Abstract
The cystathionine β-synthase module of OpuA in conjunction with an anionic membrane surface acts as a sensor of internal ionic strength, which allows the protein to respond to osmotic stress. We now show by chemical modification and cross-linking studies that CBS2-CBS2 interface residues are critical for transport activity and/or ionic regulation of transport, whereas CBS1 serves no functional role. We establish that Cys residues in CBS1, CBS2, and the nucleotide-binding domain are more accessible for cross-linking at high than low ionic strength, indicating that these domains undergo conformational changes when transiting between the active and inactive state. Structural analyses suggest that the cystathionine β-synthase module is largely unstructured. Moreover, we could substitute CBS1 by a linker and preserve ionic regulation of transport. These data suggest that CBS1 serves as a linker and the structured CBS2-CBS2 interface forms a hinge point for ionic strength-dependent rearrangements that are transmitted to the nucleotide-binding domain and thereby affect translocation activity.
Keywords: ABC Transporter, Chemical Modification, Membrane Reconstitution, Protein Cross-linking, Protein Engineering, CBS Domain, Ionic Strength Sensing, Osmotic Stress, Structure Analysis
Introduction
Cell volume regulation is an essential function of any cell to overcome the consequences of osmotic stress. Under hypo-osmotic conditions, prokaryotes rapidly release large amounts of osmolytes via mechanosensitive channels to avoid the cell from lysing (1). Under hyper-osmotic conditions, a cell accumulates or synthesizes compatible solutes to prevent shrinkage and ultimately plasmolysis (2). Osmoregulatory transporters respond to hyper-osmotic stress by taking up compatible solutes such as glycine betaine, carnitine, trimethylamine N-oxide, or proline (3). Three osmoregulatory transporters have been studied in great detail as follows: ProP in Escherichia coli (4–8), BetP in Corynebacterium glutamicum (9–10), and OpuA in Lactococcus lactis (11–13). ProP and BetP are secondary transporters driven by the proton and sodium electrochemical gradient across the membrane, respectively, whereas OpuA is a primary transporter driven by ATP. Each of these systems has been shown to sense the intracellular concentration of ions (and hydration in case of ProP (14)). Whereas ProP and OpuA seem indifferent to the type of ion, BetP senses K+ ions preferentially (9, 14–16). For each of the systems, the C-terminal domain is involved in ionic sensing but is not sufficient for ionic regulation (5, 12, 13, 17–19). For BetP and OpuA, it has been shown that ionic regulation is dependent on anionic lipids in the membrane (11, 20). In fact, we have shown that the C-terminal CBS2 module of OpuA interacts directly or indirectly with the anionic membrane surface (12) and that raising the ionic strength above threshold values screens the electrostatic interactions and unlocks the transporter.
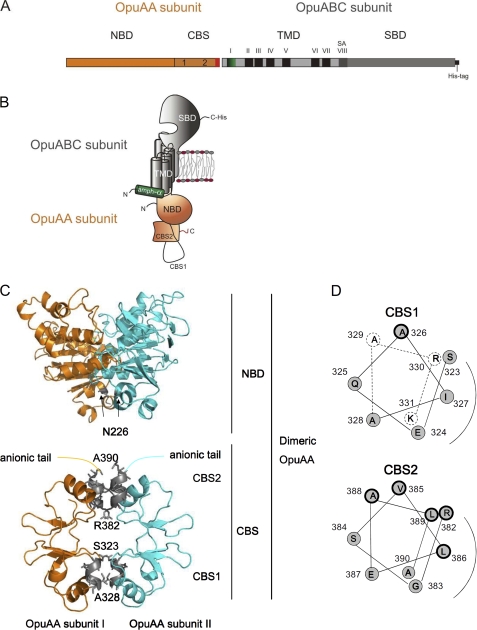
OpuA consists of two different polypeptides (Fig. 1, A and B) as follows: OpuABC with the substrate-binding domain fused C-terminal to the transmembrane domain (TMD), and OpuAA with tandem CBS domains fused C-terminal to the nucleotide-binding domain (NBD) (21). The active OpuA complex consists of two OpuABC subunits and two OpuAA subunits. The structure of the substrate-binding domain has been solved at 1.9 Å resolution, and glycine betaine binding to OpuA has been shown to be independent of ionic strength (22). Homology models are available for the NBD and CBS domains, but there is no high resolution structure of a homologue of the TMD. The tandem CBS domains each have predicted β-α-β-β-α secondary structure, and crystal structures of analogous proteins suggest a dimeric state, consistent with the dimeric structure of OpuA (23–27). CBS domains are not highly conserved in sequence, and the molecular basis for regulation by CBS is poorly understood, although over 30,000 CBS-containing proteins are known to date. In some cases, conformational changes at the CBS dimer interface have been reported upon the binding of nucleotides (28, 29) or Mg2+ ions (30, 31).
FIGURE 1.
Dimer structure of CBS domains and nucleotide-binding domain in OpuA. A, OpuAA consists of the nucleotide-binding domain and tandem CBS domains connected to the anionic tail (∼20 amino acids, indicated in red). OpuABC consists of the transmembrane domain (I is amphipathic helix, indicated in green; II–VII are transmembrane segments, shown in black; and SA is the signal anchor sequence, shown in gray), connected to the substrate-binding domain (SBD). B, shows the organization of the subunits in the membrane; the differences in structure of the CBS1 and CBS2 domains are emphasized in the schematic. The functional protein is a dimer of the complex shown in B. C, CBS tandem domain and nucleotide-binding domain of OpuAA were modeled on the crystal structure of MgtE in the Mg2+-bound state (Protein Data Bank code 2YVX) and MalK in the ATP-bound state (Protein Data Bank code 1Q12), respectively, using Swiss model. Each monomer was then assembled with reference to the atomic coordinates of MgtE or the MalK dimer structure using PyMOL (70). The positions of the Cys mutations are indicated in gray (residues at the beginning and end of α2 are numbered). D, helical wheel projection of the residues in α2. The putative dimer interface on the basis of the MgtE crystal structure is indicated by the bent line. Well conserved residues between OpuAA, MgtE, and CmCLC are circled by a thick line (supplemental Fig. 2A). A more complete alignment of CBS domains in OpuAA homologs is presented in supplemental Fig. 7. Residues indicated by dotted lines have not been examined.
Guided by these studies, we have now engineered the CBS module of OpuA and probed the effects of domain modifications and cross-linking on the ionic regulation of transport. Each of the modified proteins was purified and reconstituted into liposomes of defined lipid composition, and the transport activity was determined as a function of the ionic strength and other parameters. We show ionic strength-dependent conformational changes at CBS1 and CBS2. However, only the domain interactions at the CBS2-CBS2 interface are critical for transport; CBS1 merely serves as a linker domain without a specific functionality.
EXPERIMENTAL PROCEDURES
Plasmid Construction
Site-specific mutations in the CBS or NBD part of the opuAA gene were made via the QuikChange site-directed mutagenesis method (32). Briefly, pRE-OpuA with opuAA and opuABC in the pRE vector, containing an E. coli origin of replication and the β-lactamase resistance gene (33), was used as template for the PCR and amplification by Pfu DNA polymerase (Promega) (see supplemental Table 1 for specification of the primers). Following digestion by DpnI, the DNA was transformed to E. coli, and plasmids were isolated and verified by DNA sequencing. The exchange of the vector backbone of pRE-OpuA to pERL, containing an L. lactis origin of replication, was performed as described earlier (33), resulting in pNZOpuA(X###C)His (X = residue in wild type OpuAA; ### = position of the residue in the protein sequence; C = cysteine). The plasmids were electroporated into L. lactis Opu401, a derivative of NZ9000 in which the opuA genes were deleted (12). For the CBS1 deletion mutant, KpnI restriction sites were introduced before and after the CBS1 coding region, using pRE-OpuA and QuikChange site-directed mutagenesis as described above and primer pairs with KpnI sites (AK1 and AK2; see supplemental Table 1). The plasmid was digested by KpnI and ligated after removing the DNA fragment encoding the CBS1 domain, resulting in pRE-OpuA(ΔCBS1). For substitution of CBS1 by a flexible linker (protein sequence GGGSGGGSGGGSGGGSAAAQL), KpnI and PstI restriction sites were introduced before and after the CBS1 coding region as described above, using primer pairs with KpnI sites (AK1) and PstI sites (AK3; see supplemental Table 1). Next, the plasmid was digested by KpnI and PstI, and a synthetic DNA fragment coding for the flexible linker was inserted (linker DNA sequence, 5′-GGGGTACCTTAGGTGGTGGTTCAGGTGGTGGTTCAGGTGGTGGTTCAGGTGGTGGTTCAGCTGCTGCTCAACTGCAGAACCAATGCATTGG), and the resulting plasmid was named pRE-OpuA(LΔCBS1)). The plasmid was electroporated into L. lactis Opu401 after changing the vector backbone as described above.
The gene fragment encoding the tandem CBS domain was cloned into pBADnLIC (33), using pNZOpuAhis (34) as a template, with the primer pair CBS (see supplemental Table S1). The resulting plasmid, pBADnLIC-CBS, was transformed into E. coli MC1061 cells.
Bacterial Strains, Growth Conditions, and Vesicle Preparation
L. lactis strain Opu401 (12) was cultivated semi-anaerobically at 30 °C in a medium containing 2% (w/v) gistex LS (Strik BV, Eemnes, Netherlands) and 65 mm sodium phosphate (NaPi), pH 6.5, supplemented with 1.0% (w/v) glucose and 5 μg/ml chloramphenicol when carrying pNZOpuAHis or derivatives. For isolation of membrane vesicles, cells were grown in a 2-liter pH-regulated bioreactor to an A600 of 2, after which transcription from the nisA promoter was switched on by the addition of 0.1% (v/v) culture supernatant of the nisin A-producing strain NZ9700 (35). The cells were harvested after 2 h (same temperature for induction as for initial growth). The cells were lysed by high pressure disruption (Constant Systems Ltd., UK; two passes at 39,000 p.s.i.). Unbroken cells and cell debris were removed by centrifugation at 267,000 × g, for 30 min at 4 °C, and then the membrane vesicles were collected by ultracentrifugation at 280,000 × g, for 1 h at 4 °C, and resuspended in 50 mm KPi plus 20% (v/v) glycerol. The protein concentration was determined using the DC protein assay (Bio-Rad).
For isolation of the tandem CBS domains (hereafter referred to as the CBS module), E. coli MC1061/pBADnLIC-CBS was grown aerobically at 37 °C in LB medium to an A600 of 0.5–0.6. Following induction with 0.002% (w/v) l-arabinose, the cells were grown for 3 h at 25 °C before harvesting and resuspension in 50 mm Tris-HCl, pH 8.0, 200 mm NaCl, plus 10% (v/v) glycerol; the cells were stored at −80 °C.
Purification of the CBS Module
The E. coli cells were lysed by high pressure disruption with two passes at 15,000 p.s.i. Unbroken cells and cell debris was removed by centrifugation at 267,000 × g, for 1 h at 4 °C. After addition of 10 mm imidazole, pH 8.0, 10 ml of cell extract (∼2.5 mg of protein) was incubated with 0.5 ml of nickel-Sepharose for 1 h at 4 °C and subsequently packed in a column. The column was washed with 20 column volumes of 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 5% (v/v) glycerol, plus 50 mm imidazole. The protein was eluted with 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, plus 500 mm imidazole. The protein was then loaded onto a Superdex 200 10/300GL size exclusion column, pre-equilibrated with 20 mm Tris-HCl, pH 8.0, plus 150 mm NaCl. For CD and NMR experiments, the buffer was exchanged to 100 mm KPi, pH 7.0, using a NAP10 column (GE Healthcare), and the protein was concentrated using a 10-kDa molecular mass cutoff Vivaspin concentrator to ∼580 μm.
Light-scattering Measurements
An aliquot of 200 μl of CBS module (∼0.5 mg/ml) was run at a flow rate of 0.5 ml/min on a Superdex 200 10/300GL column (GE Healthcare) in 20 mm Tris-HCl, pH 8.0, plus 150 mm NaCl, using an Agilent 1200 series isocratic pump at room temperature. Detectors were used for absorbance at 280 nm (Agilent), static light-scattering (miniDawn TREOS Wyatt), and differential refractive index (Optilab Rex Wyatt). For data analysis, the ASTRA software package version 5.3.2.10 was used (Wyatt), with a value for the refractive index increment (dn/dc) protein of 0.179 ml/mg (36, 37).
Circular Dichroism
CD measurements were done on a circular dichroism spectrometer; model 62A DS (Aviv Lakewood). Typically, 300 μl of protein sample at a protein concentration of 0.1 mg/ml in 0.1 m KPi, pH 7.0, was used in a 300-μl cuvette with a 1-mm path length. The protein concentration was based on the extinction coefficient of the tandem CBS domain (2980 m−1 cm−1), on samples that were concentrated to 3–5 mg/ml and subsequently diluted to 0.1 mg/ml for the CD measurement. The protein concentrations determined in this way corresponded well to the values measured using BCA assay. Samples without guanidine HCl (GdnHCl) were measured between 250 and 190 nm at 25 °C. Because of the dynode voltage limit of the detector, the samples with GdnHCl added were measured between 250 and 210 nm. Measurements were done in triplicate; GdnHCl was added from a 6 m stock solution. The secondary structure of the protein was estimated from spectra as shown in the inset of Fig. 5B by comparison with an expanded reference set, using the programs CONTIN, SELCON, and CDSSTR (38).
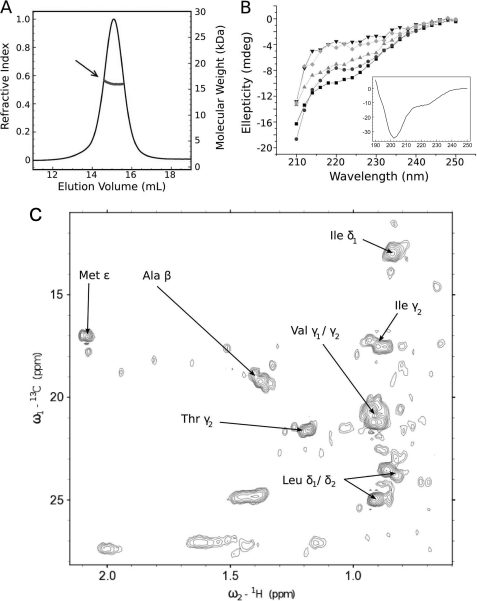
FIGURE 5.
Structural analysis of the soluble tandem CBS domain. A, by size-exclusion chromatography coupled to light scattering analysis of the tandem CBS domain of OpuA. The calculated molecular mass of the CBS domain throughout the peak is indicated. B, CD spectra of the tandem CBS domain in the absence and different concentrations of GdnHCl to unfold the protein; without GdnHCl (■), 0.4 m (●), 1 m (▴), 1.8 m (▾), 2.5 m GdnHCl (♦). The full spectrum without GdnHCl is shown in the inset. C, inset of the 1H-13C HSQC NMR spectrum of the tandem CBS domains showing cross-peaks for all methyl-containing amino acids. Although a number of discrete peaks can be observed in the spectra, the majority of the peaks superimpose, which is typical of a protein with relatively large unstructured domains.
NMR Spectroscopy
For NMR measurements, the sample contained 580 μm of unlabeled CBS module in 100 mm KPi, pH 7.0, in 93/7% (v/v) H2O/D2O. NMR experiments were performed at 26 °C on a Varian Unity Inova spectrometer operating at 600 MHz and equipped with a triple resonance room temperature probe. The 1H-13C HSQC experiment was recorded with 128 (13C) × 1024 (1H) complex data points (10.7 × 127.9 ms maximum evolution times, respectively) averaging 32 scans per FID, giving rise to an acquisition time of 160 min.
MalPEG Labeling and Cross-linking by Bis-methanethiosulfonate
5 mg of total protein in membrane vesicles were solubilized with 0.5% (w/v) DDM, and OpuA was bound to nickel-Sepharose (GE Healthcare) in Buffer A: 50 mm KPi, pH 8.0, 200 mm KCl, 20% glycerol, plus 1 mm DTT. After removal by washing the nontagged proteins and DTT (20 column volumes of Buffer B: 50 mm KPi, pH 7.0, 50 mm imidazole, 200 mm KCl, 20% glycerol, plus 0.04% DDM), OpuA(X###C) mutants were labeled with 50 μm MalPEG (Sigma), overnight at 4 °C. Following washing with 5 column volumes of Buffer B without imidazole, MalPEG-labeled OpuA was eluted in Buffer C: 50 mm KPi, pH 7.0, 200 mm imidazole, 200 mm KCl, 20% glycerol, plus 0.04% DDM. The cross-linking of Cys mutants with bis-methanethiosulfonate reagents was done similarly except that 2.5 μm MTS-3-MTS, MTS-6-MTS, or MTS-17-MTS (Toronto Research Chemicals Inc., approximate S–S distances are 5, 9, and 22 Å, respectively) was used instead of 50 μm MalPEG. We note that this cross-linking protocol was obtained after extensive testing of reaction conditions (reagent and protein concentration and reaction time), e.g. supplemental Fig. 4. Following elution, 4 mg of protein was resuspended in nonreducing sample buffer (3% (w/v) Tris-HCl, 50% (v/v) glycerol, 0.005% (v/v) bromphenol blue, plus 1% (v/v) SDS) and subjected to 12.5% SDS-PAGE. The labeling or cross-linking efficiency was calculated as a ratio from the intensity of MalPEG-labeled or cross-linked OpuAA over total OpuAA. Protein band intensities on SDS-polyacrylamide gels stained with Coomassie Brilliant Blue were measured with a densitometer, using the Scion Image beta 4.02 software (National Institutes of Health).
Cross-linking of OpuA in Membrane Vesicles
Cross-linking of the Cys mutants in membrane vesicles by the MTS cross-linker was carried out under low (50 mm KPi, pH 7.0, plus 50 mm KCl) or high (50 mm KPi, pH 7.0, plus 300 mm KCl) ionic strength conditions (low or high salt buffer). The membrane vesicles at a concentration of 2 mg/ml of protein were washed by ultracentrifugation (280,000 × g, 4 °C, 15 min) and resuspended in low or high salt buffer. The membranes were incubated with 4 μm MTS-3-MTS, MTS-6-MTS, or MTS-17-MTS for 1 min at 4 °C. The cross-linking reaction was stopped by the addition of 10 mm N-ethylmaleimide. The OpuA protein was subsequently solubilized and purified as described previously (34). About 10 μg of protein was resuspended in nonreducing sample buffer (3% (w/v) Tris-HCl, 50% (v/v) glycerol, 0.005% (v/v) bromphenol blue, plus 1% (v/v) SDS) and subjected to 12.5% SDS-PAGE.
Membrane Reconstitution of OpuA
Purified OpuA and mutant derivatives were reconstituted in liposomes composed of synthetic lipids (50 mol % DOPE, 38 mol % DOPG, plus 12 mol % DOPC) as described previously (39). The final protein to lipid ratio was 1:100 (w/w).
Transport Assays
For ATP-driven uptake of glycine betaine in proteoliposomes, the ATP-regenerating system was enclosed in two cycles of freezing in liquid N2 and thawing at room temperature (39); ATP-regenerating system corresponds to 10 mm Na2-ATP + 10 mm MgSO4 + 24 mm Na2-creatine phosphate + 2.4 mg/ml creatine kinase in 50 mm KPi, pH 7.0. After extrusion of the proteoliposomes through a polycarbonate filter (200 nm pore size), the proteoliposomes were washed twice with 90 mm KPi, pH 7.0 (osmolality of ∼ 240 mosmol/kg), by centrifugation (280,000 × g; for 15 min at 4 °C) and resuspended in the same buffer to a concentration of 80 mg of lipid/ml. For osmotically activated transport, the proteoliposomes were diluted to a lipid concentration of 6 mg/ml into assay buffer (90 mm KPi, pH 7.0, supplemented with different concentrations of KCl). Following incubation for 2 min at 30 °C, the transport reaction was initiated by the addition of [14C]glycine betaine to a final concentration of 51 μm (more than 10-fold above the Km for transport and KD for binding) (40). At given time intervals, 26-μl samples were taken and diluted with 2 ml of ice-cold isotonic assay buffer. The samples were filtered rapidly through 0.45-μm pore-size cellulose nitrate filters (Schleicher & Schuell) and washed twice with 2 ml of ice-cold assay buffer. The radioactivity on the filters was determined by liquid scintillation counting. Radiolabeled [N-methyl-14C]choline chloride (55 mCi/mmol) was obtained from Amersham Biosciences and converted to [N-methyl-14C]glycine betaine as described (41). Osmotic activation curves were fitted with the equation: V = Amax·In/(K0.5n + In), in which V is the initial rate of uptake; Amax is the maximal rate of uptake; I is the KCl concentration of the outside buffer; K0.5 is the concentration of KCl at which Amax is 50%, and n is the activation coefficient (slope at K0.5). A summary of all parameter for each of the mutants and analyzed under different conditions is given in supplemental Table 3.
Blue Native Electrophoresis
OpuA wild type and L386C were purified and labeled with MalPEG as described above. Blue native electrophoresis was performed as described previously (42), using the protocol of Ref. 43. 10 μg of protein was applied to a linear 8–16% (w/v) polyacrylamide gradient gel. The electrophoresis was started at 100 V for 45 min and continued for 3 h at 200 V at room temperature. The gel was stained with Coomassie Brilliant Blue.
Nucleotide Binding
Binding to OpuA of the fluorescent ATP analog TNP-ATP was analyzed as described previously (44). TNP-ATP fluorescence was first determined in the absence of protein by measuring the fluorescence emission at 540 nm and an excitation wavelength of 409 nm for TNP-ATP concentrations in the range of 0–20 μm, in steps of 1 μm. The same experiment was conducted in the presence of 0.5 μm purified OpuA; the change in fluorescence (ΔF) was calculated after subtracting the fluorescence change in the absence of OpuA. The fluorescence change as a function of TNP-ATP concentration was fitted to a simple hyperbola as shown in Equation 1 to determine the dissociation constant (KD),
where [S] is the TNP-ATP concentration and ΔFmax is the maximal change in fluorescence intensity and related to the maximal number of binding sites.
RESULTS
Cys Engineering at the Putative Dimer Interface of the CBS Module of OpuA
CBS domains have been observed as dimeric structures with various interfaces, e.g. face-to-face or back-to-back configurations of CBS1:CBS1-CBS2:CBS2 and CBS1:CBS2-CBS2:CBS1 (supplemental Fig. 1) (23–27, 45–47). It is possible that some of these are crystallization artifacts as most structures were obtained from isolated domains rather than full-length proteins. The complete structures of membrane proteins with tandem CBS domains are from the Mg2+-regulated transporter MgtE (30) and the CmCLC chloride transporter (48). In MgtE, helix 2 (α2) of one CBS domain packs onto the corresponding domain of the other subunit, forming face-to-face CBS1:CBS1-CBS2:CBS2 structures (supplemental Fig. 2B). CmCLC, on the other hand, forms a back-to-back β-sheet CBS2-CBS2 interface (supplemental Fig. 2B). The primary sequence of the CBS domain in OpuA is more similar to that of MgtE (supplemental Fig. 2A), and therefore, we initially modeled the OpuA CBS module on the basis of the crystal structure of MgtE (Protein Data Bank code 2VYX), taking advantage of multiple sequence alignments to position the conserved residues (Fig. 1C; helical wheel representations of α2 are shown in Fig. 1D). We used this model to project the amino acid substitutions in CBS1 and CBS2, which we subsequently analyzed by accessibility and cross-linking studies.
Effect of Single Cys Substitutions on Transport Activity
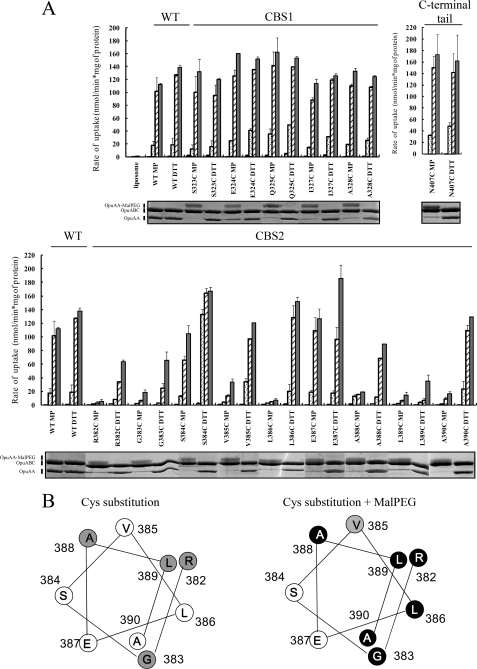
We determined the activity of wild type OpuA and of each of the Cys mutants in 90 mm KPi, pH 7.0, at low (no KCl added), intermediate (100 mm KCl), and high (200 and 300 mm KCl) ionic strength. Under these conditions and with 38 mol % of anionic lipid in the membrane, wild type OpuA is inactive at low ionic strength and maximally active at or above 200 mm salt. None of the CBS1 mutants was significantly affected in its transport activity or ionic regulation upon DTT treatment to avoid the spontaneous cross-linking of Cys residues. On the contrary, R382C, G383C, A388C, and L389C in CBS2 were to differing degrees reduced in activity (Fig. 2A), pointing to critical contacts made by these residues.
FIGURE 2.
Effect of MalPEG modification on the activity and ionic activation of OpuA. A, uptake of [14C]glycine betaine by wild type OpuA and Cys mutants was assayed in 90 mm KPi, pH 7.0, supplemented with 0, 100, 200, or 300 mm KCl. Purified unlabeled or MalPEG-labeled OpuA was reconstituted in proteoliposomes composed of 50 mol % DOPE, 12 mol % DOPC, plus 38 mol % DOPG and supplemented with 1 mm DTT; in A, unlabeled protein is indicated by DTT; MalPEG-labeled by MP. The ATP-regenerating system was enclosed inside the proteoliposomes. The average rates and standard deviations of four time points in the linear range (0–120 s) of the uptake curve are shown. 0 mm KCl, white bar; 100 mm KCl, dotted bar; 200 mm KCl, slashed bar; 300 mm KCl, gray bar. SDS-PAGE of the proteins and mobility shifts as a result of MalPEG labeling are shown below the top panel; 750 μg of proteoliposomes (lipid weight, corresponding to 7.5 μg of protein) were treated with nonreducing sample buffer with 4% (v/v) SDS and loaded per lane on a 12.5% SDS-polyacrylamide gel. B, helical wheel projection of the Cys residues in putative α-helix 2 of CBS2. Mutants with more than 80% activity are depicted by a white circle; mutants with 20–80% activity by a gray circle; and mutants with less than 20% activity by black circles.
MalPEG Modification of OpuA
We then tested the effect of MalPEG labeling on the transport activity of each of the Cys mutants. MalPEG has a large 5-kDa polyethyleneglycol moiety, which is predicted to perturb interactions at the dimer interface but may not have much effect when the residue is surface-exposed or the region is disordered. In agreement, MalPEG labeling of the C-terminal tail at Cys-407 (N407C), which is predicted to be unstructured (13), did not significantly affect the activation profile of OpuA (Fig. 2A) despite a high labeling efficiency (supplemental Table 2). As shown in the gel panels of Fig. 2A, a high degree of labeling was achieved for each of the mutants except A326C (data not shown) (supplemental Table 2). The extra mass of the MalPEG moiety resulted in a significant retardation of the OpuAA mobility on SDS-PAGE. The shift was larger for the polyethyleneglycol moiety linked to CBS1 than CBS2 (Fig. 2A). Strikingly, modification of the CBS1 domain with MalPEG had no effect on the transport activity or ionic regulation of OpuA. On the contrary, several but not all of the CBS2 Cys mutants were highly compromised in their transport activity (Fig. 2A). Fig. 2B projects the effects on transport activity of the Cys substitutions and MalPEG labeling of CBS2 in the form of a color scale: white, <20%; gray, 20–80%; and black >80% inhibition. The data indicate a clear periodicity in the inhibition of transport. On the contrary, the binding of the ATP analog TNP-ATP was not affected in the MalPEG-labeled mutants (supplemental Fig. 3A). Next, we examined whether MalPEG labeling would affect the oligomeric state of OpuA, using blue native electrophoresis. The MalPEG-labeled inactive L386C mutant had the same apparent mass as unlabeled, fully active, L386C, and wild type OpuA (supplemental Fig. 3B). Thus, MalPEG modification of one particular face of α2 in the CBS2 domain interferes with the translocation cycle of OpuA but does not affect the ATP binding or oligomeric state of the protein. We have previously shown that deletion of the CBS module of OpuA results in a transporter that is no longer regulated by ionic strength but is otherwise fully functional (13). Taken together, the data indicate that the accessory CBS module imposes a control mechanism onto the catalytic cycle of OpuA.
Cross-linking of the CBS Domains of OpuA
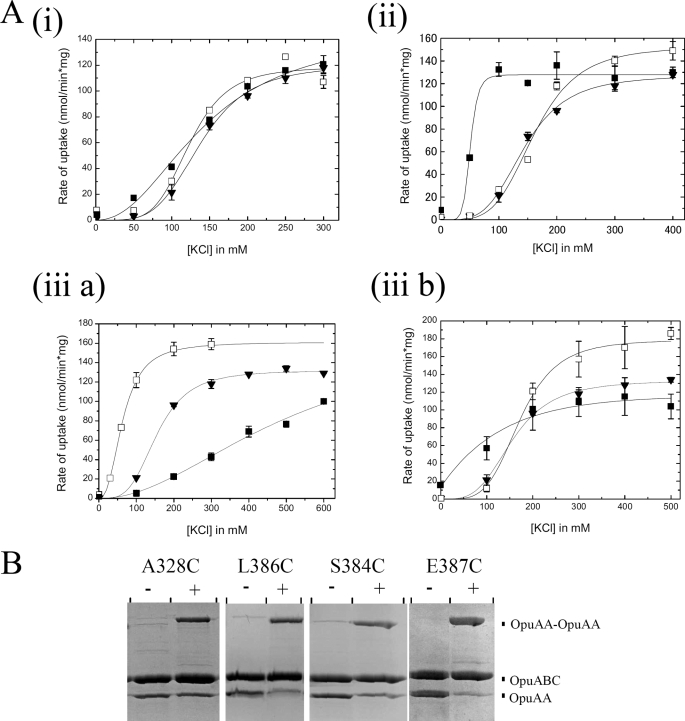
Because the integrity of putative interface residues of CBS2 domain proved important for a high activity of OpuA, we tested whether the CBS2-CBS2 domains would reversibly associate and dissociate as part of the catalytic cycle of OpuA, i.e. much like the NBDs do upon binding and hydrolyzing ATP and releasing of ADP plus Pi (49). Alternatively, it seemed possible that such association-dissociation would form the basis of the ionic gating mechanism rather than being intrinsic to the catalytic cycle of the protein. We thus examined the effect of cross-linking of the CBS domains on the transport activity of OpuA. The Cys mutants were cross-linked, following purification of the proteins, and then reconstituted in liposomes. The cross-linking by MTS-3-MTS was optimized with respect to reagent concentration and reaction time (supplemental Fig. 4). The cross-linking efficiency was optimal at 1 μm (i.e. a 1.3-fold excess of linker over OpuA); at higher concentrations of MTS-3-MTS (or other cross-linker), individual protein thiols became labeled with the reagent, preventing intersubunit cross-linking (data not shown). On average, the cross-linking efficiency was 60–80%, except for E324C, I327C, and L389C (Fig. 3B, supplemental Fig. 5L, and supplemental Table 2). Fig. 3A, supplemental Fig. 5, A–K, and supplemental Table 3 show the diverse effects of cross-linking on the ionic activation profile of OpuA; see under “Experimental Procedures” for the definition of the salt dependence parameter K0.5 and the activation coefficient n. We observed the following: (i) no significant change in the activation profile or maximal activity for E324C, Q325C, I327C, and A328C (all in CBS1); (ii) a shift to lower salt concentrations (i.e. a lower K0.5) for S323C, L386C, L389C, and A390C; and (iii) a lower activation coefficient (n) for A326C, G383C, S384C, and E387C. Within category iii, the K0.5 of E387C (Fig. 3A, panel iii b) is shifted to lower values and that of G383C and S384C (Fig. 3A, panel iii a) to higher values of ionic strength. We note that in case of R382C, G383C, and L389C (all in CBS2), a pronounced shift to higher salt concentrations was already observed upon substitution of the native residue to Cys (in the presence of DTT). Thus, the most pronounced effects of cross-linking on the ionic activation are seen in S384C, E387C (positions that are little or not affected by MalPEG labeling), L386C, and L389C, all in CBS2.
FIGURE 3.
Effect of CBS cross-linking on the ionic activation of OpuA. A, Cys mutants bound to nickel-Sepharose were treated with MTS-3-MTS or DTT as described under “Experimental Procedures.” Purified OpuA was reconstituted in liposomes composed of 50 mol % DOPE, 12 mol % DOPC, plus 38 mol:% DOPG. The ATP-regenerating system was enclosed inside the proteoliposomes. Uptake of [14C]glycine betaine was assayed in 90 mm KPi, pH 7.0, with or without KCl as indicated on the x axis. WT, ▾; DTT-treated Cys mutant, □; MTS-3-MTS cross-linked Cys-mutant, ■. Panel i, A328C; panel ii, L386C; panel iii a, S384C; panel iii b, E387C. B, visualization of the reconstituted proteins by SDS-PAGE (see legend to Fig. 2 for details). Left lanes, DTT-treated; right lanes, MTS-3-MTS treated; the positions of monomeric and cross-linked subunits are indicated. The error bars indicate the S.E. derived from multiple measurements (n = 3).
Overall, the effects of cross-linking (MTS reagents) in CBS1 were small or not significant, whereas each of the CBS2 mutants was significantly affected. We note that the effects of cross-linking of CBS2 are moderate compared with MalPEG labeling. The interactions across the CBS2-CBS2 interface tune the salt dependence of transport, but they are not intrinsic to the ionic sensing mechanism. The effects of MalPEG labeling as presented in Fig. 2 must represent disruption of the dimeric CBS or NBD-CBS structure by the large polyethyleneglycol moiety.
Cross-linking as a Function of Ionic Strength
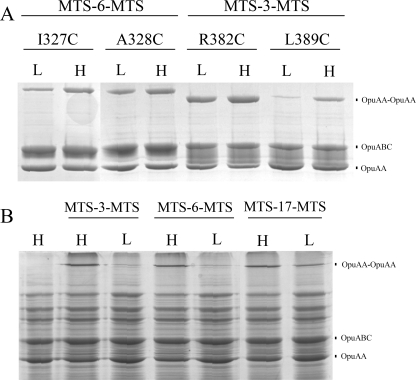
In membranes with 38 mol % of anionic lipids (e.g. DOPG), OpuA is inactive at an ionic strength (Iin) of 0.3 (i.e. 90 mm KPi plus 50 mm KCl) and maximally active at Iin of 0.8 (i.e. 90 mm KPi plus 300 mm KCl) (12). We used these conditions to probe differences between the inactive and active state of OpuA, using native membrane vesicles and MTS-3-MTS as cross-linker. Subsequently, the cross-linked proteins were solubilized, purified, and analyzed on SDS-polyacrylamide gels. The cross-linking efficiency with MTS-3-MTS was very low for each of the CBS1 mutants (data not shown) and both at low and high salt conditions, suggesting that the α2 helices of CBS1 are relatively distantly spaced. However, if we used the MTS-6-MTS reagent, which has a longer spacer arm (S–S distance is ∼10 Å) than MTS-3-MTS, we could detect significant cross-linking for I327C and A328C, which increased with ionic strength (Fig. 4A). The cross-linking of CBS2 by MTS-3-MTS was significant, and in case of R382C and L389C, it was much higher at high than low salt conditions (Fig. 4A). These data confirm that these Cys residues in the α2 helix of CBS2 are in close proximity and positioned differently at low and high salt. Control experiments with surface-exposed Cys residues show that the reactivity of the MTS reagents is similar at low and high salt concentrations (data not shown). Our data thus indicate that there is a conformational rearrangement at the CBS-CBS interface upon activation of OpuA with salt.
FIGURE 4.
Salt dependence of cross-linking. A, 2 mg of membrane vesicle protein was resuspended in 50 mm KPi, pH 7.0, 20% glycerol, plus 50 or 300 mm KCl. The Cys mutants (as indicated in the figure) were incubated with 1 mm MTS-3-MTS or MTS-6-MTS for 1 min on ice. Then 10 mm N-ethylmaleimide (NEM) was added to stop the cross-linking reaction. Cross-linked Cys mutants were purified by nickel-Sepharose resin and applied to nonreducing 12.5% SDS-PAGE. B, OpuA N226C was treated as described above. After stopping the cross-linking reaction, membrane vesicles were applied to 12.5% SDS-PAGE. N226C without cross-linker treatment is shown in 1st lane. H, refers to high salt (50 mm KPi, pH 7.0, plus 300 mm KCl) and L to low salt (50 mm KPi, pH 7.0, plus 50 mm KCl) conditions.
Because the tandem CBS domains are connected to the NBD, conformational changes in one of the domains are likely transmitted to the other. To examine whether the structure of the NBD is affected by ionic strength, we constructed N226C using a homology model of the NBD part of OpuAA on the basis of the crystal structure of MalK (50). The equivalent residue in MalK is located at the dimer interface (Fig. 1C). Contrary to the proximity of the residues in the MalK dimer structure, N226C did not form a disulfide bridge spontaneously (Fig. 4B, 1st lane), and we did not detect cross-linking in the presence of MTS-3-MTS or MTS-6-MTS at low salt conditions (Fig. 4B, 3rd and 5th lanes). Cross-linked OpuAA was observed with both MTS reagents at high salt concentration (Fig. 4B, 2nd and 4th lanes; with MTS-17-MTS, the cross-linking was stimulated by a high ionic strength), suggesting that the NBD dimer structure is affected by ionic strength, presumably as a result of conformational changes in the ion-sensing CBS domains.
Structural Analysis of the CBS Module from OpuA
The cross-linking and chemical modification data indicate that CBS1 is not critical for activity or ionic strength sensing of OpuA. In fact, the lack of any significant effect of MalPEG labeling points toward a partially unstructured CBS1 domain, consistent with the crystal structure of CBS1 from P. horikoshii (supplemental Fig. 1D). To substantiate this notion, we characterized the purified CBS module by size-exclusion chromatography coupled to light scattering, circular dichroism (CD), and nuclear magnetic resonance (NMR) spectroscopy. Monomeric CBS has a calculated molecular mass of 17.3 kDa, but the protein elutes on a size-exclusion column with an apparent molecular mass of ∼40 kDa (using standard globular proteins as size markers, data not shown). However, when analyzed using static light-scattering coupled to refractive index and UV absorption (by size-exclusion chromatography coupled to light scattering), the molecular mass of the purified protein is ∼16 kDa over the entire peak (Fig. 5A). This indicates that the CBS module in solution is monomeric and has an elongated structure. We then estimated the content of secondary structure elements by CD and NMR. The CD spectra show that the protein has some secondary structure (Fig. 5B, inset), mainly α-helical, but less than would be expected from the crystal structures of CBS domains. Analyses of the CD spectra, using the CDPro software package, indicate that ∼60% of the CBS module adopts an extended conformation. The two-dimensional 1H-13C HSQC spectrum confirms this assertion, with the majority of the amino acids residing in the regions typical for intrinsically disordered proteins (Fig. 5C) (51, 52). Fig. 5B shows that addition of denaturant (GdnHCl) completely unfolds the CBS module; in the presence of GdnHCl, only the 250–210-nm region of the CD spectrum could be recorded (see under “Experimental Procedures” for details). The LS-SEC, CD, and NMR data together with the chemical modification of the CBS module in OpuA strongly suggest that part(s) of the protein are intrinsically disordered, and most likely compose the CBS1 domain.
Replacing CBS1 by an Artificial Linker Region
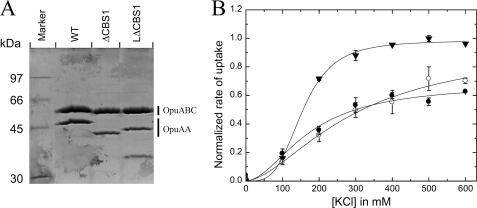
The previously described experiments point toward a critical role of CBS2 in ion sensing and/or ionic regulation of OpuA, whereas CBS1 merely fulfills a role as a linker domain. If true, one should be able to replace CBS1 by a flexible linker and still observe ionic strength-dependent transport. We thus constructed an OpuA mutant lacking CBS1, in which the NBD and CBS2 were connected by an 11- (OpuA(ΔCBS1)) or 34 (OpuA(LΔCBS1))-amino acid long linker domain. The OpuA(ΔCBS1) and OpuA(LΔCBS1) proteins were somewhat less stable than wild type OpuA as can be inferred from the reduced OpuAA/OpuABC ratio (Fig. 6A). However, the transporter was still under ionic strength control (Fig. 6B). These data show that CBS2 is necessary and sufficient for ionic regulation of OpuA and that CBS1 merely serves as a linker domain.
FIGURE 6.
Ionic activation profiles of CBS1 deletion mutants. A, visualization of the reconstituted proteins by SDS-PAGE; 7.5 μg of protein was loaded (see legend to Fig. 2A). B, uptake of [14C]glycine betaine by wild type OpuA, OpuA(ΔCBS1), and OpuA(LΔCBS1). Assay conditions were the same as in the legend to Fig. 2. Data were normalized by correcting for the amount of OpuAA subunit (i.e. the lower OpuAA/OpuABC ratio in the case of OpuA(ΔCBS1) and OpuA(LΔCBS1)). The error bars indicate the standard error derived from multiple measurements (n = 3).
DISCUSSION
Proteins with CBS domains are widespread in genomes from all kingdoms of life. At present more than 30,000 sequences and several dozens of crystal structures of CBS domains are available. Despite this wealth of genomic and structural information, relatively little is known about the regulatory mechanisms and the functions of the CBS domains. CBS domains have been found associated with ABC transporters (12, 53), chloride channels of the CLC family (54–58), magnesium transporters (30, 31), and water-soluble enzymes, including sugar (d-arabinose 5-phosphate) isomerases (59), AMP-activated protein kinase (60–63), inosine 5′-monophosphate dehydrogenase (60, 61, 64), metalloproteases (65), pyrophosphatases (29), and many proteins with unknown functions.
The crystal structures of more than 70 CBS domains/proteins have been determined, predominantly as isolated domains, but most of the structural data is not supported by functional data. Despite a low sequence identity (the primary sequences of CBS domains are generally not considered homologous), the secondary structure elements seem well conserved. On the basis of these crystal structures different types of association between CBS tandems have been reported as shown in supplemental Fig. 1 and as follows: head-to-head (CBS1-CBS1 and CBS2-CBS2) (A–D), head-to-tail (CBS1-CBS2 and CBS2-CBS1) (E), CBS1-CBS2 (F), CBS2-CBS2 (G), back-to-back (CBS1-CBS1 and CBS2-CBS2) (H), and back-to-back (CBS1-CBS2 and CBS2-CBS1) (I). So far, we have not succeeded in obtaining well diffracting crystals of OpuA, and we thus used the structure of MgtE to guide the engineering of the CBS module of OpuA (Fig. 1). We tested the model by chemical modification and cross-linking studies, using purified protein as well as native membranes. The cross-linking data confirm the head-to-head structure of the tandem CBS of OpuA. The head-to-tail but also the back-to-back structures can be ruled out on the basis of the long distance between α2 in CBS2 in these structures (supplemental Fig. 1, panels E–I). For the head-to-head structures, four different conformational states are available as shown in supplemental Fig. 1, A–D and as follows: α2 in CBS1 and CBS2 are close to each other (A-type); the CBS2 domains are further apart than CBS1 (B-type); the CBS1 domains are further apart than CBS2 (C-type); and CBS1 is partially disordered (D-type). Our cross-linking data indicate that the CBS2 domains do not have to move away from each other upon transiting from an inactive to active species, making the B-type head-to-head state a less relevant conformation. The MalPEG labeling data suggest that the CBS1 domains may be in an open conformation (C-type) or partly unstructured (D-type). The structural studies of the isolated CBS module are most consistent with the CBS1 being partly disordered (D-type). The data collectively point to a head-to-head structure of the dimeric tandem CBS domains with CBS1 serving as linker region between the NBD and CBS2.
Comparison of the CBS domains of the OTCN family of ABC transporters shows that CBS2 domains are better conserved than CBS1 domains (supplemental Fig. 6). In fact, a subset of proteins only has the CBS2 and lacks a full CBS1 domain (16), which coincides with our observations on OpuA(ΔCBS1) and OpuA(LΔCBS1). For example, YheX of the osmotic stress-inducible YheZYXW complex of E. coli contains only a CBS2 domain (66). Thus, for simple CBS-mediated ionic strength-dependent regulation of transport, CBS2 is critical, but CBS1 can be lost during evolution without adverse physiological consequences.
So far, MgtE (30) and CmCLC (48) are the only two transporters with CBS domains for which a crystal structure is available. In these systems, the CBS2 domains are closest to the TMD, and a linker region connects the TMD with the CBS1. In case of CmCLC, the CBS2 directly contacts helices that extend into the ion pathway of the protein. In fact, the majority of disease-causing mutations in human CLCs are found at this TM-CBS2 interface (48). In the tandem CBS domains of CLC-5 whose structure is consistent with CmCLC, a nucleotide-binding site is present, and mutagenesis of this region greatly affects the channel function (46). Interactions between the TMD and CBS2 would be important for transmitting conformational changes upon binding of nucleotides. In MgtE, the CBS2 is connected to the TMD via a “connecting helix” and indirectly controls access to the ion transport pathway. Four putative Mg2+ ions are bound at the interface of the connecting helices and the TMD and CBS domains and stabilize the closed state of the pore. Without Mg2+ (low intracellular concentrations), the CBS2 domains are repelled from each other by the negatively charged residues at the dimer interface, which favors the open conformation (30, 31). Overall, these findings support the idea that the tandem CBS domains sense intracellular ligands and elicit conformational changes in the associated protein domains. Because MalPEG labeling of CBS1 does not affect OpuA, whereas the transporter is sensitive to substitutions and modifications in CBS2, we propose a domain organization similar to that in MgtE and CmCLC, with CBS2 transmitting the conformational changes to the NBD. Contrary to MgtE, we do not propose a specific ion/effector-binding site on the CBS module, rather CBS2 directly or indirectly interacts in an ionic strength-dependent manner with the anionic surface of the membrane, whereas CBS1 serves a linker function.
The crystal structures of ABC transporters ModBC (67), MetNI (68), and MalFGK2E (69) have revealed a C-terminal regulatory domain, and the binding of ligand (e.g. methionine in MetD and molybdate or tungstate in ModC) traps the system in the open conformation and thereby provides a negative feedback to the transporter (67, 68). Although the CBS domains are structurally unrelated to these accessory domains, the regulatory mechanism has similar consequences (21). Thus, salt-dependent conformational changes in CBS2 are likely to affect the NBD structure (i.e. their opening and closing), and thereby affect the transport reaction. Importantly, not only the CBS module but also anionic lipids are critical for activity. Here, a high ionic strength is needed to prevent a too strong interaction of the CBS module with the membrane, either directly or indirectly (via another site on the protein). Because mutations on the surface of the CBS module (Lys and Arg to neutral residues) reduce the salt dependence of transport (main contribution from K377V on CBS2 (13)), we conclude that salt does not act directly at the CBS2-CBS2 interface like Mg2+ does in MgtE and nucleotides are thought to do in CmCLC. Rather surface interactions with the CBS2 affect the NBD-NBD interface and thereby the hydrolysis of ATP. Consistent with this notion are the observations that cross-linking of the CBS domains and NBDs of OpuA from L. lactis is salt-dependent (this work) and that, in solution, the monomer-dimer equilibrium of the OpuAA subunit from B. subtilis is nucleotide- and salt concentration-dependent (49).
In conclusion, the CBS domains in OpuA form a dimeric structure with head-to-head interactions, mostly if not only via CBS2. The CBS1 domain may be largely unstructured and merely serve as a linker between the NBD and CBS2. CBS2-CBS2 interactions are critical for transport activity and influence the ionic activation of OpuA. In contrast to MgtE and CmCLC, the CBS domains in the ABC transporter do not bind specific ligands, rather the CBS module transmits ionic strength-dependent interactions with the anionic surface of the membrane to conformational changes in the NBD dimer.
This work was supported by Netherlands Organization for Scientific Research Top Subsidy Grant 700.56.302 (to B. P.) and the European Union EDICT Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
- CBS
- cystathionine β-synthase
- ABC
- ATP-binding cassette
- NBD
- nucleotide-binding domain
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine
- DOPG
- 1,2-dioleoyl-sn-glycero-3-phosphatidylglycerol
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine
- MalPEG
- methoxy-polyethylene glycol maleimide 5000
- MTS-3-MTS
- 1,3-propanediyl bismethanethiosulfonate
- MTS-6-MTS
- 1,6-hexanediyl bismethanethiosulfonate
- MTS-17-MTS
- 3,6,9,12,15-pentaoxaheptadecane-1,17-diyl bis-methanethiosulfonate
- TNP-ATP
- 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate
- DDM
- n-dodecyl-β-d-maltoside
- TMD
- transmembrane domain
- GdnHCl
- guanidine HCl
- CD
- circular dichroism.
REFERENCES
- 1. Hamill O. P., Martinac B. (2001) Physiol. Rev. 81, 685–740 [DOI] [PubMed] [Google Scholar]
- 2. Wood J. M., Bremer E., Csonka L. N., Kraemer R., Poolman B., van der Heide T., Smith L. T. (2001) Comp. Biochem. Physiol. 130, 437–460 [DOI] [PubMed] [Google Scholar]
- 3. Bolen D. W. (2001) Methods Mol. Biol. 168, 17–36 [DOI] [PubMed] [Google Scholar]
- 4. Racher K. I., Culham D. E., Wood J. M. (2001) Biochemistry 40, 7324–7333 [DOI] [PubMed] [Google Scholar]
- 5. Tsatskis Y., Khambati J., Dobson M., Bogdanov M., Dowhan W., Wood J. M. (2005) J. Biol. Chem. 280, 41387–41394 [DOI] [PubMed] [Google Scholar]
- 6. Culham D. E., Vernikovska Y., Tschowri N., Keates R. A., Wood J. M., Boggs J. M. (2008) Biochemistry 47, 13584–13593 [DOI] [PubMed] [Google Scholar]
- 7. Culham D. E., Romantsov T., Wood J. M. (2008) Biochemistry 47, 8176–8185 [DOI] [PubMed] [Google Scholar]
- 8. Keates R. A., Culham D. E., Vernikovska Y. I., Zuiani A. J., Boggs J. M., Wood J. M. (2010) Biochemistry 49, 8847–8856 [DOI] [PubMed] [Google Scholar]
- 9. Rübenhagen R., Morbach S., Krämer R. (2001) EMBO J. 20, 5412–5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ziegler C., Bremer E., Krämer R. (2010) Mol. Microbiol. 78, 13–34 [DOI] [PubMed] [Google Scholar]
- 11. van der Heide T., Stuart M. C., Poolman B. (2001) EMBO J. 20, 7022–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biemans-Oldehinkel E., Mahmood N. A., Poolman B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahmood N. A., Biemans-Oldehinkel E., Poolman B. (2009) J. Biol. Chem. 284, 14368–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Culham D. E., Henderson J., Crane R. A., Wood J. M. (2003) Biochemistry 42, 410–420 [DOI] [PubMed] [Google Scholar]
- 15. Schiller D., Rübenhagen R., Krämer R., Morbach S. (2004) Biochemistry 43, 5583–5591 [DOI] [PubMed] [Google Scholar]
- 16. Mahmood N. A., Biemans-Oldehinkel E., Patzlaff J. S., Schuurman-Wolters G. K., Poolman B. (2006) J. Biol. Chem. 281, 29830–29839 [DOI] [PubMed] [Google Scholar]
- 17. Hillar A., Culham D. E., Vernikovska Y. I., Wood J. M., Boggs J. M. (2005) Biochemistry 44, 10170–10180 [DOI] [PubMed] [Google Scholar]
- 18. Schiller D., Krämer R., Morbach S. (2004) FEBS Lett. 563, 108–112 [DOI] [PubMed] [Google Scholar]
- 19. Ott V., Koch J., Späte K., Morbach S., Krämer R., (2008) Biochemistry 47, 12208–12218 [DOI] [PubMed] [Google Scholar]
- 20. Ozcan N., Ejsing C. S., Shevchenko A., Lipski A., Morbach S., Krämer R. (2007) J. Bacteriol. 189, 7485–7496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biemans-Oldehinkel E., Doeven M. K., Poolman B. (2006) FEBS Lett. 580, 1023–1035 [DOI] [PubMed] [Google Scholar]
- 22. Wolters J. C., Berntsson R. P., Gul N., Karasawa A., Thunnissen A. M., Slotboom D. J., Poolman B. (2010) Plos One 5, e10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bateman A. (1997) Trends Biochem. Sci. 22, 12–13 [DOI] [PubMed] [Google Scholar]
- 24. Miller M. D., Schwarzenbacher R., von Delft F., Abdubek P., Ambing E., Biorac T., Brinen L. S., Canaves J. M., Cambell J., Chiu H. J., Dai X., Deacon A. M., DiDonato M., Elsliger M. A., Eshagi S., Floyd R., Godzik A., Grittini C., Grzechnik S. K., Hampton E., Jaroszewski L., Karlak C., Klock H. E., Koesema E., Kovarik J. S., Kreusch A., Kuhn P., Lesley S. A., Levin I., McMullan D., McPhillips T. M., Morse A., Moy K., Ouyang J., Page R., Quijano K., Robb A., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., Vincent J., Wang X., West B., Wolf G., Xu Q., Hodgson K. O., Wooley J., Wilson I. A. (2004) Proteins 57, 213–217 [DOI] [PubMed] [Google Scholar]
- 25. Ragunathan P., Kumarevel T., Agari Y., Shinkai A., Kuramitsu S., Yokoyama S., Ponnuraj K. (2008) Biochem. Biophys. Res. Commun. 375, 124–128 [DOI] [PubMed] [Google Scholar]
- 26. Proudfoot M., Sanders S. A., Singer A., Zhang R., Brown G., Binkowski A., Xu L., Lukin J. A., Murzin A. G., Joachimiak A., Arrowsmith C. H., Edwards A. M., Savchenko A. V., Yakunin A. F. (2008) J. Mol. Biol. 375, 301–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gómez-García I., Oyenarte I., Martínez-Cruz L. A. (2010) J. Mol. Biol. 399, 53–70 [DOI] [PubMed] [Google Scholar]
- 28. Lucas M., Encinar J. A., Arribas E. A., Oyenarte I., García I. G., Kortazar D., Fernández J. A., Mato J. M., Martínez-Chantar M. L., Martínez-Cruz L. A. (2010) J. Mol. Biol. 396, 800–820 [DOI] [PubMed] [Google Scholar]
- 29. Tuominen H., Salminen A., Oksanen E., Jämsen J., Heikkilä O., Lehtiö L., Magretova N. N., Goldman A., Baykov A. A., Lahti R. (2010) J. Mol. Biol. 398, 400–413 [DOI] [PubMed] [Google Scholar]
- 30. Hattori M., Tanaka Y., Fukai S., Ishitani R., Nureki O. (2007) Nature 448, 1072–1075 [DOI] [PubMed] [Google Scholar]
- 31. Ishitani R., Sugita Y., Dohmae N., Furuya N., Hattori M., Nureki O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 15393–15398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng L., Baumann U., Reymond J. L. (2004) Nucleic Acids Res. 32, e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Geertsma E. R., Poolman B. (2007) Nat. Methods 4, 705–707 [DOI] [PubMed] [Google Scholar]
- 34. van der Heide T., Poolman B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7102–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Ruyter P. G., Kuipers O. P., de Vos W. M. (1996) Appl. Environ. Microbiol. 62, 3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Folta-Stogniew E., Williams K. R. (1999) J. Biomol Tech. 10, 51–63 [PMC free article] [PubMed] [Google Scholar]
- 37. Slotboom D. J., Duurkens R. H., Olieman K., Erkens G. B. (2008) Methods 46, 73–82 [DOI] [PubMed] [Google Scholar]
- 38. Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 39. Geertsma E. R., Nik Mahmood N. A., Schuurman-Wolters G. K., Poolman B. (2008) Nat. Protoc. 3, 256–266 [DOI] [PubMed] [Google Scholar]
- 40. Biemans-Oldehinkel E., Poolman B. (2003) EMBO J. 22, 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Landfald B., Strøm A. R. (1986) J. Bacteriol. 165, 849–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schägger H., Cramer W. A., von Jagow G. (1994) Anal. Biochem. 217, 220–230 [DOI] [PubMed] [Google Scholar]
- 43. Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H., Poolman B. (2002) J. Mol. Biol. 317, 591–600 [DOI] [PubMed] [Google Scholar]
- 44. Poolman B., Doeven M. K., Geertsma E. R., Biemans-Oldehinkel E., Konings W. N., Rees D. C. (2005) Methods Enzymol. 400, 429–459 [DOI] [PubMed] [Google Scholar]
- 45. Meyer S., Dutzler R. (2006) Structure 14, 229–307 [DOI] [PubMed] [Google Scholar]
- 46. Meyer S., Savaresi S., Forster I. C., Dutzler R. (2007) Nat. Struct. Mol. Biol. 14, 60–67 [DOI] [PubMed] [Google Scholar]
- 47. Sharpe M. L., Gao C., Kendall S. L., Baker E. N., Lott J. S. (2008) J. Mol. Biol. 383, 822–836 [DOI] [PubMed] [Google Scholar]
- 48. Feng L., Campbell E. B., Hsiung Y., MacKinnon R. (2010) Science 330, 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Horn C., Bremer E., Schmitt L. (2003) J. Mol. Biol. 334, 403–419 [DOI] [PubMed] [Google Scholar]
- 50. Lu G., Westbrooks J. M., Davidson A. L., Chen J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17969–17974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Meinema A. C., Laba J. K., Hapsari R. A., Otten R., Mulder F. A., Kralt A., van den Bogaart G., Lusk C. P., Poolman B., Veenhoff L. M. (2011) Science 333, 90–93 [DOI] [PubMed] [Google Scholar]
- 52. Mulder F. A., Lundqvist M., Scheek R. M. (2010) in Nuclear Magnetic Resonance Spectroscopy Applied to (Intrinsically) Disordered Proteins (Uversky V. N., Longhi S.) pp. 59–87, John Wiley & Sons, Inc., Hoboken, NJ [Google Scholar]
- 53. Chen C., Beattie G. A. (2007) J. Bacteriol. 189, 6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schmidt-Rose T., Jentsch T. J. (1997) J. Biol. Chem. 272, 20515–20521 [DOI] [PubMed] [Google Scholar]
- 55. Maduke M., Williams C., Miller C. (1998) Biochemistry 37, 1315–1321 [DOI] [PubMed] [Google Scholar]
- 56. Hebeisen S., Biela A., Giese B., Müller-Newen G., Hidalgo P., Fahlke C. (2004) J. Biol. Chem. 279, 13140–13147 [DOI] [PubMed] [Google Scholar]
- 57. Garcia-Olivares J., Alekov A., Boroumand M. R., Begemann B., Hidalgo P., Fahlke C. (2008) J. Physiol. 586, 5325–5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bennetts B., Rychkov G. Y., Ng H. L., Morton C. J., Stapleton D., Parker M. W., Cromer B. A. (2005) J. Biol. Chem. 280, 32452–32458 [DOI] [PubMed] [Google Scholar]
- 59. Tan L., Darby C. (2006) Mol. Microbiol. 61, 861–870 [DOI] [PubMed] [Google Scholar]
- 60. Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. (2004) J. Clin. Invest. 113, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ignoul S., Eggermont J. (2005) Am. J. Physiol. Cell Physiol. 289, C1369–C1378 [DOI] [PubMed] [Google Scholar]
- 62. Viana R., Towler M. C., Pan D. A., Carling D., Viollet B., Hardie D. G., Sanz P. (2007) J. Biol. Chem. 282, 16117–16125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Townley R., Shapiro L. (2007) Science 315, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 64. Colby T. D., Vanderveen K., Strickler M. D., Markham G. D., Goldstein B. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3531–3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou R., Cusumano C., Sui D., Garavito R. M., Kroos L. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16174–16179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Checroun C., Gutierrez C. (2004) FEMS Microbiol. Lett. 236, 221–226 [DOI] [PubMed] [Google Scholar]
- 67. Gerber S., Comellas-Bigler M., Goetz B. A., Locher K. P. (2008) Science 321, 246–250 [DOI] [PubMed] [Google Scholar]
- 68. Kadaba N. S., Kaiser J. T., Johnson E., Lee A., Rees D. C. (2008) Science 321, 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen J., Lu G., Lin J., Davidson A. L., Quiocho F. A. (2003) Mol. Cell 12, 651–661 [DOI] [PubMed] [Google Scholar]
- 70. Delano W. L. (2008) The PyMOL Molecular Graphics System, Version 0.99rc6, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]