Abstract
In Saccharomyces cerevisiae, non-coding RNAs, including cryptic unstable transcripts (CUTs), are subject to degradation by the exosome. The Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex in S. cerevisiae is a nuclear exosome cofactor that recruits the exosome to degrade RNAs. Trf4/5 are poly(A) polymerases, Mtr4 is an RNA helicase, and Air1/2 are putative RNA-binding proteins that contain five CCHC zinc knuckles (ZnKs). One central question is how the TRAMP complex, especially the Air1/2 protein, recognizes its RNA substrates. To characterize the function of the Air1/2 protein, we used random mutagenesis of the AIR1/2 gene to identify residues critical for Air protein function. We identified air1-C178R and air2-C167R alleles encoding air1/2 mutant proteins with a substitution in the second cysteine of ZnK5. Mutagenesis of the second cysteine in AIR1/2 ZnK1–5 reveals that Air1/2 ZnK4 and -5 are critical for Air protein function in vivo. In addition, we find that the level of CUT, NEL025c, in air1 ZnK1–5 mutants is stabilized, particularly in air1 ZnK4, suggesting a role for Air1 ZnK4 in the degradation of CUTs. We also find that Air1/2 ZnK4 and -5 are critical for Trf4 interaction and that the Air1-Trf4 interaction and Air1 level are critical for TRAMP complex integrity. We identify a conserved IWRXY motif in the Air1 ZnK4-5 linker that is important for Trf4 interaction. We also find that hZCCHC7, a putative human orthologue of Air1 that contains the IWRXY motif, localizes to the nucleolus in human cells and interacts with both mammalian Trf4 orthologues, PAPD5 and PAPD7 (PAP-associated domain containing 5 and 7), suggesting that hZCCHC7 is the Air component of a human TRAMP complex.
Keywords: Protein-Protein Interactions, RNA Processing, RNA Turnover, RNA-binding Proteins, Yeast Genetics, Air1, RNA Quality Control, TRAMP Complex, Trf4, Zinc Knuckle Protein
Introduction
Production of mature RNAs in eukaryotes requires a complex set of processing steps, including 5′-end capping, splicing, 3′-end cleavage, polyadenylation, nucleolytic cleavage/trimming, and base modifications, by numerous processing components. Incorrectly processed RNAs are rapidly degraded by RNA quality control machinery to prevent deleterious effects on the cell. Processing and degradation of multiple classes of RNA are performed by RNA endo/exoribonucleases that are recruited to their RNA substrates by specific protein cofactors. These nucleases and their cofactors are highly regulated and evolutionarily conserved.
In Saccharomyces cerevisiae, non-coding RNAs (ncRNAs),2 including precursors of rRNAs, snoRNAs, and snRNAs, are processed and/or degraded by the nuclear exosome, an evolutionarily conserved ringlike riboexonuclease complex containing two active 3′-5′-riboexonucleases, Rrp44/Dis3 and Rrp6 (1–8). Hypomodified initiator tRNAs (tRNAiMet) from cells with an impaired tRNA methyltransferase and aberrant pre-mRNAs from cells with defective 3′-end processing, splicing, or nuclear export factors are also degraded by the nuclear exosome (9–13). More recently, a novel class of small (250–300-nucleotide) intergenic RNA polymerase II transcripts, termed cryptic unstable transcripts (CUTs), was discovered in cells that lack the RRP6 gene (14, 15). CUTs are polyadenylated and normally rapidly degraded by the nuclear exosome, but they are readily stabilized in rrp6Δ cells (14). CUTs initiate from nucleosome-free 5′ promoter regions or 3′-ends of protein-coding genes, and some CUTS regulate gene expression (16–19). Importantly, exosome depletion from human cells has revealed the presence of short, polyadenylated, unstable RNAs arising upstream (∼1 kb) of promoters, termed promoter upstream transcripts, indicating that exosome-targeted degradation of such ncRNAs is conserved in humans (20).
In S. cerevisiae, the nuclear exosome is recruited to ncRNAs and aberrant RNAs through the evolutionarily conserved Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) complex, a cofactor and critical RNA surveillance component that adds a short poly(A) tail to RNA substrates and promotes their degradation by stimulating the exonucleolytic activity of the exosome (13, 14, 21–23). In particular, the TRAMP complex enhances RNA degradation by Rrp6 (24). The TRAMP complex is composed of a non-canonical poly(A) polymerase (Trf4 or Trf5), a putative RNA-binding protein (Air1 or Air2), and an RNA helicase (Mtr4) (14, 21, 22). In S. cerevisiae, there are two types of defined TRAMP complex, TRAMP4 (Trf4-Air1/2-Mtr4) and TRAMP5 (Trf5-Air1-Mtr4), which each function in nuclear RNA quality control (14, 21–23). In a basic model for TRAMP complex function, Air1/2 proteins recognize the RNA substrate, Mtr4 unwinds/remodels the RNA, and Trf4/5 poly(A) polymerases polyadenylate the RNA, which presents an unstructured substrate for the exosome to degrade (14, 21, 22, 25, 26). An outstanding question in nuclear RNA surveillance is how the TRAMP complex can recognize its RNA substrates and distinguish between “good” properly processed and “bad” incorrectly processed RNA. Characterizing the putative RNA-binding proteins, Air1 and Air2, is thus paramount.
Trf4 and Trf5 (DNA topoisomerase I-related function) are nuclear poly(A) polymerases of the Pol-β-like nucleotidyltransferase family that includes the canonical poly(A) polymerase, Pap1 (14, 21, 22, 27–30). Unlike Pap1, which processively adds 70–90 adenosines to an RNA substrate, Trf4, in the context of TRAMP4, distributively adds a short poly(A) tail to an RNA substrate (21, 22, 31). Notably, Trf4 is inactive by itself and requires Air1/2 to actively polyadenylate RNA (14, 21, 22). Mutations in TRF4 and RRP44 stabilize unstable hypomodified tRNAiMet that lacks a methyl group in cells with a defective tRNA methyltransferase, and Trf4 overexpression causes polyadenylation of this hypomodified, but not wild type, tRNAiMet in these mutant cells, indicating that Trf4 polyadenylates and marks hypomodified tRNAiMet for degradation by the exosome (13). Critically, budding yeast-derived TRAMP4 complex or recombinant Trf4-Air1/2 complex exclusively polyadenylates hypomodified tRNAiMet, suggesting that the TRAMP4 complex recognizes the structure/folding of an RNA substrate (22). Besides aberrant tRNAs, Trf4 also polyadenylates and stimulates the degradation of snRNAs, snoRNAs, rRNAs, and CUT RNAs, including the prototypic CUT, NEL025c (14, 21, 32, 33).
Trf4 and Trf5 (48% identical) are functionally redundant because trf4Δ or trf5Δ single mutants are viable, but trf4Δ trf5Δ double mutants are inviable (21, 34). Surprisingly, a catalytically inactive mutant of Trf4, trf4-DADA, supports the degradation of most Trf4 RNA substrates in trf4Δ cells and rescues the lethality of trf4Δ trf5Δ cells, suggesting that Trf4 can target RNAs to the exosome in a polyadenylation-independent manner (14, 34, 35). Trf4 orthologues are present in Schizosaccharomyces pombe (Cid14), Drosophila (TRF4-1), and humans (hTrf4–1/PAPD7 and hTrf4–2/PAPD5) (25, 26, 29, 30, 36–39). Mtr4 (mRNA transport, also known as Dob1) is an essential nuclear DEXH RNA helicase in S. cerevisiae that is required for the processing/degradation ncRNAs in vivo and preferentially binds to poly(A) RNA and unwinds RNA duplexes in vitro (1, 3, 23, 40–44).
Air1 and Air2 (arginine methyltransferase-interacting RING finger) are nuclear zinc knuckle proteins required for Trf4-mediated polyadenylation and degradation of RNA substrates, including ncRNAs, hypomodified tRNAiMet, and CUTs, such as NEL025c, in S. cerevisiae (14, 21, 22, 45). Air1 and Air2 are functionally redundant because air1Δ and air2Δ single deletion mutant cells grow similarly to wild type cells, but air1Δ air2Δ double mutant cells exhibit a strong growth defect (45). Both Air1 and Air2 copurify with Trf4, but only Air1 copurifies with Trf5 (14, 21–23). A single functional Air1 orthologue exists in S. pombe (SPBP35G2.08c) (38) and two human proteins, hZCCHC7 and hZCCHC9, have been proposed to be the putative human Air1 orthologue based on sequence similarity (25, 26, 46), but no evidence has been published to support the idea that either protein is a functional Air1 protein.
Air1 and Air2 (45% identical) are predicted to bind RNA because recombinant Air1/2 is essential for Trf4-mediated polyadenylation of hypomodified tRNAiMet in vitro, and air1Δ air2Δ cells exhibit loss of polyadenylation of ncRNAs in vivo (14, 21, 22). Air1/2 proteins contain five adjacent CX2CX4HX4C zinc knuckles (ZnK1–5; Fig. 1A) that could interact with nucleic acids or bind protein (22, 45). In retroviral nucleocapsid proteins, one or two CX2CX4HX4C zinc knuckles form metal-coordinated reverse turns that typically contact viral single-stranded tetraloop RNA via the variable (C)X2 and (H)X4 residues (14, 22, 47, 48). A recent crystal structure of Trf4 in complex with an Air2 fragment shows that Air2 zinc knuckles 4 and 5 interact with the Trf4 central domain (49). In addition, recombinant Trf4 in complex with Air2 ZnK1–5 preferentially polyadenylates aberrant tRNA in vitro, indicating that Air2 zinc knuckles can also recognize RNA (49). A key question is therefore which zinc knuckles of the Air1/2 proteins contribute to RNA recognition and Air protein function in vivo. At present, studies of Air zinc knuckle function have only been carried out in vitro using recombinant protein, and only mutants of ZnK1, ZnK2, and ZnK3 have been generated in the context of Air2 ZnK1–5 (49). Trf4 complexed with an Air2 ZnK1 mutant, but not a ZnK2 or ZnK3 mutant, exhibits impaired polyadenylation of mutant tRNA, suggesting that Air2 ZnK1 is important for RNA recognition (49). An Air2 ZnK4-5 fragment also supports weak Trf4-mediated polyadenylation of aberrant tRNA, indicating that ZnK4 and ZnK5 also have the capacity to recognize RNA (49). However, these conclusions are all based on biochemical experiments, and the contributions of the individual zinc knuckles to the in vivo function of the Air proteins have not been addressed.
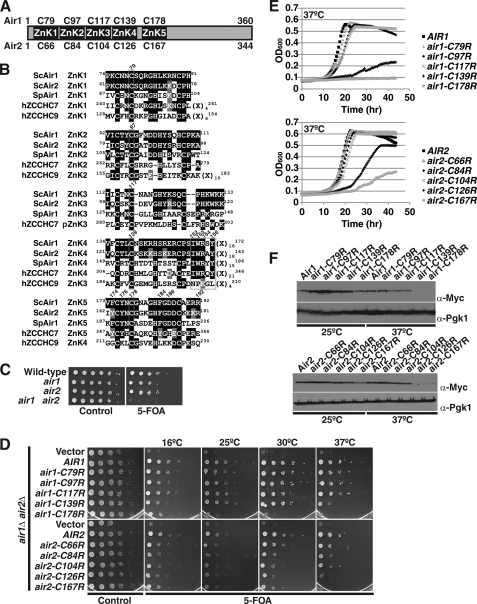
FIGURE 1.
Air1/2 zinc knuckle 4 and 5 are functionally important. A, schematic of Air1 and Air2 proteins depicting the five CCHC zinc knuckles (ZnK1–5). The amino acid positions of the second cysteine within each zinc knuckle of Air1 (above) and Air2 (below) that was changed to arginine are shown. B, peptide alignment of the CCHC zinc knuckles 1–5 (ZnK1–5) of S. cerevisiae Air1 and Air2 (ScAir1 and ScAir2), S. pombe Air1 (SpAir1), hZCCHC7 proteins, and ZnK1–4 of hZCCHC9 protein. hZCCHC7 contains four intact ZnKs (ZnK1-2 and ZnK4-5) and a pseudo-ZnK3 (pZnK3) in which the spacing of the histidine is altered. Identical residues are shaded in black, and similar residues are shaded in gray. Amino acid positions of the zinc knuckles within each protein are shown in superscript. Positions of residues substituted in Air1 zinc knuckles are depicted as slanted numbers above the Air1 sequence. The second cysteines of each ZnK and IWRXY motif are boxed. The length of nondepicted linker residues (X) is shown in subscript. C, air1Δ and air2Δ single mutant cells show no growth defects, but air1Δ air2Δ double mutant cells exhibit a severe growth defect. Wild type, air1Δ, air2Δ, and air1Δ air2Δ cells containing an AIR2 URA3 plasmid were grown to saturation, serially diluted in 10-fold dilutions, and spotted on control and 5-FOA plates. D, air1 ZnK4 and -5 mutants (air1-C139R and -C178R) and air2 ZnK4 and -5 mutants (air2-C126R and -C167R) exhibit temperature-sensitive growth. air1Δ air2Δ cells containing AIR2 URA3 maintenance plasmid and vector, AIR1, air1 ZnK1–5 mutant (air1-C79R, -C97R, -C117R, -C139R, or -C178R), AIR2, or air2 ZnK1–5 mutant (air2-C66R, -C84R, -C104R, -C126R, or -C167R) LEU2 test plasmids were grown to saturation, serially diluted, and spotted on control and 5-FOA plates, and grown at the indicated temperatures. E, growth curve analysis of air1/2 ZnK4 and -5 mutants confirms that they grow more slowly than AIR1/AIR2 cells. air1Δ air2Δ cells carrying only AIR1, air1 ZnK1–5 mutant, AIR2, or air2 ZnK1–5 mutant plasmids were grown to saturation and diluted, and their optical density was measured at A600 for 40 h at 37 °C as described under “Experimental Procedures.” F, steady-state protein levels of air1 ZnK4 and -5 mutant proteins (air1-C139R and -C178R) and air2 ZnK4 and -5 mutant proteins (air2-C126R and -C167R) are profoundly decreased at 37 °C. Whole cell lysates from air1Δ air2Δ cells expressing Myc-tagged Air1, air1 ZnK1–5 mutant, Air2, or air2 ZnK1–5 mutant protein and grown at 25 and 37 °C were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody (α-Myc) to detect Air1/2-Myc proteins and an anti-Pgk1 antibody (α-Pgk1) to detect 3-phosphoglycerate kinase as a loading control.
Here, we present the first in vivo functional analysis of full-length Air1/2 zinc knuckle 1–5 mutants. We find that Air1/2 ZnK4 and ZnK5 are functionally important. In particular, Air1/2 ZnK4 and ZnK5 mutants exhibit temperature-sensitive growth and reduced binding to Trf4. In addition, air1 ZnK4 mutant cells exhibit the highest levels of NEL025c CUT RNA relative to AIR1 cells and the other air1 ZnK mutants, suggesting that Air1 ZnK4 may help facilitate RNA recognition. We also find that Air1/2 and Trf4, but not Trf5 or Mtr4, suppress the temperature-sensitive growth of the air1 ZnK5 mutant. In addition, Air1 interaction with Trf4 and Air1 level are critical for the stability of TRAMP complex components and the integrity of the TRAMP complex. Importantly, our studies also identify a key evolutionarily conserved IWRXY motif in the Air1 ZnK4-5 linker region that is important for Air1 function and interaction with Trf4. A putative human orthologue of the Air proteins, hZCCHC7, which contains the IWRXY motif, localizes to the nucleolus and binds to both mammalian Trf4-like proteins, PAPD5 and PAPD7, providing evidence that hZCCHC7 is the Air protein component of a human TRAMP complex. Together, the data suggest that Air1/2 interacts with Trf4 predominantly via the ZnK4-5 linker and ZnK5 and facilitates recognition of RNA via ZnK1–4.
EXPERIMENTAL PROCEDURES
Plasmids, Strains, and Chemicals
All DNA manipulations were performed according to standard methods (50), and all media were prepared by standard procedures (51). All chemicals were obtained from Sigma-Aldrich, U.S. Biological (Swampscott, MA), or Fisher unless otherwise noted. S. cerevisiae strains and plasmids used are described in Table 1. The TRF4 gene was subcloned from pCB727 (a gift from Michael F. Christman) into pRS426 to create URA3 2μ TRF4 plasmid (pAC2147). The TRF5 gene was subcloned from pCB557 (a gift Michael F. Christman) into pRS426 to create URA3 2μ TRF5 plasmid (pAC2931). The MTR4 gene was amplified by polymerase chain reaction (PCR) from S. cerevisiae genomic DNA with oligonucleotides (Integrated DNA Technologies) and cloned into pRS426 to create URA3 2μ MTR4 plasmid (pAC2897). The URA3 2μ trf4-DADA mutant plasmid (pAC2710) and trf4–378 mutant plasmid (pAC3048) were generated by site-directed mutagenesis with trf4-DADA oligonucleotides encoding D236A and D238A residue substitutions and trf4–378 oligonucleotides encoding E378A and E381A residue substitutions, TRF4 (pAC2147) plasmid template, and the QuikChange site-directed mutagenesis kit (Stratagene). C-terminally Myc-tagged TRF4 (pAC2910), trf4-DADA mutant (pAC2914), and trf4–378 mutant (pAC3049) were constructed by PCR amplification of TRF4 using oligonucleotides and TRF4 (pAC2147), trf4-DADA (pAC2710), or trf4–378 (pAC3048) template and cloning into pRS415, followed by insertion of 2xMyc PCR product. C-terminally Myc-tagged TRF5 (pAC3050) and MTR4 (pAC3051) were constructed by PCR amplification of TRF5 and MTR4 using oligonucleotides and TRF5 (pAC2931) and MTR4 (pAC2897) template and cloning into pRS415 followed by insertion of 2xMyc PCR product. Wild type AIR1 and AIR2 genes were amplified by PCR from S. cerevisiae genomic DNA with oligonucleotides and cloned into pRS416 to create URA3 CEN AIR1 plasmid (pAC1613) and URA3 CEN AIR2 (pAC1614) and cloned into pRS415 to create LEU2 CEN AIR1 plasmid (pAC1856) and LEU2 CEN AIR2 (pAC1857). Untagged air1 zinc knuckle 1–5 mutants (pAC2727–2730 and pAC2012) and untagged air2 zinc knuckle 1–5 mutants (pAC2731–2734 and pAC2013) with cysteine to arginine substitutions in the second cysteine of zinc knuckles 1–5 were generated by site-directed mutagenesis using oligonucleotides encoding the amino acid substitutions, AIR1 (pAC1856) or AIR2 (pAC1857) plasmid template, and the QuikChange site-directed mutagenesis kit (Stratagene). Additional untagged air1 zinc knuckle 4-5 linker and zinc knuckle 5 mutants (pAC2837–2840, pAC2845–2848, and pAC2884) with alanine substitutions were also generated by site-directed mutagenesis. C-terminally GFP-tagged AIR1/2 and air1/2 zinc knuckle mutants (pAC2825–2836, pAC2841–2844, and pAC2863–2866) and C-terminally Myc-tagged AIR1/2 and air1/2 zinc knuckle mutants (pAC2885–2896 and pAC2902–2909) were constructed by PCR amplification of AIR1/2 genes using oligonucleotides and AIR1/2 or air1/2 templates (pAC1856, pAC2727–2734, pAC2012–2013, pAC2837–2840, pAC2845–2848, and pAC2884) and cloning into pRS415, followed by insertion of GFP or 2xMyc PCR products. C-terminally Myc-tagged murine PAPD5 (mPAPD5) and human PAPD7 (hPAPD7) were constructed by PCR using oligonucleotides and pCR4-TOPO-mPAPD5 (IMAGE 40131040, accession number BC145737; OpenBiosystems) and pCMV6-AC-hPAPD7-GFP (accession number NM_006999.3; OriGene) templates and cloning into pcDNA3 (Invitrogen). All constructs were sequenced to ensure the presence of each desired mutation and the absence of any additional mutations.
TABLE 1.
Yeast strains and plasmids
| Strain/Plasmid | Description | Source |
|---|---|---|
| Strain | ||
| W303 wild type (ACY233) | MATα ura3Δ leu2Δ trp1Δ his3Δ | |
| BY4741 wild type (ACY402) | MATa leu2Δ ura3Δ his3Δ TRP1 met15Δ | Research Genetics |
| air1Δ (ACY1090) | MATa leu2Δ ura3Δ his3Δ TRP1 AIR1::KAN | Research Genetics |
| air2Δ (ACY1091) | MATa leu2Δ ura3Δ his3Δ TRP1 AIR2::KAN | Research Genetics |
| rrp6Δ (ACY1641) | MATa leu2Δ ura3Δ his3Δ TRP1 RRP6::KAN | Research Genetics |
| trf4Δ (ACY2149) | MATa leu2Δ ura3Δ his3Δ TRP1 TRF4::KAN | Research Genetics |
| air1Δ air2Δ (ACY1095) | MATa leu2Δ ura3Δ his3Δ TRP1 met15Δ AIR1::KAN AIR2::KAN (pAC1614) | This study |
| air1-C178R air2Δ (ACY2020) | MATα ura3Δ leu2Δ trp1Δ his3Δ air1-C178R AIR2::NAT | This study |
| air1Δ air2ΔTRF4-TAP (ACY2131) | ura3Δ leu2Δ trp1Δ his3Δ AIR1::NAT AIR2::HPH TRF4-TAP:Sphis5+ | This study |
| AIR1-TAP (ACY1061) | MATa ura3Δ leu2Δ his3Δ met15Δ TRP1 AIR1-TAP:Sphis5+ | Open Biosystems |
| Plasmid | ||
| pRS416 | CEN, URA3, AMPR | Ref. 65 |
| pAC1613 | AIR1, CEN, URA3, AMPR | This study |
| pAC1614 | AIR2, CEN, URA3, AMPR | This study |
| pRS415 | CEN, LEU2, AMPR | Ref. 65 |
| pAC1856 | AIR1, CEN, LEU2, AMPR | This study |
| pAC2727 | air1-C79R, CEN, LEU2, AMPR | This study |
| pAC2728 | air1-C97R, CEN, LEU2, AMPR | This study |
| pAC2729 | air1-C117R, CEN, LEU2, AMPR | This study |
| pAC2730 | air1-C139R, CEN, LEU2, AMPR | This study |
| pAC2012 | air1-C178R, CEN, URA3, AMPR | This study |
| pAC2845 | air1-I152A, CEN, LEU2, AMPR | This study |
| pAC2837 | air1-W153A, CEN, LEU2, AMPR | This study |
| pAC2884 | air1-R154A, CEN, LEU2, AMPR | This study |
| pAC2846 | air1-Y156A, CEN, LEU2, AMPR | This study |
| pAC2847 | air1-F174A, CEN, LEU2, AMPR | This study |
| pAC2838 | air1-Y176A, CEN, LEU2, AMPR | This study |
| pAC2848 | air1-F184A, CEN, LEU2, AMPR | This study |
| pAC2839 | air1-D186A, CEN, LEU2, AMPR | This study |
| pAC2840 | air1-R192A, CEN, LEU2, AMPR | This study |
| pAC2825 | AIR1-GFP, CEN, LEU2, AMPR | This study |
| pAC2826 | air1-C79R-GFP, CEN, LEU2, AMPR | This study |
| pAC2827 | air1-C97R-GFP, CEN, LEU2, AMPR | This study |
| pAC2828 | air1-C117R-GFP, CEN, LEU2, AMPR | This study |
| pAC2829 | air1-C139R-GFP, CEN, LEU2, AMPR | This study |
| pAC2830 | air1-C178R-GFP, CEN, URA3, AMPR | This study |
| pAC2863 | air1-I152A-GFP, CEN, LEU2, AMPR | This study |
| pAC2841 | air1-W153A-GFP, CEN, LEU2, AMPR | This study |
| pAC2944 | air1-R154A-GFP, CEN, LEU2, AMPR | This study |
| pAC2864 | air1-Y156A-GFP, CEN, LEU2, AMPR | This study |
| pAC2865 | air1-F174A-GFP, CEN, LEU2, AMPR | This study |
| pAC2842 | air1-Y176A-GFP, CEN, LEU2, AMPR | This study |
| pAC2866 | air1-F184A-GFP, CEN, LEU2, AMPR | This study |
| pAC2843 | air1-D186A-GFP, CEN, LEU2, AMPR | This study |
| pAC2844 | air1-R192A-GFP, CEN, LEU2, AMPR | This study |
| pAC2885 | AIR1–2xMyc, CEN, LEU2, AMPR | This study |
| pAC2886 | air1-C79R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2887 | air1-C97R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2888 | air1-C117R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2889 | air1-C139R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2890 | air1-C178R-2xMyc, CEN, URA3, AMPR | This study |
| pAC2906 | air1-I152A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2902 | air1-W153A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2937 | air1-R154A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2907 | air1-Y156A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2908 | air1-F174A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2903 | air1-Y176A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2909 | air1-F184A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2904 | air1-D186A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2905 | air1-R192A-2xMyc, CEN, LEU2, AMPR | This study |
| pAC1857 | AIR2, CEN, LEU2, AMPR | This study |
| pAC2731 | air2-C66R, CEN, LEU2, AMPR | This study |
| pAC2732 | air2-C84R, CEN, LEU2, AMPR | This study |
| pAC2733 | air2-C104R, CEN, LEU2, AMPR | This study |
| pAC2734 | air2-C126R, CEN, LEU2, AMPR | This study |
| pAC2013 | air2-C167R, CEN, LEU2, AMPR | This study |
| pAC2831 | AIR2-GFP, CEN, LEU2, AMPR | This study |
| pAC2832 | air2-C66R-GFP, CEN, LEU2, AMPR | This study |
| pAC2833 | air2-C84R-GFP, CEN, LEU2, AMPR | This study |
| pAC2834 | air2-C104R-GFP, CEN, LEU2, AMPR | This study |
| pAC2835 | air2-C126R-GFP, CEN, LEU2, AMPR | This study |
| pAC2836 | air2-C167R-GFP, CEN, LEU2, AMPR | This study |
| pAC2891 | AIR2–2xMyc, CEN, LEU2, AMPR | This study |
| pAC2892 | air2-C66R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2893 | air2-C84R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2894 | air2-C104R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2895 | air2-C126R-2xMyc, CEN, LEU2, AMPR | This study |
| pAC2896 | air2-C167R-2xMyc, CEN, LEU2, AMPR | This study |
| pRS426 | 2μ, URA3, AMPR | Ref. 66 |
| pCB727 | TRF4, 2μ, TRP1, AMPR | M. F. Christman |
| pCB557 | TRF5, 2μ, TRP1, AMPR | M. F. Christman |
| pAC2147 | TRF4, 2μ, URA3, AMPR | This study |
| pAC2710 | trf4-DADA, 2μ, URA3, AMPR | This study |
| pAC3048 | trf4–378, 2μ, URA3, AMPR | This study |
| pAC2931 | TRF5, 2μ, URA3, AMPR | This study |
| pAC2897 | MTR4, 2μ, URA3, AMPR | This study |
| pAC2910 | TRF4–2xMyc, 2μ, URA3, AMPR | This study |
| pAC2914 | trf4-DADA-2xMyc, 2μ, URA3, AMPR | This study |
| pAC3049 | trf4–378-2xMyc, 2μ, URA3, AMPR | This study |
| pAC3050 | TRF5–2xMyc, 2μ, URA3, AMPR | This study |
| pAC3051 | MTR4–2xMyc, 2μ, URA3, AMPR | This study |
| pRS423 | 2μ, HIS3, AMPR | Ref. 66 |
| pAC2940 | TRF4, 2μ, HIS3, AMPR | This study |
| pAC2946 | trf4-DADA, 2μ, HIS3, AMPR | This study |
| pAC2998 | pcDNA3-mPAPD5–2xMyc, AMPR | This study |
| pAC2999 | pcDNA3-hPAPD7–2xMyc, AMPR | This study |
Cell Growth Assays
To assess the growth of double deletion mutant air1Δ air2Δ cells compared with single mutant air1Δ or air2Δ cells, wild type (BY4741), air1Δ (ACY1090), air2Δ (ACY1091), and air1Δ air2Δ cells (ACY1095) containing an AIR2 URA3 maintenance plasmid (pAC1614) were grown overnight at 30 °C to saturation in Ura− minimal media with 2% glucose. Cell number was then normalized by A600; serially diluted and spotted onto control Ura− minimal media or minimal media containing 5-fluoroorotic acid (5-FOA), which selects for cells that have lost the AIR2 URA3 maintenance plasmid (52); and grown at 25 °C. In vivo functional analysis of air1 and air2 zinc knuckle mutants was carried out through a standard plasmid shuffle assay combined with serial dilution and spotting or growth curves. To generate single mutants of air1 or air2, air1Δ air2Δ cells (ACY1095), containing AIR2 URA3 maintenance plasmid (pAC1614), were transformed with AIR1 (pAC1856), air1 mutant (pAC2727–2730 and pAC2012), AIR2 (pAC1857), air2 mutant (pAC2731–2734 and pAC2013) or vector (pRS415) LEU2 plasmids and selected on Leu−Ura− minimal media with 2% glucose. air1Δ air2Δ cells (ACY1095) containing AIR2 URA3 maintenance plasmid (pAC1614) were similarly transformed with additional air1 zinc knuckle 4-5 linker and zinc knuckle 5 mutant (pAC2837–2840, pAC2845–2848, and pAC2884) LEU2 plasmids. Cells were grown overnight at 25 °C to saturation in Leu−Ura− minimal media with 2% glucose. Cell concentrations were normalized by A600, and cultures were serially diluted in sterile H2O to obtain ∼10,000, 1000, 100, 10, or 1 cell per 3-μl volume. These dilutions were spotted onto control Leu−Ura− minimal media, where the AIR2 URA3 maintenance plasmid is maintained, or Leu− minimal media containing 5-FOA, which selects for cells that have lost the AIR2 URA3 maintenance plasmid. Growth of air1Δ air2Δ cells harboring AIR1/2 or air1/2 zinc knuckle mutants as the sole copy of air1/2 was examined at 16, 25, 30, and 37 °C.
For growth curve analysis, air1Δ air2Δ cells harboring air1/2 mutants as the sole copy of Air1/2 were grown overnight at 25 °C to saturation in Leu− media with 2% glucose. Cell concentrations were normalized by A600, diluted 100-fold in 100 μl of Leu− media with 2% glucose, and added to wells of a MicroWell F96 microtiter plate (Nunc). Samples were loaded in quintuplicate. Cells in plate wells were grown at 37 °C with shaking, and absorbance at A600 was measured every 30 min for 40 h in an ELX808 Ultra microplate reader with KCjunior software (Bio-Tek Instruments, Inc.). Quintuplicate sample absorbances for time points were averaged and plotted using Microsoft Excel for Mac 2008 (Microsoft Corp.).
Random Mutagenesis of AIR1/2
To generate random mutants of AIR1/2, the AIR1 and AIR2 genes were subjected to random PCR mutagenesis using the AIR1 and AIR2 template plasmids (pAC1856 and pAC1857), oligonucleotides, 0.4 mm dGTP (0.2 mm dCTP, dATP, and dTTP), 2 mm MgCl2, and Taq polymerase (Qiagen). The air mutant PCR products were introduced into linearized AIR1 and AIR2 LEU2 plasmid (pAC1856 and pAC1857) via homologous recombination by transformation into air1Δ air2Δ cells (ACY1095) containing an AIR2 URA3 maintenance plasmid (pAC1614) and selection on Ura−Leu− minimal medium plates with 2% glucose at 30 °C. Temperature-sensitive air1 and air2 mutants were then identified by replica plating to Leu− minimal media containing 5-FOA to select for cells that have lost the AIR2 URA3 maintenance plasmid. Colonies that grew at 25 °C but not at 37 °C were selected. Plasmids containing the air1 and air2 mutants were recovered and fully sequenced to identify the air1 and air2 mutations.
Localization of GFP-tagged Proteins
To localize Air1/2-GFP proteins, air1Δ air2Δ cells (ACY1095) harboring an AIR2 URA3 maintenance plasmid (pAC1614) and expressing GFP-tagged Air protein (pAC2825–2836, pAC2841–2844, and pAC2863–2866) were grown in Ura−Leu− media with 2% glucose overnight at 25 °C, transferred to Ura−Leu− media with 2% glucose, and grown to log phase at 25 °C. GFP fusion proteins were visualized by direct fluorescence microscopy using an Olympus BX60 direct fluorescence microscope equipped with a photometric Quantix digital camera from Roper Scientific (Tucson, AZ) and filters from Chroma Technology (Brattleboro, VT). All images were captured using IP Lab Spectrum software.
Total RNA Isolation
To prepare S. cerevisiae total RNA from cell pellets of 50-ml cultures grown to A600 = 0.5, 2–3 scoops of glass beads were added to each cell pellet in a 2-ml screw cap tube, 1 ml of TRIzol (Invitrogen) was added, and the cell sample was vigorously disrupted in a Mini Bead Beater 16 cell disrupter (Biospec) for 2 min at 25 °C. For each sample, 100 μl of 1-bromo-3-chloropropane was added, and the sample was vortexed for 15 s and incubated at 25 °C for 2 min. Each sample was then centrifuged at 16,300 × g for 8 min at 4 °C, and the upper layer was transferred to a fresh Eppendorf tube. RNA was precipitated with 500 μl of isopropyl alcohol, and the sample was vortexed for 10 s to mix. Total RNA was pelleted by centrifugation at 16,300 × g for 8 min at 4 °C. Supernatant was decanted, and 1 ml of 75% ethanol was added to wash the RNA pellet. Sample was centrifuged at 16,300 × g for 5 min at 4 °C. Supernatant was decanted, remaining ethanol was removed, and the RNA pellet was air-dried for 15 min. Total RNA was resuspended in 50 μl of diethylpyrocarbonate (Sigma)-treated water and stored at −80 °C.
Quantitative RT-PCR to Detect CUT RNAs
To measure the levels of CUTs, total RNA was isolated from cells and reverse transcribed to cDNA, and cDNA of CUTs was detected by real-time quantitative PCR. Wild type (BY4741), air2Δ (ACY1091), trf4Δ (ACY2149), rrp6Δ (ACY1641), and air1Δ air2Δ cells (ACY1095) harboring AIR1 (pAC1856) or air1 mutant (pAC2727–2730 and pAC2012) LEU2 plasmids as the sole copy of air1 were inoculated into 2 ml of YEPD (yeast extract, peptone, dextrose) media and grown overnight at 25 °C. An aliquot of 100 μl of each overnight culture was used to inoculate 50 ml of YEPD media and grown overnight at 25 °C. Cultures were grown to an A600 = 0.5. Cells were centrifuged at 2,163 × g, transferred to fresh tubes, centrifuged at 16,000 × g, and stored at −80 °C. Following total RNA isolation from each cell pellet, 1 μg of RNA was reverse transcribed to first strand cDNA using the QuantiTect® reverse transcription kit (Qiagen) according to the manufacturer's protocol. Relative CUT cDNA levels were detected by quantitative PCR on triplicate samples of 50 ng of cDNA using either 2.5 μm NEL025c CUT primers or control PGK1 primers and QuantiTect® SYBR Green Master Mix (Qiagen) on an iCycler iQTM real-time PCR detection system (Bio-Rad). Results were analyzed using the iCycler optical system version 3.0a software (Bio-Rad), and data were normalized by the ΔΔCt method to a control PGK1 transcript (53).
High Copy Suppression
To assess suppression of air1-C178R temperature-sensitive growth by TRAMP components, air1-C178R air2Δ cells (ACY2020) transformed with vector (pRS426), AIR1 (pAC1613), AIR2 (pAC1614), TRF4 (pAC2147), trf4-DADA (pAC2710), TRF5 (pAC2931), MTR4 (pAC2897), or trf4–378 (pAC3048) 2μ URA3 plasmid or cells transformed with control vector (pRS415), AIR1 (pAC1856), or air1 mutant (pAC2727–2730 and pAC2012) CEN LEU2 plasmid were serially diluted and spotted onto minimal medium plates and grown at 25, 30, and 37 °C.
Trf4-TAP and Air1-TAP Binding Assay
To assess binding between Trf4 and air1/2 mutant proteins, air1Δ air2Δ TRF4-TAP cells (ACY2131) containing AIR1 URA3 maintenance plasmid (pAC1613) and expressing Myc-tagged wild type Air1/2 (pAC2885 and pAC2891) or air1/2 mutants (pAC2886–2890, pAC2892–2896, pAC2902–2909, and pAC2937) were inoculated into 50 ml of Ura−Leu− minimal media and grown overnight at 25 °C to an A600 = 2. To assay binding between Air2 and trf4 mutant proteins, AIR1-TAP cells (ACY1061) overexpressing Myc-tagged wild type Trf4 (pAC2910), trf4-DADA (pAC2914), or trf4–378 (pAC3049) were similarly grown. Cells were pelleted by centrifugation at 2,163 × g and transferred to fresh tubes. Each cell pellet was resuspended in 1 ml of IPP150 buffer (10 mm Tris-HCl, pH 8, 150 mm NaCl, 0.1% Nonidet P-40) supplemented with protease inhibitors (1 mm phenylmethanesulfonyl fluoride (PMSF), 3 ng/ml pepstatin A, leupeptin, aproptinin, and chymostatin), 10 scoops of glass beads were added, and sample was vigorously disrupted in a Mini Bead Beater 16 cell disrupter (Biospec) for 4 × 2 min at 25 °C. Cell debris was pelleted by centrifugation at 16,000 × g for 30 min at 4 °C. Protein supernatant was transferred to a fresh Eppendorf tube and centrifuged at 16,000 × g for an additional 20 min at 4 °C. Protein supernatant was transferred to a new Eppendorf tube using a 1-ml syringe. Protein lysate concentration was determined by a Bradford assay using the Bio-Rad protein assay. Lysate (2–5 mg) in 1 ml of IPP150 buffer was incubated with 25 μl of IgG-SepharoseTM beads (GE Healthcare) overnight at 4 °C with mixing. Beads were pelleted by centrifugation at 2,300 × g for 1 min, and unbound supernatant was transferred to fresh Eppendorf tube. Beads were then washed three times with 1 ml of IPP150 buffer for 10 min each. Input lysate (50 μg), unbound supernatant (50 μg), and all bound Trf4-TAP-bead or Air1-TAP-bead sample were analyzed by SDS-PAGE and immunoblotting with anti-Myc monoclonal antibody to detect Air1/2-Myc proteins or Trf4-Myc proteins and loading control anti-Pgk1 monoclonal antibody to detect 3-phosphoglycerate kinase.
Analysis of Protein Expression Levels
For analysis of Myc-tagged air1/2 mutant protein levels, air1Δ air2Δ cells (ACY1095) harboring an AIR2 URA3 maintenance plasmid (pAC1614) and expressing Myc-tagged air1/2 protein (pAC2885–2896) were grown in minimal media overnight at 25 °C, the optical density of the cells was measured at 600 nm, and 10-ml cultures with an A600 = 0.5 were prepared and grown for 4 h at 25 °C. The cultures were then split, with one set of 5-ml cultures maintained at 25 °C and the other set shifted to 37 °C for an additional 2 h. For analysis of Myc-tagged Trf4, trf4 mutant, Trf5, and Mtr4 protein levels, wild type cells (W303) or air1-C178R air2Δ cells (ACY2020) overexpressing Myc-tagged Trf4 (pAC2910), trf4-DADA (pAC2914), trf4–378 (pAC3049), Trf5 (pAC3050), or Mtr4 (pAC3051) protein were grown in minimal media overnight at 25 °C, and 10-ml cultures with an A600 = 0.5 were prepared and grown for 4 h at 25 °C and then split, with one set of cultures maintained at 25 °C and the other set shifted to 30 °C for an additional 2 h. For analysis of Myc-tagged air1-C178R protein levels upon overexpression of Trf4 protein, air1Δ air2Δ cells expressing Myc-tagged air1-C178R (pAC2890) and containing vector (pRS423) or overexpressing Trf4 (pAC2940) or trf4-DADA (pAC2946) were grown in minimal media overnight at 25 °C, and then 10-ml cultures with an A600 = 0.5 were prepared and grown for 6 h at 25 °C. Whole cell lysates of cells were then prepared, and 40–50 μg of protein was analyzed by immunoblotting with an anti-Myc monoclonal antibody to detect Myc-tagged proteins.
TRAMP Protein Stability Assay
To determine the stability of TRAMP proteins, wild type (W303) cells and air1-C178R air2Δ (ACY2020) cells overexpressing Trf4 (pAC2910), trf4-DADA (pAC2914), Trf5 (pAC3050), or Mtr4 (pAC3051) were grown in 10 ml of minimal media overnight at 25 °C, the optical density of the cells was measured at 600 nm, and 60-ml cultures with an A600 = 0.5 were prepared and grown for 3 h at 25 °C. Cycloheximide (Sigma) was added to the cultures at a final concentration of 100 μg/ml. Samples (10 ml) were taken immediately after the cycloheximide addition at the zero time point. Cultures were then grown at 25 °C, and 10-ml samples were collected at 30, 60, 90, 120, and 150 min time points after the cycloheximide addition. Protein lysate from each cell sample time point was prepared by resuspension of the cells in 0.5 ml of Lysis Buffer (PBS, 0.1% Nonidet P-40, protease inhibitors (1 mm PMSF, 3 ng/ml pepstatin A, leupeptin, aproptinin, and chymostatin)), the addition of three scoops of glass beads, disruption in a Mini Bead Beater 16 cell disrupter (Biospec) for 4 × 1 min at 25 °C, and centrifugation to remove cell debris at 16,000 × g for 20 min at 4 °C. Protein lysate concentration was determined by a Bradford assay using the Bio-Rad protein assay. Protein lysate (30 μg) from each time point was analyzed by SDS-PAGE and immunoblotting with an anti-Myc monoclonal antibody to detect Myc-tagged TRAMP proteins and an anti-Pgk1 monoclonal antibody to detect 3-phosphoglycerate kinase as a loading control.
SDS-PAGE and Immunoblotting
To detect Myc-tagged Air1/2, Trf4, Trf5, and Mtr4 proteins in cell lysates, and Myc-tagged Air1/2 and Trf4 proteins in input/unbound lysates, and bound IgG-Sepharose bead protein samples from the Trf4-TAP or Air1-TAP binding assay, protein samples were resolved on Criterion 4–20% gradient gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad), and membranes were probed with anti-Myc monoclonal antibody 9B11 (1:2000; Cell Signaling) and peroxidase-conjugated anti-mouse antibody (1:3000; Jackson ImmunoResearch Laboratories) to detect Air1/2-Myc or Trf4-Myc proteins. As a loading control, 3-phosphoglycerate kinase (Pgk1) protein levels in lysate samples were detected using an anti-Pgk1 monoclonal antibody (1:15,000; Invitrogen) and peroxidase-conjugated anti-mouse antibody (1:3000; Jackson ImmunoResearch Laboratories). To detect hZCCHC7 in HeLa cell lysates and unbound and bound Myc-agarose bead protein samples from mPAPD5/hPAPD7-Myc immunoprecipitations, protein samples were resolved on TGX Mini 4–15% gradient gels (Bio-Rad) and transferred to nitrocellulose membranes (Bio-Rad), and membranes were probed with anti-hZCCHC7 rabbit polyclonal antibody (1:2000; HPA021088, Sigma) and peroxidase-conjugated anti-rabbit antibody (1:4000; Jackson ImmunoResearch Laboratories). Membranes were also probed with anti-Myc monoclonal antibody 9B11 (1:2000; Cell Signaling) to detect Myc-tagged PAPD5 and PAPD7.
Quantitation of Immunoblots
The protein band intensities/areas from all immunoblots were quantitated using ImageJ version 1.4 software (National Institute of Health, Bethesda, MD), and relevant percentages of protein were calculated in Microsoft Excel for Mac 2008 (Microsoft Corp.). In binding assays, all bound air1/2-Myc intensities were normalized to Trf4-TAP intensities, bound trf4-Myc intensities were normalized to Air1-TAP intensities, and input air1/2/trf4-Myc intensities were normalized to Pgk1 intensities. In TRAMP protein level and stability assays, all Myc-tagged Trf4, trf4-DADA, Trf5, and Mtr4 intensities were normalized to Pgk1. In the Trf4-TAP and Air1-TAP binding assays, to quantitate the percentage of bound air1/2-Myc or trf4-Myc proteins relative to the amount of input protein and wild type Air1/2 or Trf4, the normalized bound air1/2/trf4 intensities were divided by the normalized input air1/2/trf4 intensities, and these values were divided by the wild type Air1/2/Trf4 intensity. For the percentage of bound and input air1 ZnK1–5 mutants, the S.E. value (S.D. divided by the square root of n, where n = 3) from triplicate binding experiments was calculated. To quantitate the percentage of input air1/2/trf4 proteins relevant to wild type Air1/2/Trf4, the normalized input air1/2/trf4 intensities were divided by the input wild type Air1/2/Trf4 intensities. To quantitate the percentage decrease in Myc-tagged TRAMP proteins in wild type and air1-C178R air2Δ cells at 30 °C compared with 25 °C, the normalized TRAMP protein intensities at 30 °C were divided by normalized intensities at 25 °C. To quantitate the -fold increase in air1-C178R in cells overexpressing Trf4 or trf4-DADA relevant to cells containing vector alone, the air1-C178R intensities in vector alone and Trf4/trf4 samples were normalized to Pgk1 intensities, and the normalized air1-C178R intensities in Trf4/trf4 samples were divided by that in the vector alone sample. In the TRAMP protein stability assay, to quantitate the percentages of Myc-tagged TRAMP proteins in wild type and air1-C178R air2Δ cells at each time point relevant to time 0, the normalized TRAMP protein intensities at each time point were divided by the normalized TRAMP protein intensity at time 0. These percentages were graphed using Microsoft Excel. To generate the graphs with least-squares fit lines through the log-transformed TRAMP protein intensities at time points 0, 60, and 90 min, the natural logarithms (ln) of normalized TRAMP protein intensities at each time point were graphed, and the best fit lines were calculated using Microsoft Excel. All best fit lines had good correlation coefficients (R2 = 0.98–0.99). The slopes of the best fit lines were calculated to determine the decay rate constants (k) for the TRAMP proteins, and the half-lives (T½) of the TRAMP proteins were calculated using the equation, T½ = ln(2)/k as previously used to determine protein half-lives (54).
Localization of hZCCHC7
Endogenous hZCCHC7 was visualized in HeLa cells by indirect immunofluorescence microscopy using an anti-hZCCHC7 polyclonal rabbit antibody (HPA021088, Sigma) according to the protocol from the Human Protein Atlas (available on the World Wide Web) (55). Cells were fixed with 4% formaldehyde (EM Science) in growth medium supplemented with 10% fetal bovine serum (FBS) for 15 min, permeabilized with 0.1% Triton X-100 in PBS for 3 × 5 min, washed with PBS, and incubated at 4 °C overnight with primary rabbit anti-hZCCHC7 antibody (1:1000; HPA021088, Sigma) and mouse anti-fibrillarin antibody (1:1000; EnCore) and in PBS supplemented with 4% FBS. Cells were then washed with PBS for 4 × 5 min and incubated with secondary anti-rabbit fluorescein-conjugated antibody or anti-mouse Texas Red-conjugated antibody (1:1000; Jackson ImmunoResearch Laboratories). Cells were incubated with Hoechst stain to mark nuclear DNA. Images were obtained using an Olympus IX81 microscope with a 0.3 numerical aperture, ×60 Zeiss Plan-Neofluor objective. Images were captured using a Hamamatsu digital camera with Slidebook software (version 5) and globally processed for brightness using Adobe Photoshop.
Immunoprecipitation of mPAPD5/hPAPD7 and hZCCHC7
To immunoprecipitate mPAPD5/hPAPD7, HEK293 cells were transiently transfected with Myc-tagged mPAPD5 (pAC2998) or Myc-tagged hPAPD7 (pAC2999) plasmid using Lipofectamine 2000 (Invitrogen), and cells were collected 48 h after transfection. Control untransfected HEK293 cells or cells containing Myc-tagged mPAPD5 or hPAPD7 were lysed in binding buffer (50 mm Tris-HCl, pH 8, 100 mm NaCl, 0.3% Nonidet P-40, 32 mm NaF, protease inhibitors (1 mm PMSF, 3 ng/ml pepstatin A, leupeptin, aprotinin, and chymostatin)). Cell lysates (0.5 mg) in 0.5 ml of binding buffer were incubated with 40 μl of agarose-conjugated monoclonal mouse c-Myc (9E10) antibody beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 2 h at 4 °C with mixing. Beads were pelleted by centrifugation, unbound supernatant was removed, and beads were washed three times with 0.5 ml of binding buffer for 5 s each. Input, unbound (25 μg), and bound Myc bead (one-quarter the total amount) samples were analyzed by SDS-PAGE and immunoblotting with anti-hZCCHC7 polyclonal antibody (Sigma) to detect hZCCHC7 protein and anti-Myc monoclonal antibody (Cell Signaling) to detect mPAPD5 and hPAPD7. To immunoprecipitate hZCCHC7, HEK293 cells were transiently transfected with Myc-tagged mPAPD5 or hPAPD7 and lysed in binding buffer as before. HEK293 cell lysates (0.5 mg) containing Myc-tagged mPAPD5 or hPAPD7 in 0.5 ml of lysis buffer were incubated with 5 μl of anti-hZCCHC7 polyclonal antibody (Sigma) or a control rabbit IgG antibody (GE Healthcare) alone overnight at 4 °C with mixing. Lysates were further incubated with 20 μl of Dynabeads® coupled to Protein G (Invitrogen) for 2 h at 4 °C with mixing. Beads were then washed three times as before, and input, unbound, and bound hZCCHC7-bead or IgG bead samples were analyzed by SDS-PAGE and immunoblotting with anti-Myc monoclonal antibody (Cell Signaling) to detect mPAPD5 and hPAPD7.
RESULTS
Functional Analysis of Air1/2 Zinc Knuckle Mutants
The Air1 and Air2 proteins of the TRAMP complex are thought to mediate RNA recognition via CCHC zinc knuckle domains (Fig. 1A). The Air1 and Air2 zinc knuckle domains contain five CCHC-type zinc knuckles that are highly similar to one another (67% identity, 80% similarity; Fig. 1B). To identify temperature-sensitive mutants of AIR1/2 and thus pinpoint regions within the Air proteins that are functionally important in an unbiased manner, we performed random PCR mutagenesis of the AIR1/2 open reading frame and screened for temperature-sensitive mutants in air1Δ air2Δ cells at 37 °C as described under “Experimental Procedures.” The screen was performed in cells deleted for both AIR1 and AIR2 because air1Δ or air2Δ single deletion mutants showed no significant growth defects, but an air1Δ air2Δ double deletion mutant exhibited a severe growth defect compared with wild type cells (Fig. 1C) (14, 45). This screen led to the identification of an air2 mutant containing a single cysteine to arginine amino acid change in the second cysteine of zinc knuckle 5 (air2-C167R) that showed significantly impaired growth at 37 °C (Fig. 1D). An air1 mutant with an analogous amino acid substitution in zinc knuckle 5 (air1-C178R), generated by site-directed mutagenesis, also exhibited profoundly slow growth at 37 °C (Fig. 1D). This result suggests that zinc knuckle 5 of the Air proteins is functionally important.
Given that random mutagenesis of AIR1/2 identified a mutation in zinc knuckle 5 (ZnK5) that confers temperature-sensitive growth, we systematically altered the corresponding second cysteine in each of the other four zinc knuckles (ZnK1–4) of Air1/2 to arginine (Fig. 1A) and assessed the impact on protein function by examining the growth of air1Δ air2Δ cells expressing each air mutant as the sole cellular copy of the Air protein. Cell growth for each air mutant was monitored at 16, 25, 30, and 37 °C. The air1 and air2 ZnK1–3 mutants showed growth comparable with control AIR1/2 cells at all temperatures tested (Fig. 1D; summarized in Table 2). In contrast, air1 ZnK4 mutant (air1-C139R) and air2 ZnK4 mutant (air2-C126R) are temperature-sensitive and exhibit severely impaired growth at 37 °C compared with AIR1/2 wild type cells (Fig. 1D; summarized in Table 2). We confirmed these results by measuring the optical density of the cells in liquid culture over time and generating quantitative growth curves for the cells (Fig. 1E). The growth analysis highlights the fact that growth of the air1/2 ZnK4 mutants is not as severely impaired as that of the air1/2 ZnK5 mutants at 37 °C. In addition, the curves indicate that air1 ZnK4/5 mutants are more growth-impaired/thermosensitive than the air2 ZnK4/5 mutants at 37 °C. Growth analysis of air1/2 zinc knuckle 1–5 mutants therefore suggests that both zinc knuckle 4 and 5 of the Air proteins are functionally important.
TABLE 2.
Summary of air1 mutant phenotypes
+++, strong/high; ++, intermediate; +, weak/low; −, no growth; ND, not determined.
| Growth at |
Levels of NEL025c CUT | Interaction with Trf4 | ||||
|---|---|---|---|---|---|---|
| 16 °C | 25 °C | 30 °C | 37 °C | |||
| Air1 wild type | +++ | +++ | +++ | +++ | + | +++ |
| air1 ZnK1–5 mutants | ||||||
| C79R | +++ | +++ | +++ | +++ | ++ | +++ |
| C97R | +++ | +++ | +++ | +++ | ++ | +++ |
| C117R | +++ | +++ | +++ | +++ | ++ | +++ |
| C139R | +++ | +++ | +++ | + | +++ | ++ |
| C178R | +++ | + | − | − | ++ | + |
| air1 IWRXY mutants | ||||||
| I152A | +++ | +++ | +++ | ++ | ND | ++ |
| W153A | +++ | +++ | + | − | ND | + |
| R154A | +++ | +++ | +++ | + | ND | ++ |
| Y156A | +++ | +++ | + | − | ND | + |
| air1 ZnK5 mutants | ||||||
| F174A | +++ | +++ | +++ | +++ | ND | ++ |
| Y176A | +++ | +++ | +++ | − | ND | + |
| F184A | +++ | +++ | +++ | − | ND | + |
| D186A | +++ | +++ | +++ | − | ND | + |
| R192A | +++ | +++ | +++ | +++ | ND | ++ |
To further characterize the Air1/2 zinc knuckle mutants, we examined the steady-state protein levels of C-terminally Myc-tagged wild type Air1/2 and air1/2 ZnK1–5 mutant proteins. air1Δ air2Δ cells containing AIR2 URA3 maintenance plasmid and expressing Myc-tagged wild type Air1/2 protein or air1/2 ZnK1–5 mutant protein were grown at 25 and 37 °C, and whole cell lysates prepared from these cells were analyzed by SDS-PAGE and immunoblotting with anti-Myc antibody. At 25 °C, air1/2 ZnK1–4 mutants were expressed at a level similar to that of wild type Air1/2, whereas the air1/2 ZnK5 mutant showed decreased expression compared with wild type Air1/2 (Fig. 1F). At 37 °C, the expression of air1/2 ZnK1–3 mutants was similar to that of wild type Air1/2; however, the expression of air1/2 ZnK4 and -5 mutants was profoundly decreased relative to wild type Air1/2 (Fig. 1F). These data indicate that the air1/2 ZnK4 and -5 mutant proteins are unstable at 37 °C and suggest that the thermosensitive growth of the air1/2 ZnK4 and -5 mutants could result from a decrease in the steady-state level of the mutant proteins at 37 °C. However, unlike the air1/2 ZnK5 mutants, the steady-state level of the air1/2 ZnK4 mutant proteins is not greatly reduced at 25 °C.
To examine the cellular localization of the air1/2 zinc knuckle mutants, we localized C-terminally GFP-tagged Air1/2 and air1/2 ZnK1–5 mutants in live cells by direct fluorescence microscopy. Like wild type Air1, air1 ZnK1–5 mutants localized predominantly to the nucleolus and to a lesser extent to the nucleoplasm, whereas, similarly to wild type Air2, air2 ZnK1–5 mutants localized mostly to the nucleoplasm with a degree of nucleolar localization (supplemental Fig. S1). These results indicate that the air1/2 zinc knuckle mutant proteins were correctly targeted to the nucleus.
Residues in the Air1 Zinc Knuckle 4-5 Linker and Zinc Knuckle 5 Are Critical for Air1 Function
Our results with air1/2 ZnK1–5 mutants indicate that ZnK4 and ZnK5 play a role in the structural integrity of the Air proteins, which complicates analysis but does not exclude the possibility that ZnK4 and ZnK5 also play an important functional role. To gain further insight into the specific functional requirements of the ZnK4-5 region, we targeted residues within the linker region between Air1 ZnK4 and ZnK5 that would be less likely to impact the structural integrity of the protein. To identify conserved residues in the Air1 ZnK domain, we performed a BLAST search of the NCBI RefSeq protein data base with the Air1 ZnK1–5 amino acid sequence to identify putative Air1 orthologues and aligned the ZnK domains of these Air1 proteins (Fig. 1B). The alignment of ZnK domains from S. cerevisiae Air1 and Air2, S. pombe Air1, and human hZCCHC7 and hZCCHC9 highlights conserved residues within and flanking Air1 ZnK5 (Fig. 1B). In addition, the alignment reveals a conserved IWRXY motif adjacent to Air1 ZnK4 within the Air1 ZnK4-5 linker region (Fig. 1B). Notably, one putative human Air1 orthologue, hZCCHC7, a 542-residue protein with four intact CCHC zinc knuckles (35% identity to Air1 ZnK1–5), contains the IWRXY motif, but the other potential human Air1 orthologue, hZCCHC9, a 271-residue protein with four CCHC zinc knuckles (19% identity to Air1 ZnK1–5), lacks this IWRXY motif (Fig. 1B).
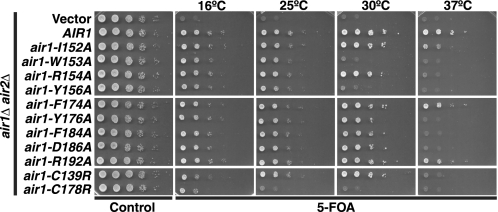
Given the importance of Air1 ZnK4 and 5, we wished to determine if the IWRXY motif in the Air1 ZnK4-5 linker region and the evolutionarily conserved residues in Air1 ZnK5 are functionally important. To this end, we mutated the IWRXY motif and ZnK5 residues to alanine in Air1 and tested the function of these air1 mutants by examining the growth of air1Δ air2Δ cells expressing each air mutant at different temperatures as described under “Experimental Procedures.” The air1 IWRXY mutants, air1-W153A and air1-Y156A, grew slowly at 30 °C and exhibited severely impaired growth at 37 °C compared with AIR1 cells (Fig. 2; summarized in Table 2). The slow growth of air1-W153A and air-Y156A cells at 30 and 37 °C was similar to that of air1-C178R cells, but, unlike air1-C178R cells, air1-W153A and air-Y156A cells grow normally at 25 °C. The air1 IWRXY mutants air1-I152A and air1-R154A grew similarly to AIR1 cells at 30 °C but showed slower growth at 37 °C relative to AIR1 cells. These results confirm that the IWRXY motif residues in the Air1 ZnK4-5 linker, particularly Trp-153 and Tyr-156, are functionally important. The air1 ZnK5 mutant air1-F174A and air1-R192A cells grew similarly to AIR1 cells at all temperatures, suggesting that Air1 residues Phe-174 and Arg-192 that flank the Air1 ZnK5 are not critical for function and importantly do not impact protein stability (Fig. 2; summarized in Table 2). In contrast, air1 ZnK5 mutants air1-Y176A, air1-F184A and air1-D186A grew similarly to AIR1 cells at 30 °C but exhibited profoundly temperature-sensitive growth at 37 °C (Fig. 2; summarized in Table 2). The slow growth of these air1 ZnK5 mutants at 37 °C was similar to that of air1 ZnK4 mutant air1-C139R cells. This result suggests that Air1 residues Tyr-176, Phe-184, and Asp-186 within Air1 ZnK5 are also functionally important. Critically, these air1 IWRXY and ZnK5 mutants predominantly localize, like wild type Air1, to the nucleolus and to a lesser extent to the nucleoplasm (supplemental Fig. S1).
FIGURE 2.
Air1 zinc knuckle 4-5 linker IWRXY motif and zinc knuckle 5 residues are functionally important. air1 IWRXY motif mutants (air1-I152A, -W153A, -R154A, and -Y156A) and air1 ZnK5 mutants (air1-Y176A, -F184A, and -D186A) exhibit temperature-sensitive growth. air1Δ air2Δ cells containing AIR2 URA3 maintenance plasmid and vector, AIR1, air1 IWRXY mutant (air1-I152A, -W153A, -R154A, or Y156A), air1 ZnK5 mutant (air1-F174A, -Y176A, -F184A, -D186A, or -R192A), or air1 ZnK4 and -5 mutant (air1-C139R or -C178R) LEU2 test plasmids were grown to saturation, serially diluted, and spotted on control and 5-FOA plates at the indicated temperatures.
Air1 Zinc Knuckle Mutants Exhibit Elevated CUT RNA Levels
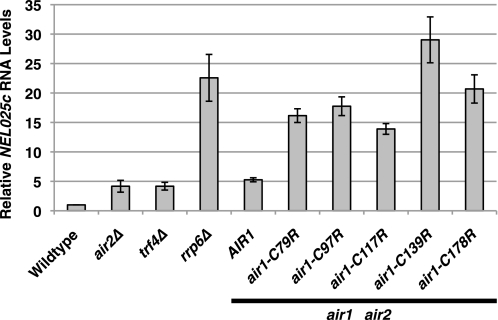
Because retroviral CCHC type zinc knuckles bind to single-stranded tetraloop RNA and Air1/2 protein is required for Trf4-mediated polyadenylation of tRNA and RNA oligomers in vitro (14, 21, 22, 47, 48), Air1/2 CCHC zinc knuckles are hypothesized to facilitate recognition of RNA. In support of a key role for Air proteins in mediating TRAMP function, CUTs, normally polyadenylated by TRAMP and degraded by the exosome, are stabilized in air1Δ air2Δ cells (14). To determine whether CUTs are stabilized in air1 zinc knuckle 1–5 mutants, we analyzed CUT RNA levels by quantitative RT-PCR. Total RNA was isolated from wild type cells, control TRAMP/exosome mutant cells (air2Δ and trf4Δ/rrp6Δ), and air1Δ air2Δ cells containing only AIR1 or air1 ZnK1–5 mutant plasmids grown at 25 °C and reverse transcribed, and cDNA was measured by quantitative PCR using gene-specific primers for the CUT NEL025c. Relative to wild type cells, the air2Δ and trf4Δ TRAMP mutants exhibited a 3-fold increase in NEL025c RNA levels, whereas the rrp6Δ exosome mutant showed a 22-fold increase in NEL025c RNA levels (Fig. 3). Like air2Δ cells, air1Δ air2Δ cells expressing AIR1 exhibited a 4-fold increase in NEL025c RNA levels relative to wild type cells. In contrast, air1Δ air2Δ cells expressing air1 ZnK1–3 and air1 ZnK5 mutants showed a 13–20-fold increase in NEL025c RNA levels, whereas the air1Δ air2Δ cells expressing air1 ZnK4 mutant cells showed a 28-fold increase in NEL025c RNA levels relative to wild type cells (Fig. 3; summarized in Table 2). Because air1 ZnK1–4 mutant proteins were expressed at levels similar to those of wild type Air1 protein at 25 °C (Fig. 1E), these results suggest that Air1 zinc knuckles 1–4 are important for the recognition/degradation of CUT RNA. In particular, Air1 zinc knuckle 4, which when mutated causes the greatest increase in NEL025c RNA levels, may be critical for CUT RNA recognition/degradation.
FIGURE 3.
Air1 zinc knuckles 1–5 are important for the degradation of NEL025c CUT RNA. air1 ZnK1–5 mutants (air1-C79R, -C97R, -C117R, -C139R, and -C178R) show elevated levels of NEL025c CUT RNA relative to wild type cells. Total RNA from wild type, air2Δ, trf4Δ, rrp6Δ, and air1Δ air2Δ cells containing only AIR1 or air1 ZnK1–5 mutant plasmid grown at 25 °C was measured by quantitative RT-PCR using gene-specific primers for the CUT NEL025c as described under “Experimental Procedures.” Relative NEL025c RNA levels were measured in triplicate biological samples, normalized to a control PGK1 transcript by the ΔΔCt method, averaged, and graphically shown as -fold increase relative to wild type cells. Error bars, S.E.
Air1/2 Zinc Knuckles 4 and 5 and ZnK4-5 Linker IWRXY Motif Are Required for Proper Interaction with Trf4
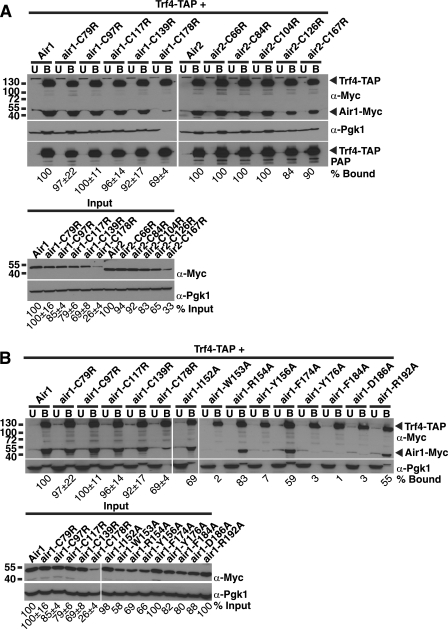
The recently reported Trf4-Air2 structure shows that Air2 ZnK4 and ZnK5 interact with Trf4 (49), which confirms a previous yeast two-hybrid screen with Trf4 that identified Air1 ZnK4 and Air2 ZnK2–5 peptides as interacting with Trf4 (22). To determine if air1/2 ZnK1–5 mutants show decreased interaction with Trf4, we examined binding of Myc-tagged air1/2 ZnK mutants to TAP-tagged Trf4 expressed in S. cerevisiae cells as described under “Experimental Procedures.” The air1/2 ZnK1, -2, and -3 mutants showed binding to Trf4 that was comparable with wild type Air1/2 (96–100% bound; Fig. 4A; summarized in Table 2). In contrast, air1/2 ZnK4 and -5 mutants showed decreased binding to Trf4 compared with wild type Air1/2 (84–92% ZnK4 mutant bound, 69–90% ZnK5 mutant bound; Fig. 4A; summarized in Table 2), confirming that Air1/2 zinc knuckles 4 and 5 are important for binding to Trf4.
FIGURE 4.
Air1 zinc knuckle 4 and 5 and zinc knuckle 4-5 linker IWRXY motif are important for interaction with Trf4. A, air1 ZnK4 and -5 mutants (air1-C139R and -C178R) and air2 ZnK4 and -5 mutants (air2-C126R and -C167R) show decreased interaction with Trf4 relative to wild type Air1. TAP-tagged Trf4 was precipitated from lysates of TRF4-TAP cells expressing Myc-tagged Air1, air1 ZnK1–5 mutants (air1-C79R, -C97R, -C117R, -C139R, and -C178R), Air2, or air2 ZnK1–5 mutants (air1-C66R, -C84R, -C104R, -C126R, and -C167R) and bound (B), unbound (U), and input fractions were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody (α-Myc) to detect Air1/2-Myc proteins and an anti-Pgk1 antibody (α-Pgk1) to detect 3-phosphoglycerate kinase as a loading control. Unbound and bound fractions were also probed with peroxidase anti-peroxidase (PAP) antibody to detect Trf4-TAP proteins. B, air1 IWRXY mutants (air1-W153A and -Y156A) and air1 ZnK5 mutants (air1-Y176A, -F184A, and -D186A) show decreased interaction with Trf4 relative to wild type Air1. TAP-tagged Trf4 was precipitated from lysates of TRF4-TAP cells expressing Myc-tagged Air1, air1 ZnK1–5 mutants (air1-C79R, -C97R, -C117R, -C139R, and -C178R), air1 IWRXY mutants (air1-I152A, -W153A, -R154A, and -Y156A), and air1 ZnK5 mutants (air1-F174A, -Y176A, -F184A, -D186A, and -R192A), and bound (B), unbound (U), and input fractions were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody to detect Air1/2-Myc proteins and an anti-Pgk1 antibody to detect 3-phosphoglycerate kinase as a loading control. The percentage of bound air1 protein relative to the amount of input protein and wild type Air1 (% Bound) is shown below the bound fractions. For the percentage of bound and input air1 ZnK1–5 mutants, S.E. values from three binding experiments are shown. The percentage of input air1 protein relative to the amount of wild type Air1 protein (% Input) is shown below the input fractions. The percentages of protein were calculated by measuring the intensities of the protein bands as described under “Experimental Procedures.”
We next analyzed whether any of the air1 IWRXY ZnK4-5 linker mutant proteins or air1 ZnK5 mutant proteins exhibit altered interactions with Trf4 relative to wild type Air1. The expression levels of air1 IWRXY mutant air1-I152A and ZnK5 mutants air1-F174A and air1-R192A were similar to wild type Air1 (98–100%), whereas the levels of air1 IWRXY mutants air1-W153A, air1-R154A, and air1-Y156A and ZnK5 mutants air1-Y176A, air1-F184A, and air1-D186A were decreased relative to wild type Air1 (58–88%; Fig. 4B, Input). Importantly, the levels of air1 IWRXY mutants (58–98%) are still greater than air1 ZnK5 mutant, air1-C178R (26%). As observed previously, the air1 ZnK1–3 mutants bound to the same extent as wild type Air1, but the air1 ZnK4 and -5 mutant showed decreased binding to Trf4 compared with wild type Air1 (Fig. 4B; summarized in Table 2). The air1 IWRXY mutants also bound more weakly to Trf4 relative to wild type Air1 (Fig. 4B; summarized in Table 2). The interaction of air1-I152A and air1-R154A with Trf4 was partially decreased compared with Air1 (69–83% bound), whereas air1-W153A and air1-Y156A binding to Trf4 was profoundly decreased relative to Air1 (2–7% bound). These results indicate that the Air1 IWRXY motif residues Trp-153 and Tyr-156 are critical for interaction with Trf4. The air1 ZnK5 mutants also bound more weakly to Trf4 compared with wild type Air1 (Fig. 4B; summarized in Table 2). The interaction of air1-F174A and air1-R192A with Trf4 was partially decreased compared with Air1 (55–59% bound); however, air1-Y176A, air1-F184A and air1-D186A binding to Trf4 was greatly decreased (1–3% bound). These results suggest that Air1 residues Tyr-176, Phe-184, and Asp-186, within ZnK5, are critical for interaction with Trf4. In contrast, Air1 residues Phe-174 and Arg-192, which flank ZnK5, may not be as critical for binding to Trf4. Combined, the data argue that ZnK4, the ZnK4-5 linker IWRXY motif, and ZnK5 in Air1 are required to mediate interactions with Trf4.
Levels of TRAMP Complex Components Are Reduced in air1 Zinc Knuckle 5 Mutant Cells
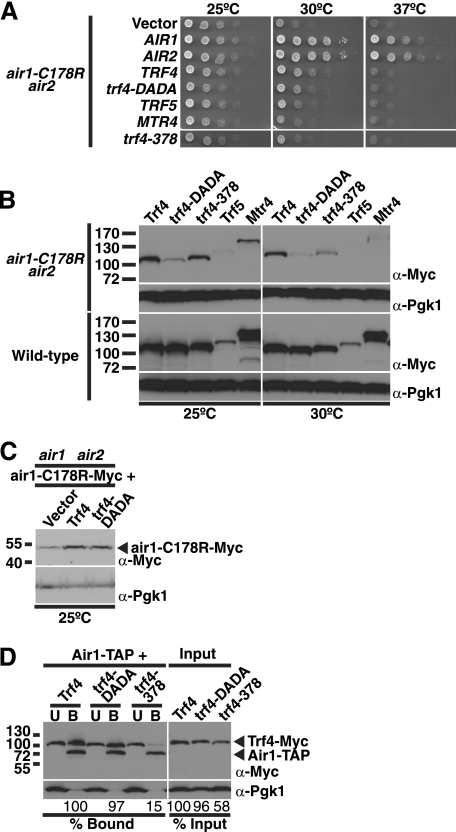
The previous data reveal that air1 ZnK5 mutant air1-C178R cells are temperature-sensitive at 37 °C. To examine genetic interactions with air1-C178R, we integrated the air1-C178R allele at the genomic AIR1 locus in air2Δ cells to create an air1-C178R air2Δ mutant strain. Like air1Δ air2Δ cells harboring air1-C178R on a plasmid, the integrated air1-C178R air2Δ strain was highly thermosensitive and exhibited slow growth at 30 °C and profoundly impaired growth at 37 °C (Fig. 4A). To determine whether components of the TRAMP complex can suppress the temperature-sensitive growth of the air1-C178R mutant, we overexpressed Air1/2, Trf4/5, trf4-DADA, a catalytically inactive mutant of Trf4 (D236A/D238A in the catalytic domain; see Fig. 8) (22), and Mtr4 in the integrated air1-C178R air2Δ cells and examined cell growth at different temperatures as described under “Experimental Procedures.” We also overexpressed trf4–378 (E378A/E381A in the central domain; see Fig. 8), a nonfunctional mutant of Trf4 containing alanine substitutions of Glu-378 and Glu-381 residues (56), which are close to Air2 residues in the Trf4-Air2 structure and could mediate interactions with the Air proteins (49). As expected, overexpression of Air1 or Air2 completely suppressed the slow growth of air1-C178R cells at 30 °C and 37 °C (Fig. 5A). In addition, we overexpressed air1 ZnK1–5 mutants in air1-C178R cells and found that air1 ZnK1–4 mutants, but not the air1 ZnK5 mutant, fully suppress the slow growth of air1-C178R cells at 30 °C (supplemental Fig. S2). Overexpression of Trf4 partially suppressed the slow growth of air1-C178R cells at 30 °C but not at 37 °C (Fig. 4A). Interestingly, overexpression of the catalytically inactive Trf4 mutant, trf4-DADA, suppressed the slow growth of air1-C178R cells at 30 °C only weakly if at all and had no effect at 37 °C, suggesting that the catalytic function of Trf4 is required or that the expression level of the trf4-DADA mutant is reduced in air1-C178R cells (Fig. 4A). Overexpression of Trf5 suppressed the slow growth of air1-C178R cells at 30 °C only weakly and had no effect at 37 °C (Fig. 5A). Overexpression of Mtr4 or trf4–378, however, did not suppress the slow growth of air1-C178R cells at 30 °C or 37 °C (Fig. 5A). These data indicate that Trf4, but not Trf5 or Mtr4, can suppress air1-C178R thermosensitive cell growth.
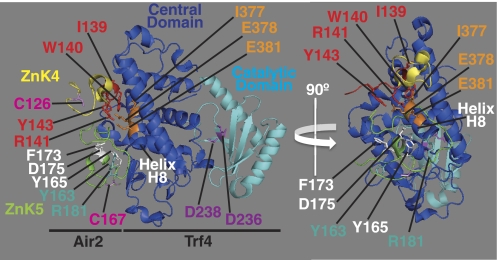
FIGURE 8.
Trf4-Air2 structural model that highlights Air2 ZnK4, ZnK5, and IWRXY motif residues and Trf4 central and catalytic domain residues. A structural model of the Trf4-Air2 complex (Protein Data Bank code 3NYB) based on the recent work of by Hamill et al. (49) is shown to highlight positions of key amino acid residues analyzed in this study. Two views of the Trf4-Air2 complex rotated 90° about the vertical axis relative to each other are shown. Air2 ZnK4 (yellow), Air2 ZnK5 (green), Trf4 central domain (blue), and Trf4 catalytic domain (cyan) are depicted. Air2 ZnK4 residue Cys-126 and ZnK5 Cys-167 (pink), equivalent to Air1 Cys-139 and Cys-178, are labeled. Air2 ZnK5 residues Tyr-165, Phe-173, and Asp-175 (white), equivalent to Air1 ZnK5 residues Tyr-176, Phe-184, and Asp-186, and Air2 ZnK5 residues Tyr-163 and Arg-181 (teal), equivalent to Air1 ZnK5 residues Phe-174 and Arg-192, are labeled. Air2 IWRXY motif residues Ile-139, Trp-140, Arg-141, and Tyr-143 (red), equivalent to Air1 IWRXY residues Ile-152, Trp-153, Arg-154, and Tyr-156, are labeled. Trf4 central domain helix H8 (white) and helix H8 residues Ile-377, Glu-378, and Glu-381 (orange) are labeled. Trf4 catalytic residues Asp-236 and Asp-238 (magenta) are labeled. The structural model of the crystal structure of Trf4-Air2 fragment complex was reproduced from the Protein Data Bank file (entry 3NYB) of the atomic coordinates and structure factors (49) using MacPyMOL software (64) and altered and annotated using Adobe Photoshop and Illustrator CS4 (Adobe).
FIGURE 5.
Overexpression of TRAMP complex components can suppress the thermosensitive growth of air1-C178R ZnK5 mutant cells, and TRAMP component levels are reduced in air1-C178R cells. A, overexpression of Air1/2 and Trf4, but not Trf5 or Mtr4, can effectively suppress air1-C178R thermosensitive growth. air1-C178R air2Δ cells containing vector, AIR1, AIR2, TRF4, trf4-DADA mutant, TRF5, MTR4, or trf4–378 mutant 2μ URA3 plasmids were grown to saturation, serially diluted and spotted on plates, and grown at the indicated temperatures. B, steady-state levels of TRAMP components are reduced in air1-C178R cells relative to wild-type cells. Lysates of wild type and air1-C178R air2Δ cells overexpressing Myc-tagged Trf4, trf4-DADA mutant, trf4–378 mutant, Trf5, or Mtr4 grown at 25 and 30 °C were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody (α-Myc) to detect Trf4-Myc proteins and anti-Pgk1 antibody (α-Pgk1) to detect 3-phosphoglycerate kinase as a loading control. In air1-C178R air2Δ cells, relative to the amounts of the TRAMP proteins at 25 °C, the amounts of the TRAMP proteins at 30 °C show the following percentage decreases: Trf4 (23%), trf4-DADA (50%), trf4–378 (63%), Trf5 (100%), and Mtr4 (79%). In wild type cells, relative to the amounts of the TRAMP proteins at 25 °C, the amounts of the TRAMP proteins at 30 °C show the following percentage decreases: Trf4 (0%), trf4-DADA (8%), trf4–378 (18%), Trf5 (8%), and Mtr4 (0%). C, overexpression of Trf4 increases the steady-state level of air1-C178R mutant protein. Lysates of air1Δ air2Δ cells expressing Myc-tagged air1-C178R protein and containing vector alone or overexpressing Trf4 or trf4-DADA grown at 25 °C were analyzed by immunoblotting with an anti-Myc antibody to detect air1-C178R-Myc protein and anti-Pgk1 antibody to detect 3-phosphoglycerate kinase as a loading control. Relative to cells containing vector alone, cells overexpressing Trf4 show a 3.8-fold increase in air1-C178R, whereas cells overexpressing trf4-DADA show a 3.1-fold increase in air1-C178R protein. D, nonfunctional Trf4 mutant, trf4–378, shows decreased binding to Air1 relative to wild-type Trf4. TAP-tagged Air1 was precipitated from lysates of AIR1-TAP cells expressing Myc-tagged Trf4 or trf4 mutants, trf4-DADA or trf4–378, and bound (B), unbound (U), and input fractions were analyzed by immunoblotting with an anti-Myc antibody to detect Air1/2-Myc proteins and an anti-Pgk1 antibody to detect 3-phosphoglycerate kinase as a loading control. The percentage of bound trf4 protein relative to the amount of input protein and wild type Trf4 (% Bound) is shown below the bound fractions. The percentage of input trf4 protein relative to the amount of wild type Trf4 protein (% Input) is shown below the input fractions. The percentages of protein were calculated by measuring the intensities of the protein bands as described under “Experimental Procedures.”
The variation in suppression of air1-C178R cell growth by the TRAMP components could reflect differences in the expression levels of the proteins. To address this point, we examined the expression levels of Myc-tagged Trf4, trf4-DADA, trf4–378, Trf5, and Mtr4 proteins in air1-C178R cells grown at 25 and 30 °C by immunoblotting. As a control, we also examined the levels of these proteins in wild-type cells. Surprisingly, the level of the trf4-DADA mutant was greatly reduced compared with wild-type Trf4 in air1-C178R cells at 25 °C but not in wild-type cells (Fig. 5B). In addition, the levels of Trf5 and Mtr4 are reduced in air1-C178R cells at 25 °C compared with wild type cells (Fig. 5B). Moreover, the levels of trf4-DADA, trf4–378, Trf5, and Mtr4 were reduced (23–100%) in air1-C178R cells shifted to 30 °C compared with the same cells at 25 °C (Fig. 5B). These data indicate that the steady-state levels of the trf4-DADA mutant, Trf5, and Mtr4 are decreased in air1-C178R cells and suggest that the integrity of the TRAMP complex is compromised in these mutant cells because they express reduced levels of Air1 protein.
The most logical explanation for suppression of air1-C178R thermosensitive cell growth by Trf4 is that Trf4 stabilizes the air1-C178R mutant protein. To address this possibility, we overexpressed Trf4 and trf4-DADA in air1Δ air2Δ cells expressing Myc-tagged air1-C178R protein and examined the level of air1-C178R protein by immunoblotting. Compared with cells containing vector alone, cells overexpressing Trf4 and trf4-DADA showed a 3.1–3.8-fold increase in the level of the air1-C178R mutant protein (Fig. 5C). This result indicates that Trf4 can stabilize the air1 ZnK5 mutant.
As mentioned, the trf4–378 mutant, which did not suppress the air1-C178R thermosensitive cell growth (Fig. 5A) and could not support trf4Δ trf5Δ cell growth (56) (data not shown), contains substitutions in Glu-378 and Glu-381 residues that could mediate interactions with the Air proteins. To test whether Trf4 residues Glu-378 and Glu-381 are important for interaction with Air1, we examined the binding of Myc-tagged Trf4, trf4-DADA, and trf4–378 to TAP-tagged Air1 expressed in S. cerevisiae cells as described under “Experimental Procedures.” Compared with wild type Trf4, the trf4–378 mutant showed greatly decreased binding to Air1 (15% bound), whereas the trf4-DADA mutant showed similar binding to Air1 (97% bound; Fig. 5D). This result indicates that Trf4 residues Glu-378 and Glu-381 are important for interaction with Air1.
TRAMP Complex Proteins Are Unstable in air1 Zinc Knuckle 5 Mutant Cells
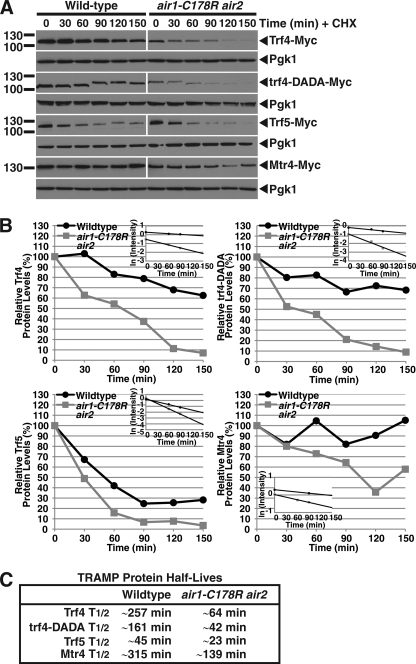
The previous results show that the steady-state levels of TRAMP proteins are reduced in air1-C178R air2Δ cells relative to wild type cells. One potential explanation for the decreased TRAMP protein levels in air1 ZnK5 mutant cells could be that the TRAMP proteins are more unstable in these cells. To address the question of TRAMP protein stability in a rigorous manner, we measured the levels of Myc-tagged Trf4, trf4-DADA, Trf5, and Mtr4 in wild type and air1-C178R air2Δ cells over time following translation inhibition with cycloheximide. We found that the TRAMP proteins were indeed more unstable in air1 ZnK5 mutant cells compared with wild type cells (Fig. 6). Quantitation of the immunoblots (Fig. 6A) showed that the half-lives of TRAMP proteins were reduced 2–4-fold in air1 ZnK5 mutant cells compared with wild type cells (Fig. 6B). Notably, trf4-DADA had a shorter half-life than Trf4 in air1 ZnK5 mutant cells and wild type cells (Fig. 6C). Together, these results indicate that the stability of the TRAMP complex proteins are reduced in air1 ZnK5 mutant cells and suggest that the integrity of the TRAMP complex is compromised in cells with lower levels of Air1 protein.
FIGURE 6.
TRAMP components are unstable in air1-C178R ZnK5 mutant cells. A, protein levels of Trf4, trf4-DADA, Trf5, and Mtr4 decrease more rapidly over time in air1-C178R air2Δ cells relative to wild type cells following cycloheximide treatment. Wild type and air1-C178R air2Δ cells overexpressing Myc-tagged Trf4, trf4-DADA, Trf5, or Mtr4 were treated with translation inhibitor cycloheximide, and cell samples were collected every 30 min. Cell sample lysates from each time point were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody to detect Myc-tagged proteins (Trf4-Myc, trf4-DADA-Myc, Trf5-Myc, and Mtr4-Myc) and an anti-Pgk1 antibody (Pgk1) to detect 3-phosphoglycerate kinase as a loading control. B, the immunoblots shown in A were quantitated to plot the percentage of Trf4, trf4-DADA, Trf5, and Mtr4 proteins at each time point relative to inhibition of translation at time 0 in wild type and air1-C178R air2Δ cells. The inset graphs show the log-transformed intensities of Trf4, trf4-DADA, Trf5, and Mtr4 protein bands at time points 0, 60, and 90 min fitted with linear least-squares fit lines. These best fit lines were used to determine the decay rate constant (k) for each protein. C, the table shows the half-lives (T½) of the TRAMP proteins in wild type and air1-C178R air2Δ cells that were calculated from each decay rate constant using the equation T½ = ln(2)/k (54). Further details on the measurement of the percentages of protein using protein band intensities and calculation of the protein half-lives are described under “Experimental Procedures.”
Endogenous hZCCHC7 Localizes to the Nucleolus and Interacts with Mammalian PAPD5 and PAPD7
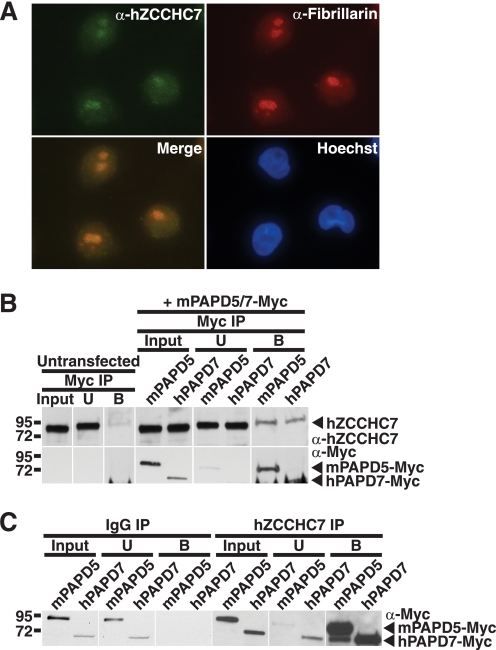
Like S. cerevisiae Air1, a putative human Air1 orthologue should possess CCHC zinc knuckles, localize to the nucleus, and have the capacity to interact with a Trf4-like protein. The human protein, hZCCHC7, which possesses four intact CCHC zinc knuckles, contains the conserved Air1 ZnK4-5 linker IWRXY motif residues and Air1 ZnK5 residues that are important for interaction with Trf4 (see Fig. 1B). To examine endogenous hZCCHC7, we immunoblotted HeLa cell lysates with a commercially available anti-hZCCHC7 polyclonal antibody raised to the N terminus of hZCCHC7. This antibody detected a single band of ∼85 kDa in HeLa cell lysate (supplemental Fig. S3). We localized endogenous hZCCHC7 in HeLa cells by indirect immunofluorescence with the anti-hZCCHC7 antibody as described under “Experimental Procedures.” This immunolocalization revealed that hZCCHC7 colocalized with the nucleolar protein fibrillarin to the nucleolus in HeLa cells (Fig. 7A) similar to the observed nucleolar localization of Air1 in S. cerevisiae cells.
FIGURE 7.
Endogenous hZCCHC7 localizes to the nucleolus in HeLa cells and interacts with both mammalian Trf4 orthologues, PAPD5 and PAPD7, in HEK293 cells. A, endogenous hZCCHC7 localizes to the nucleolus in HeLa cells. hZCCHC7 was localized in HeLa cells by indirect immunofluorescence with anti-hZCCHC7 antibody (α-hZCCHC7). The nucleolar protein fibrillarin was detected with an anti-fibrillarin antibody (α-Fibrillarin) to show the position of the nucleolus. The merge shows that hZCCHC7 colocalizes with fibrillarin in the nucleolus. Hoechst dye was used to stain DNA and show the position of the nucleus. B, endogenous hZCCHC7 interacts with both mammalian Trf4 homologues, PAPD5 and PAPD7. Myc-tagged mPAPD5 and Myc-tagged hPAPD7 were immunoprecipitated (IP) from lysates of HEK293 cells expressing these tagged proteins. As a control, Myc beads alone were used with untransfected HEK293 cells. Input, unbound (U), and bound (B) fractions were analyzed by SDS-PAGE and immunoblotting with an anti-hZCCHC7 antibody. hZCCHC7 binds to both mPAPD5 and hPAPD7, but no binding to Myc beads alone is detected. C, both PAPD5 and PAPD7 interact with endogenous hZCCHC7. Endogenous hZCCHC7 was immunoprecipitated from lysates of HEK293 cells expressing Myc-tagged mPAPD5 (mPAPD5) or Myc-tagged hPAPD7 (hPAPD7), and input, unbound (U) and bound (B) fractions were analyzed by SDS-PAGE and immunoblotting with an anti-Myc antibody. As a control, IgG beads alone were also used for immunoprecipitation. Both mPAPD5 and hPAPD7, but not IgG beads alone, bind to hZCCHC7.
To determine if hZCCHC7 interacts with one or both of the putative mammalian Trf4 orthologues, PAPD5 and PAPD7, we immunoprecipitated Myc-tagged murine PAPD5, which exhibits 94% sequence identity to human PAPD5 and Myc-tagged human PAPD7, from lysates of HEK293 cells expressing these tagged proteins and examined the binding to endogenous hZCCHC7 by immunoblotting with an anti-hZCCHC7 antibody. Endogenous hZCCHC7 bound to both mPAPD5 and hPAPD7 but showed little or no binding to control Myc beads alone (Fig. 7B). To confirm the interactions between hZCCHC7 and PAPD5/7, we immunoprecipitated endogenous hZCCHC7 from lysates of HEK293 cells expressing Myc-tagged mPAPD5 or hPAPD7 and immunoblotted the samples with an anti-Myc antibody. As observed previously, both mPAPD5 and hPAPD7 bound to hZCCHC7 but did not bind to control IgG beads (Fig. 7C). Nucleolar hZCCHC7 thus has the capacity to bind both putative mammalian Trf4 orthologues, PAPD5 and PAPD7, and could be the Air component of a human TRAMP complex.
DISCUSSION
Here we present the first in vivo analysis of full-length Air1 and Air2 zinc knuckle mutant proteins, which reveals that Air1/2 zinc knuckles 4 and 5 are critical for cell function. In particular, our data indicate that Air1/2 ZnK5 is important for interaction with Trf4 and suggest that Air1 ZnK4 may help facilitate recognition of RNA targets. Furthermore, interaction of Air1 with Trf4 and the level of Air1 are critical for the integrity of the TRAMP complex. We also identify a key evolutionarily conserved IWRXY motif in the Air1 ZnK4-5 linker region that is important for function and Trf4 interaction. The presence of the IWRXY motif, a potential signature for Air orthologues, in the human CCHC zinc knuckle protein, hZCCHC7, strongly suggests that this is the human orthologue of S. cerevisiae Air1. In support of the idea, we find that hZCCHC7 localizes to the nucleolus in human cells and binds to both mammalian Trf4 orthologues, PAPD5 and PAPD7, consistent with it being a component of a human TRAMP complex that performs nuclear RNA surveillance. Taken together, the data suggest that Air1/2 forms a stable complex with Trf4 via the ZnK4-5 linker and ZnK5 and targets CUT RNAs for degradation using the ZnK1–4.
Our results support the recent structural model of Trf4 in complex with an Air2 fragment reported by Hamill and colleagues (49), which shows that Air2 ZnK4 and -5 interact with the Trf4 central domain. In Fig. 8, we use the Trf4-Air2 structural model (49) to highlight the positions of the Air2 ZnK4, ZnK5, and IWRXY motif residues and Trf4 central and catalytic domain residues. Importantly, the structural model reveals that Air2 ZnK5 is buried against the surface of Trf4, but Air2 ZnK4 makes little contact with the Trf4 surface and remains tethered to Trf4 via an adjacent linker (49). Our data showing that several Air1 mutants of the ZnK4-5 linker-located IWRXY motif and ZnK5 bind more weakly to Trf4 compared with Air1 ZnK4 mutants are consistent with these structural data. In particular, we note that the Air1 IWRXY motif residues Trp-153 and Tyr-156 and the ZnK5 CX2 residue Tyr-176 and HX4 residues Phe-184 and Asp-186 (in CX2CX4HX4C) are critical for interaction with Trf4 (1–7% bound, Fig. 4B). Importantly, in the Trf4-Air2 structure, the equivalent Air2 IWRXY residues Trp-140 and Tyr-143 and ZnK5 residues Tyr-165, Phe-173, and Asp-175 are located close to Trf4 residues and thus have the potential to interact with Trf4. The Trf4-Air2 structure highlights an interaction between Air2 and the Trf4 helix H8 (residues 373–386) in the central domain (49) (Fig. 8). The proximity of Air2 IWRXY residues to Trf4 helix H8 residues, especially Ile-377, Glu-378, and Glu-381, suggests that they could interact. In particular, Air2 IWRXY residues Trp-140, Arg-141, and Tyr-143 could interact with Trf4 helix H8 residues Ile-377, Glu-381, and Glu-378, respectively, based on interresidue distances of 2.5–4 Å (Fig. 8). In support of this idea, we find that the Trf4 mutant, trf4–378, containing alanine substitutions of helix H8 residues Glu-378 and Glu-381, which is nonfunctional (56) and cannot suppress the air1-C178R thermosensitive cell growth (Fig. 5A), shows decreased binding to Air1 (15% bound; Fig. 5D). These results indicate that Trf4 residues Glu-378 and Glu-381 are functionally important and that altering these residues can impair the Trf4-Air interaction.
The data presented here indicate that the Air1-Trf4 interaction and the steady-state level of the Air1 protein contribute to the stability of TRAMP complex proteins and integrity of the TRAMP complex. The air1-C178R ZnK5 mutant protein shows decreased binding to Trf4 (69 ± 4%) and a lower steady-state level (26 ± 4%) compared with wild-type Air1 protein (Fig. 4A). In air1 ZnK5 mutant cells, Trf4, trf4-DADA, Trf5, and Mtr4 proteins all exhibit reduced stability (T½ reduced 2–4-fold) compared with wild-type cells (Fig. 6). Such instability of the TRAMP proteins in air1 ZnK5 mutant cells may account for the lower steady-state levels of TRAMP proteins in these cells (Fig. 5B). However, we note that the initial (time 0) TRAMP protein levels are reduced in air1 ZnK5 cells relative to wild type cells (Fig. 6), suggesting that additional mechanisms operate to regulate TRAMP protein levels. One possibility is that TRF4/5/MTR4 RNA levels are regulated by the TRAMP complex itself in a feedback mechanism. Thus, in air1 ZnK5 cells, decreased TRAMP complex could result in reduced TRF4/5/MTR4 RNA levels, leading to reduced TRAMP protein levels. Consistent with this proposal, TRF4 RNA levels are reduced in trf5Δ cells (34).
The reduced stability of the air1/2 ZnK5 mutant proteins could, in part, be caused by disruption of the Air1/2-Trf4 interaction. In support, cysteine to arginine substitutions within the zinc-liganding residues in these air1/2 ZnK mutants should disrupt ZnK5, and Air2 ZnK5 is buried against the surface of Trf4 in the structure (49) (Fig. 8). In addition, overexpression of Trf4 causes a 3.8-fold increase in the steady-state level of air1-C178R protein (Fig. 5C). Furthermore, Trf4 protein and trf4-DADA mutant protein, which contains alanine substitutions of catalytic domain residues Asp-236 and Asp-238 that are quite distant from the Trf4 Air1/2-interacting central domain (Fig. 8), are more unstable in air1 ZnK5 cells compared with wild-type cells (T½ reduced 4-fold; Fig. 6), suggesting that Trf4 proteins are normally stabilized by interaction with Air1/2 in wild type cells. The combined results thus suggest that Air1/2 is probably in constant partnership with Trf4/5 in S. cerevisiae cells, consistent with biochemical purification of the complex, and, furthermore, that the Air-Trf4/5 interaction plays an important role in the integrity of the TRAMP complex.
Interestingly, we find that overexpression of Trf4, but not Trf5 or Mtr4, can effectively suppress the thermosensitive growth of air1-C178R ZnK5 mutant cells (Fig. 5A). Trf4 most likely suppresses air1-C178R cell growth by stabilizing the air1-C178R protein because Trf4 overexpression increases the steady-state level of air1-C178R protein (Fig. 5C). The observation that Trf5 only weakly suppresses air1-C178R cell growth is somewhat surprising, given the functional redundancy between Trf5 and Trf4; however, the steady-state level of Trf5 is 4-fold less than Trf4 in wild type cells (57, 58), suggesting that a lower Trf5 level may not be sufficient to stabilize the air1-C178R mutant. The weak suppression by Trf5 may also be explained by the fact that Trf5 is more unstable in air1-C178R cells compared with wild-type cells and exhibits the shortest half-life of the TRAMP components (Fig. 6). Surprisingly, Mtr4 does not suppress air1-C178R cell growth. Like Trf5, the reduced stability of Mtr4 in air1-C178R cells relative to wild-type cells may contribute to the lack of suppression (Fig. 6). In addition, Mtr4 may not directly contact Air1 to stabilize the air1-C178R mutant protein.
The data presented here begin to address the question of which Air1/2 zinc knuckles are important for recognition of RNA. We find that, relative to AIR1 cells, air1 ZnK1–3 and air1 ZnK5 mutant cells show a 2–3-fold increase in NEL025 CUT RNA levels, whereas air1 ZnK4 mutant cells show a 5-fold increase in NEL025 RNA levels (Fig. 3). These results suggest that Air1 ZnK1–5 are important for the recognition/degradation of CUT RNA and suggest an important role for ZnK4 in CUT RNA recognition because the air1 ZnK4 mutant shows the greatest level of NEL025c RNA. Recent work by Hamill et al. (49) using recombinant air2 ZnK1, ZnK2, or ZnK3 mutants in Trf4 polyadenylation assays showed that Air2 ZnK1 is important for polyadenylation of tRNA, suggesting that ZnK1 helps to recognize RNA. This study reported that recombinant Air2 ZnK4-5 alone can weakly support Trf4 polyadenylation of tRNA, suggesting that Air2 ZnK4 and/or ZnK5 may help recognize RNA (49). Although the Trf4-Air2 structure showed that Air2 ZnK4 does not make many contacts with Trf4 and would be available to interact with RNA (Fig. 8), recombinant air2 ZnK4 mutant could not be made to directly test the contribution of ZnK4 in Trf4 polyadenylation. Our observation that the level of NEL025c CUT RNA is increased to the greatest extent in air1-C139R ZnK4 mutant cells is consistent with the idea that Air1/2 ZnK4 facilitates recognition of RNA. Potentially, one or more Air1/2 ZnKs may interact with the RNA substrate nonspecifically, and another Air1/2 ZnK, such as ZnK4, may recognize the RNA substrate specifically. Further Air1/2-RNA binding studies will be required to confirm which zinc knuckles interact directly with RNA.
Our combined results support the notion that the human CCHC zinc knuckle protein, hZCCHC7, is a putative human Air1 orthologue. First, hZCCHC7 ZnK1–5, which show 35% sequence identity to Air1 ZnK1–5, contain the ZnK4-5 linker-located IWRXY motif and key ZnK5 residues (e.g. CX2 residue Tyr-350) that are conserved in the yeast Air1/2 ZnK domains and important for Trf4 interaction (see Fig. 1B). Second, like Air1, hZCCHC7 localizes to the nucleolus in HeLa cells, consistent with hZCCHC7 serving a nuclear function, such as RNA processing/degradation. Third, hZCCHC7 interacts with both putative mammalian Trf4 orthologues, PAPD5 and PAPD7, suggesting that hZCCHC7 could be the Air component in a human TRAMP complex. Although the human CCHC zinc knuckle protein, hZCCHC9, has also been proposed to be a putative human Air1 orthologue, based on the fact that its four zinc knuckles show 19% sequence identity to Air1 ZnK1–5 and it localizes to the nucleolus (46, 59), this seems less likely because hZCCHC9 lacks the conserved IWRXY motif and ZnK5 residues important for Trf4 interaction (see Fig. 1B). In addition, hZCCHC9 ZnK1 has less sequence similarity within Air1 ZnK1, which may be important for tRNA interaction (49), and the hZCCHC9 ZnK3–4 linker (13 residues) is much shorter than the Air1 ZnK4-5 linker (25 residues) (see Fig. 1B).
Future analysis of the function of hZCCHC7, hPAPD5/7, and the human TRAMP complex will be intriguing. Because promoter upstream transcripts are polyadenylated and degraded in an exosome-dependent manner in human cells (20) and aberrant pre-rRNAs have been shown to be polyadenylated by the Trf4 orthologue, PAPD5, in mouse cells (60), it seems highly likely that a human TRAMP complex will act as an exosome cofactor in the processing/degradation of ncRNAs in human cells. Very recently, hZCCHC7 and hPAPD5 were identified in hMTR4 and hRRP6 precipitates and shown to interact with each other and play a role in the polyadenylation of pre-rRNAs (61), providing strong support for a human TRAMP complex containing ZCCHC7 and PAPD5 and also supporting the data presented here. Notably, ZCCHC7 is disrupted in t(9;20) chromosomal translocations in acute lymphoblastic leukemia, and ZCCHC7 expression is up-regulated in early B-cells (62, 63), suggesting that hZCCHC7 plays roles in lymphoblast and B-cell development. hZCCHC7 and the human TRAMP complex may thus help target ncRNAs for degradation to regulate gene expression and cell development.
Acknowledgments
We are most grateful to members of the Corbett laboratory for helpful discussions and comments. We also thank Michael F. Christman for generously providing the TRF4 (pCB727) and TRF5 (pCB557) plasmids.
This work was supported, in whole or in part, by a National Institutes of Health grant (to A. H. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ncRNA
- non-coding RNA
- mRNA
- messenger RNA
- rRNA
- ribosomal RNA
- snRNA
- small nuclear RNA
- snoRNA
- small nucleolar RNA
- tRNA
- transfer RNA
- tRNAiMet
- initiator tRNA
- CUT
- cryptic unstable transcript
- TRAMP
- Trf4/5-Air1/2-Mtr4 polyadenylation
- ZnK
- zinc knuckle
- DAPI
- 4′,6-diamidino-2-phenylindole-dihydrochloride
- 5-FOA
- 5-fluoroorotic acid
- DIC
- differential interference contrast
- TAP tag
- tandem affinity purification tag
- 5-FOA
- 5-fluoroorotic acid
- PAPD5/7
- PAP-associated domain 5/7
- mPAPD5
- murine PAPD5
- hPAPD5
- hPAPD7, hZCCHC7, and hZCCHC9, human PAPD5, PAPD7, ZCCHC7, and ZCCHC9, respectively.
REFERENCES
- 1. Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. (1999) EMBO J. 18, 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T. H. (2001) Nature 413, 538–542 [DOI] [PubMed] [Google Scholar]
- 3. van Hoof A., Lennertz P., Parker R. (2000) Mol. Cell Biol. 20, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuai L., Fang F., Butler J. S., Sherman F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. (1997) Cell 91, 457–466 [DOI] [PubMed] [Google Scholar]
- 6. Dziembowski A., Lorentzen E., Conti E., Séraphin B. (2007) Nat. Struct. Mol. Biol. 14, 15–22 [DOI] [PubMed] [Google Scholar]
- 7. Burkard K. T., Butler J. S. (2000) Mol. Cell Biol. 20, 604–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. (1999) Genes Dev. 13, 2148–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bousquet-Antonelli C., Presutti C., Tollervey D. (2000) Cell 102, 765–775 [DOI] [PubMed] [Google Scholar]
- 10. Das B., Butler J. S., Sherman F. (2003) Mol. Cell Biol. 23, 5502–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T. H. (2002) Mol. Cell Biol. 22, 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Torchet C., Bousquet-Antonelli C., Milligan L., Thompson E., Kufel J., Tollervey D. (2002) Mol. Cell 9, 1285–1296 [DOI] [PubMed] [Google Scholar]
- 13. Kadaba S., Krueger A., Trice T., Krecic A. M., Hinnebusch A. G., Anderson J. (2004) Genes Dev. 18, 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wyers F., Rougemaille M., Badis G., Rousselle J. C., Dufour M. E., Boulay J., Régnault B., Devaux F., Namane A., Séraphin B., Libri D., Jacquier A. (2005) Cell 121, 725–737 [DOI] [PubMed] [Google Scholar]
- 15. Davis C. A., Ares M., Jr. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu Z., Wei W., Gagneur J., Perocchi F., Clauder-Münster S., Camblong J., Guffanti E., Stutz F., Huber W., Steinmetz L. M. (2009) Nature 457, 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neil H., Malabat C., d'Aubenton-Carafa Y., Xu Z., Steinmetz L. M., Jacquier A. (2009) Nature 457, 1038–1042 [DOI] [PubMed] [Google Scholar]
- 18. Martens J. A., Laprade L., Winston F. (2004) Nature 429, 571–574 [DOI] [PubMed] [Google Scholar]
- 19. Camblong J., Iglesias N., Fickentscher C., Dieppois G., Stutz F. (2007) Cell 131, 706–717 [DOI] [PubMed] [Google Scholar]
- 20. Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M. S., Mapendano C. K., Schierup M. H., Jensen T. H. (2008) Science 322, 1851–1854 [DOI] [PubMed] [Google Scholar]
- 21. LaCava J., Houseley J., Saveanu C., Petfalski E., Thompson E., Jacquier A., Tollervey D. (2005) Cell 121, 713–724 [DOI] [PubMed] [Google Scholar]
- 22. Vanácová S., Wolf J., Martin G., Blank D., Dettwiler S., Friedlein A., Langen H., Keith G., Keller W. (2005) PLoS Biol. 3, e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houseley J., Tollervey D. (2006) EMBO Rep. 7, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Callahan K. P., Butler J. S. (2010) J. Biol. Chem. 285, 3540–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Houseley J., Tollervey D. (2008) Biochim. Biophys. Acta 1779, 239–246 [DOI] [PubMed] [Google Scholar]
- 26. Anderson J. T., Wang X. (2009) Crit. Rev. Biochem. Mol. Biol. 44, 16–24 [DOI] [PubMed] [Google Scholar]
- 27. Sadoff B. U., Heath-Pagliuso S., Castaño I. B., Zhu Y., Kieff F. S., Christman M. F. (1995) Genetics 141, 465–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Z., Castaño I. B., De Las Peñas A., Adams C., Christman M. F. (2000) Science 289, 774–779 [DOI] [PubMed] [Google Scholar]
- 29. Aravind L., Koonin E. V. (1999) Nucleic Acids Res. 27, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walowsky C., Fitzhugh D. J., Castaño I. B., Ju J. Y., Levin N. A., Christman M. F. (1999) J. Biol. Chem. 274, 7302–7308 [DOI] [PubMed] [Google Scholar]
- 31. Brown C. E., Sachs A. B. (1998) Mol. Cell Biol. 18, 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Egecioglu D. E., Henras A. K., Chanfreau G. F. (2006) RNA 12, 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kadaba S., Wang X., Anderson J. T. (2006) RNA 12, 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. San Paolo S., Vanacova S., Schenk L., Scherrer T., Blank D., Keller W., Gerber A. P. (2009) PLoS Genet. 5, e1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rougemaille M., Gudipati R. K., Olesen J. R., Thomsen R., Seraphin B., Libri D., Jensen T. H. (2007) EMBO J. 26, 2317–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Win T. Z., Draper S., Read R. L., Pearce J., Norbury C. J., Wang S. W. (2006) Mol. Cell Biol. 26, 1710–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bühler M., Haas W., Gygi S. P., Moazed D. (2007) Cell 129, 707–721 [DOI] [PubMed] [Google Scholar]
- 38. Keller C., Woolcock K., Hess D., Bühler M. (2010) RNA 16, 1124–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakamura R., Takeuchi R., Takata K., Shimanouchi K., Abe Y., Kanai Y., Ruike T., Ihara A., Sakaguchi K. (2008) Mol. Cell Biol. 28, 6620–6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang S., Hitomi M., Hu Y. H., Liu Y., Tartakoff A. M. (1996) Mol. Cell Biol. 16, 5139–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de la Cruz J., Kressler D., Tollervey D., Linder P. (1998) EMBO J. 17, 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Houseley J., Kotovic K., El Hage A., Tollervey D. (2007) EMBO J. 26, 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X., Jia H., Jankowsky E., Anderson J. T. (2008) RNA 14, 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bernstein J., Patterson D. N., Wilson G. M., Toth E. A. (2008) J. Biol. Chem. 283, 4930–4942 [DOI] [PubMed] [Google Scholar]
- 45. Inoue K., Mizuno T., Wada K., Hagiwara M. (2000) J. Biol. Chem. 275, 32793–32799 [DOI] [PubMed] [Google Scholar]
- 46. Sanudo M., Jacko M., Rammelt C., Vanacova S., Stefl R. (2011) Biomol. NMR Assign. 5, 19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. De Guzman R. N., Wu Z. R., Stalling C. C., Pappalardo L., Borer P. N., Summers M. F. (1998) Science 279, 384–388 [DOI] [PubMed] [Google Scholar]
- 48. D'Souza V., Summers M. F. (2004) Nature 431, 586–590 [DOI] [PubMed] [Google Scholar]
- 49. Hamill S., Wolin S. L., Reinisch K. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 15045–15050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 51. Adams A., Gottschling D. E., Kaiser C. A., Stearns T. (1997) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 52. Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 53. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 54. Belle A., Tanay A., Bitincka L., Shamir R., O'Shea E. K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uhlén M., Björling E., Agaton C., Szigyarto C. A., Amini B., Andersen E., Andersson A. C., Angelidou P., Asplund A., Asplund C., Berglund L., Bergström K., Brumer H., Cerjan D., Ekström M., Elobeid A., Eriksson C., Fagerberg L., Falk R., Fall J., Forsberg M., Björklund M. G., Gumbel K., Halimi A., Hallin I., Hamsten C., Hansson M., Hedhammar M., Hercules G., Kampf C., Larsson K., Lindskog M., Lodewyckx W., Lund J., Lundeberg J., Magnusson K., Malm E., Nilsson P., Odling J., Oksvold P., Olsson I., Oster E., Ottosson J., Paavilainen L., Persson A., Rimini R., Rockberg J., Runeson M., Sivertsson A., Sköllermo A., Steen J., Stenvall M., Sterky F., Strömberg S., Sundberg M., Tegel H., Tourle S., Wahlund E., Waldén A., Wan J., Wernérus H., Westberg J., Wester K., Wrethagen U., Xu L. L., Hober S., Pontén F. (2005) Mol. Cell Proteomics 4, 1920–1932 [DOI] [PubMed] [Google Scholar]
- 56. Wang Z., Castaño I. B., Adams C., Vu C., Fitzhugh D., Christman M. F. (2002) Genetics 160, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 58. Reis C. C., Campbell J. L. (2007) Genetics 175, 993–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou A., Zhou J., Yang L., Liu M., Li H., Xu S., Han M., Zhang J. (2008) J. Genet. Genomics 35, 467–472 [DOI] [PubMed] [Google Scholar]
- 60. Shcherbik N., Wang M., Lapik Y. R., Srivastava L., Pestov D. G. (2010) EMBO Rep. 11, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lubas M., Christensen M. S., Kristiansen M. S., Domanski M., Falkenby L. G., Lykke-Andersen S., Andersen J. S., Dziembowski A., Jensen T. H. (2011) Mol. Cell 43, 624–637 [DOI] [PubMed] [Google Scholar]
- 62. An Q., Wright S. L., Moorman A. V., Parker H., Griffiths M., Ross F. M., Davies T., Harrison C. J., Strefford J. C. (2009) Haematologica 94, 1164–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hystad M. E., Myklebust J. H., Bø T. H., Sivertsen E. A., Rian E., Forfang L., Munthe E., Rosenwald A., Chiorazzi M., Jonassen I., Staudt L. M., Smeland E. B. (2007) J. Immunol. 179, 3662–3671 [DOI] [PubMed] [Google Scholar]
- 64. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrodinger, LLC, New York [Google Scholar]
- 65. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. (1992) Gene 110, 119–122 [DOI] [PubMed] [Google Scholar]