Background: Altered function has been reported when cannabinoid CB1 and orexin OX1 receptors are co-expressed.
Results: Direct physical interactions between these receptors were observed.
Conclusion: The CB1-OX1 heteromer is a selective target for orexin A.
Significance: Co-regulation of this heteromer may alter wakefulness and feeding behavior.
Keywords: Seven-helix Receptor, Cannabinoid Receptors, Fluorescence Resonance Energy Transfer (FRET), G Protein-coupled Receptors (GPCR), Peptide Hormones, Pharmacology
Abstract
Agonist-induced internalization was observed for both inducible and constitutively expressed forms of the cannabinoid CB1 receptor. These were also internalized by the peptide orexin A, which has no direct affinity for the cannabinoid CB1 receptor, but only when the orexin OX1 receptor was co-expressed along with the cannabinoid CB1 receptor. This effect of orexin A was concentration-dependent and blocked by OX1 receptor antagonists. Moreover, the ability of orexin A to internalize the CB1 receptor was also blocked by CB1 receptor antagonists. Remarkably, orexin A was substantially more potent in producing internalization of the CB1 receptor than in causing internalization of the bulk OX1 receptor population, and this was true in cells in which the CB1 receptor was maintained at a constant level, whereas levels of OX1 could be varied and vice versa. Both co-immunoprecipitation and cell surface, homogenous time-resolved fluorescence resonance energy transfer based on covalent labeling of N-terminal “SNAP” and “CLIP” tags present in the extracellular N-terminal domain of the receptors confirmed the capacity of these two receptors to heteromultimerize. These studies confirm the capacity of the CB1 and OX1 receptors to interact directly and demonstrate that this complex has unique regulatory characteristics. The higher potency of the agonist orexin A to regulate the CB1-OX1 heteromer compared with the OX1-OX1 homomer present in the same cells and the effects of CB1 receptor antagonists on the function of orexin A suggest an interplay between these two systems that may modulate appetite, feeding, and wakefulness.
Introduction
It is now widely recognized that many and indeed perhaps all members of the G protein-coupled receptor (GPCR)2 superfamily are able to form homodimers or homomultimers (1–4). There are also a substantial number of reports of heterointeractions between co-expressed pairs of GPCRs (5–9). Although the functional significance and consequences of a number of such pairings, including those between dopamine D1 and D2 (10–12) and μ- and δ-opioid (13–16) receptor subtypes, have been explored, the relevance of other pairings has been studied less extensively. With notable exceptions such as interactions between adenosine and dopamine receptor subtypes (17, 18), this is particularly true of pairings between GPCRs for which the endogenous agonist ligands are distinct. Despite this, a number of commentators have discussed the potential for such heteromers to respond to ligands in unique ways and to offer the potential as novel sets of drug targets (3, 19–22). In substantial part, this reflects a growing understanding of the mechanisms of G protein activation via GPCR dimers (23) and the appreciation that interactions between protomeric elements of GPCR homomers and heteromers must cause allosteric effects upon one another (23–26). The consequences of such allosteric effects are likely to be unique for different GPCR pairs and indeed potentially for different ligands. One means to explore such effects is via ligand binding studies (6, 27). Although potentially powerful, in a number of cases, this may be limited by the availability of suitable, particularly agonist radiolabeled probes. As such, measures of ligand function at GPCR multimers have been more widely explored. For example, the potency of the hallucinogenic serotonin 5-HT2A agonist 2,5-dimethoxy-4-iodoamphetamine to activate Gi family G proteins is increased more than 100-fold when the 5-HT2A receptor is co-expressed alongside and interacts with the metabotropic glutamate receptor 2 (mGluR2), and this is reversed upon co-addition of an mGluR2 agonist (28). A further pairing in which alterations in potency of agonist ligands has been observed following co-expression is the cannabinoid CB1 receptor and the orexin OX1 receptor (29, 30). Here, Hilairet et al. (29) observed that the potency of the peptide agonist orexin A to activate the mitogen-activated protein kinase pathway via the OX1 receptor was increased 100-fold in the presence of the CB1 receptor, whereas Ellis et al. (30) noted that the CB1 receptor antagonist/inverse agonist SR141716A caused a decrease in potency of orexin A to activate the mitogen-activated protein kinases ERK1/2 only in cells co-expressing the two receptors. There have also been a number of reports of coordinated trafficking of pairs of GPCRs by individual ligands that are expected only to occupy and activate one of the pair (31). These studies are at least consistent with the concept of GPCR heteromultimerization (5–9).
In recent times, the ability to monitor cell surface multimerization of GPCRs in intact cells has been enhanced greatly by the development of SNAP and CLIP tag variants of the enzyme O6-alkylguanine-DNA-alkyltransferase. These allow the covalent attachment of a variety of fluorophores and other probes to such modified receptors (32–36). This tagging approach has also proved valuable in monitoring ligand-induced receptor trafficking (37). Herein, we used a range of such approaches to confirm heteromultimerization between the CB1 receptor and the orexin OX1 receptor and show a substantially greater potency of orexin A to promote internalization of this heteromer compared with the OX1 receptor homomer. The concept that an endogenously produced peptide ligand has higher affinity/potency at a receptor heteromer than at the supposed primary monomeric/homomeric target is both novel and fascinating and provides new insights into both physiological signaling processes and the prospects for drug discovery.
EXPERIMENTAL PROCEDURES
Materials
Lipofectamine transfection reagent was from Invitrogen. SB334867 (N-(2-methyl-6-benzoxazolyl)-N′-1,5-naphthyridin-4-yl urea), SB408124 (N-(6,8-difluoro-2-methyl-4-quinolinyl)-N′-[4-(dimethylamino)phenyl]urea), CP55940 ((−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl)phenyl]-trans-4-(3-hydroxypropyl)cyclohexanol), AM251 (N-(piperidin-1-yl)-5-(4-iodophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide), and O2050 ((6aR,10aR)-3-(1-methanesulphonylamino-4-hexyn-6-yl)-6a,7,10,10a-tetrahydro-6,6,9-trimethyl-6H-dibenzo[b,d]pyran) were from Tocris Biosciences (Avonmouth, UK). Orexin A was from Bachem (UK) Ltd. (St. Helens, UK). WIN55212-2 mesylate ((R)-(+)-[2,3-dihydro-5-methyl-3-(4-morpholinylmethyl)pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl]-1-naphthalenylmethanone mesylate) was from Sigma-Aldrich. Oligonucleotides were from ThermoElectron (Ulm, Germany), and all materials for tissue culture were from Invitrogen with the exception of fetal bovine serum, which was from PAA Laboratories Ltd. (Yeovil, UK). [3H]SB674042 (1-(5-(2-fluorophenyl)-2-methylthiazol-4-yl)-1-((S)-2-(5-phenyl-(1,3,4)oxadiazol-2-ylmethyl)-pyrrolidin-1-yl)methanone) and [3H]SR141716A were from PerkinElmer Life Sciences. SR141716A (rimonabant; 5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-N-(piperidin-1-yl)-1H-pyrazole-3-carboxamide) was a gift from Stephen Rees, Screening and Compound Profiling, GlaxoSmithKline (Stevenage, UK). Antibodies to epitope tags were obtained from Insight Biotechnology Ltd. (Wembley, UK) (anti-VSV-G) and Roche Diagnostics (anti-HA). SNAP- and CLIP-specific labels were supplied by New England Biolabs (Hitchin, UK), and Tag-liteTM reagents were supplied by Cisbio Bioassays (Bagnols-sur-Cèze, France). All other reagents were obtained from Fisher Scientific or Sigma-Aldrich.
DNA Constructs
Constructs expressing the SNAP/CLIP and epitope-tagged receptors (VSV-G-SNAP-OX1, VSV-SNAP-CB1, HA-CLIP-OX1, and HA-CLIP-CB1) were made as described (36, 37).
Generation and Maintenance of stable Flp-InTM T-RExTM 293 Cells
To generate Flp-In T-REx 293 cells (36–40) able to inducibly express the VSV-G-SNAP-OX1/CB1 or HA-CLIP-OX1/CB1 constructs, cells were co-transfected with the plasmids pOG44 and pcDNA5/FRT/TO (Invitrogen) containing the desired cDNA at a ratio of 9:1 using Lipofectamine. After 48 h, the medium was supplemented with 200 μg·ml−1 hygromycin to select for stably transfected cells. Pools of cells were established and tested for inducible expression by the addition of 1 μg·ml−1 doxycycline for 48 h followed by screening for VSV-G-, HA-, or SNAP/CLIP-tagged protein expression by Western blotting. To constitutively and stably express a second receptor-tag combination in these cells, cDNA constructs based upon pSEMS1-26m (VSV-G-SNAP) or pCEMS1-CLIP10m (HA-CLIP) with either OX1 or CB1 were transfected into the inducible stable cell lines to make a double stable cell line with the “opposite” combination of receptor and tag. Thus, for example, cells inducibly expressing VSV-G-SNAP-OX1 were transfected with HA-CLIP-CB1 as the constitutive component and vice versa. After transfection, the cells were subjected to selection with 1 mg·ml−1 G418, and resistant colonies were picked, amplified, and screened first by Western blotting with anti-VSV-G and anti-HA antibodies and then staining with SNAP/CLIP fluorescent dyes that were visualized by microscopy.
Co-immunoprecipitation
The double stable clones were grown up in 75-cm2 flasks with and without 10 ng·ml−1 doxycycline induction for 24 h. Cells were harvested and resuspended in lysis buffer (150 mm NaCl, 0.01 mm Na3PO4, 2 mm EDTA, 0.5% n-dodecyl β-d-maltoside, and 5% glycerol plus protease inhibitor mixture tablets, pH 7.4) before incubation on a rotating wheel for 30 min at 4 °C. Samples were then centrifuged for 15 min at 10,000 × g at 4 °C, and the supernatant was transferred to fresh tubes and incubated with anti-VSV-G-agarose beads (Sigma-Aldrich) for 2 h at 4 °C on a rotating wheel. The samples were then washed five times with lysis buffer, and the bound receptors were eluted by the addition of 100 μl of SDS-PAGE sample buffer followed by heating at 65 °C for 5 min. The samples were subjected to SDS-PAGE analysis using 4–12% Bis-Tris gels (NuPAGE, Invitrogen) and MOPS buffer. After separation, the proteins were electrophoretically transferred to nitrocellulose membrane, which was then blocked (5% fat-free milk powder in PBS with 0.1% Tween 20 (PBS-Tween)) at 4 °C on a rotating shaker overnight. The membrane was incubated for 3 h with appropriate primary antibody (see figure legends) in 2% fat-free milk powder in PBS-Tween, washed (3 × 10 min with PBS-Tween), and then incubated for 3 h with appropriate secondary antibody (horseradish peroxidase (HRP)-linked sheep anti-mouse or goat anti-rat HRP; GE Healthcare) diluted 1:10,000 in 2% fat-free milk powder in PBS-Tween. After washing, proteins were detected by enhanced chemiluminescence (Pierce) according to the manufacturer's instructions.
Cell Membrane Preparation
Pellets of cells were frozen at −80 °C for a minimum of 1 h, thawed, and resuspended in ice-cold 10 mm Tris, 0.1 mm EDTA, pH 7.4 (TE buffer) supplemented with Complete protease inhibitor mixture (Roche Diagnostics). Cells were homogenized on ice by 40 strokes of a Teflon-glass homogenizer followed by centrifugation at 1000 × g for 5 min at 4 °C to remove unbroken cells and nuclei. The supernatant fraction was removed and passed through a 25-gauge needle 10 times before being transferred to ultracentrifuge tubes and subjected to centrifugation at 50,000 × g for 30 min at 4 °C. The resulting pellets were resuspended in ice-cold TE buffer. Protein concentration was assessed, and membranes were stored at −80 °C until required.
[3H]SB674042 Binding Assays
Saturation binding curves were established by the addition of 5 μg of membrane protein to assay buffer (25 mm HEPES, 0.5 mm EDTA, 2.5 mm MgCl2, pH 7.4 supplemented with 0.3% bovine serum albumin (BSA)) containing varying concentrations of [3H]SB674042 (37, 41) (0.4–20 nm). Nonspecific binding was determined in the presence of 3 μm SB408124. Reactions were incubated for 90 min at 25 °C, and bound ligand was separated from free ligand by vacuum filtration through GF/C filters (Brandel Inc., Gaithersburg, MD). The filters were washed twice with cold 1× PBS (120 mm NaCl, 25 mm KCl, 10 mm Na2HPO4, 3 mm KH2PO4, pH 7.4), and bound ligand was estimated by liquid scintillation spectrometry.
[3H]SR141716A Binding Assays
Saturation binding curves for [3H]SR141716A (37) were determined as in the previous section but with the following detailed differences. 30 μg of membrane protein was added to assay buffer composed of 50 mm Tris-HCl, 3 mm MgCl2, 1 mm EDTA, 0.3% BSA, pH 7.4 containing varying concentrations of [3H]SR141716A (0.5–14 nm). Nonspecific binding was determined by the addition of 10 μm AM251, and unbound ligand was separated by washing with cold 1× PBS supplemented with 0.1% poly(ethyleneimine).
Dual Fluorescent Labeling of Cells
Cells stably expressing the human OX1 receptor N-terminally tagged with CLIP or the human CB1 receptor N-terminally tagged with SNAP were grown on coverslips that had been sterilized and treated with 0.1 mg·ml−1 poly-d-lysine. The cells were rinsed with growth medium and then transferred into fresh culture medium containing CLIP-Surface 488 (5 μm) (New England Biolabs). The cells were then incubated in this medium at 37 °C for 30 min before being washed three times with growth medium and then transferred into fresh culture medium containing 2.5 μm SNAP-Surface 549 (New England Biolabs). After incubation at 37 °C for 30 min, the cells were washed three times with growth medium and then rinsed in Hanks' balanced salt solution (Invitrogen) prior to confocal imaging.
Confocal Imaging of Fluorescently Labeled Cells
A Zeiss 510 PASCAL Exciter laser-scanning confocal inverted microscope equipped with a 63× oil immersion Plan Fluor Apochromat objective lens (1.4 numerical aperture) was used to visualize cells labeled with CLIP-Surface 488 and SNAP-Surface 549. Using the appropriate laser lines and emission filters, a set of sequential images were acquired to determine the total fluorescence emission intensity associated with each cell prior to treatment with orexin A (1 μm) or vehicle. The effect of these reagents upon the distribution of receptors between the cell surface and cytoplasm was assessed by time lapse microscopy. Sequential images were acquired at 15-min intervals for a 45–60-min time period. Merged images for each time point were then created to determine the degree of receptor co-localization.
Quantitation of Distribution of Fluorescently Labeled Receptors
The level of background fluorescence in the CLIP-Surface 488 or SNAP-Surface 549 channel images was determined by drawing a region of interest adjacent to groups of fluorescently labeled cells. Background fluorescence was then subtracted from each pixel in each channel and segmented using Otsu's threshold algorithm. Segmented images were exported into AutoQuant and AutoDeblur/AutoVisualize software (version 9.6.3, Media Cybernetics, Inc., Silver Spring, MD). Using the AutoQuant image algebra module, each segmented image was duplicated, and binary masks were then generated by dividing each segmented image with its duplicate. The binary masks were then used to mask out the segmented regions from each channel image. The AutoQuant manual segmentation paintbrush tool, which allows the selection of pixels as it is moved across the raw image, was then used to distinguish regions of cell surface receptor fluorescence from receptor fluorescence located in the cytosol. This region was then displayed as a yellow mask over the grayscale raw image. A new image was then created in which all pixels of the raw image that are not part of the segmentation mask are set to zero and removed. This manual segmentation method was used to quantify the mean total fluorescence intensity values corresponding to SNAP-Surface 488- or SNAP-Surface 549-labeled receptors located at the membrane surface and within the cytoplasm of the cell. The total fluorescence pixel intensity from each region was expressed as a percentage of the total fluorescent SNAP-Surface 488 or SNAP-Surface 549 intensity. These values were exported into Prism 5.2 (GraphPad Software, San Diego, CA), and all data were expressed as the mean ± S.E. Measurements were made of six cells in each group.
Enzyme-linked Immunosorbent Assay
Flp-In T-REx 293 cells able to express appropriate tagged receptors were seeded into poly-d-lysine-coated, clear, 96-well tissue culture plates at a density of 50,000 cells/well. After incubation for 24 h, the medium was removed, replaced with 100 μl/well warmed primary antibody solution (normal medium with 1:1000 dilution of mouse anti-VSV-G), and incubated at 37 °C for 30 min. The primary antibody solution was removed, and the wells were washed with 100 μl/well warm DMEM HEPES (13.4 g·liter−1 Dulbecco's modified Eagle's medium-high glucose, 20 mm HEPES, pH 7.4, filter-sterilized). 100 μl/well warmed secondary antibody solution was added (1:2000 dilution sheep anti-mouse secondary antibody linked to horseradish peroxidase and 1:1000 dilution Hoechst stain (Hoechst 33342 trihydrochloride trihydrate, Molecular Probes, Eugene, OR) in normal medium). Following incubation at 37 °C for 30 min, the secondary antibody solution was removed, and the wells were washed with 2 × 100 μl/well warm PBS. During the second PBS wash, the Hoechst staining was measured at 460 nm using a Victor2 1420 multilabel counter (PerkinElmer Life Sciences). The PBS was completely removed and replaced with 100 μl/well warmed 3,3′,5,5′-tetramethylbenzine substrate (SureBlue ReserveTM TMB peroxidase substrate, Insight Biotechnology, Wembley, UK). After a 5-min incubation in the dark at room temperature, the absorption at 620 nm was measured, and these values were then corrected for cell number using the Hoechst staining values.
SNAP/CLIP-Lumi4Tb Binding Studies
Cells expressing combinations of the OX1 receptor containing the SNAP tag or the CB1 receptor tagged with CLIP were seeded at 100,000 cells/well in solid black 96-well plates (Greiner BioOne) that had been treated with 0.1 mg·ml−1 poly-d-lysine. Following overnight growth, the cells were subjected to the required ligand treatments. The growth medium was replaced with 50 μl of either 10 nm Tag-lite SNAP-Lumi4Tb or 20 nm CLIP-Lumi4Tb in 1× labeling medium (all from Cisbio Bioassays). Plates were incubated for 1 h (or 30 min in the case of postligand treatment to allow comparison with the ELISA measurements) at 37 °C in 5% CO2 in a humidified atmosphere. The plates were subsequently washed four times in 100 μl/well labeling medium, and a final 100 μl/well labeling medium was added. After excitation at 337 nm, emission at 620 nm was determined using a PHERAstar FS homogeneous time-resolved fluorescence-compatible reader (BMG Labtechnologies, Offenburg, Germany) (37).
Homogenous Time-resolved Fluorescence Resonance Energy Transfer (htrFRET) Studies
Clone B6 double stable cells (VSV-G-SNAP-OX1 (inducible)/HA-CLIP-CB1 (constitutive)) (Fig. 1) were seeded at 100,000 cells/well in solid black 96-well plates (Greiner BioOne) that had been treated with 0.1 mg·ml−1 poly-d-lysine. The growth medium was replaced with 50 μl of a mixture containing the fixed optimal concentrations of donor alone (Tag-lite SNAP-Lumi4Tb or CLIP-Lumi4Tb), acceptor alone (Tag-lite SNAP-Red or CLIP-Red), or optimized mixtures of donor and acceptor in 1× labeling medium (all from Cisbio Bioassays). Plates were incubated for 1 h at 37 °C in 5% CO2 in a humidified atmosphere and subsequently washed four times in labeling medium. 100 μl of labeling medium was added to each well, and the plates were read using a PHERAstar FS homogeneous time-resolved fluorescence-compatible reader (BMG Labtechnologies). The emission signal from the Tag-lite SNAP/CLIP-Lumi4Tb cryptate (620 nm) and the htrFRET signal resulting from the acceptor Tag-lite SNAP/CLIP-Red (665 nm) were recorded (36).
FIGURE 1.
Organization of receptor expression in clonal cell lines. In all the clonal cell lines used in the studies reported, one GPCR is expressed constitutively, whereas expression of the second can be induced by addition of doxycycline. In the schematics, the GPCR that is constitutively expressed is shown in white, whereas the inducible component is shown in black. In A, a specific clone nomenclature (A8, C2, B6, and D10) is provided that is then used consistently throughout. In B and in subsequent figures, the GPCR that is being measured in individual assays is highlighted with a black arrow. In C specifically and in Fig. 7, the black and white arrows define studies on the detection of heteromers (where arrows identify different receptors) and homomers (where arrows identify the same receptor). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
Biotinylation Protection Assay
Cells were grown and induced overnight with doxycycline in 6-well plates treated with 0.1 mg·ml−1 poly-d-lysine. The plates were put on ice, the medium was removed, and the cells were washed with 2 ml/well ice-cold PBS. 1 ml/well 0.3 mg·ml−1 sulfo-NHS-SS-biotin (sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithiopropionate; Pierce, Thermo Scientific) in PBS was added, and the plates were incubated on ice in the dark for 30 min. The biotin was removed, the cells were washed as above, 1 ml of prewarmed cell growth medium/well was added, and the plates were incubated at 37 °C in 5% CO2 for 15 min. Ligand treatments were performed by adding 1 ml/well 2× concentrated ligand or 1 ml of unsupplemented medium as appropriate before incubation at 37 °C in 5% CO2 for 1 h. The plates were placed on ice, the medium was removed, and the cells were washed as above. 2 ml/well stripping solution (50 mm glutathione, 0.3 m NaCl, 75 mm NaOH, 1% fetal bovine serum) was added followed by incubation on ice in the dark for 30 min. The stripping solution was removed, and the cells were washed with 2 × 2 ml of cold PBS to ensure complete removal of the stripping solution prior to cell lysis. Cells were lysed by the addition of 400 μl of ice-cold radioimmunoprecipitation assay buffer (50 mm HEPES, 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 10 mm NaF, 5 mm EDTA, 10 mm NaH2PO4, 5% ethylene glycol, pH 7.4) supplemented with Complete protease inhibitor mixture (Roche Diagnostics), scraped from the plate, and transferred to Eppendorf tubes. The lysates were incubated on a rotating wheel at 4 °C for 15 min and then spun at 18,000 × g for 10 min at 4 °C, and the protein concentrations of the supernatants were determined and equalized (lysate samples were taken for later analysis). 100 μl/tube streptavidin-agarose resin (Pierce, Thermo Scientific) was added, and tubes were incubated on a rotating wheel at 4 °C overnight before being spun at 2000 × g for 2 min at 4 °C. The supernatant was removed, and the resin was washed with 3 × 500 μl of radioimmunoprecipitation assay buffer. 100 μl of SDS-PAGE sample buffer containing 5% 2-mecaptoethanol was added, and tubes were incubated at 37 °C for 1 h. The cell lysates and biotinylated samples were subjected to SDS-PAGE analysis and Western blotting as above.
Data Analysis
Data were quantified, grouped, and analyzed using Prism 5.2 (GraphPad Software). Data are expressed as means ± S.E. Statistical analysis was carried out by one-way analysis of variance.
RESULTS
Orexin A Causes Internalization of Cannabinoid CB1 Receptor but Only When Orexin OX1 Receptor Is Co-expressed
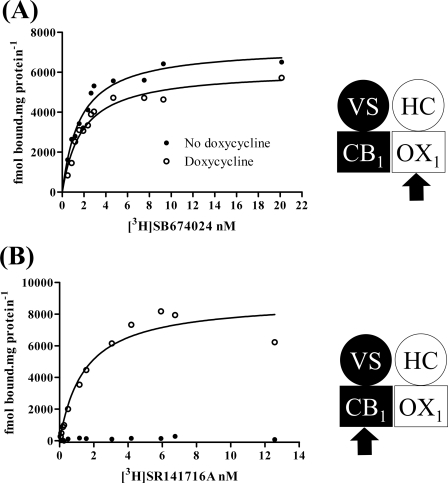
A form of the cannabinoid CB1 receptor N-terminally tagged with both the VSV-G epitope tag and the SNAP variant of O6-alkylguanine-DNA-alkyltransferase (37) was cloned into the Flp-In locus of Flp-In T-REx 293 cells. Populations of cells harboring this construct were isolated as described previously (37). Flp-In T-REx 293 cells allow the production of protein from DNA located at the Flp-In locus upon addition of the antibiotic tetracycline or its analog doxycycline (36–40). These cells were further transfected with a form of the orexin OX1 receptor N-terminally tagged with a combination of the HA epitope tag and the CLIP variant of O6-alkylguanine-DNA-alkyltransferase (37). Individual clones constitutively expressing HA-CLIP-OX1 were then isolated by imaging fluorescence following addition of the CLIP tag-specific dye substrate CLIP-505 (37). Clone B6 is an example of such a cell line (Fig. 1). Both with and without induction of expression of VSV-G-SNAP-CB1, membranes isolated from these cells bound the highly selective OX1 receptor antagonist [3H]SB674024 with high affinity (Fig. 2A and Table 1) (Kd = 1.62 ± 0.39 and 1.72 ± 0.17 nm with and without doxycycline addition, respectively). Neither the affinity nor the extent of specific binding of [3H]SB674024 was altered significantly following doxycycline-induced expression of VSV-G-SNAP-CB1 (Fig. 2A and Table 1). However, although specific binding of the CB1-specific antagonist [3H]SR141716 was absent in membranes of uninduced cells (Fig. 2B and Table 1), this ligand bound with high affinity (Kd = 1.54 ± 0.28 nm) to membranes of doxycycline-induced cells (Fig. 2B and Table 1).
FIGURE 2.
CB1 receptor expression does not alter expression levels of OX1 receptor or its affinity to bind [3H]SB674024. Clone B6 (see Fig. 1) Flp-In T-REx 293 cells that constitutively express HA-CLIP-OX1 and harbor VSV-G-SNAP-CB1 at the Flp-In locus were untreated (filled symbols) or treated with doxycycline (10 ng·ml−1 for 24 h) (open symbols). Membranes prepared from these cells were used in saturation [3H]SB674024 (OX1 receptor antagonist) (A) or [3H]SR141716A (CB1 receptor antagonist) (B) binding studies. Representative experiments of n = 3 are shown. VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
TABLE 1.
Ligand binding data for cell lines used
The data are presented as mean values of three independent experiments ±S.E. A8, B6, C2, and D10 refer to the clones used and are described in Fig. 1. Induction with 10 ng·ml−1 doxycycline for 24 h is present (+) or absent (−) as indicated.
| [3H]SB674042 (OX1 receptor) |
[3H]SR141716A (CB1 receptor) |
|||
|---|---|---|---|---|
| Bmax | Kd | Bmax | Kd | |
| pmol·mg protein−1 | nm | pmol·mg protein−1 | nm | |
| A8 − | Not detected | 1.52 ± 0.11 | 1.13 ± 0.25 | |
| A8 + | 7.32 ± 0.71 | 1.70 ± 0.15 | 1.61 ± 0.97 | 1.44 ± 0.02 |
| B6 − | 7.05 ± 0.60 | 1.62 ± 0.39 | Not detected | |
| B6 + | 6.37 ± 0.38 | 1.72 ± 0.17 | 9.54 ± 2.70 | 1.54 ± 0.28 |
| C2 − | Not detected | 3.70 ± 0.88 | 1.30 ± 0.44 | |
| C2 + | 8.65 ± 1.07 | 1.59 ± 0.19 | 3.76 ± 0.77 | 1.49 ± 0.32 |
| D10 − | 5.25 ± 0.88 | 1.76 ± 0.04 | Not detected | |
| D10 + | 5.05 ± 1.00 | 1.88 ± 0.46 | 1.51 ± 0.10 | 1.12 ± 0.35 |
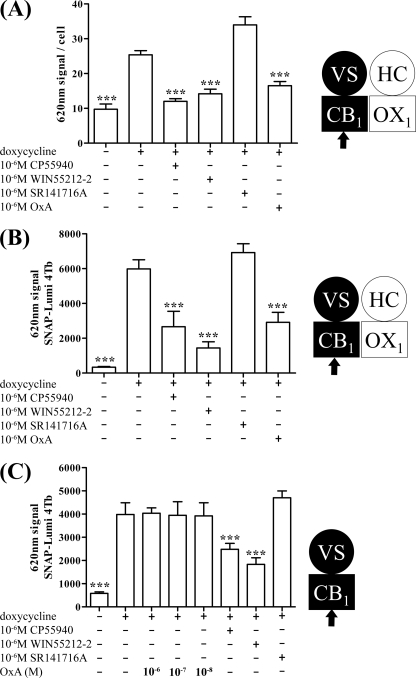
Ligand binding studies performed on cell membrane preparations do not provide direct information on the cellular location of the receptors of interest. To address this, following induction of VSV-G-SNAP-CB1 expression by these cells, substantial binding of both an anti-VSV-G antibody (Fig. 3A) and SNAP-Lumi4Tb (Fig. 3B) was detected on intact cells. Because the N-terminal domain of plasma membrane-localized GPCRs is extracellular, these results demonstrated the presence at steady state of a population of cell surface VSV-G-SNAP-CB1. For both labels, binding to uninduced cells was much lower (Fig. 3, A and B) with the signal to background ratio of induced versus uninduced cells being substantially greater when using SNAP-Lumi4Tb (Fig. 3, compare A with B). As anticipated from previous studies (37), addition for 40 min of either of the CB1 receptor agonists CP55940 and WIN55212-2 (10−6 m) prior to addition of anti-VSV-G (Fig. 3A) or SNAP-Lumi4Tb (Fig. 3B) resulted in a substantial reduction in cell labeling by these reagents. This is consistent with enhanced internalization of the CB1 receptor construct by these agonists. By contrast, over a 40-min time period, the CB1 receptor antagonist SR141716A had no statistically significant effect on detection of the receptor by either anti-VSV-G (Fig. 3A) or SNAP-Lumi4Tb (Fig. 3B), although there was a trend toward increased receptor detection. Unexpectedly, however, treatment of these cells with 10−7 m orexin A (an OX1 receptor agonist with no direct affinity for the CB1 receptor; see Ref. 30) also resulted in a large reduction in levels of anti-VSV-G (Fig. 3A) and SNAP-Lumi4Tb (Fig. 3B) labeling. This effect required the presence of the OX1 receptor because in cells able to induce expression of VSV-G-SNAP-CB1 but in which HA-CLIP-OX1 was not expressed equivalent treatment with orexin A was completely unable to modulate SNAP-Lumi4Tb binding (Fig. 3C). This was despite both CP55940 and WIN55212-2 being able to reduce SNAP-Lumi4Tb binding in these cells in a fashion consistent with enhanced CB1 receptor internalization (Fig. 3C).
FIGURE 3.
Both CB1 receptor agonists and orexin A produce internalization of CB1 receptor: effect of orexin A requires OX1 receptor. Cells of clone B6 (A and B) were uninduced (− doxycycline) or induced to express VSV-G-SNAP-CB1 (+ doxycycline). These were challenged with the identified ligands (CP55940 and WIN55212-2 are CB1 receptor agonists, whereas SR141716A is an antagonist at this receptor. Orexin A (OxA) is an agonist of the OX1 receptor) for 40 min. Cell surface VSV-G-SNAP-CB1 was detected via anti-VSV-G ELISA (A) or the binding of SNAP-Lumi4Tb (B). C, similar studies were performed on cells able to express inducible VSV-G-SNAP-CB1 but that lack expression of HA-CLIP-OX1, and cell surface VSV-G-SNAP-CB1 was detected by the binding of SNAP-Lumi4Tb. ***, p < 0.001 (different from doxycycline). Combined data from n = 3 experiments (means ± S.E.) are shown. VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
Effects of Orexin A on Cannabinoid CB1 Receptor Are Produced No Matter Which Receptor Is Inducible and Which Is Expressed Constitutively
We next assessed whether this effect of orexin A would also be observed in cells in which the CB1 receptor was present constitutively, whereas the OX1 receptor could be induced on demand. In clone C2 (Fig. 1), HA-CLIP-OX1 is located at the Flp-In locus. In these cells, specific and high affinity (Kd = 1.59 ± 0.19 nm) binding of [3H]SB674024 was only observed following treatment with doxycycline (supplemental Fig. 1A and Table 1). By contrast, these cells, which were clonally selected for inducible expression of HA-CLIP-OX1 and constitutive expression of VSV-G-SNAP-CB1 (Fig. 1), displayed specific, high affinity binding of [3H]SR141716 in the absence of doxycycline (Kd = 1.30 ± 0.44 nm), and this was unchanged (Kd = 1.49 ± 0.32 nm) by induction of HA-CLIP-OX1 (supplemental Fig. 1B and Table 1). As expected in this configuration, binding of SNAP-Lumi4Tb was high in the absence of doxycycline treatment and was unaffected by the addition of orexin A (supplemental Fig. 1C). Furthermore, steady-state binding of SNAP-Lumi4Tb was not affected by doxycycline induction of HA-CLIP-OX1 (supplemental Fig. 1C). However, after induction of HA-CLIP-OX1 expression, addition of orexin A was as effective as CP55940 or WIN55212-2 in reducing cell surface binding of SNAP-Lumi4Tb (supplemental Fig. 1C).
To ensure that the basic observations were not related in some way to the identity of the N-terminal tags added to the CB1 receptor, we reversed the tags and organization of the receptor pairing. In clone D10 (Fig. 1), VSV-G-SNAP-OX1 was expressed constitutively, whereas HA-CLIP-CB1 could be induced by addition of doxycycline, and in clone A8 (Fig. 1), HA-CLIP-CB1 was expressed constitutively, whereas VSV-G-SNAP-OX1 could be induced. This was again confirmed via saturation binding experiments with the selective 3H-antagonists (supplemental Fig. 2A and Table 1). As before, in both these cases, the CB1 agonists CP55940 and WIN55212-2 promoted reduction in cell surface CB1 receptor levels in this case measured by the binding of CLIP-Lumi4Tb (supplemental Fig. 2B). By contrast, orexin A only caused reduction in CLIP-Lumi4Tb binding and therefore enhanced internalization of the CB1 receptor when both the CB1 receptor and the OX1 receptor construct were present (supplemental Fig. 2B).
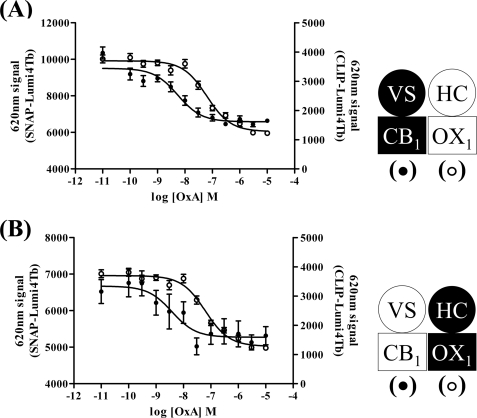
Orexin A Is More Potent in Causing Internalization of CB1 Receptor than OX1 Receptor
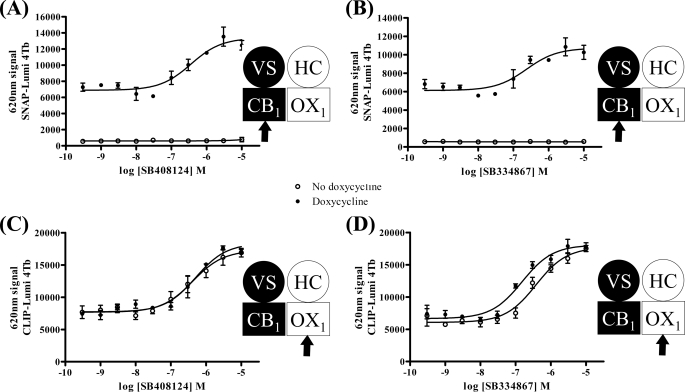
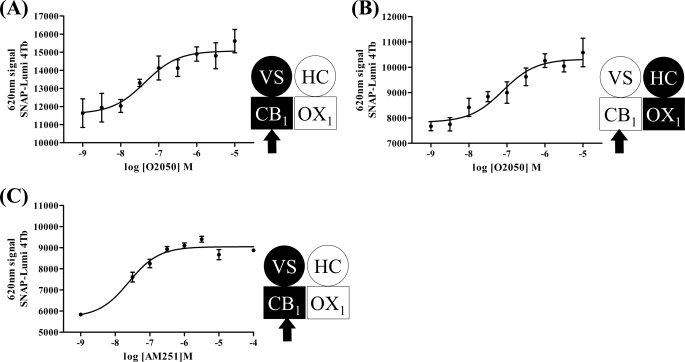
The ability of orexin A to promote internalization of forms of the CB1 receptor was concentration-dependent. Using clones B6 (Fig. 4A) and C2 (Fig. 4B) as exemplars, the pEC50 values for orexin A were 8.26 ± 0.13 and 8.42 ± 0.35, respectively. Remarkably, this was ∼10-fold more potent (B6, p < 0.001; C2, p < 0.0025) than the ability of orexin A to cause internalization of the OX1 receptor (clone B6, pEC50 = 7.20 ± 0.07; clone C2, pEC50 = 7.38 ± 0.13) (Fig. 4, A and B). Furthermore, the ability of 5 × 10−7 m orexin A to cause internalization of the CB1 receptor was blocked in a concentration-dependent manner by the OX1 receptor antagonists SB408124 (Fig. 5A) and SB334867 (Fig. 5B). The potency (SB408124, pIC50 = 6.45 ± 0.21; SB334867, pIC50 = 6.63 ± 0.25) of these two antagonists to prevent this effect of orexin A, however, was the same as their potency to block orexin A-mediated internalization of the OX1 receptor (SB408124, pIC50 = 6.38 ± 0.09 and 6.3 ± 0.13, minus and plus doxycycline, respectively; SB334867, pIC50 = 6.42 ± 0.11 and 6.82 ± 0.12, minus and plus doxycycline, respectively) (Fig. 5, C and D). Potentially even more interestingly, orexin A-mediated internalization of the CB1 receptor constructs was blocked in a concentration-dependent fashion by the CB1 receptor antagonist O2050 (Fig. 6). This was true in both cells in which expression of VSV-G-SNAP-CB1 was induced in the presence of HA-CLIP-OX1 (clone B6, pIC50 = 7.4 ± 0.18) (Fig. 6A) or in which expression of HA-CLIP-OX1 was induced in the face of constitutive expression of VSV-G-SNAP-CB1 (clone C2, pIC50 = 7.1 ± 0.2) (Fig. 6B). Importantly, this effect was not restricted to a single CB1 receptor antagonist: a second antagonist, AM251, also inhibited the effect of orexin A in a concentration-dependent fashion (clone B6, pIC50 = 7.6 ± 0.11) (Fig. 6C).
FIGURE 4.
Orexin A is more potent in causing internalization of CB1 receptor than OX1 receptor. Cells of clones B6 (A) and C2 (B) were induced to express the harbored receptor by treatment with doxycycline. The ability of different concentrations of orexin A to cause internalization of CB1 (filled symbols) and OX1 receptors (open symbols) was assessed in parallel by measuring binding of either SNAP-Lumi4Tb or CLIP-Lumi4Tb to intact cells. In both cases, orexin A (OxA) was more potent in producing internalization of the CB1 receptor than the OX1 receptor. Data are means ± S.E. (n = 3 in each case). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
FIGURE 5.
OX1 receptor antagonists block effects of orexin A at both OX1 and CB1 receptors. Clone B6 was untreated (open symbols) or treated with doxycycline (10 ng·ml−1 for 24 h) (filled symbols) to induce expression of VSV-G-SNAP-CB1. Cell surface VSV-G-SNAP-CB1 (A and B) and HA-CLIP-OX1 (C and D) were measured, respectively, by the binding of SNAP-Lumi4Tb (A and B) and CLIP-Lumi4Tb (C and D) after treatment of cells for 40 min with 5 × 10−7 m orexin A and varying concentrations of the OX1 receptor antagonists SB408124 (A and C) and SB334867 (B and D). Data are means ± S.E. (n = 3). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
FIGURE 6.
Orexin A-induced CB1 receptor internalization is blocked by CB1 receptor antagonists. Clone B6 (A and C) or clone C2 (B) cells were induced to express the harbored receptor (VSV-G-SNAP-CB1 (A and C) or HA-CLIP-OX1 (B)). The effects of varying concentrations of the CB1 antagonists O-2050 (A and B) and AM251 (C) to modulate internalization of the CB1 receptor mediated by treatment with 1 × 10−6 m orexin A for 40 min were assessed by the binding of SNAP-Lumi4Tb. Data are means ± S.E. (n = 3). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
Identification of CB1-OX1 Receptor Heteromers
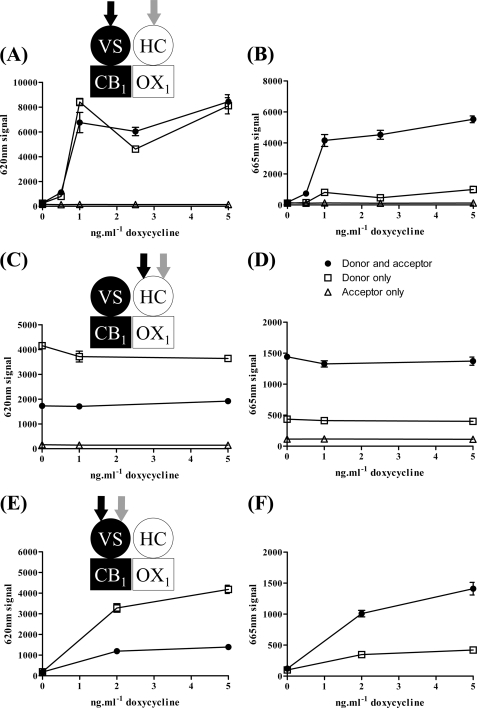
All of the above observations are at least consistent with the potential of the CB1 and OX1 receptors to exist within heterodimers or heteromultimers. To assess this more directly, we used htrFRET based on the potential for resonance energy transfer between energy donors and acceptors linked to SNAP- and CLIP-tagged substrates that were added individually or co-added to cells (clone B6) in which CB1 receptor levels could be regulated. Initial optimization studies determined the most appropriate energy donor (SNAP-Lumi4Tb) to energy acceptor (CLIP-Red) ratio to detect the presence of CB1-OX1 heteromers (supplemental Fig. 3). These cells were uninduced or induced with varying concentrations of doxycycline and then labeled with donor + acceptor, donor alone, or acceptor alone, and emission at 620 nm (donor binding) (Fig. 7A) and 665 nm (htrFRET signal) (Fig. 7B) was determined after excitation at 337 nm. Although binding of donor and htrFRET signal were absent without induction of VSV-G-SNAP-CB1 (zero doxycycline), substantial signal of each was detected at all concentrations of doxycycline above 0.5 ng·ml−1 (Fig. 7, A and B). Such observations indicate that a proportion of the cell surface OX1 and CB1 receptors are in proximity sufficient to be considered a heterodimeric/heteromultimeric complex. In parallel, we also assessed the potential presence of both CB1-CB1 homomers and OX1-OX1 homomers. To detect OX1-OX1 interactions, cells were labeled with combinations of CLIP-Lumi4Tb and CLIP-Red (Fig. 7, C and D). Because the CLIP-tagged OX1 receptor is expressed constitutively in these cells, emission at 620 nm, representing binding of CLIP-Lumi4Tb to the receptor, was present even without doxycycline treatment and was unaffected by addition of the antibiotic (Fig. 7C). When added alone, binding of CLIP-Lumi4Tb was higher than when CLIP-Red was also added concurrently because CLIP-Red competes with CLIP-Lumi4Tb for the receptor (Fig. 7C). The combination of CLIP-Lumi4Tb and CLIP-Red also generated htrFRET emission at 665 nm consistent with the presence of OX1-OX1 homomers (Fig. 7D). Equivalent studies using combinations of SNAP-Lumi4Tb and SNAP-Red allowed detection of interactions consistent with the presence of CB1-CB1 homomers also (Fig. 7, E and F). However, as anticipated, binding of both SNAP-Lumi4Tb and htrFRET signal was absent without prior doxycycline treatment of the cells (Fig. 7, D and F).
FIGURE 7.
htrFRET detects each of cell surface CB1-OX1 receptor heteromultimers, CB1-CB1 homomers, and OX1-OX1 homomers. Cells of clone B6 were untreated or treated for 24 h with varying concentrations of doxycycline to induce VSV-G-SNAP-CB1 expression. A and B, SNAP-Lumi4Tb (donor alone; open squares) (to label VSV-G-SNAP-CB1), CLIP-Red (acceptor alone; open triangles) (to label HA-CLIP-OX1), or an optimal mixture (10 nm SNAP-Lumi4Tb and 350 nm CLIP-Red) (see supplemental Fig. 3) of SNAP-Lumi4Tb and CLIP-Red (filled circles) (to allow detection of potential CB1-OX1 heteromers) was added. In A, the signal at 620 nm (as a measure of VSV-G-SNAP-CB1 expression) was assessed. In B, the signal at 665 nm (as a measure of heteromer detection) was assessed. C–F, equivalent studies explored OX1-OX1 interactions (C and D) via separate addition of CLIP-Lumi4Tb (open squares) or CLIP-Red (open triangles) or their co-addition (filled circles) and CB1-CB1 interactions (E and F) via separate addition of SNAP-Lumi4Tb (open squares) or co-addition of SNAP-Lumi4Tb and SNAP-Red (filled circles). Data are means ± S.E. (n = 3). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
To extend the studies on CB1-OX1 heteromers, we performed a series of co-immunoprecipitation experiments. B6 cells were maintained with and without doxycycline and lysed, and samples were precipitated with anti-VSV-G antibody-coated agarose beads. Bound proteins were separated by SDS-PAGE, transferred, and probed with anti-HA antibody (Fig. 8). Anti-HA immunoreactivity corresponding to HA-CLIP-OX1 was only detected in anti-VSV-G immunoprecipitates from cells induced with doxycycline. Similar studies were then performed with the three other cell lines described above (A8, C2, and D10) (Fig. 8). In each case, anti-HA immunoreactivity was present only in the anti-VSV-G immunoprecipitates from doxycycline-treated cells with HA-CLIP-OX1 migrating somewhat further in such gels than HA-CLIP-CB1 (Fig. 8).
FIGURE 8.

Co-expression of CB1 and OX1 receptors allows their co-immunoprecipitation. Clones A8, B6, C2, and D10 were untreated (−) or induced (+) with doxycycline. Lysates from these cells were immunoprecipitated with anti-VSV-G. Subsequently, these samples were resolved by SDS-PAGE and immunoblotted (IB) with anti-HA. In all cases, co-expression of the CB1 and OX1 receptors was both required and sufficient to allow co-immunoprecipitation (CO-IP). Representative experiments of n = 3 are shown. VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
Orexin A Produces Internalization of CB1-OX1 Receptor Heteromers
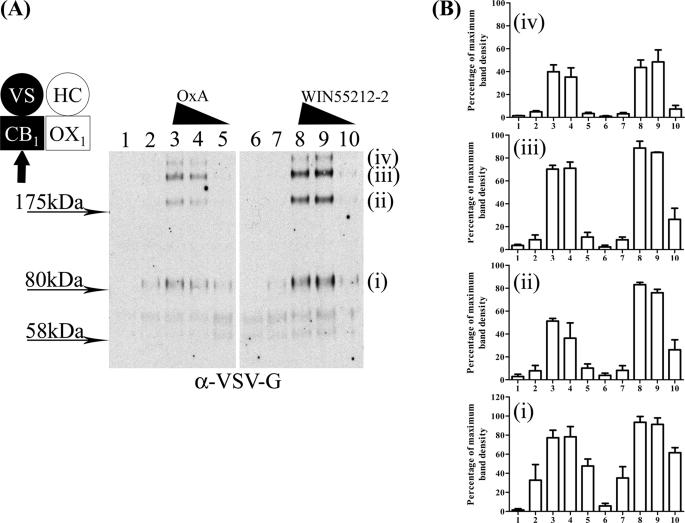
To further explore the consequences of heterointeractions between the CB1 and OX1 receptors, we performed a series of “biotinylation protection” assays. In such experiments, proteins at the cell surface are labeled with biotin, and those that are internalized over the period of the assay are “protected” from subsequent cleavage of the biotin label (42). In B6 cells that were induced to express VSV-G-SNAP-CB1, treatment with WIN55212-2 resulted in the appearance of protected forms of VSV-G-SNAP-CB1 identified following SDS-PAGE separation, and this effect of WIN55212-2 was concentration-dependent (Fig. 9A). As well as a protected form of VSV-G-SNAP-CB1 migrating at a position consistent with the resolved monomeric CB1 receptor construct (calculated molecular mass of 76.6 kDa), a series of anti-VSV-G-reactive species were also observed to migrate with lower mobility (Fig. 9A), and these presumably reflect incompletely dissociated multimeric receptor complexes. Equivalent studies using orexin A again resulted in the protection and therefore internalization of biotinylated forms of the CB1 receptor in a manner dependent upon the concentration of orexin A used (Fig. 9A). Again, a substantial fraction of the anti-VSV-G immunoreactivity migrated as species with lower mobility than predicted for a monomer of the CB1 receptor (Fig. 9). The combined quantification for the major species is shown in Fig. 9B.
FIGURE 9.
Orexin A-induced internalization of CB1-OX1 heteromer. Clone B6 cells were uninduced (lanes 1 and 6) or induced with doxycycline (10 ng·ml−1 for 24 h) (all other lanes). Following cell surface biotinylation, cells were treated with vehicle (lanes 2 and 7) or either orexin A (OxA) (lanes 3–5) or WIN55212-2 (lanes 8–10) (10−6 m (lanes 3 and 8), 10−7 m (lanes 4 and 9), or 10−8 m (lanes 5 and 10)) for 40 min. Biotin was cleaved from proteins remaining at the cell surface, and the internalized and therefore protected CB1 receptor was detected following SDS-PAGE. A representative experiment of n = 3 is shown in A. Individual blots were scanned, and polypeptides were quantified and normalized. Combined results are displayed as means ± S.E. in B. VS, VSV-G + SNAP tag; HC, HA + CLIP tag. Polypeptides with apparent mass labeled (i–iv) in A were quantitated in B across lanes 1–10.
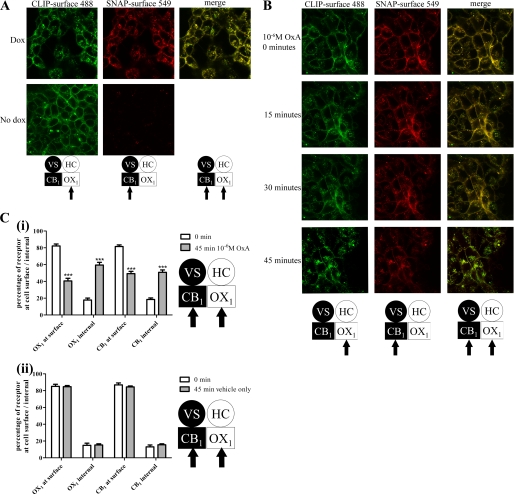
The ability to selectively link various cell-impermeant fluorophores to the SNAP and CLIP tags in a covalent fashion offered an alternate means to visualize the presence of the two receptors and their co-regulation in intact cells. In the absence of doxycycline, addition of CLIP-Surface 488 to clone B6 cells allowed identification of HA-CLIP-OX1 (Fig. 10A), whereas as anticipated, SNAP-Surface 549 did not label the cells in a specific manner (Fig. 10A). However, following doxycycline-induced expression of VSV-G-SNAP-CB1, both CLIP-Surface 488 and SNAP-Surface 549 labeled the surface of cells effectively, and merging of such images indicated strong overlap of the signals (Fig. 10A). Most noticeably in this situation, however, a proportion of both the CLIP-Surface 488 and SNAP-Surface 549 labels was now located in punctate intracellular but non-nuclear locations (Fig. 10A). Again, merging of such images indicated strong overlap at these locations. As these fluorophores are cell-impermeant, this must indicate that both the VSV-G-SNAP-CB1 and HA-CLIP-OX1 were labeled at the cell surface but subsequently became internalized (Fig. 10A). Although small numbers of individual “red” and “green” pixels could be observed in the merged images and may represent non-co-localized VSV-G-SNAP-CB1 and HA-CLIP-OX1 receptors, this may also reflect a degree of nonspecific staining of the CLIP-Surface 488 and SNAP-Surface 549 fluorophores as small patches of SNAP-Surface 549 in cells lacking expression of VSV-G-SNAP-CB1 and as larger, apparently random agglomerations of CLIP-Surface 488 staining in cell images could be observed (Fig. 10A). Following addition of orexin A, the extent of the intracellular location of both the CLIP-Surface 488 and SNAP-Surface 549 labels increased in a time-dependent manner (Fig. 10B), and once again, merging of such images indicated strong co-localization of the internalized labels and their associated receptors (Fig. 10B). This effect required orexin A because addition of vehicle did not result in a time-dependent increase in intracellular levels of CLIP-Surface 488 and/or SNAP-Surface 549. Quantification of such images demonstrated that the percentage of intracellular HA-CLIP-OX1 increased from 17.8 ± 2.3 to 59.5 ± 3.3% over a 45-min exposure to 10−6 m orexin A with intracellular VSV-G-SNAP-CB1 increasing from 18.4 ± 1.9 to 50.8 ± 2.7% (Fig. 10C).
FIGURE 10.
Imaging co-localization of internalized CB1 and OX1 receptors. A, clone B6 cells that were either uninduced (No dox) or induced with doxycycline for 24 h (Dox) were imaged confocally following addition of CLIP-Surface 488 (green) and SNAP-Surface 549 (red). A “merge” of the two images is also shown for the doxycycline-induced cells. B, doxycycline-treated cells as in A were treated with orexin A (OxA) (10−6 m), and images were taken at times between 0 and 45 min. C, experiments akin to B (i) or cells treated with vehicle (ii) were quantified. Data represent means ± S.E. for six individual cells. ***, p < 0.001 (different between 45 and 0 min). VS, VSV-G + SNAP tag; HC, HA + CLIP tag.
DISCUSSION
Previous studies have either inferred physical interactions between CB1 and OX1 receptors based on alterations in function of receptor selective agonists in cells transfected to co-express this pair of GPCRs (29) or used FRET between C-terminally cyan fluorescent protein- and YFP-tagged variants to detect such interactions in intracellular structures that may represent recycling endocytic vesicles (30). A further set of observations consistent with the presence of CB1-OX1 heteromers is that selective antagonists for either receptor are able to traffic both receptors to the cell surface from an intracellular location (30). However, although consistent with such an interaction, much of the evidence for the existence of a CB1-OX1 heteromer has been indirect and/or has not addressed whether such a complex is present at the surface of cells. In recent times, the introduction of SNAP or CLIP tags into the N-terminal domain of GPCRs has become an effective way of detecting cell surface receptors because the N-terminal region is exposed to the extracellular environment, and the SNAP and CLIP tags can be labeled covalently with a variety of cell-impermeant fluorophores or other reagents (32–37). As such, cell surface receptors can be labeled and imaged, and the effects of ligands on the cellular location of the receptor can be monitored by following the location of the fluorophore because the ligand binding site of the receptor is not altered by this procedure (33, 37) unlike when using irreversible receptor ligands or those with high affinity and very slow dissociation rates (43). Although we have previously used this approach to monitor individually cell surface location and agonist-induced internalization of both the CB1 receptor and the OX1 receptor (37), the most extensive use of SNAP tagging and related technologies has been to covalently label cell surface receptors with pairs of htrFRET-competent donors and acceptors to detect protein-protein interactions at the cell surface (32, 33). In studies on heteromeric interactions, separate SNAP and CLIP tagging of a pair of proteins allows the concurrent addition of distinct FRET acceptors/donors that label the two proteins differentially and the subsequent detection of htrFRET if the proteins are located within FRET-compliant distances of one another (32). To assess the presence of CB1-OX1 heteromers, we generated sets of cell lines in which either a SNAP-tagged CB1 or OX1 receptor was expressed constitutively, whereas the complementary CLIP-tagged receptor was harbored at a doxycycline-inducible locus. In cells in which the SNAP-tagged receptor was at the inducible locus, no binding of the FRET donor SNAP-Lumi4Tb was detected until the receptor was induced. However, when induced and in the presence of the FRET acceptor CLIP-Red, the htrFRET signal reported the presence of the CB1-OX1 heteromer at the cell surface. As a more conventional approach, we also took advantage of the presence of HA and VSV-G epitope tags that were also engineered into the N-terminal domain of each receptor to show that no matter which receptor was SNAP- or CLIP-tagged or which receptor was at the inducible locus co-immunoprecipitation was achieved after addition of doxycycline to cells to cause receptor co-expression.
A further use of the SNAP/CLIP tags is to monitor receptor internalization. Only a cell surface receptor is available to be labeled with SNAP/CLIP-Lumi4Tb, and in cells expressing only SNAP-CB1, CB1 agonists substantially reduced levels of SNAP-Lumi4Tb binding over short time periods. As this construct also contained the VSV-G epitope tag, a similar effect of CB1 agonists was observed in intact cell ELISA studies using a VSV-G antibody. However, the signal to background ratio in studies using SNAP-Lumi4Tb binding was far superior to ELISA; therefore, SNAP-Lumi4Tb was used routinely in these studies. Although initially unexpected, in cells co-expressing both CB1 and OX1, maximally effective concentrations of orexin A were as effective at producing internalization of the CB1 receptor as CB1 receptor agonists. This is consistent with the CB1-OX1 heteromer being a stable complex and binding of only orexin A causing the entire complex to be internalized. Importantly, when exploring the potency of orexin A to produce this effect, we noted in different cell lines that orexin A was some 10-fold more potent in producing internalization of the CB1 receptor than in causing internalization of the co-expressed OX1 receptor. This may seem contrary to the concept of internalization of a stable CB1-OX1 heteromer, but it may be anticipated that only a proportion of the expressed OX1 receptor is within such a heteromeric complex. As such, the most obvious but also most interesting interpretation is that orexin A has substantially higher affinity for the CB1-OX1 heteromer than for the OX1-OX1 homomer (or OX1 monomer). This would be consistent with the earlier observation that orexin A is substantially more potent in promoting ERK mitogen-activated protein kinase phosphorylation when the OX1 receptor is co-expressed with the CB1 receptor (29) and the concept that receptor heteromers are unique species.
To assess whether OX1-OX1 homomers were also present in SNAP-CB1/CLIP-OX1-co-expressing cells, we added a combination of CLIP-Lumi4Tb and CLIP-Red, which can label only the OX1 receptor population, and again were able to detect htrFRET consistent with such an OX1-OX1 interaction. Equivalent studies using the equivalent pair of SNAP labels also identified CB1-CB1 interactions. Although certainly consistent with mixtures of homodimers and heterodimers, these results are also potentially consistent with the CB1 and OX1 receptors co-existing in larger complexes such as tetramers, and there is growing evidence for such oligomeric GPCR complexes (44–46), including tetramers (47–50). Although requiring further analysis, this would also be consistent with the ability of orexin A to internalize similar proportions of the CB1 receptor as the cannabinoid agonists.
Co-internalization of the CB1 receptor along with the OX1 receptor in response to orexin A was also observed in cell surface biotinylation protection experiments that were designed to provide a biochemical correlate of the htrFRET studies. Equally, by labeling the receptors with cell-impermeant fluorescent SNAP and CLIP tag substrates, we were able to image the co-internalization of both OX1 and CB1 receptors in response to addition of orexin A. This approach also demonstrated, as shown previously using a pair of receptors C-terminally tagged with autofluorescent proteins (30), that the simple presence of the CB1 receptor alters directly the cellular location profile of the OX1 receptor even in the absence of receptor ligands. Co-internalization of co-expressed receptors has been observed in a substantial number of cases. For example, the μ-opioid receptor agonist [d-Ala2,N-MePhe4,Gly-ol]enkephalin is able to cause internalization of the mGluR5 as well as the μ-opioid receptor when the two receptors are co-expressed, whereas the non-competitive mGluR5 antagonist 2-methyl-6-(phenylethynyl)pyridine limits [d-Ala2,N-MePhe4,Gly-ol]enkephalin-induced internalization of the μ-opioid receptor (51). Similarly, interactions between purinergic P2Y11 and P2Y1 receptors are reported to allow agonist internalization of P2Y11, although this receptor is not generally able to be internalized in response to agonist when expressed in isolation (52).
It is noteworthy that the CB1 receptor appears to be able to form heteromers with distinct properties with a variety of other receptors (53). For example, interactions with the angiotensin AT1 receptor (54) results in CB1 blockers limiting mitogenic signaling by the AT1 receptor, and although not observed in all studies (40), interactions of the CB1 receptor with the μ-opioid receptor (56, 57) have also been reported to modulate function. Further interactions with the dopamine D2 receptor (58, 59) and with the β2-adrenoreceptor (60) have also been reported to have functional sequelae. By contrast, there is little information on other heteromers that incorporate the OX1 receptor. However, this may simply reflect the much more extensive literature on cannabinoid receptors than on the OX1 receptor rather than a more limited propensity to make such interactions.
Although initial focus on the OX1 receptor centered on potential roles in appetite and feeding, much recent work has concentrated on the capacity of orexin receptor antagonists potentially targeting the orexin OX2 receptor as well as the OX1 receptor to limit wakefulness and hence treat insomnia (61, 62). By contrast, until recently, there have been very limited efforts to identify small molecule orexin receptor agonists, and because of this, the current studies have been limited to using the native peptide agonist orexin A as an activator of the OX1 receptor. This may change with indications that agonists at the OX1 receptor could be effective in the treatment of colon cancer (63, 64). The CB1 receptor has been a target of great interest not least because of the psychotropic effects of the active ingredient of cannabis produced via activation of this receptor. The association of increased feeding with cannabis use resulted in the development of CB1 receptor antagonists/inverse agonists for weight loss and the approval (later retracted because of the potential for psychiatric side effects) of the ligand rimonabant (65). Despite this, the contribution of CB1 receptors to energy balance and fat storage in the periphery has suggested that peripherally restricted CB1 receptor antagonists/inverse agonists might well have clinical potency without the central nervous system liabilities (65–67). Peripherally restricted CB1 agonists have also been promoted as potential therapeutic agents in both inflammatory and neuropathic pain (55). The effects of heteromeric interactions between the CB1 and OX1 receptors on such end points remain uncertain, but the substantial effects on ligand potency reported previously (29, 30) and the effects of orexin A and presumably synthetic OX1 receptor agonists as they are developed on CB1 receptor trafficking and function will need to be explored.
This work was supported by Medical Research Council Grant G0900050.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- GPCR
- G protein-coupled receptor
- htrFRET
- homogenous time-resolved FRET
- VSV-G
- vesicular stomatitis virus G
- 5-HT
- 5hydroxytryptamine
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- mGluR
- metabotropic glutamate receptor.
REFERENCES
- 1. Milligan G., Canals M., Pediani J. D., Ellis J., Lopez-Gimenez J. F. (2006) Ernst Schering Found. Symp. Proc. 2, 145–161 [DOI] [PubMed] [Google Scholar]
- 2. Milligan G. (2008) Br. J. Pharmacol. 153, Suppl. 1, S216–S229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dalrymple M. B., Pfleger K. D., Eidne K. A. (2008) Pharmacol. Ther. 118, 359–371 [DOI] [PubMed] [Google Scholar]
- 4. Pin J. P., Comps-Agrar L., Maurel D., Monnier C., Rives M. L., Trinquet E., Kniazeff J., Rondard P., Prézeau L. (2009) J. Physiol. 587, 5337–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Milligan G. (2009) Br. J. Pharmacol. 158, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birdsall N. J. (2010) Trends Pharmacol. Sci. 31, 499–508 [DOI] [PubMed] [Google Scholar]
- 7. Pin J. P., Neubig R., Bouvier M., Devi L., Filizola M., Javitch J. A., Lohse M. J., Milligan G., Palczewski K., Parmentier M., Spedding M. (2007) Pharmacol. Rev. 59, 5–13 [DOI] [PubMed] [Google Scholar]
- 8. Rozenfeld R., Devi L. A. (2010) Trends Pharmacol. Sci. 31, 124–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ferré S., Navarro G., Casadó V., Cortés A., Mallol J., Canela E. I., Lluís C., Franco R. (2010) Prog. Mol. Biol. Transl. Sci. 91, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hasbi A., Fan T., Alijaniaram M., Nguyen T., Perreault M. L., O'Dowd B. F., George S. R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21377–21382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rashid A. J., So C. H., Kong M. M., Furtak T., El-Ghundi M., Cheng R., O'Dowd B. F., George S. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 654–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pei L., Li S., Wang M., Diwan M., Anisman H., Fletcher P. J., Nobrega J. N., Liu F. (2010) Nat. Med. 16, 1393–1395 [DOI] [PubMed] [Google Scholar]
- 13. van Rijn R. M., Whistler J. L., Waldhoer M. (2010) Curr. Opin. Pharmacol. 10, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milan-Lobo L., Whistler J. L. (2011) J. Pharmacol. Exp. Ther. 337, 868–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He S. Q., Zhang Z. N., Guan J. S., Liu H. R., Zhao B., Wang H. B., Li Q., Yang H., Luo J., Li Z. Y., Wang Q., Lu Y. J., Bao L., Zhang X. (2011) Neuron 69, 120–131 [DOI] [PubMed] [Google Scholar]
- 16. Gomes I., Ijzerman A. P., Ye K., Maillet E. L., Devi L. A. (2011) Mol. Pharmacol. 79, 1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borroto-Escuela D. O., Romero-Fernandez W., Tarakanov A. O., Ciruela F., Agnati L. F., Fuxe K. (2011) J. Mol. Biol. 406, 687–699 [DOI] [PubMed] [Google Scholar]
- 18. Soriano A., Ventura R., Molero A., Hoen R., Casadó V., Cortés A., Fanelli F., Albericio F., Lluís C., Franco R., Royo M. (2009) J. Med. Chem. 52, 5590–5602 [DOI] [PubMed] [Google Scholar]
- 19. Casadó V., Cortés A., Mallol J., Pérez-Capote K., Ferré S., Lluis C., Franco R., Canela E. I. (2009) Pharmacol. Ther. 124, 248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Milligan G. (2006) Drug Discov. Today 11, 541–549 [DOI] [PubMed] [Google Scholar]
- 21. Panetta R., Greenwood M. T. (2008) Drug Discov. Today 13, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 22. Filizola M. (2010) Life Sci. 86, 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rovira X., Pin J. P., Giraldo J. (2010) Trends Pharmacol. Sci. 31, 15–21 [DOI] [PubMed] [Google Scholar]
- 24. Smith N. J., Milligan G. (2010) Pharmacol. Rev. 62, 701–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Springael J. Y., Urizar E., Costagliola S., Vassart G., Parmentier M. (2007) Pharmacol. Ther. 115, 410–418 [DOI] [PubMed] [Google Scholar]
- 26. Milligan G., Smith N. J. (2007) Trends Pharmacol. Sci. 28, 615–620 [DOI] [PubMed] [Google Scholar]
- 27. Rovira X., Vivó M., Serra J., Roche D., Strange P. G., Giraldo J. (2009) Br. J. Pharmacol. 156, 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. González-Maeso J., Ang R. L., Yuen T., Chan P., Weisstaub N. V., López-Giménez J. F., Zhou M., Okawa Y., Callado L. F., Milligan G., Gingrich J. A., Filizola M., Meana J. J., Sealfon S. C. (2008) Nature 452, 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hilairet S., Bouaboula M., Carrière D., Le Fur G., Casellas P. (2003) J. Biol. Chem. 278, 23731–23737 [DOI] [PubMed] [Google Scholar]
- 30. Ellis J., Pediani J. D., Canals M., Milasta S., Milligan G. (2006) J. Biol. Chem. 281, 38812–38824 [DOI] [PubMed] [Google Scholar]
- 31. Milligan G. (2010) Curr. Opin. Pharmacol. 10, 23–29 [DOI] [PubMed] [Google Scholar]
- 32. Maurel D., Comps-Agrar L., Brock C., Rives M. L., Bourrier E., Ayoub M. A., Bazin H., Tinel N., Durroux T., Prézeau L., Trinquet E., Pin J. P. (2008) Nat. Methods 5, 561–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alvarez-Curto E., Ward R. J., Pediani J. D., Milligan G. (2010) J. Biol. Chem. 285, 23318–23330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Albizu L., Cottet M., Kralikova M., Stoev S., Seyer R., Brabet I., Roux T., Bazin H., Bourrier E., Lamarque L., Breton C., Rives M. L., Newman A., Javitch J., Trinquet E., Manning M., Pin J. P., Mouillac B., Durroux T. (2010) Nat. Chem. Biol. 6, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monnier C., Tu H., Bourrier E., Vol C., Lamarque L., Trinquet E., Pin J. P., Rondard P. (2011) EMBO J. 30, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu T. R., Ward R. J., Pediani J. D., Milligan G. (2011) Biochem. J. 439, 171–183 [DOI] [PubMed] [Google Scholar]
- 37. Ward R. J., Pediani J. D., Milligan G. (2011) Br. J. Pharmacol. 162, 1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bennett K. A., Langmead C. J., Wise A., Milligan G. (2009) Mol. Pharmacol. 76, 802–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smith N. J., Stoddart L. A., Devine N. M., Jenkins L., Milligan G. (2009) J. Biol. Chem. 284, 17527–17539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Canals M., Milligan G. (2008) J. Biol. Chem. 283, 11424–11434 [DOI] [PubMed] [Google Scholar]
- 41. Langmead C. J., Jerman J. C., Brough S. J., Scott C., Porter R. A., Herdon H. J. (2004) Br. J. Pharmacol. 141, 340–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martini L., Waldhoer M., Pusch M., Kharazia V., Fong J., Lee J. H., Freissmuth C., Whistler J. L. (2007) FASEB J. 21, 802–811 [DOI] [PubMed] [Google Scholar]
- 43. Hern J. A., Baig A. H., Mashanov G. I., Birdsall B., Corrie J. E., Lazareno S., Molloy J. E., Birdsall N. J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2693–2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lopez-Gimenez J. F., Canals M., Pediani J. D., Milligan G. (2007) Mol. Pharmacol. 71, 1015–1029 [DOI] [PubMed] [Google Scholar]
- 45. Klco J. M., Lassere T. B., Baranski T. J. (2003) J. Biol. Chem. 278, 35345–35353 [DOI] [PubMed] [Google Scholar]
- 46. Guo W., Urizar E., Kralikova M., Mobarec J. C., Shi L., Filizola M., Javitch J. A. (2008) EMBO J. 27, 2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pisterzi L. F., Jansma D. B., Georgiou J., Woodside M. J., Chou J. T., Angers S., Raicu V., Wells J. W. (2010) J. Biol. Chem. 285, 16723–16738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Raicu V., Stoneman M. R., Fung R., Melnichuk M., Jansma D. B., Pisterzi LF, Rath S., Fox M., Wells J. W., Saldin D. K. (2009) Nat. Photonics 3, 107–111 [Google Scholar]
- 49. Fung J. J., Deupi X., Pardo L., Yao X. J., Velez-Ruiz G. A., Devree B. T., Sunahara R. K., Kobilka B. K. (2009) EMBO J. 28, 3315–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Comps-Agrar L., Kniazeff J., Nørskov-Lauritsen L., Maurel D., Gassmann M., Gregor N., Prézeau L., Bettler B., Durroux T., Trinquet E., Pin J. P. (2011) EMBO J. 30, 2336–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schröder H., Wu D. F., Seifert A., Rankovic M., Schulz S., Höllt V., Koch T. (2009) Neuropharmacology 56, 768–778 [DOI] [PubMed] [Google Scholar]
- 52. Ecke D., Hanck T., Tulapurkar M. E., Schäfer R., Kassack M., Stricker R., Reiser G. (2008) Biochem. J. 409, 107–116 [DOI] [PubMed] [Google Scholar]
- 53. Hudson B. D., Hébert T. E., Kelly M. E. (2010) Mol. Pharmacol. 77, 1–9 [DOI] [PubMed] [Google Scholar]
- 54. Rozenfeld R., Gupta A., Gagnidze K., Lim M. P., Gomes I., Lee-Ramos D., Nieto N., Devi L. A. (2011) EMBO J. 30, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yu X. H., Cao C. Q., Martino G., Puma C., Morinville A., St-Onge S., Lessard E., Perkins M. N., Laird J. M. (2010) Pain 151, 337–344 [DOI] [PubMed] [Google Scholar]
- 56. Rios C., Gomes I., Devi L. A. (2006) Br. J. Pharmacol. 148, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hojo M., Sudo Y., Ando Y., Minami K., Takada M., Matsubara T., Kanaide M., Taniyama K., Sumikawa K., Uezono Y. (2008) J. Pharmacol. Sci. 108, 308–319 [DOI] [PubMed] [Google Scholar]
- 58. Marcellino D., Carriba P., Filip M., Borgkvist A., Frankowska M., Bellido I., Tanganelli S., Müller C. E., Fisone G., Lluis C., Agnati L. F., Franco R., Fuxe K. (2008) Neuropharmacology 54, 815–823 [DOI] [PubMed] [Google Scholar]
- 59. Przybyla J. A, Watts V. J. (2010) J. Pharmacol. Exp. Ther. 332, 710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hudson B. D., Hébert T. E., Kelly M. E. (2010) Br. J. Pharmacol. 160, 627–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Scammell T. E., Winrow C. J. (2011) Annu. Rev. Pharmacol. Toxicol. 51, 243–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Coleman P. J., Cox C. D., Roecker A. J. (2011) Curr. Top. Med. Chem. 11, 696–725 [DOI] [PubMed] [Google Scholar]
- 63. Laburthe M., Voisin T. (2011) Br. J. Pharmacol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Voisin T., El Firar A., Fasseu M., Rouyer-Fessard C., Descatoire V., Walker F., Paradis V., Bedossa P., Henin D., Lehy T., Laburthe M. (2011) Cancer Res. 71, 3341–3351 [DOI] [PubMed] [Google Scholar]
- 65. Christopoulou F. D., Kiortsis D. N. (2011) J. Clin. Pharm. Ther. 36, 10–18 [DOI] [PubMed] [Google Scholar]
- 66. Wu Y. K., Yeh C. F., Ly T. W., Hung M. S. (2011) Curr. Top. Med. Chem. 11, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 67. Tam J., Vemuri V. K., Liu J., Bátkai S., Mukhopadhyay B., Godlewski G., Osei-Hyiaman D., Ohnuma S., Ambudkar S. V., Pickel J., Makriyannis A., Kunos G. (2010) J. Clin. Investig. 120, 2953–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]