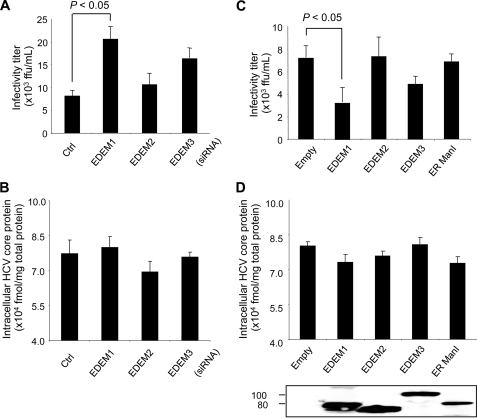
Given the involvement of EDEMs in the turnover of HCV glycoproteins, we investigated whether EDEMs affect the replication and production of infectious virus particles. EDEMs were knocked down in HCV-infected HuH-7 cells by siRNA transfection, and the production of infectious particles was then monitored by measuring the extracellular infectivity titer. Knocking down of EDEM1 and EDEM3 in the infected cells resulted in ∼3.1-fold (p < 0.05) and ∼2.3-fold increases in virus production, respectively, compared with control cells. No effect on virus production was observed following EDEM2 gene silencing (Fig. 4A). On the other hand, no significant differences were observed with regard to intracellular HCV core protein levels among mock- and EDEM siRNA-transfected cells (Fig. 4B), which indicates that replication of the viral genome is not affected by EDEM proteins. To examine further whether this effect on virus production was due to turnover of HCV envelope proteins, we performed loss-of-EDEM-function experiments in HuH-7 cells carrying HCV subgenomic replicons. Because the replicons do not require envelope proteins, they should be insensitive to the expression levels of genes involved in the ERAD pathway. As expected, siRNA-mediated knockdown of EDEMs resulted in little to no change in genome replication (supplemental Fig. S4A). To investigate further the participation of EDEMs in the HCV life cycle, HCV-infected cells were examined 48 h after transfection with an expression plasmid for either EDEM1, EDEM2, or EDEM3. As expected, exogenous expression of EDEM1 in the infected cells led to a 2.4-fold decrease in virus production compared with mock-transfected cells (p < 0.05) (Fig. 4C). A moderate decrease of 1.7-fold was observed in the cells overexpressing EDEM3 protein. Ectopic expression of EDEMs and ER ManI did not cause any change in intracellular HCV core protein levels (Fig. 4D). Similarly, little or no change was observed in genome replication when plasmids carrying EDEMs were introduced into HCV subgenomic replicon cells (supplemental Fig. S4B). These results indicate that EDEM1 and EDEM3, particularly EDEM1, regulate virus production, possibly through post-translational control of HCV glycoproteins.
FIGURE 4.
Role of EDEMs in HCV replication and production of infectious virus particles. A, HCV production in HuH-7 cells transfected with EDEM siRNAs. Cells were infected with JFH-1 at a m.o.i. of 1. Twenty-four hours later, the cells were transfected with the indicated siRNAs at a final concentration of 10 nm. The culture medium was harvested 48 h later and was used to infect naïve HuH-7.5.1 cells seeded in a 96-well plate. Immunostaining using anti-HCV core antibodies was performed at 72 h after infection, and focus-forming units were counted. B, siRNA-transfected and HCV-infected cells described in A harvested at 48 h after infection. Intracellular HCV core protein was measured. The values were normalized to total protein in the cell lysate samples. C, HCV production in HuH-7 cells transfected with plasmids carrying EDEM1-HA, EDEM2-HA, EDEM3-HA, or ER ManI-HA genes. D, intracellular HCV core protein within the cells described in C. Expression levels of the EDEMs and ER ManI were determined by anti-HA immunoblotting. The mean ± S.D. (error bars) of three independent experiments are shown in all of the panels.
Chemical Inhibition of the ERAD Pathway Increases HCV Production
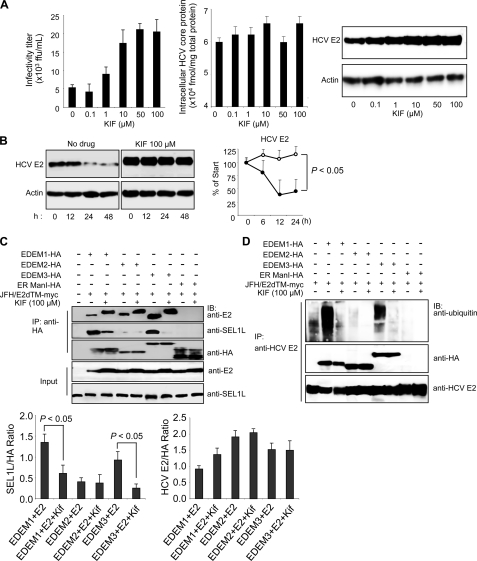
KIF, a potent inhibitor of ER mannosidase, is reported to inhibit the ERAD pathway. When HCV-infected cells were treated with KIF, virus production increased in the culture medium in a dose-dependent manner (Fig. 5A, left), and the steady-state level of E2 in the cells increased accordingly (Fig. 5A, right). No change was observed in intracellular HCV core protein levels after KIF treatment (Fig. 5A, center). Kinetic analyses showed that E2 was stabilized dramatically in KIF-treated cells (Fig. 5B), whereas the fate of HCV core protein, a nonglycoprotein, was not affected by KIF treatment (supplemental Fig. S5). No effect on virus replication was observed when the cells harboring JFH-1 subgenomic replicons were treated with KIF (data not shown).
FIGURE 5.
Effect of KIF on HCV production and stability of E2. A, extracellular HCV titer, intracellular HCV core protein expression, and steady-state level of HCV E2 in HuH-7 cells treated with different concentrations of KIF. B, CHX-based HCV protein stability assay of HCV E2 protein in KIF-treated cells as described in Fig. 3E. E2 protein levels normalized to actin levels are shown in the graph on the right. The open and filled circles indicate KIF-treated and nontreated cells, respectively. The mean ± S.D. (error bars) of two independent experiments are shown. C, binding of EDEMs and ER ManI with HCV E2 and SEL1L in 293T cells in the absence or presence of KIF. 293T cells were seeded in 6-well plates at a density of 3 × 105 cells/well. After overnight incubation, the cells were co-transfected with plasmids carrying HCV E2-myc (1 μg) and EDEM1-HA, EDEM2-HA, EDEM3-HA, or ER ManI-HA proteins (1 μg each). After 6 h, the culture medium was replaced with fresh or KIF-containing medium (100 μm). Forty-eight hours later, the cells were harvested and immunoprecipitated (IP) with anti-HA antibodies, after which Western blotting (IB) was performed with the indicated antibodies. Specific signals were quantified by densitometry, and the ratio between HCV E2 and HA (right graph) and between SEL1L and HA (left graph) in the same lanes is plotted on the graphs. The mean ± S.D. of three independent experiments are shown. D, EDEM protein-mediated ubiquitylation of HCV E2 protein in 293T cells in the absence or presence of KIF. The experimental procedure was the same as that described in Fig. 5C, except that immunoprecipitation was performed with anti-HCV E2 antibodies.
On the basis of these findings, one may hypothesize that KIF contributes to the stabilization of HCV glycoprotein(s) by interfering with the interaction between (i) EDEMs and viral proteins, or (ii) EDEMs and SEL1L. To address this, HCV E2 was co-expressed in 293T cells with EDEM1, EDEM2, EDEM3, or ER ManI in the presence or absence of KIF, followed by immunoprecipitation (Fig. 5C). E2 was shown to interact with EDEM1, EDEM2, and EDEM3, analogous to the data shown in Fig. 3A, and KIF did not block the interactions. Decreased electrophoretic mobility of E2 was detected in KIF-treated cells, possibly due to a change in glycan composition caused by inhibition of mannosidase activity. These findings led us to investigate whether the glycans on HCV glycoproteins are required for binding to EDEMs. We generated E1 and E2 mutants by replacing their N-glycosylation sites with glutamine residues and analyzed their interaction with EDEMs. Removal of the glycans did not inhibit the binding of E1 and E2 proteins to EDEM, demonstrating that N-glycans on the surface of viral proteins are not indispensible for an interaction between EDEMs and HCV glycoproteins to occur (supplemental Fig. S6). The effect of KIF on the association of EDEMs with downstream ERAD machinery was examined further. In cells co-expressing E2 and EDEMs, the interaction of SEL1L with EDEM1 and EDEM3 was significantly reduced in the presence of KIF (p < 0.05) (Fig. 5C). Consistent with these results, KIF abrogated the EDEM1- and EDEM3-mediated ubiquitylation of HCV E2 protein (Fig. 5D). This inhibitory effect of KIF on the SEL1L-EDEM interaction was also observed in HuH-7 cells (supplemental Fig. S7). These results suggest that KIF stabilizes HCV glycoproteins by interfering with the SEL1L-EDEM interaction and thus leads to an increase in virus production.
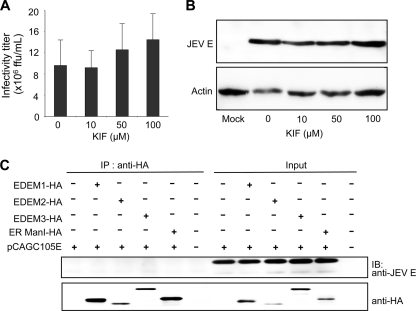
Role of ERAD in the Life Cycle of JEV
This study demonstrates involvement of the ERAD pathway in HCV production. However, the role of this pathway in the production of other viruses is still unknown. To this end, we examined its role in the life cycle of JEV, another member of the Flaviviridae family. In contrast to HCV, KIF treatment had little effect on JEV production in infected cells (Fig. 6A) or the steady-state level of viral E glycoprotein (Fig. 6B). Interaction of EDEMs with JEV E was analyzed further. Neither EDEMs nor ER ManI was found to interact with JEV E in cells (Fig. 6C), indicating no significant role of the ERAD pathway in the JEV life cycle. Altogether, these results strongly suggest that the ERAD pathway is involved in the quality control of glycoproteins of specific viruses, possible through an interaction with EDEM(s), and subsequent regulation of virus production.
FIGURE 6.
Binding of JEV envelope glycoprotein with EDEMs and effect of KIF on JEV production. A, JEV production in HuH-7 cells treated with KIF. The mean ± S.D. (error bars) of three independent experiments are shown. B, effect of KIF on the steady-state level of JEV envelope protein. C, binding of EDEMs with the JEV envelope.
DISCUSSION
Accumulating evidence points to a role of the ERAD pathway in the pathogenesis of different genetic and degenerative diseases. However, the involvement of ERAD in the life cycle of viruses and infectious diseases remains poorly understood. Until recently, an experimental HCV cell culture infection system has been lacking such that studies evaluating the effect of HCV infection on the ERAD pathway were performed by either using HCV subgenomic replicons which lack structural proteins or by ectopic expression of one or multiple structural proteins (21, 22). However, this problem was solved by identification of an HCV clone, JFH-1, capable of replicating and assembling infectious virus particles in cultured hepatocytes (15). In the present study, we used JFH-1 to examine the effect of HCV infection on activation of the ERAD pathway and its role in the virus life cycle. Our results show that the ERAD pathway is activated in HCV-infected cells, as evidenced by the maturation of XBP1 mRNA to its active form and up-regulation of EDEM1 (Fig. 1, A–D). Knocking down IRE1 reversed the induction of EDEM1, indicating that HCV infection-induced activation of the ERAD pathway is mediated through IRE1 (Fig. 1F). Loss- and gain-of-function analyses indicated that EDEM1 and EDEM3, particularly EDEM1, are involved in the post-translational control of HCV glycoproteins by which viral production is down-regulated (Figs. 3, D and E, and 4A). Our results suggest that EDEM1 and EDEM3 play a role in delivery of viral glycoproteins to the SEL1L-containing ubiquitin-ligase complex. It has recently been reported that coronavirus infection causes an accumulation of EDEM1 in membrane vesicles which are sites of viral replication, but that EDEM1 is not required for coronavirus replication (23). To our knowledge, the present study is the first to demonstrate regulation of the viral life cycle by ERAD machinery through interaction of EDEMs with viral glycoproteins.
We propose that the mechanisms described here are important during the early stages of establishing persistent HCV infection. ER stress caused by high levels of HCV infection during the acute phase presumably results in activation of the ERAD pathway. Induced EDEMs enhance the degradation of HCV envelope proteins, thereby reducing virus production. Maintenance of moderately low levels of HCV in the infected liver may contribute to the persistence of HCV infection, often associated with a lengthy asymptomatic phase that can last for decades. A range of viruses, including flaviviruses such as JEV, dengue virus, and West Nile virus, have been reported to induce XBP1 mRNA splicing triggered by ER stress (2, 3, 24). However, we demonstrate here that, in contrast to HCV, the envelope protein of JEV, which causes acute encephalitis, is not recognized by EDEMs, and the ERAD pathway does not control JEV production.
N-Linked glycoproteins displaying the glycan precursor Glc1Man9GlcNAc2 bind ER chaperones, such as calnexin or calreticulin, which facilitates protein folding. Removal of the terminal Glc from glycans disrupts this interaction with chaperones leading to Man trimming and delivery to ERAD machinery. A glucosyltransferase can transfer the terminal Man-linked Glc back to glycans, thereby allowing the “calnexin cycle” to continue until the glycoproteins are properly folded (for review, see Ref. 25). During this cycle, the decision of when to abandon additional folding attempts for immature polypeptides and to direct them instead toward the degradation pathway appears to be a crucial element of protein quality control. The basis by which this occurs, however, is not fully understood. Here, we demonstrate that stabilization of HCV envelope proteins and increased virus production occurs with KIF treatment (Fig. 5, A and B) and with gene silencing of either EDEM1 or EDEM3 (Figs. 3, D and E, and 4A). It is generally accepted that ERAD functions to eliminate proteins that are unable to adopt their native structure after translocation into the ER. From our results, however, one could argue that, during the HCV life cycle, at least a fraction of the competently folded viral glycoprotein intermediates may be released from the calnexin cycle before maturation and thereby be recognized as ERAD substrates. As suggested previously, the processes of protein folding and ERAD compete to some extent for newly synthesized polypeptides (26, 27). Under conditions in which high concentrations of ERAD-related factors are found in the ER due to induction of ER stress by viral infection, activated ERAD machinery may efficiently capture protein intermediates with folding/refolding capacity and cause premature termination of chaperone-assisted protein folding.
EDEM1 has recently been found to bind SEL1L, which is involved in the translocation of ERAD substrates from the ER to the cytoplasm (20). Our results demonstrate efficient binding of EDEM1 and EDEM3 to SEL1L, whereas EDEM2 exhibits only residual binding. In agreement with these results, increased ubiquitylation of HCV E2 protein was observed in cells overexpressing EDEM1 and EDEM3, but not in cells overexpressing the EDEM2 ortholog (Fig. 3B). Furthermore, KIF inhibited the binding of EDEM1 and EDEM3 with SEL1L, thus abrogating the ubiquitylation and enhancing the stability of HCV E2 protein (Fig. 5, B and D). It has been reported that KIF inhibits the interaction between EDEM1 and SEL1L, thus stabilizing ERAD substrates (4). Therefore, our results confirm previous findings and show that, along with EDEM1, KIF inhibits the binding of SEL1L to EDEM3. Furthermore, we have been the first to show that HCV E2 is a virus-derived ERAD substrate that can be used to analyze the mechanisms of this pathway. Taken together, our results indicate that EDEM1 and EDEM3, but not EDEM2, might be involved in targeting ERAD substrates to the translocation machinery, which may partly explain the different roles of the three EDEMs in HCV production. Although both EDEM1 and EDEM3 bind SEL1L and HCV envelope proteins, EDEM1 appears to have a larger role in regulation of HCV production than EDEM3. This is supported further by the finding that enhanced ubiquitylation of HCV E2 occurs in the presence of EDEM1 overexpression (Figs. 3B and 5D). In EDEM3-knockdown cells, EDEM1 may take over the function of delivering ERAD substrates to the translocation machinery. We also speculate that EDEM1 may function as a helper for EDEM3. This is supported by the observation that EDEM1 and EDEM3 synergistically increase HCV production when knocked down together (data not shown). HCV glycoproteins are a suitable means by which to investigate differences and redundancies pertaining to the role of EDEMs in the ERAD pathway.
HCV-infected and TM-treated cells demonstrated the greatest activation of EDEM1 transcript production among EDEMs (Fig. 1, C and D, and supplemental Fig. S1). Although it is known that XBP1 binds to specific ER stress-responsive cis-acting elements to induce EDEMs (28, 29), the exact mechanism of transcriptional regulation is not fully understood. It will be interesting to examine regulatory mechanism(s) specific to individual EDEM homologs in an ER stress-dependent or -independent manner.
These findings highlight the crucial role of the ERAD pathway in the HCV life cycle. Further studies are needed to clarify the details of this complex pathway. The data generated in this work, however, further contribute to our understanding of the mechanisms that govern the maturation and fate of viral glycoproteins in the ER.
Acknowledgments
We thank Dr. F. V. Chisari for the HuH-7.5.1 cells, Drs. N. Hosokawa and K. Nagata for the EDEM expression plasmids, Dr. K. Mori for the reporter plasmids of GRP78 and GRP94, and Drs. C. K. Lim and T. Takasaki for the anti-JEV antibody. We thank Drs. Chia-Yi Yu and Yi-Ling Lin for valuable advice and T. Date, M. Kaga, M. Sasaki, and T. Mizoguchi for assistance.
This work was supported by grants-in-aid from the Ministry of Health, Labor and Welfare, and from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
D. Akazawa, N. Nakamura, and T. Wakita, unpublished data.
- ER
- endoplasmic reticulum
- CHX
- cycloheximide
- EDEM
- ER degradation-enhancing α-mannosidase-like protein
- ERAD
- ER-associated degradation
- HCV
- hepatitis C virus
- JEV
- Japanese encephalitis virus
- KIF
- kifunensine
- ManI
- mannosidase I
- m.o.i.
- multiplicity of infection
- TM
- tunicamycin
- XBP1
- X-box-binding protein 1
- IRE
- inositol-requiring enzyme.
REFERENCES
- 1. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu C. Y., Hsu Y. W., Liao C. L., Lin Y. L. (2006) J. Virol. 80, 11868–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barry G., Fragkoudis R., Ferguson M. C., Lulla A., Merits A., Kohl A., Fazakerley J. K. (2010) J. Virol. 84, 7369–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Isler J. A., Skalet A. H., Alwine J. C. (2005) J. Virol. 79, 6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 6. Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., Moremen K. W. (2005) Glycobiology 15, 421–436 [DOI] [PubMed] [Google Scholar]
- 7. Hirao K., Natsuka Y., Tamura T., Wada I., Morito D., Natsuka S., Romero P., Sleno B., Tremblay L. O., Herscovics A., Nagata K., Hosokawa N. (2006) J. Biol. Chem. 281, 9650–9658 [DOI] [PubMed] [Google Scholar]
- 8. Bartenschlager R., Lohmann V. (2000) J. Gen. Virol. 81, 1631–1648 [DOI] [PubMed] [Google Scholar]
- 9. Reed K. E., Rice C. M. (2000) Curr. Top. Microbiol. Immunol. 242, 55–84 [DOI] [PubMed] [Google Scholar]
- 10. Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D. R., Wieland S. F., Uprichard S. L., Wakita T., Chisari F. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao Z., Date T., Li Y., Kato T., Miyamoto M., Yasui K., Wakita T. (2005) J. Gen. Virol. 86, 2209–2220 [DOI] [PubMed] [Google Scholar]
- 12. Tani H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., Matsuura Y. (2010) J. Virol. 84, 2798–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yoshida H., Haze K., Yanagi H., Yura T., Mori K. (1998) J. Biol. Chem. 273, 33741–33749 [DOI] [PubMed] [Google Scholar]
- 14. Murakami K., Kimura T., Osaki M., Ishii K., Miyamura T., Suzuki T., Wakita T., Shoji I. (2008) J. Gen. Virol. 89, 1587–1592 [DOI] [PubMed] [Google Scholar]
- 15. Wakita T., Pietschmann T., Kato T., Date T., Miyamoto M., Zhao Z., Murthy K., Habermann A., Kräusslich H. G., Mizokami M., Bartenschlager R., Liang T. J. (2005) Nat. Med. 11, 791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim C. K., Takasaki T., Kotaki A., Kurane I. (2008) Virology 374, 60–70 [DOI] [PubMed] [Google Scholar]
- 17. Takeuchi T., Katsume A., Tanaka T., Abe A., Inoue K., Tsukiyama-Kohara K., Kawaguchi R., Tanaka S., Kohara M. (1999) Gastroenterology 116, 636–642 [DOI] [PubMed] [Google Scholar]
- 18. Deng L., Adachi T., Kitayama K., Bungyoku Y., Kitazawa S., Ishido S., Shoji I., Hotta H. (2008) J. Virol. 82, 10375–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Masaki T., Suzuki R., Saeed M., Mori K., Matsuda M., Aizaki H., Ishii K., Maki N., Miyamura T., Matsuura Y., Wakita T., Suzuki T. (2010) J. Virol. 84, 5824–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cormier J. H., Tamura T., Sunryd J. C., Hebert D. N. (2009) Mol. Cell 34, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tardif K. D., Mori K., Kaufman R. J., Siddiqui A. (2004) J. Biol. Chem. 279, 17158–17164 [DOI] [PubMed] [Google Scholar]
- 22. Chan S. W., Egan P. A. (2005) FASEB J. 19, 1510–1512 [DOI] [PubMed] [Google Scholar]
- 23. Reggiori F., Monastyrska I., Verheije M. H., Calì T., Ulasli M., Bianchi S., Bernasconi R., de Haan C. A., Molinari M. (2010) Cell Host Microbe 7, 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Medigeshi G. R., Lancaster A. M., Hirsch A. J., Briese T., Lipkin W. I., Defilippis V., Früh K., Mason P. W., Nikolich-Zugich J., Nelson J. A. (2007) J. Virol. 81, 10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Molinari M. (2007) Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- 26. Eriksson K. K., Vago R., Calanca V., Galli C., Paganetti P., Molinari M. (2004) J. Biol. Chem. 279, 44600–44605 [DOI] [PubMed] [Google Scholar]
- 27. Wu Y., Swulius M. T., Moremen K. W., Sifers R. N. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8229–8234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olivari S., Galli C., Alanen H., Ruddock L., Molinari M. (2005) J. Biol. Chem. 280, 2424–2428 [DOI] [PubMed] [Google Scholar]
- 29. Yoshida H., Matsui T., Hosokawa N., Kaufman R. J., Nagata K., Mori K. (2003) Dev. Cell 4, 265–271 [DOI] [PubMed] [Google Scholar]