Background: Ethanol-induced neuronal apoptosis causes brain shrinkage and cognitive defects.
Results: Exposure to ethanol (0.5% v/v for 72 h) in SH-SY5Y cells induced expression of miR-497 and miR-302b and down-regulated expression of BCL2 and/or cyclin D2.
Conclusion: Ethanol-induced neuronal apoptosis follows both the mitochondria-mediated and non-mitochondria-mediated pathways.
Significance: Our study shows that miRNAs are involved in regulation of ethanol neurotoxicity.
Keywords: Apoptosis, Gene Regulation, Gene Silencing, MicroRNA, Neurotoxin, Dicer, Ethanol, miR-302b, miR-497
Abstract
In chronic alcoholism, brain shrinkage and cognitive defects because of neuronal death are well established, although the sequence of molecular events has not been fully explored yet. We explored the role of microRNAs (miRNAs) in ethanol-induced apoptosis of neuronal cells. Ethanol-sensitive miRNAs in SH-SY5Y, a human neuroblastoma cell line, were identified using real-time PCR-based TaqMan low-density arrays. Long-term exposure to ethanol (0.5% v/v for 72 h) produced a maximum increase in expression of miR-497 (474-fold) and miR-302b (322-fold). Similar to SH-SY5Y, long-term exposure to ethanol induced miR-497 and miR-302b in IMR-32, another human neuroblastoma cell line. Using in silico approaches, BCL2 and cyclin D2 (CCND2) were identified as probable target genes of these miRNAs. Cotransfection studies with 3′-UTR of these genes and miRNA mimics have demonstrated that BCL2 is a direct target of miR-497 and that CCND2 is regulated negatively by either miR-302b or miR-497. Overexpression of either miR-497 or miR-302b reduced expression of their identified target genes and increased caspase 3-mediated apoptosis of SH-SY5Y cells. However, overexpression of only miR-497 increased reactive oxygen species formation, disrupted mitochondrial membrane potential, and induced cytochrome c release (mitochondria-related events of apoptosis). Moreover, ethanol induced changes in miRNAs, and their target genes were substantially prevented by pre-exposure to GSK-3B inhibitors. In conclusion, our studies have shown that ethanol-induced neuronal apoptosis follows both the mitochondria-mediated (miR-497- and BCL2-mediated) and non-mitochondria-mediated (miR-302b- and CCND2-mediated) pathway.
Introduction
MicroRNAs (miRNAs)4 are small regulatory RNA molecules that down-regulate the expression of protein coding genes in a sequence-specific manner. MiRNAs regulate the expression of several genes that are important for apoptosis, senescence, aging, neuronal differentiation, neurogenesis, and neurodegeneration (1, 2). Knockout studies of dicer (the endoribonuclease responsible for the formation of mature miRNAs) have shown that miRNAs are required for the survival of neural progenitor cells and differentiation and maturation of postmitotic neurons (3, 4). Ethanol is a well known neurotoxin for both the developing and the adult brain (5). In the developing brain, only a single dose of ethanol can induce widespread apoptotic neurodegeneration (6). Over-expression of antiapoptotic BCL2 can protect the neonatal cerebellum from ethanol-induced apoptosis (7). In adults, chronic use of ethanol decreases neurogenesis of the hippocampus, which is the major site of adult neurogenesis and ethanol neurotoxicity (8–10).
Recent studies have clearly shown the role of miRNAs in adult and embryonic neurogenesis (11, 12). Levels of cyclin D2 (CCND2), which plays a crucial role in the maintenance of the neurogenesis potential of the adult brain, are directly regulated by miR-302b (13, 14). Pietrzykowski et al. (15) have shown that neuronal adaptation to ethanol is regulated by miR-9. Studies have also shown that competing interactions between a set of miRNAs can decide the fate of ethanol-exposed neural progenitor cells (16). Ethanol-induced neurotoxicity can be prevented substantially by pre-exposure to glycogen synthase kinase 3 β (GSK-3Β) inhibitors (17). The GSK-3 isoforms, GSK-3A and GSK-3B are constitutively active serine/threonine protein kinases of the WNT/β-catenin signaling cascade. GSK-3 regulates protein translation, promotion of mitochondrial apoptosis, and levels of other signaling elements like cyclin D1 and D2 by activation of different transcription factors (18–20). In the brain, GSK-3B plays a crucial role in the regulation of neurogenesis, neuronal survival, and neurite outgrowth (21). Moreover, exposure to GSK-3B inhibitors (LiCl3 or valproate) has been shown to alter the expression profile of selective miRNAs (down-regulated: let-7b, let-7c, miR-128a, miR-24a, miR-30c, miR-34a, and miR-221; up-regulated: miR-144) that target proteins involved in neurite outgrowth; neurogenesis; and the phosphatase and tensin homolog (PTEN), extracellular signal-regulated kinases (ERK), and Wnt/β-catenin pathways (22).
Studies were initiated to explore the involvement of miRNAs in ethanol-induced neuronal death by silencing dicer in SH-SY5Y, a human neuroblastoma cell line. Increased sensitivity of dicer-silenced SH-SY5Y cells toward ethanol exposure has prompted us to identify ethanol-sensitive miRNAs. Further studies were concentrated on two miRNAs that showed maximum alterations after long-term exposure to ethanol. Potential target genes of these miRNAs were identified and validated by 3′-UTR binding assays, and their role in ethanol-induced neuronal apoptosis was investigated. Moreover, the effect of pre-exposure to two well known GSK-3Β inhibitors (lithium and DZD-8) was also studied on ethanol-induced alterations in miRNAs and their target genes.
EXPERIMENTAL PROCEDURES
Chemicals
A TaqMan low-density array (TLDA), individual miRNA assays, an RT kit, preamplification master mix, TaqMan universal master mix., miRVana, and other reagents required for real-time PCR were procured from Applied Biosystems, Inc. TDZD-8, LiCl3, and 2′,7′-dichlorofluorescein diacetate were procured from Sigma. siRNA for dicer and miRNA mimics were obtained from Qiagen, and the non-targeting control (NTC) was procured from Dharmacon Research. The Cell Line Nucleofector® Kit V was procured from Lonza. Anti-BCL2 and anti-cytochrome c were procured from Chemicon. Anti-CCND2 and anti-β-actin were procured from Sigma. Polyclonal anti-VDAC-1 was procured from Abcam. DMEM-F12, fetal bovine serum, and antibiotic-antimycotic were purchased from Invitrogen. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, a tetrazole) and neutral red dyes were procured from SRL, India. Apoptosis assays were performed with the FITC- annexin V apoptosis detection kit from BD Pharmingen. The Dual-Glo luciferase assay kit was procured from Promega.
Cell Culture and Exposure to Chemicals
SH-SY5Y cells were grown in a 1:1 mixture of Eagle's minimum essential medium with non-essential amino acids and Ham's F12 medium. IMR-32 human neuroblastoma cells were grown in Eagle's minimum essential medium with 2 mm l-glutamine and non-essential amino acids. The growth media of both cell lines were supplemented with 10% fetal bovine serum and 1% antibiotic and antimycotic solution. Cells were kept in 5% CO2 95% atmosphere with high humidity at 37 °C. Considering the volatile properties of ethanol, exposure to ethanol in a 96-well plate was given as described earlier (23). For short-term exposure studies, cells were exposed to 0.5, 1.5, 2.5, 3.5, 4.5, or 5.5% v/v ethanol for 4 h. For long-term exposure studies, cells were exposed to 0.125, 0.25, 0.5, 1.0, 2.0, or 4.0% v/v ethanol for 72 h. A non-cytotoxic concentration of ethanol was selected by carrying out MTT and neutral red uptake (NRU) assays as described in our earlier paper (24). GSK-3B inhibition was carried out by exposing cells to either TDZD-8 (10 μm) or LiCl3 (20 mm) as reported earlier (17, 25).
Dicer Silencing Studies
SH-SY5Y cells were transfected with siRNA for dicer (target sequence: anti-Dicer 1, ACCACTGATTCTGCATATGAA or anti-Dicer 2, AATGTGCTATCTGGATCCTAG) or NTC siRNA using the Cell Line Nucleofactor Kit V. After 24 h of transfection, a decrease of more than 80% in expression of dicer mRNA and protein was achieved by using anti-Dicer 1 siRNA (supplemental Fig. S1). After 24 h of transfection, cells were exposed with different concentrations of ethanol, and cell viability was assayed by MTT and NRU assays.
Expression Profiling of miRNAs
The total RNA, which also contained small RNAs, was isolated by means of the mirVana miRNA isolation kit as described by the manufacturer. The expression of 667 unique miRNAs was studied by TaqMan low-density arrays pool A and pool B (TaqMan® Human MicroRNA Set Cards v2.0, Part no. 4400238). Primers of 667 miRNAs were present on a set of two plates named pool A (containing primers for 380 unique miRNAs) and pool B (containing primers for 287 unique miRNAs). After RNA isolation, RT was carried out with megaplex RT primers of pool A (part no. 4399966) and pool B (part no. 4399968), which are a set of two predefined pools containing stem-looped reverse transcription primers for mature miRNAs. RT was performed with the TaqMan miRNA RT kit from ABI as described by the manufacturer. In brief, the reaction mixture contained 1× megaplex RT primers, 2.6 mm deoxynucleotide triphosphates (dNTPs) with deoxythymidine triphosphate (dTTP), 75 units of MultiScribe reverse transcriptase, 1× buffer, 3 mm MgCl2, 2 units of RNase inhibitor, and 100 ng of total RNA with a final volume of 7.5 μl. Thermal cycling conditions used for reverse transcription involved 40 cycles of 16 °C for 2 min, 42 °C for 1 min, and 50 °C for 1 s. Finally, the reaction mixture was denatured by incubating it at 85 °C. After reverse transcription, preamplification was carried out by means of the TaqMan preamp master mix and megaplex preamp primers of pool A (part no. 4399233) and pool B (4399201). 12.5 μl of preamp master mixture was mixed with 2.5 μl of preamp primers and 2.5 μl of reverse transcription product with a total volume of 25 μl. Thermal cycling conditions used for preamplification were 10 min at 95 °C, 2 min at 55 °C, 2 min at 72 °C, and 12 cycles of 15 s at 95 °C, and 4 min at 60 °C. Finally, the preamp products were incubated for 10 min at 99.9 °C. The preamplification products were diluted up to four times with 0.1 × Tris-EDTA buffers. For running TLDA plates, 450 μl of the TaqMan universal PCR master mixture was mixed with 9 μl of the diluted preamplification product in a total volume of 900 μl. In each port of the TLDA plates, 100 μl from the above-mentioned master mixture was added, and after sealing and spinning, the plates were loaded on a thermal cycler. Relative quantification was done by means of the -ΔΔ cycle threshold method, considering the levels of mammalian U6 RNA as endogenous control. The expression of miR-497 and miR-302b were studied using individual TaqMan miRNA assays.
Overexpression of miRNA Mimics
SH-SY5Y and IMR-32 cells were transfected with miRNA mimics, Syn-miR-497 (mature miRNA sequence 5′-CAGCAGCACACUGUGGUUUGU-3′) or Syn-miR-302b (mature miRNA sequence 5′-UAAGUGCUUCCAUGUUUUAGUAG-3′), or NTC siRNA by means of the Cell Line Nucleofactor Kit V.
Luciferase Assay
The effect of miR-497 or miR-302b binding on 3′-UTR of BCL2 or CCND2 was studied by cotransfection of 3′-UTR and miRNA mimics. 3′-UTR of BCL2 (1–2751 bp after the stop codon of NM_000633) and CCND2 (536–2137 bp after the stop codon of NM_001759) were cloned in the pEZX-MTO1 vector, which contains firefly luciferase as the reporter gene (controlled by the SV40 promoter) and Renilla luciferase as the tracking gene (controlled by the CMV promoter). Both firefly and Renilla luciferase activities were measured by means of the Dual-Glo luciferase assay kit at 10-min intervals. Maximal luciferase activity was calculated by normalizing firefly luciferase activity with Renilla luciferase activity.
Flow Cytometry Studies
Cell Death
Cell death in SH-SY5Y and IMR-32 cells was measured by means of FITC-labeled annexin V. Cells were transfected with either NTC or syn-miR-497 or syn-miR-302b. After 72 h of transfection, the cells were detached from the culture plate, washed twice with PBS, and suspended in 100 μl of binding buffer. 5 μl of annexin V was added, and the cells were incubated in the dark for 15 min. Subsequently, 5 μl of propidium iodide (PI) was added, and cell death was measured using flow cytometry (BD FACScanto II).
Reactive Oxygen Species (ROS) and Mitochondrial Membrane Potential (MMP) Measurement in SH-SY5Y
ROS formation was measured using 2′,7′-dichlorodihydrofluorescein diacetate, a non-fluorescent probe that is converted into a highly fluorescent (2′,7′-dichlorofluorescein) molecule in the presence of ROS. In brief, cells were transfected with miRNA mimics or NTC and cultured for 72 h. Subsequently, cells were detached from wells and incubated in phosphate buffer saline containing 20 μm 2′,7′-dichlorodihydrofluorescein diacetate in the dark at 37 °C for 30 min. After 30 min. the cells were pelleted, resuspended in PBS, and analyzed by flow cytometry. Changes in MMP were studied using JC-1, a cationic dye that shows potential dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green to red. In brief, cells were detached from 6-well plates and incubated in an incomplete medium containing 10 μm JC-1 in the dark at 37 °C for 20 min. After 20 min, cells were pelleted and resuspended in PBS and analyzed by flow cytometry or a fluorescence microscope.
Caspase 3 Activity and Cytochrome c Release
Total cell lysates of SH-SY5Y cells transfected with miRNA mimics or NTC were prepared using Cell Lytic M reagent. Caspase 3 activity was measured in cell lysates using fluorogenic substrate Ac-DEVD-AMC of caspase 3 from BD Pharmingen as described by the manufacturer. Cytochrome c release was studied by immunoblotting of cytochrome c in the cytosolic and mitochondrial fraction of SH-SY5Y cells and transfected with miRNA mimics or NTC. Mitochondrial fractions were prepared using mitochondrial protein isolation buffer from AMRESCO supplemented with protease inhibitor mixture and DTT. Prior to immunoblotting of cytochrome c, immunoblotting of β-actin and VDAC-1 were performed to establish the purity of mitochondrial and cytosolic fractions. Levels of β-actin and VDAC-1 were also used for normalization of protein loading.
Real-time PCR of BCL2, CCND2, and β-Actin
RT was performed with the high-capacity cDNA kit from ABI. The reaction mixture contains 1× RT buffer, 8 mm dNTP mix, 1× RT random primers, and 1 μl each of MultiScribe reverse transcriptase and RNase inhibitor, with a total volume of 20 μl, which also includes 1 μg of total RNA. The thermal cycling conditions used for RT were 10 min at 25 °C, 120 min at 37 °C, and 5 s at 85 °C. Real-time PCR was carried out using SYBR Green chemistry. In brief, the reaction mixture contained 1× SYBR Green master mixture, 1 μl of cDNA, and 0.50 μm of forward and reverse primers, with a total volume of 20 μl. Primers of BCL2 and CCND2 were designed with Primer Express software from ABI. The sequence of primers used was as follows. BCL2: forward, 5′-CTGAGTACCTGAACCGGCACC-3′ and reverse, 5′-GAGCAGAGTCTTCAGAGACAG-3′. CCND2: forward, 5′-GTTCCTGGCCTCCAAACTCA-3′ and reverse, 5′-CTTGATGGAGTTGTCGGTGTAAAT-3′.
Immunoblotting
The total cell lysates were prepared with CellLytic M cell lysis reagent from Sigma and supplemented with a protease inhibitor mixture and DTT. Immunoblotting was performed as described in our earlier paper (26). In brief, protein samples were subjected to SDS-PAGE (3% acrylamide stacking gel and 12 or 15% acrylamide separating gel). After electrophoresis, proteins were transferred to a nitrocellulose membrane. The membrane was incubated overnight with a primary antibody (1:1000 dilution) at 4 °C and for 1 h at room temperature. HRP-conjugated secondary antibodies were used at a 1:5000 dilution for protein detection. Blots were developed with the Pierce Supersignal West Pico horse-peroxidase reagent.
Statistical Analysis
Where comparisons had to be made between only two groups, Student's t test was employed to calculate the statistical significance. p < 0.05 in Student's t test was considered significant. A one-way analysis of variance followed by a post hoc Tukey test were conducted for pairwise comparisons made between more than two groups.
RESULTS
Dicer Silencing Increased Ethanol Toxicity in SH-SY5Y Cells
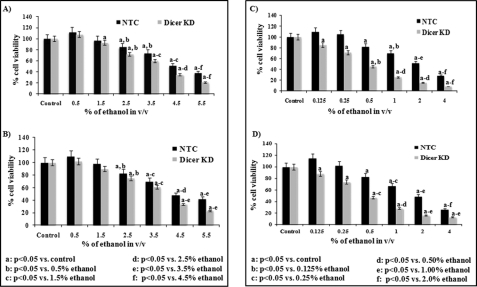
Dicer-silenced cells showed a higher rate of cell death compared with cells transfected with NTC after either short-term or long-term exposure to ethanol (Fig. 1). In NTC-transfected cells, short-term exposure to 5.5% v/v ethanol and long-term exposure to 4 and 2% of ethanol reduced cell viability below 50% (Fig. 1). In dicer-silenced cells, short-term exposure to 5.5 and 4.5% v/v ethanol and long-term exposure to 4, 2, 1, and 0.5% v/v ethanol reduced cell viability below 50% (Fig. 1, A–D). For further studies, 2.5% (for 4 h) and 0.5% (for 72 h) of ethanol were selected as short-term and long-term exposure concentrations, respectively (Fig. 1).
FIGURE 1.
Effect of either short-term (4 h, A and B) or long-term (72 h, C and D) ethanol exposure on viability of SH-SY5Y cells transfected with NTC siRNA or dicer siRNA. Cell viability is measured by MTT (A and C) and NRU (B and D) assays. The viability of cells transfected with NTC siRNA and exposed to different concentration of ethanol (either short-term or long-term) is compared with the viability of cells transfected with NTC siRNA and not exposed to ethanol (control = 100%). Similarly, the viability of cells transfected with dicer siRNA and exposed to ethanol (either short-term or long-term) is compared with the viability of cells transfected with dicer siRNA and not exposed to ethanol. All values are mean of four different experiments, and the error bars represent + S.E. of the mean. Statistical significance between control and different ethanol exposed groups was calculated by one-way analysis of variance followed by a post hoc Tukey test. Values showing p < 0.05 in the Tukey test are considered statistically significant and are marked a, b, c, d, e, and f. (Dicer KD: dicer knockdown (KD)).
Identification of Ethanol-responsive miRNAs in Neuronal Cells
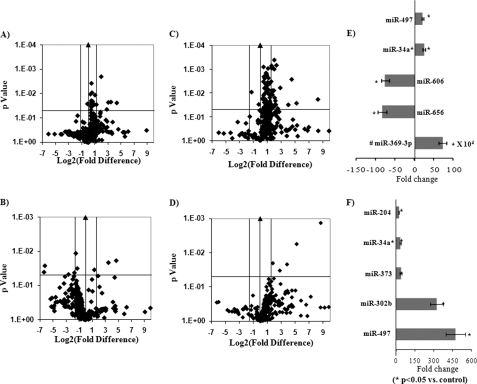
Ethanol-responsive miRNAs were identified by global profiling of miRNAs in SH-SY5Y cells exposed to ethanol for either short-term (2.5% v/v × 4 h) or long-term (0.5% v/v × 72 h). A volcano plot drawn between log2 of the fold difference and the p value shows that long-term exposure to ethanol produces more significant (p < 0.05) alterations in the expression pattern of miRNAs than short-term exposure to ethanol (Fig. 2, A–D). Short-term exposure to ethanol significantly increased the expression of seven miRNAs more than 5-folds (Fig. 2E and supplemental Table 1). Maximum increase was found in miR-369–3p (731,234-fold; undetected in control and in undetected samples, Ct is taken as 35, which is equal to the number of cycles) followed by miR-34a* (25-fold) and miR-493 (20-fold) (Fig. 2E and supplemental Table 1). Significant down-regulation in the expression of three miRNAs (5-fold) were also observed after short-term exposure. After short-term exposure to ethanol, the maximum decrease was found in miR-656 (82-fold), followed by miR-606 (75-fold) and miR-27a* (6-fold) (Fig. 2E). Long-term exposure to ethanol up-regulated the expression of 11 miRNAs more than 5-fold (supplemental Table 2). After long-term exposure to ethanol, miR-497 showed the maximum increase (474-fold), followed by miR-302b (322-fold), miR-373 (37-fold), miR-34a* (36-fold), and miR-204 (23-fold) (Fig. 2F). However, after long-term exposure significant down-regulation was found only in expression of miR-33b (2.38-fold) (supplemental Table 2). Similar to SH-SY5Y, long-term exposure to ethanol significantly (p < 0.05) induced the expression of miR-497 (128-fold) and miR-302b (74-fold) in IMR-32 (Fig. 3A).
FIGURE 2.
Global expression profiling of miRNAs by real-time PCR using TLDA arrays in SH-SY5Y cells. A, volcano plot of miRNA expression in SH-SY5Y cells after either short-term exposure (A and B) or long-term exposure (C and D) to ethanol. Fig. 1, A and C, represents the set of miRNAs present in pool A of the TLDA plates. Fig. 1, B and D, represents the set of miRNAs present in pool B of the TLDA plates. For short-term exposure, cells are exposed to 2.0% v/v ethanol for 4 h, and for long-term exposure, cells were exposed to 0.5% v/v ethanol for 72 h. The volcano plot is plotted between log2 of the fold changes versus its p value obtained from the Student's t test. The black line with the arrowhead in the center indicates a fold change in the gene expression of 1. Parallel to the central line, two vertical lines, one on each side, indicate a + 3-fold change in gene expression. The single horizontal line inside the boxes indicates the threshold for the p value of the t test, set as 0.05. E, the top five miRNAs altered after short-term exposure of ethanol. F, the top five miRNAs altered after long-term exposure to ethanol. Cells not exposed to ethanol are considered as control and are used for calculation of the fold change with the -ΔΔ cycle threshold method (control = 1). #, MiR-369–3p had not amplified in control sample, so for calculations, their Ct is taken as 35 (final number of the cycle). In the bar diagram, fold change is represented by multiplying it from 104. All values are the mean of three individual experiments. Significant changes are calculated by Student's t test. *, p < 0.05.
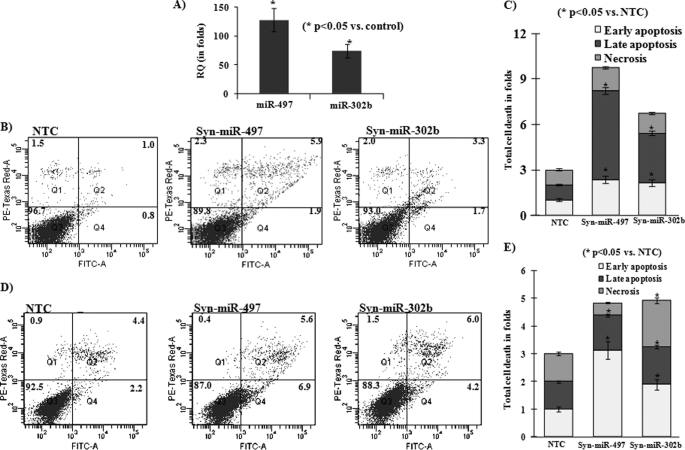
FIGURE 3.
Induction of miR-497 and miR-302b by ethanol exposure in IMR-32 cells (A) and effect of ectopic expression of these miRNAs on apoptosis of SH-SY5Y (B and C) and IMR-32 (D and E) cells. A, real-time PCR of miR-497 and miR-302b in control and ethanol-exposed (long-term exposure, 0.5% v/v × 72 h) IMR-32 cells. B, dot plots of cell death analysis by FITC annexin V staining followed by flow cytometry analysis in SH-SY5Y cells transfected with either NTC, Syn-miR-497, or Syn-mir-302b. C, bar diagram analysis of total cell death in SH-SY5Y cells as observed by flow cytometry analysis. D, dot plots of cell death analysis by FITC annexin V staining followed by flow cytometry analysis in IMR-32 cells transfected with either NTC, Syn-miR-497, or Syn-mir-302b. E, bar diagram analysis of total cell death of IMR-32 cells as observed by flow cytometry analysis. Dot plots are plotted between FITC-A and PE-Texas Red. Q1, quadrant I (necrotic cells); Q2, quadrant 2 (late apoptotic cells); Q3, quadrant 3 (living cells); Q4, quadrant 4 (early apoptotic cells). For calculating fold change in total cell death, fold change in early apoptosis, late apoptosis, and necrosis is calculated individually and added. Cells transfected with NTC are considered as control and used for calculating fold change in early apoptosis, late apoptosis, or necrosis of cells transfected with Syn-miR-497 or Syn-miR-302b. All values are the mean of three individual experiments. Significant changes are calculated by Student's t test. *, p < 0.05. RQ, relative quantification.
Ectopic Expression of miR-497- and miR-302b-induced Apoptosis and Caspase 3 Activity
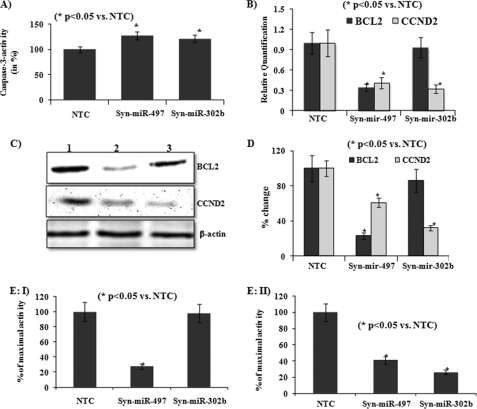
In SH-SY5Y cells, overexpression of miR-497 increased early apoptosis, late apoptosis, and necrosis up to 2.3-, 5.9-, and 1.5-fold, respectively (Fig. 3, B and C). Similarly, overexpression of miR-302b increased early apoptosis, late apoptosis, and necrosis up to 2.1-, 3.3-, and 1.3-fold, respectively (Fig. 3, B and C). In IMR-32 cells, overexpression of miR-497 increased early apoptosis and late apoptosis up to 1.2- and 3.13-fold, respectively (Fig. 3, D and E). Similarly, overexpression of miR-302b increased early apoptosis and late apoptosis up to 1.9- and 1.3-fold, respectively (Fig. 3, D and E). In SH-SY5Y, overexpression of either miR-497 or miR-302b produced the maximum increase in late apoptosis (Fig. 3, B and C), whereas in IMR-32 cells, overexpression of either miR-497 or miR-302b produced the maximum increase in early apoptosis (D and E). Moreover, overexpression of miR-497 and miR-302b in SH-SY5Y cells increased caspase 3 activity up to 27.7% and 21.2%, respectively (Fig. 4A).
FIGURE 4.
Ectopic expression of miR-497 or miR-302b induced caspase 3 activity (A) and down-regulated expression of BCL2 and/or CCND2 (B–D) by targeting 3′-UTR of these genes (E-I and E-II). A, caspase 3 activity measured in lysates of cells transfected with NTC (control = 100%), Syn-miR-497, or Syn-miR-302b using the fluorogenic substrate Ac-DEVD-AMC. Fluorescence was measured using a multiple-well plate reader at an excitation wavelength of 380 nm and an emission wavelength of 450 nm. B, effect of ectopic expression of miR-497, miR-302b, or NTC (control = 1) on transcript levels of BCL2 or CCND2 as measured by real-time PCR. Expression of BCL2 and CCND2 in cells transfected with Syn-miR-497 or Syn-miR-302b is compared with their expression in cells transfected with NTC (control = 1). C, effect of ectopic expression of miR-497, miR-302b, or NTC on protein levels of BCL2 or CCND2 measured by immunoblotting in cell lysates of SH-SY5Y cells. Lane 1, NTC; lane 2, Syn-miR-497; lane 3, Syn-miR-302b. D, densitometry of immunoblots of BCL2 and CCND2 shown in B. E, luciferase assays in SH-SY5Y cells cotransfected with the pEZX-MT01 vector with 3′-UTR of BCL2 (I) or CCND2 (II) and miRNA mimics or NTC (control = 100%). When cells were transfected with NTC siRNAs, the empty pEZX-MT01 vector was used for cotransfection. Both firefly luciferase and Renilla luciferase activities were measured as described under “Experimental Procedures,” and firefly luciferase activity was normalized with Renilla luciferase activities in the same well. All the values are the mean of three individual experiments. Significant changes are calculated by Student's t test. *, p < 0.05.
Identification of BCL2 and CCND2 as Potential Target Genes Involved in Ethanol-induced Neuronal Apoptosis
After observing a significant increase (p < 0.05) in apoptosis by ectopic expression of miR-497 or miR-302b, we generated a list of potential target genes of miR-497 and mir-302b. After scanning of in silico-predicted targets, we identified BCL2 and CCND2 as possible regulators of ethanol-induced neuronal apoptosis. Interestingly, scanning of 3′-UTR of BCL-2 and CCND2 (supplemental Fig. 2) showed the presence of target sites for several miRNAs (BCL2: miR-497, miR-204, miR-181, miR-182, miR-449, and miR-365; CCND2: miR-497, miR-302, miR-204, miR-182, miR-503, and let 7) that were induced at least 2-fold by long-term exposure to ethanol. Transfection of the miR-497 mimic in SH-SY5Y cells reduced the levels of BCL2 and CCND2 mRNAs up to 34 and 41%, respectively (Fig. 4B). While transfection of the miR-302b mimic in SH-SY5Y cells reduced the expression of CCND2 by up to 32%, no significant change was observed in expression of BCL2 (Fig. 4B). In line with real-time PCR results, ectopic expression of miR-497 reduced the levels of BCL2 and CCND2 proteins, and ectopic expression of miR-302b reduced the levels of CCND2 protein (Fig. 4, C and D). Moreover, in SH-SY5Y cells, cotransfection of Syn-miR-497 with 3′-UTR of BCL2 significantly decreased (up to 72%) maximal luciferase activity in comparison to cells cotransfected with NTC (Fig. 4, E-I). As expected, cotransfection of Syn-miR-302b with 3′-UTR of BCL2 did not produce a significant change in maximal luciferase activity (Fig. 4, E-I). However, cotransfection of 3′-UTR of CCND2 with mimics of either miR-497 or miR-302b significantly reduced maximal luciferase activity by up to 41 and 26%, respectively (Fig. 4, E-II).
Overexpression of miR-497 Increased ROS Formation, Disrupted MMP, and Induced Cytochrome c Release
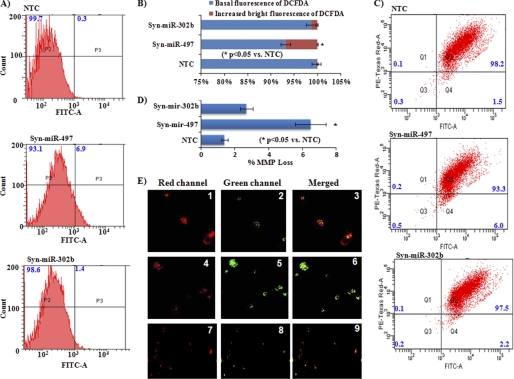
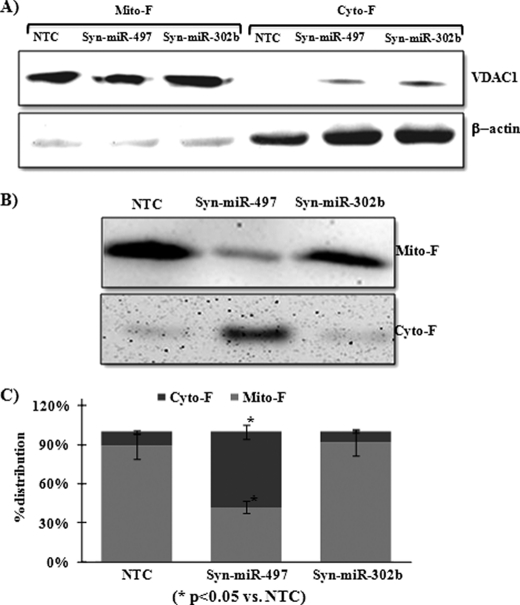
Flow cytometry analysis of SH-SY5Y cells transfected with the miR-497 mimic showed a significant increase (6.6%) in ROS formation (Fig. 5, A and B). In accordance with ROS formation, overexpression of miR-497 also increased the MMP loss by up to 6%, which was 1.5% in cells transfected with NTC (Fig. 5, C and D). Fluorescence imaging of cells transfected with mimics of miR-497 clearly showed the presence of the green JC-1 dye, and this validated our flow cytometry results (Fig. 5E). Overexpression of miR-302b in SH-SY5Y cells did not produce any significant change in ROS formation or MMP loss (Fig. 5, A–E). Similarly, although overexpression of only miR-497 induced the release of cytochrome c from mitochondria to the cytosol, overexpression of miR-302b did not produce any significant change in the localization of cytochrome c (Fig. 6, B and C).
FIGURE 5.
Effect of ectopic expression of miR-497, miR-302b, or NTC (control) on ROS formation (A and B) and MMP loss (C–E) in SH-SY5Y cells. A, ROS formation was measured by flow cytometry in cells transfected with Syn-miR-497, Syn-miR-302b, or NTC (control) as described under “Experimental Procedures. P2, basal fluorescence; P3, increased bright fluorescence. B, % change in ROS formation as measured by flow cytometry in A. C, flow cytometry analysis of MMP loss using JC-1, a dual-emission potential-sensitive probe in SH-SY5Y cells transfected with Syn-miR-497, Syn-miR-302b, or NTC (control). D, bar diagram for representing percent of MMP loss observed after flow cytometry in C. E, fluorescence imaging of cells incubated with JC-1 for validating MMP loss induced by ectopic expression of miRNAs. 1-3, cells transfected with NTC. 4-6, cells transfected with Syn-miR-497. 7-9, cells transfected with Syn-miR-302b. All values are the mean of three individual experiments. Significant changes are calculated by Student's t test. *, p < 0.05.
FIGURE 6.
Effect of ectopic expression of miR-497, miR-302b, or NTC (control) on cytochrome c release in SH-SY5Y cells. A, levels of VDAC-1 (mitochondrial loading control) and β-actin (cytosolic loading control) in mitochondrial fraction (Mito-F) and cytosolic fraction (Cyto-F) of cells transfected with NTC, Syn-miR-497, or Syn-miR-302b. B, immunoblotting of cytochrome c in Mito-F and Cyto-F of cells transfected with NTC, Syn-miR-497, or Syn-miR-302b. C, percent distribution of cytochrome c between mitochondrial and cytosolic fraction calculated by densitometry of cytochrome c immunoblots. After densitometry, the amount of cytochrome c detected in the mitochondrial and cytosolic fraction of NTC-, Syn-miR-497-, or Syn-miR-302b-transfected cells is added separately and considered as 100%. All values are the mean of three individual experiments. Significant changes are calculated by Student's t test. *, p < 0.05.
Effect of GSK-3Β Inhibition on Ethanol-induced Apoptosis of SH-SY5Y Cells
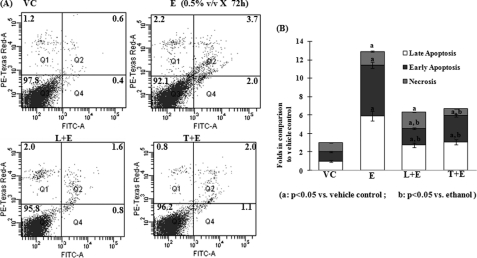
Pre-exposure with LiCl3 or TDZD-8 significantly (p < 0.05) prevented ethanol-induced apoptosis (Fig. 7, A and B). Long-term exposure to ethanol (0.5% × 72 h) induced early and late apoptosis up to 5.0- and 3.3-fold, respectively. Pre-exposure with LiCl3 and TDZD-8 reduced early apoptosis to 2.0- and 2.7-fold, respectively (Fig. 7, A and B). Similarly, pre-exposure to LiCl3 and TDZD-8 reduced late apoptosis to 2.6- and 3.3-fold, respectively (Fig. 7, A and B).
FIGURE 7.
Effect of pre-exposure of GSK-3B inhibitors on ethanol-induced death of SH-SY5Y cells. A, cells were exposed to ethanol (E, 0.5% × 72 h), lithium (L, 20 mm) + E, TDZD-8 (T, 10 μm) + E, or vehicle control (VC). The cell death assay was carried out by FITC annexin V staining followed by flow cytometry analysis. Data are presented as dot plots (FITC-A plotted against PE-Texas Red). Q1, quadrant I (necrotic cells); Q2, quadrant 2 (early apoptotic cells); Q3, quadrant 2 (living cells); Q4, quadrant 4 (late apoptotic cells). B, bar diagram analysis of total cell death as observed by flow cytometry analysis in A. For calculating fold change in total cell death, the fold change in early apoptosis, late apoptosis, and necrosis is calculated individually and added. The fold change in early apoptosis, late apoptosis, and necrosis of E-, L+E-, or T+E-exposed groups is calculated against the vehicle control. Significant changes are calculated by one-way analysis of variance followed by a pairwise Tukey test. Values showing p < 0.05 in the Tukey test are considered statistically significant. a, p < 0.05 when compared with vehicle control; b, p < 0.05 when compared with ethanol-exposed cells).
Effect of GSK-3Β Inhibition on Ethanol-mediated Regulation of miR-497, miR-302b, and Their Target Genes in SH-SY5Y Cells
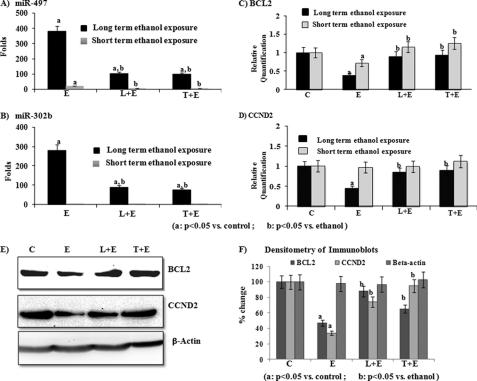
Pre-exposure to GSK-3Β inhibitors significantly (p < 0.05) prevented ethanol-mediated up-regulation of miR-497 and miR-302b (Fig. 8, A and B). Short-term and long-term exposure to ethanol produced a 20- and 380-fold increase in miR-497 expression, respectively. Pre-exposure with LiCl3 or TDZD-8 prevented approximately 90% of the increase found in expression of miR-497 after either short-term exposure (20-fold was reduced to 3-fold) or long-term exposure (380-fold was reduced to 40-fold) to ethanol (Fig. 8A). Although long-term exposure to ethanol induced the expression of miR-302b up to 280-fold, short-term exposure did not alter the expression of miR-302b (Fig. 8B). Pre-exposure with LiCl3 or TDZD-8 decreased the ethanol-induced expression of miR-302b up to 22-fold (LiCl3) and 27-fold (TDZD-8) (Fig. 8B). In accordance with induction of miR-497, expression of BCL2 (the target gene of miR-497) decreased by 72 and 38% after long-term or short-term exposure to ethanol, respectively (Fig. 8C). Pre-exposure with LiCl3 and TDZD-8 significantly prevented the ethanol-mediated decrease in mRNA expression of BCL2 after either short-term (increased from 72 to 89%) or long-term (increased from 38 to 93%) exposure to ethanol (Fig. 8C). Similarly, when cells were pre-exposed with TDZD-8, expression of BCL2 decreased significantly after either short-term or long-term exposure to ethanol (Fig. 8C). Short-term exposure to ethanol did not produce any significant alteration in expression of CCND2, but long-term exposure reduced the expression of CCND2 up to 45%. When these cells were pre-exposed with LiCl3 or TDZD-8, expression of CCND2 decreased up to 85 and 90%, respectively (Fig. 8D). In line with real-time PCR results, our immunoblotting studies also showed a significant decrease in the expression of BCL2 (53%) and CCND2 (66%) after long-term exposure to ethanol (Fig. 8, E and F). Further pre-exposure to LiCl3 and TDZD-8 significantly (p < 0.05) prevented an ethanol-meditated decrease in the levels of BCL2 and CCND2 (Fig. 8, E and F).
FIGURE 8.
Effect of pre-exposure of GSK-3B inhibitors (L, lithium or T, TDZD-8) on ethanol-induced alterations in expression of miR-497, miR-302b, BCL2, or CCND2. A–D, expression of miR497 (A), miR-302b (B), BCL2 (C), and CCND2 (D) measured by real-time PCR in SH-SY5Y cells exposed with GSK-3B inhibitors and ethanol. E, immunoblotting of BCL2, CCND2, and β-actin in total cell lysates of SH-SY5Y cells exposed with GSK-3B inhibitors and ethanol. F, densitometry of immunoblots. The concentrations used for exposure are ethanol (E), 0.5% × 72 h; lithium (L), 20 mm; and TDZD-8 (T), 10 μm. The expression of miR-497 and miR-302b was measured by using TaqMan assays, whereas expression of BCL2 and CCND2 was measured using SYBR Green chemistry as mentioned under “Experimental Procedures.” The fold change in expression of miR-497 and miR-302b was calculated by considering their levels as 1-fold in cells exposed to vehicle control. Significant changes are calculated by one-way analysis of variance followed by a pairwise Tukey test. a, p < 0.05 versus vehicle control; b, p < 0.05 versus ethanol.
DISCUSSION
Increased Sensitivity of dicer-silenced SH-SY5Y cells to ethanol indicated the involvement of miRNAs in ethanol neurotoxicity. In dicer-silenced SH-SY5Y cells, long-term exposure to low dosages of ethanol has shown more toxicity than short-term exposure to higher dosages of ethanol. Similarly, long-term exposure to a low dose of ethanol (0.5% v/v for 72 h) altered the expression of more miRNAs (28 miRNAs, 3-fold and p < 0.05) than short-term exposure (13 miRNAs, 3-fold and p < 0.05) of a high dose (2.5% v/v for 4 h) of ethanol in SH-SY5Y cells. MiR-369–3p, miR-493, miR-518f, and miR-370 were found to be specifically up-regulated by short-term exposure to ethanol, whereas expression of miR-302b, miR-373, miR204, miR-208b, miR-432, miR-375, miR-936, and miR-483–5p were up-regulated specifically by long-term exposure to ethanol. Long-term exposure to ethanol also induced the expression of miR-204 (23.17-fold) and miR-375 (7.4-fold), which are known to express at higher levels in the brain (27, 28). Sathyan et al. (16) have shown that ethanol at levels found in alcoholics alters the expression of miR-21, miR-335, miR-9, and miR-153 in neurospheres derived from neuroepithelial cells of a fetal mouse cerebral cortex at gestational day 12.5. Similar to SH-SY5Y cells, long-term exposure to ethanol substantially induced expression of miR-497 and miR-302b in IMR-32 cells. In SH-SY5Y cells, mir-497 was found to be particularly sensitive to ethanol exposure and exhibited a 21- and 474-fold increase in expression after short-term and long-term exposure, respectively.
Ectopic Expression of miR-497 in SH-SY5Y cells increased the rate of early and late apoptosis up to 2.37- and 5.90-fold, respectively. Similarly, ectopic expression of miR-302b increased the rate of early and late apoptosis up to 2.12- and 3.3-fold, respectively. Moreover, overexpression of either miR-497 or miR-302b in SH-SY5Y cells induced the activity of caspase 3, an effector caspase of apoptosis. Interestingly, several miRNAs (miR-497, miR-449a, miR-192, miR-139–5p, miR-146a, miR-328, let-7b, and miR-342–3p) that were induced by ethanol have been reported previously to target apoptosis-related genes in other systems or cells (29, 30). Using the TargetScan web portal, we generated list of potential target genes of miR-497 or miR-302b and selected BCL2 and CCND2 as possible candidate genes involved in the regulation of ethanol-induced neuronal apoptosis. The presence of target sites for several miRNAs in 3′-UTR of BCL2 (miR-497, miR-204, miR-181, miR-182, miR-449, and miR-365) and CCND2 (miR-497, miR-302, miR-204, miR-182, miR-503, and let 7) that are induced by long-term exposure to ethanol have supported our selection. A significant decrease in maximal luciferase activity in neuronal cells cotransfected with 3′-UTR of BCL2 and Syn-miR-497 provided further evidence that BCL2 is directly targeted by miR-497. The maximum luciferase activity of plasmids containing 3′-UTR of CCND2 decreased because of cotransfection of either miR-497 or miR-302b mimics. Also, the decreased expression of BCL2 mRNAs and proteins in cells transfected with miR-497 mimics has confirmed the direct role of miR-497 in down-regulation of BCL2. Similarly, the decreased expression of CCND2 mRNA and protein in SH-SY5Y cells transfected with mimics of either miR-497 or miR-302b has proved that levels of CCND2 are regulated by both miR-497 and miR302b.
Overexpression of miR-497, which targets BCL2, also induced ROS formation and MMP loss. Down-regulation of antiapoptotic protein BCL2 along with increased ROS formation and MMP loss indicates involvement of a mitochondria-mediated pathway of apoptosis (31). Also, the increased release of cytochrome c in cells overexpressing miR-497 confirms the involvement of a mitochondrial pathway of apoptosis. Studies carried out by Young et al. (32) have shown that ethanol-induced neuronal apoptosis is an intrinsic pathway-mediated phenomenon that involves disruption of mitochondrial membrane potential, cytochrome c release, and caspase 3 activation. Similar to our results, a recent study has shown that induction of miR-497 in response to ischemia and oxygen-glucose deprivation induces neuronal cell death by down-regulating BCL2 (33). BCL2-mediated neuronal apoptosis is also known to be regulated by miR-34a (34, 35). Although no alteration was observed in expression of miR-34a, a significant and substantial increase was found in the expression of miR-34a* (a minor strand of pre-miR-34) after either short-term (25.11-fold) or long-term (36.9-fold) exposure to ethanol. Overexpression of miR-302b also induced the caspase 3-mediated apoptosis of SH-SY5Y cells. However, overexpression of miR-302b did not produce significant alterations in ROS formation, MMP loss, and cytochrome c localization, which indicates that miR-302b-induced neuronal apoptosis is not mitochondria-mediated.
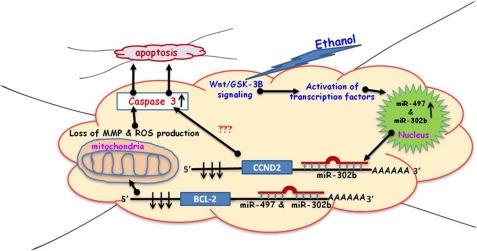
FIGURE 9.
Pictorial summary of results. The arrows with ● at the bottom indicate a connection between two events. ↑ indicate an increase in activity or expression, and ↓ indicate decreased expression.
Overexpression of either miR-497 or miR-302b decreased the expression of CCND2, a cyclin also known to regulate neurogenesis in the adult brain (13). CCND2 regulates the cell cycle by exporting p27 (a cell cycle inhibitor known to control cell cycle progression at G1) from the nucleus to the cytoplasm, which results in degradation of p27 at the G0-G1 transition of the cell cycle (36). Our cotransfection studies have shown that miR-302b directly targets 3′-UTR of CCND2 in neuronal cells. Earlier studies that used gain and loss of function approaches showed that miR-302b is involved in the regulation of stemness and inhibits differentiation of neuronal cells by targeting CCND2 (14). Moreover, CCND2 is the only cyclin isoform known to express in the hippocampus and regulates neurogenesis in the hippocampus of the adult brain (13). Chronic use of ethanol is known to target the hippocampus and its associated physiological functions (37). Several studies have shown that miRNAs play an important role in the regulation of neurogenesis (11, 38, 39), and interestingly, our in silico studies (supplemental Fig. 3) have also identified neurogenesis as the primary cellular process targeted by ethanol-sensitive miRNAs. As in vitro systems alone may not be sufficient for studying adult neurogenesis, animal studies are required to further identify the role of miR-302b in ethanol-mediated alterations in adult neurogenesis.
Cellular stress in the brain modulates several events of neurogenesis by activating GSK-3B (40). In dicer knockdown mice (or in the absence of mature miRNAs), GSK-3B is inactivated because of increased phosphorylation at the serine 9 position (41). Moreover, during neuronal differentiation, ethanol inhibits neurite outgrowth by activating GSK-3B through dephosphorylation at serine 9 (42). Prevention of ethanol-induced neuronal apoptosis at substantial levels by GSK-3B inhibition supports earlier reports about the involvement of GSK-3B signaling in ethanol neurotoxicity (17). Earlier studies have also shown that inhibition of GSK-3B with LiCl3 down-regulates the expression of CCND2 (18). Prevention of ethanol-mediated induction of miRNAs and their target genes because of inhibition of GSK-3B suggests that the modulation in miRNA levels is preceded by GSK-3B activity. In line with our findings, the recent studies of Ji et al. (43) have shown that consistent activation of the Wnt/β-catenin signaling pathway by inhibition of GSK-3B induces transcriptional activation of the miR-181 family (43). Inhibition of GSK-3B can result in activation of several transcription factors that are probably involved in the induction of miRNAs (21).
In summary, our studies have shown that the absence of miRNAs in SH-SY5Y cells increases ethanol-induced apoptosis, particularly after long-term exposure to ethanol. We have identified miRNAs that are regulated by both short-term and long-term exposure to ethanol (miR-34a*, miR-523, miR-497, and miR-618) and miRNAs regulated by either short-term (miR-369–3p, miR-656, miR-606, and miR-493) or long-term (miR-302b, miR-373, miR204, miR-208b, and miR-432) exposure to ethanol. Our studies have also shown that in neuronal cells, miR-497 regulates expression of both BCL2 and CCND2, whereas miR-302b regulates expression of CCND2. Moreover, although sour experiments have demonstrated involvement of mitochondria in miR-497-induced apoptosis of neuronal cells, miR-302b-induced apoptosis is independent of mitochondria. Also, our studies with LiCl3 and TCZD-8 indicated that ethanol-induced modulation in miRNAs and their target genes are dependent on GSK-3B activity. In conclusion, our studies suggest that miR-497 and miR-302b regulate ethanol-induced neuronal apoptosis by both mitochondria-dependent and mitochondria-independent pathways, respectively. Further in vivo studies are required to explore the role of miR-302b and CCND2 in ethanol-mediated inhibition of adult neurogenesis.
Acknowledgments
We thank the Director, Indian Institute of Toxicology Research (formerly ITRC), Lucknow, for his keen interest and support in carrying out the study. We also thank Mr. Bhaskar Bhattacharji and Naveen Rana for helping with language improvement, Dr. Mukesh Srivastav for helping with statistical analysis, and Mr. Rajesh Misra for technical assistance.
This work was supported by the SIP08 and NWP34 projects of CSIR, New Delhi.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Tables 1 and 2.
- miRNA
- microRNA
- GSK-3Β
- glycogen synthase kinase 3 β
- CCND2
- cyclin D2
- TLDA
- TaqMan low-density array
- NTC
- non-targeting control
- MTT
- (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide)
- PI
- propidium iodide
- ROS
- reactive oxygen species
- NRU
- neutral red uptake
- MMP
- mitochondrial membrane potential
- RQ
- relative quantification.
REFERENCES
- 1. Slezak-Prochazka I., Durmus S., Kroesen B. J., van den Berg A. (2010) RNA 16, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li X., Jin P. (2010) Nat. Rev. Neurosci. 11, 329–338 [DOI] [PubMed] [Google Scholar]
- 3. Davis T. H., Cuellar T. L., Koch S. M., Barker A. J., Harfe B. D., McManus M. T., Ullian E. M. (2008) J. Neurosci. 28, 4322–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawase-Koga Y., Otaegi G., Sun T. (2009) Dev. Dyn. 238, 2800–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brust J. C. (2010) Int. J. Environmental Res. Public Health 7, 1540–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ikonomidou C., Bittigau P., Ishimaru M. J., Wozniak D. F., Koch C., Genz K., Price M. T., Stefovska V., Hörster F., Tenkova T., Dikranian K., Olney J. W. (2000) Science 287, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 7. Heaton M. B., Moore D. B., Paiva M., Gibbs T., Bernard O. (1999) Brain Res. 817, 13–18 [DOI] [PubMed] [Google Scholar]
- 8. Nixon K. (2006) Hippocampus 16, 287–295 [DOI] [PubMed] [Google Scholar]
- 9. Yadav S., Dhawan A., Singh R. L., Seth P. K., Parmar D. (2006) Mol. Cell. Biochem. 286, 171–180 [DOI] [PubMed] [Google Scholar]
- 10. Elibol-Can B., Jakubowska-Dogru E., Severcan M., Severcan F. (2011) Alcohol. Clin. Exp. Res. PMID: 21631543 [DOI] [PubMed] [Google Scholar]
- 11. Liu C., Zhao X. (2009) Neuromolecular Med. 11, 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freyberg Z., Ferrando S. J., Javitch J. A. (2010) Am. J. Psychiatry 167, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kowalczyk A., Filipkowski R. K., Rylski M., Wilczynski G. M., Konopacki F. A., Jaworski J., Ciemerych M. A., Sicinski P., Kaczmarek L. (2004) J. Cell Biol. 167, 209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee N. S., Kim J. S., Cho W. J., Lee M. R., Steiner R., Gompers A., Ling D., Zhang J., Strom P., Behlke M., Moon S. H., Salvaterra P. M., Jove R., Kim K. S. (2008) Biochem. Biophys. Res. Commun. 377, 434–440 [DOI] [PubMed] [Google Scholar]
- 15. Pietrzykowski A. Z., Friesen R. M., Martin G. E., Puig S. I., Nowak C. L., Wynne P. M., Siegelmann H. T., Treistman S. N. (2008) Neuron 59, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sathyan P., Golden H. B., Miranda R. C. (2007) J. Neurosci. 27, 8546–8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luo J. (2009) Mol. Neurobiol. 40, 108–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang W., Chang H. Y., Fei T., Wu H., Chen Y. G. (2007) Oncogene 26, 2471–2482 [DOI] [PubMed] [Google Scholar]
- 19. Ali A., Hoeflich K. P., Woodgett J. R. (2001) Chem. Rev. 101, 2527–2540 [DOI] [PubMed] [Google Scholar]
- 20. Jope R. S., Johnson G. V. (2004) Trends Biochem. Sci. 29, 95–102 [DOI] [PubMed] [Google Scholar]
- 21. Grimes C. A., Jope R. S. (2001) Prog. Neurobiol. 65, 391–426 [DOI] [PubMed] [Google Scholar]
- 22. Zhou R., Yuan P., Wang Y., Hunsberger J. G., Elkahloun A., Wei Y., Damschroder-Williams P., Du J., Chen G., Manji H. K. (2009) Neuropsychopharmacology 34, 1395–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mitchell J. J., Paiva M., Heaton M. B. (1998) Alcohol 15, 137–139 [DOI] [PubMed] [Google Scholar]
- 24. Siddiqui M. A., Singh G., Kashyap M. P., Khanna V. K., Yadav S., Chandra D., Pant A. B. (2008) Toxicol. In Vitro 22, 1681–1688 [DOI] [PubMed] [Google Scholar]
- 25. Chen G., Bower K. A., Ma C., Fang S., Thiele C. J., Luo J. (2004) FASEB J. 18, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 26. Yadav S., Johri A., Dhawan A., Seth P. K., Parmar D. (2006) Toxicol. Appl. Pharmacol. 217, 15–24 [DOI] [PubMed] [Google Scholar]
- 27. Zhang B., Pan X. (2009) Environ. Health Perspect. 117, 231–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Baroukh N. N., Van Obberghen E. (2009) FEBS J. 276, 6509–6521 [DOI] [PubMed] [Google Scholar]
- 29. Lizé M., Pilarski S., Dobbelstein M. (2010) Cell Death Differ. 17, 452–458 [DOI] [PubMed] [Google Scholar]
- 30. Subramanian S., Steer C. J. (2010) J. Cell. Physiol. 223, 289–298 [DOI] [PubMed] [Google Scholar]
- 31. Kuwana T., Newmeyer D. D. (2003) Curr. Opin. Cell Biol. 15, 691–699 [DOI] [PubMed] [Google Scholar]
- 32. Young C., Klocke B. J., Tenkova T., Choi J., Labruyere J., Qin Y. Q., Holtzman D. M., Roth K. A., Olney J. W. (2003) Cell Death Differ. 10, 1148–1155 [DOI] [PubMed] [Google Scholar]
- 33. Yin K. J., Deng Z., Huang H., Hamblin M., Xie C., Zhang J., Chen Y. E. (2010) Neurobiol. Dis. 38, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X., Liu P., Zhu H., Xu Y., Ma C., Dai X., Huang L., Liu Y., Zhang L., Qin C. (2009) Brain Res. Bull. 80, 268–273 [DOI] [PubMed] [Google Scholar]
- 35. Qi J., Yu J. Y., Shcherbata H. R., Mathieu J., Wang A. J., Seal S., Zhou W., Stadler B. M., Bourgin D., Wang L., Nelson A., Ware C., Raymond C., Lim L. P., Magnus J., Ivanovska I., Diaz R., Ball A., Cleary M. A., Ruohola-Baker H. (2009) Cell Cycle 8, 3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Susaki E., Nakayama K., Nakayama K. I. (2007) Mol. Cell. Biol. 27, 4626–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chin V. S., Van Skike C. E., Matthews D. B. (2010) Alcohol 44, 3–14 [DOI] [PubMed] [Google Scholar]
- 38. Shen Q., Temple S. (2009) Nat. Neurosci. 12, 369–370 [DOI] [PubMed] [Google Scholar]
- 39. Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009) Nat. Neurosci. 12, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wada A. (2009) J. Pharmacol. Sci. 110, 14–28 [DOI] [PubMed] [Google Scholar]
- 41. Hébert S. S., Papadopoulou A. S., Smith P., Galas M. C., Planel E., Silahtaroglu A. N., Sergeant N., Buée L., De Strooper B. (2010) Hum. Mol. Genet. 19, 3959–3969 [DOI] [PubMed] [Google Scholar]
- 42. Chen G., Bower K. A., Xu M., Ding M., Shi X., Ke Z. J., Luo J. (2009) Neurotox. Res. 15, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ji J., Yamashita T., Wang X. W. (2011) Cell Biosci. 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]