Background: β-Catenin mediates a wide variety of cellular processes, but the signaling pathways regulating β-catenin downstream events are not fully understood. The role of MLK3 in modulating β-catenin pathway has not been reported earlier.
Results: MLK3 can induce β-catenin stabilization but inhibit conventional β-catenin/TCF transcriptional activation.
Conclusion: These provide a new mechanism of regulating β-catenin/TCF axis.
Significance: MLK3 can be targeted in regulating the growth of β-catenin overexpressing tumors.
Keywords: AP1 Transcription Factor, Cancer Therapy, Cell Cycle, Jun N-terminal Kinase (JNK), Signal Transduction, Mixed Lineage Kinase 3, β-Catenin
Abstract
Expression of β-catenin is strictly regulated in normal cells via the glycogen synthase kinase 3β (GSK3β)- adenomatous polyposis coli-axin-mediated degradation pathway. Mechanisms leading to inactivation of this pathway (example: activation of Wnt/β-catenin signaling or mutations of members of the degradation complex) can result in β-catenin stabilization and activation of β-catenin/T-cell factor (TCF) signaling. β-Catenin-mediated cellular events are diverse and complex. A better understanding of the cellular signaling networks that control β-catenin pathway is important for designing effective therapeutic strategies targeting this axis. To gain more insight, we focused on determining any possible cross-talk between β-catenin and mixed lineage kinase 3 (MLK3), a MAPK kinase kinase member. Our studies indicated that MLK3 can induce β-catenin expression via post-translational stabilization in various cancer cells, including prostate cancer. This function of MLK3 was dependent on its kinase activity. MLK3 can interact with β-catenin and phosphorylate it in vitro. Overexpression of GSK3β-WT or the S9A mutant was unable to antagonize MLK3-induced stabilization, suggesting this to be independent of GSK3β pathway. Surprisingly, despite stabilizing β-catenin, MLK3 inhibited TCF transcriptional activity in the presence of both WT and S37A β-catenin. These resulted in reduced expression of β-catenin/TCF downstream targets Survivin and myc. Immunoprecipitation studies indicated that MLK3 did not decrease β-catenin/TCF interaction but promoted interaction between β-catenin and KLF4, a known repressor of β-catenin/TCF transcriptional activity. In addition, co-expression of MLK3 and β-catenin resulted in significant G2/M arrest. These studies provide a novel insight toward the regulation of β-catenin pathway, which can be targeted to control cancer cell proliferation, particularly those with aberrant activation of β-catenin signaling.
Introduction
The Wnt/β-catenin signaling pathway has been extensively studied in recent years due to its diverse and important roles in regulating tumorigenesis (1–4) as well as stem cell renewal (5, 6). In cells, β-catenin mediates two major functions; one in the formation of adherens junctions via its interaction with the cell adhesion molecules of the cadherin family (7), and the second is as part of the Wnt-signaling pathway via its interaction with TCF/LEF3 family of transcription factors (1). Central to the Wnt/β-catenin or canonical Wnt signaling pathway is the control of the stability of β-catenin. In the absence of Wnt signaling, free cytoplasmic β-catenin is maintained at very low levels where it is targeted to a phosphorylation-mediated degradation pathway that involves glycogen synthase kinase-3β (GSK3β), axin, and adenomatous polyposis coli (8, 9). In the presence of Wnt ligands, β-catenin degradation complex is destabilized, and β-catenin is stabilized via mechanisms that also involve phosphorylation of low density lipoprotein receptor-related proteins 5 and 6 (LRP5/6) and recruitment of axin to LRP (10). Stabilized free β-catenin then translocates to the nucleus, interacts with the TCF/LEF family of transcription factors, and activates transcription of target genes (11). The list of β-catenin/TCF target genes is still emerging and includes cyclin D1, c-myc, and c-Jun, each of which in turn promotes various cellular events (12). Interestingly, nuclear localization and interaction of β-catenin with LEF-1 are not sufficient for activating TCF-mediated transcription (13), suggesting that additional signaling mechanisms exist to modulate the transcriptional activity of this complex. Extensive information is available in the literature regarding the signaling pathways that regulate stabilization or degradation of β-catenin; however, very little is known regarding those that regulate β-catenin/TCF-mediated target gene transcription.
The serine/threonine kinase mixed lineage kinase 3 (MLK3) is a ubiquitously expressed kinase and belongs to the larger family of MLKs. In mammalian cells, MLK3 functions as a MAP3K (14, 15) and is linked closely with the activation of JNK and its downstream c-Jun transcription factors as well as other MAPK pathways (16). A prominent role of MLK3 in regulating neuronal cell apoptosis is now well established and is believed to involve downstream JNK signaling (17). In addition, recent studies have demonstrated the expression of MLK3 in cancer cells and established its involvement in mediating various pro-oncogenic pathways (18, 19). Cross-talk of Wnt/β-catenin signaling pathway with another closely linked mammalian MAP3K member TGFβ-activated kinase1 (TAK1) has been reported recently where TAK1 negatively regulates Wnt/β-catenin pathway (20). This is accomplished by phosphorylation of TCFs by TAK1 downstream kinase NLK (Nemo-like kinase), resulting in an inhibition of β-catenin/TCF DNA binding and thus β-catenin/TCF transcriptional activity. Interestingly, the Wnt pathway can in turn activate the TAK1/NLK cascade (21), and thus the TAK1-NLK axis might represent a physiological mechanism by which Wnt/β-catenin signaling is terminated in normal cells. It thus seems that MAP3Ks can antagonize the canonical Wnt/β-catenin pathway, but the mechanisms involved are still not clear.
To further elucidate the details of β-catenin pathway and to understand whether other MAP3Ks might be involved in β-catenin regulation, we designed studies in various cell types to determine the effect of MLK3 on β-catenin expression and β-catenin/TCF signaling. Our studies indicate that MLK3 can induce the expression of β-catenin in a variety of cancer cells including prostate cancer cells, an effect that requires the kinase activity of MLK3. Intense β-catenin expression is detected by immunofluorescence analysis when co-transfected with MLK3-WT associated with increased localization in the nucleus and membrane ruffles. This is achieved via post-translational stabilization of β-catenin independent of GSK3β. Surprisingly, despite increasing β-catenin expression, MLK3 antagonized β-catenin/TCF transcriptional activity, suggesting an inhibitory effect of MLK3 on canonical Wnt-pathway. This decrease in β-catenin/TCF transcriptional activity was not due to reduced interaction between β-catenin and TCF4 but was associated with an increased interaction between β-catenin and KLF4. In addition, co-expression of MLK3-WT and β-catenin resulted in cell cycle arrest at the G2/M phase.
EXPERIMENTAL PROCEDURES
Reagents
DMEM, Lipofectamine 2000, and the β-galactosidase assay kit were purchased from Invitrogen; the luciferase assay kit was from Promega (Madison, WI), and formaldehyde was from Polysciences (Warrington, PA). The antibodies utilized were obtained from the following sources: total MLK3 (Epitomics, Burlingame, CA), GAPDH (Ambion, Austin, TX), HA.11 (Covance, Berkeley, CA), β-actin, rabbit anti-HA, and M2 (anti-FLAG) (Sigma), 9E10 (anti-myc) (Roche Applied Science), β-catenin (BD Bioscience), P-c-Jun, total c-Jun, P-cdc2tyr15, total cdc2, cyclin B1, β-tubulin, and TCF4 (Cell Signaling Technology, Danvers, MA), KLF4 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), GST (Millipore-Upstate, Lake Placid, NY), fluorophore-conjugated donkey anti-mouse or anti-rabbit antibodies (Jackson ImmunoResearch, West Grove, PA). The JNK inhibitor SP600125 was from Alexis Biochemicals, Axxora (San Diego, CA), GSK3β inhibitors (AR-A014418 and Kenpaullone) and cycloheximide were from EMD Biosciences (Gibbstown, NJ), and Pan-MLK inhibitor CEP11004 was a gift from Cephalon. The pGL3OT luciferase vector was obtained from Dr. Bert Vogelstein (22), the HA-tagged GSK3β expression vectors (WT, kinase dead (K/A), and S9A) were from Dr. James R Woodgett (University of Toronto, Toronto, Canada) (23), and the KLF4 vectors (WT, ΔC, and ΔN) were from Dr. Wange Lu (University of Southern California, Los Angeles, CA (24), GFP-β-catenin was from Dr. Robert Malenka (Stanford University School of Medicine, CA) (25), and the HA-tagged β-catenin vectors (WT and S37A) were obtained from Dr. Stephen Byers (26). The M2-tagged MLK3 (WT and K/A) and GST-tagged MLK3 (WT, K/A, 1–385 WT, 1–385 K/A) vectors were utilized as described elsewhere (14, 17, 27).
Cell Culture
Human embryonic kidney cell line (HEK-293) and human cervical cancer cell line (HeLa) were maintained in DMEM supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin. Human prostate cancer cells (LNCaP, PC3, and 22Rv1) were maintained in RPMI media supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin. In the studies with various kinase inhibitors, cells were treated with the specific inhibitors after transfection followed by Western blot analysis or luciferase assays.
Transient Transfection and Luciferase Assays
Subconfluent populations of HEK-293, HeLa, LNCaP, and 22Rv1 cells were transiently transfected by various expression vectors utilizing Lipofectamine 2000 as per the manufacturer's instructions and as described previously (19). PC3 cells were transfected utilizing Amaxa nucleofection kit V. Briefly, cells were trypsinized, and 1 × 106 PC3 cells were resuspended in 100 μl of nucleofection solution, transferred to the nucleofection cuvettes, and transfected using Amaxa nucleofector device following the specific program T-019. Cells were recovered by adding 500 μl of complete pre-equilibrated media and transferred to a 6-well plate at 37 °C. After 48 h of recovery in the growth medium, the transfected cells were harvested and analyzed via Western blot analyses or luciferase assays. For the cycloheximide experiments, HeLa cells transiently transfected with the various expression vectors (or MCF-7 and MCF-7-KD cells) were treated with 50 μg/ml cycloheximide (CHX) following a 48-h recovery in growth media. Cells were harvested at various lengths of time after CHX treatment and subjected to Western blot analysis. In these experiments, because overexpression of MLK-WT increases β-catenin expression significantly, twice the amount of total protein lysates was utilized in the Western blots of β-catenin and empty vector (or MCF7-KD) samples compared with the β-catenin and MLK3-WT (or MCF7) samples to normalize β-catenin protein levels between the two sets. Luciferase assays were performed as described earlier (19). Briefly, subconfluent cells were transfected with pGL3OT-luciferase-reporter construct (28) along with β-galactosidase vector (to correct for transfection efficiency) using Lipofectamine 2000. Each transfection was performed in triplicate, and each experiment was repeated at least twice. Luciferase and β-galactosidase (β-gal) assays were performed using a luminometer (Berthold Technologies, Centro XS3 LB 960) and a plate reader (Power Wave XS, Biotek), respectively. The results obtained were calculated as the ratio of relative light units (RLU) to β-gal values (RLU/β-gal) and expressed as the % increase compared with controls.
Small Interference RNA (siRNA)
MLK3 siRNA (sense, 5′-GGGCAGUGACGUCUGGAGUUUdTdT-3′, and the corresponding antisense) (19) was synthesized by Dharmacon (Lafayette, CO). The control-siRNA was from Ambion. siRNA transfection was performed using Lipofectamine 2000 as per manufacturer's instructions following protocols described earlier (29). The MCF7 and ZR75–1 cells with stable MLK3 knockdown were created by utilizing validated MLK3-shRNA in lentiviral vector pLKO.1-Puro (Sigma) utilizing standard lentiviral procedures. MLK3-shRNA-expressing cells were selected with puromycin (100 μg/ml) and propagated, and knockdown of MLK3 expression was confirmed by PCR using validated primers (data not shown) and Western blot analysis (see Fig. 1E).
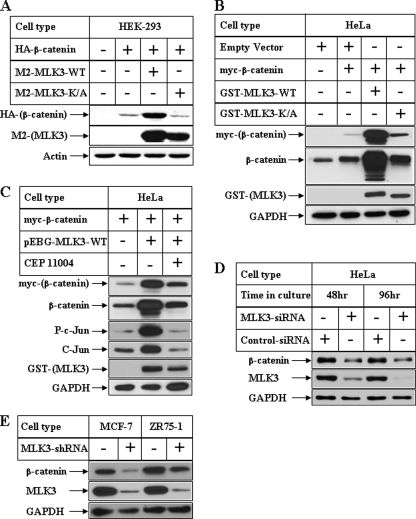
FIGURE 1.
Effect of MLK3 on β-catenin expression in various human cell lines. A, subconfluent populations of HEK-293 cells were transiently transfected with HA-tagged β-catenin alone (second lane) or in combination with M2-tagged MLK3-WT (third lane) or M2-tagged MLK3-K/A (fourth lane). Equal amounts of total protein isolated from these cells were subjected to Western blot analyses with antibodies against HA (for ectopic β-catenin) or M2 (for ectopic MLK3). The samples blotted with actin antibody served as controls. B, HeLa cells were transfected as in A utilizing myc-tagged β-catenin and GST-tagged MLK3-WT or K/A followed by Western blot analysis with antibodies against myc (for ectopic β-catenin), β-catenin (ectopic and endogenous β-catenin), GST (for ectopic MLK3), and GAPDH (as control). C, HeLa cells were transfected as in B followed by Western blot analyses with the samples utilizing the antibodies indicated. The cells in lane 3 were harvested after a pretreatment with 500 nm pan-MLK inhibitor CEP11004 for 20 h. D, HeLa cells were transiently transfected with either control-siRNA or MLK3-siRNA for the indicated periods of time followed by Western blot analysis. E, total cell extracts from control MCF-7 and ZR75–1 breast cancer cells (first and third lanes, respectively) or their stable transfectants expressing MLK3shRNA (second and fourth lanes, respectively) were analyzed by Western blots with the antibodies indicated.
Reverse Transcription-PCR Analysis
Total RNA was extracted from cells using the RNAqueous®-4PCR kit (Ambion). 500 ng of total RNA was subjected to reverse transcription reaction using the random primer and reverse transcriptase (Promega). Total gene expression was then analyzed by semiquantitative polymerase chain reaction (PCR) using high fidelity PCR system (Eppendorf, Mastercycler gradient, Hauppauge, NY). The following primers were used and obtained from IDT (San Diego, CA): β-actin, 5′-CAACCGCGAGAAGATGAC-3′ (forward) and 5′-AGGAAGGCTGGAAGAGTG-3′ (reverse); Survivin, 5′-CATTCGTCCGGTTGCGCTTTCC-3′ (forward) and 5′-GCGCACTTTCTCCGCAGTTTCC-3′ (reverse); c-myc, 5′-GGACGACGAGACCTTCATCAA-3′ (forward) and 5′-CCAGCTTCTCTGAGACGAGCTT-3′ (reverse).
Western Blot Analysis
Western blot analysis was performed utilizing procedures described previously (30). Briefly, equal amounts of total cell extracts or immunoprecipitated samples were fractionated by SDS-PAGE, transferred to PVDF membranes, and subjected to Western blot analysis utilizing various antibodies. To determine changes in β-catenin localization biochemically, detergent fractionation was carried out as follows. Cell pellets were sonicated briefly (∼10 s) in 50 mm Tris-HCl buffer, pH 7.5, containing protease inhibitors followed by centrifugation at 350,000 × g for 15 min (ultracentrifugation). The supernatant was isolated and termed the Tris-soluble fraction (or Tris), and the pellet was subjected to similar extraction and centrifugation with various detergents (in Tris-HCl buffer) in the following sequence: 1% Triton X-100 (Triton X fraction), 1% Sarkosyl (Sarkosyl fraction), and 2% SDS (SDS fraction). Tris fraction containing equal amounts of total protein along with equal volumes of Triton X, Sarkosyl, and SDS fractions (equivalent to Tris fraction) were then analyzed by SDS-PAGE.
Immunofluorescence Microscopy
HeLa cells transfected with the indicated expression constructs using Lipofectamine 2000 were allowed to adhere to fibronectin-treated glass coverslips and fixed with 3.7% formaldehyde in 0.1 m Pipes, pH 6.8, 48 h after transfection. The following primary antibodies were utilized: mouse α-myc antibody and rabbit α-HA antibody, which were secondarily labeled with fluorophore-conjugated donkey anti-mouse or anti-rabbit antibodies. They were also stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) to show the nucleus. Images were collected on a DeltaVision microscope (Applied Precision) equipped with a digital camera (CoolSNAP HQ; Photometrics) using a 60 × NA 1.42 objective lens and were deconvolved with SoftWoRx deconvolution software (Applied Precision).
Immunoprecipitation and Kinase Assay
Immunoprecipitation (IP) and GST pulldown experiments by either specific antibodies or GSH beads, respectively, were carried out as described (31). The immunoprecipitates were washed thoroughly and then processed for immunoblotting with antibodies against various interacting proteins or subjected to in vitro kinase assay in the presence of [γ-32P]ATP and GST-β-catenin as MLK3 substrate (28). In some experiments the samples were pretreated with either 400 nm CEP11004 or 40 nm SP600125 for 30 min before subjecting to kinase assay. These samples were then fractionated by SDS-PAGE, and 32P incorporation in β-catenin was quantified using a PhosphorImager (STORM 820, GE Healthcare).
Cell Cycle Analysis
HeLa cells transfected with various GFP-tagged vectors were fixed overnight at 4 °C in Fixing solution (containing 15% FBS and 15% PBS in 70% ethanol). After 2 washes in cold PBS, cells were incubated for 1 h at room temperature in propidium iodide (PI) solution containing 0.05 mg/ml PI, 0.1 mm EDTA, and 0.05 mg/ml RNase A in PBS before analyzing by flow cytometry. The percentage of cells in various stages of cell cycle in GFP-positive and GFP-negative populations were determined using FACSCanto II (BD Biosciences, San Jose, CA) and analyzed by FlowJo 7.5.5 (TreeStar Ashland OR). For Western blot analyses, cells were transfected similarly, and ∼2 × 106 GFP-positive and GFP-negative cells were sorted utilizing FACSAria (BD Biosciences) and analyzed by SDS-PAGE.
RESULTS
MLK3 Regulates β-Catenin Expression in Various Cell Types
To delineate the signaling molecules that can regulate β-catenin function, we focused on the MAP3K member MLK3, which is known to regulate various cellular pathways (18, 19). Subconfluent populations of human cell lines derived from embryonic kidney (HEK-293) and cervical carcinoma (HeLa) were transfected separately with either HA epitope-tagged β-catenin alone or in combination with M2-tagged MLK3 wild-type (WT) or K/A forms. Western blot analysis performed with antibodies against the HA tag showed a distinct induction of HA-β-catenin expression in both the cell types (Fig. 1A and supplemental Fig. 1A, HA panel, third lane). To rule out any nonspecific artifacts of overexpression, these studies were also performed with myc-epitope tagged β-catenin and GST-tagged MLK3, which showed an MLK3-mediated induction of β-catenin expression (Fig. 1B, third lane). In these studies, MLK3-mediated induction of β-catenin required the kinase activity of MLK3, since MLK3-K/A (kinase dead) was unable to induce β-catenin expression (Fig. 1, A and B, and supplemental Fig. 1, A and B, fourth lane) despite expression of significant amounts of this kinase dead form (M2 or GST panels; Fig. 1, A and B). To confirm these further, studies were also performed after pretreatment of cells with a pan-MLK inhibitor CEP11004. Pretreatment with CEP11004 antagonized MLK3/JNK-mediated c-Jun phosphorylation (Fig. 1C, phospho c-Jun panel; third lane), indicating the efficacy of CEP11004 in antagonizing the JNK axis. Similarly, CEP11004 pretreatment also antagonized MLK3-WT-induced β-catenin expression (myc and β-catenin panels), further confirming that this effect requires the kinase activity of MLK3.
To determine whether MLK3 can regulate endogenous β-catenin expression, endogenous MLK3 expression was knocked down in HeLa cells using MLK3-siRNA as described earlier (19). The results shown in Fig. 1D indicate a time-dependent decrease in MLK3 expression with MLK3-siRNA and not with control-siRNA (compare the first and second lanes and the third and fourth lanes, MLK3 panel). Knockdown of MLK3 expression also resulted in a corresponding decrease in endogenous β-catenin expression (β-catenin panel). In addition, two breast cancer cell lines (MCF-7 and ZR75–1) generated with stable overexpression of MLK3-shRNA showed a significant decrease in endogenous β-catenin expression (Fig. 1E, compare the first and second lanes and the third and fourth lanes). This was parallel to the reduced expression of endogenous MLK3 in the both cell types overexpressing MLK3-shRNA. These novel observations suggested that kinase active MLK3 can induce expression of β-catenin in various cell types and might be modulating β-catenin downstream events.
MLK3 Regulates β-Catenin Expression and Localization
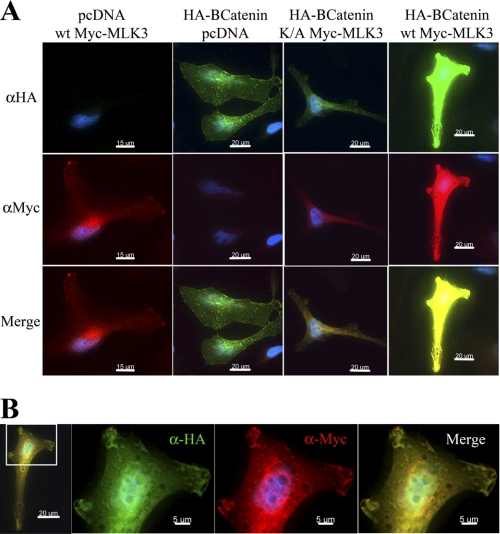
Because β-catenin can be localized in various cellular compartments, immunofluorescence studies were performed to detect any changes in β-catenin localization after co-expression with MLK3. Co-expression of β-catenin with MLK3-K/A (Fig. 2A, HA panel, third column) did not alter the level of β-catenin when compared with β-catenin alone (second column). However, when co-expressed with MLK3-WT, the expression of β-catenin was significantly enhanced such that it was impossible to determine the localization changes using the same exposure times and display settings (fourth column). When cells expressing β-catenin and MLK3-WT were viewed with display settings appropriate for their expression levels, β-catenin could be observed to localize both in the plasma membrane and in the nucleus (Fig. 2B, see HA). Both β-catenin and MLK3-WT localized in similar areas of the plasma membrane, which appear to be membrane ruffles or areas of dynamic membrane activity (see Merge). Importantly, MLK3-WT expression also seemed to be induced when transfected with HA-β-catenin (Fig. 2A, myc panel, compare the first and fourth columns). To confirm these results biochemically, cells were fractionated by sequential extraction in Tris-HCl buffer followed by Triton X-100, Sarkosyl, and SDS. These results showed that myc-β-catenin, when expressed alone, is mostly enriched in the cytoplasmic or Tris fraction (see supplemental Fig. 2, myc-β-catenin, panel D, lane 5). However, in the presence of MLK3-WT, it is significantly enriched in the SDS fraction, which is expected to be the most insoluble compartment (supplemental Fig. 2, myc-β-catenin, panel L, lane 16). The same profile was detected with anti-β-catenin antibody (supplemental Fig. 2, compare β-catenin ectopic, L panel, lane 16), whereas the endogenous β-catenin was extracted mostly in the Triton X fraction, which is where membrane-bound proteins are concentrated (see supplemental Fig. 2, compare β-cat endogenous). These studies indicate that MLK3-WT regulates β-catenin expression and localization.
FIGURE 2.
MLK3 regulates β-catenin expression and localization. A, subconfluent HeLa cells plated on fibronectin-coated cover slides were transfected with HA-epitope tagged β-catenin and myc-tagged MLK3 (WT or K/A) either alone or in combination. Immunofluorescence analysis was then performed after incubation with the appropriate primary (anti-HA or anti-myc) and fluorophore-conjugated secondary antibodies to detect ectopic β-catenin (green) and MLK3 (red), respectively. Cells were imaged using identical exposure conditions and display settings to highlight differences in expression levels. B, shown are the images obtained at a higher magnification after transfection with the HA-β-catenin and myc-MLK3-WT combination. Due to the strong intensity of the signals observed under this condition, these images were obtained with display settings appropriate for their expression levels. Areas of pronounced membrane co-localization have been enlarged in the panels on the right.
MLK3 Can Interact with β-Catenin Independent of Its Kinase Activity
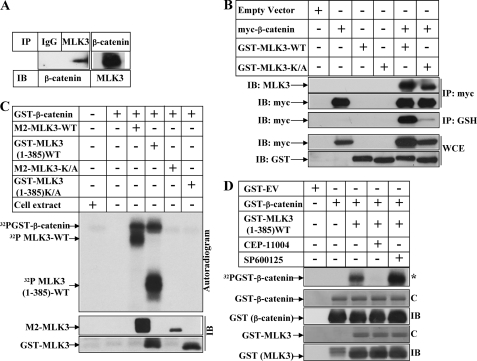
To obtain a better understanding of how MLK3 might modulate the β-catenin pathway, we determined whether they interacted with each other. Immunoprecipitation of endogenous MLK3 showed the interaction with endogenous β-catenin and vice versa (Fig. 3A). In addition, immunoprecipitation of ectopic β-catenin (with anti-myc) and immunoblotting with MLK3 antibody revealed that both WT and K/A forms of MLK3 are capable of interacting with β-catenin (Fig. 3B, fifth and sixth lanes, IP: myc and IB: MLK3 panel). A similar interaction was also detected after pulldown with GSH and immunoblotting with anti-myc antibody (IP-GSH and IB-myc panel). To confirm this interaction, similar studies were designed with HA-tagged β-catenin and M2-tagged MLK3 in HEK-293 cells. As shown in supplemental Fig. 3, immunoprecipitation with HA antibody showed the interaction with both WT and K/A MLK3 (third and fourth lanes). This suggested that MLK3 and β-catenin can interact with each other in vivo and that this interaction is independent of MLK3 kinase activity.
FIGURE 3.
MLK3 can interact with and phosphorylate β-catenin. A, equal amounts of cell extracts from HeLa cells were immunoprecipitated (IP) separately with either a control IgG (first lane) or antibodies against MLK3 (second lane) or against β-catenin (third lane). The immunoprecipitates of the first and second lanes were immunoblotted (IB) with β-catenin antibody, and that of third lane was immunoblotted with MLK3 antibody. B, subconfluent HeLa cells were transiently transfected with either empty vector (first lane), myc-tagged β-catenin (second lane), GST-tagged MLK3-WT (third lane), GST-tagged MLK3-KA (fourth lane), a combination of myc-β-catenin and GST-MLK3-WT (fifth lane), or myc-β-catenin and GST-MLK3-KA (sixth lane). IP: myc panel, equal amounts of cell extracts were immunoprecipitated with an antibody against myc and Western-blotted with an antibody against MLK3 or against myc. IP: GSH panel, equal amounts of cell extracts were subjected to pulldown with GSH beads and Western-blotted with myc antibody. WCE panel, equal amounts of whole cell extracts (WCE) were analyzed by Western blots utilizing the antibodies indicated to show expression of the respective proteins. C, M2-tagged MLK3-WT or K/A or GST-tagged MLK3-(1–385)-WT or K/A was immunoprecipitated from HEK-293 cells and subjected to in vitro kinase assays in the presence of [γ-32P]ATP using GST-β-catenin as substrate. The samples were then fractionated by SDS-PAGE, and autoradiography (top panel) or Western blot analysis with M2 or GST antibodies (bottom panel) was performed. Cell extracts (without substrate) and the substrate alone were included in the first and second lanes, respectively, as controls. D, in vitro kinase assays were carried out as described in C with GST-tagged MLK3-(1–385)-WT using GST-β-catenin as the substrate in the presence of a pretreatment with CEP11004 (fourth lane) or SP600125 (fifth lane). The asterisk (*) indicates autoradiogram; C, Coomassie Blue stain.
MLK3 Can Phosphorylate β-Catenin in Vitro
Because MLK3-mediated induction of β-catenin expression required the kinase activity of MLK3 (Fig. 1, A–C), we determined whether MLK3 was capable of phosphorylating β-catenin in vitro. To achieve this, M2-tagged MLK3-WT or K/A were transfected in HEK-293 cells. To avoid any confusion in detecting co-migrating phospho-β-catenin and phospho-MLK3 bands (due to MLK3 autophosphorylation) after kinase assay, cells were separately transfected with a truncated active GST-tagged MLK3-(1–385)-WT and K/A vector. In vitro kinase assays were then performed with immunoprecipitated ectopic MLK3 (by antibodies against M2 tag or GST tag) utilizing GST-β-catenin as a substrate as described previously (28). Results indicated that MLK3-WT and not K/A was capable of phosphorylating β-catenin in vitro (Fig. 3C, 32P-GST-β-catenin panel, compare the third and fifth lanes and the fourth and sixth lanes). To rule out the participation of any other associated kinases in the immunoprecipitates, these kinase assays were also carried out after pretreatment with CEP-11004 (pan inhibitor of MLK group of kinases) and SP600125 (inhibitor of JNK). These studies indicated increased phosphorylation of β-catenin with MLK3 (1–385)-WT, which was completely inhibited by CEP-11004 and not by SP600125 (Fig. 3D, compare the third lane with the fourth and fifth lanes). These studies indicated that this phosphorylation of β-catenin is mediated by MLK group of kinases.
MLK3 Increases β-Catenin Protein Stability
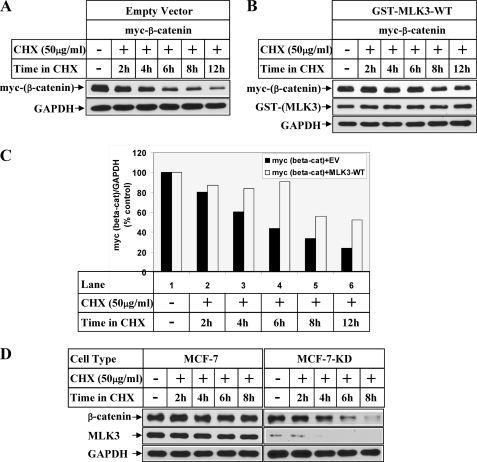
To understand whether MLK3 increased β-catenin expression at a post-translational level, cells overexpressing β-catenin and MLK3-WT were treated with cycloheximide (protein synthesis inhibitor) for different lengths of time. Western blot analysis of the samples showed a time-dependent decay of ectopic β-catenin when transfected with empty vector (Fig. 4A, myc panel) with a half-life of <6 h (Fig. 4C). However, overexpression of MLK3-WT resulted in a significant stabilization of the protein without significant decay until 12 h (Fig. 4, B and C), suggesting that MLK3 increased β-catenin expression via post-translational modification. To determine whether MLK3 also regulated endogenous β-catenin similarly, cycloheximide experiments were performed with MCF7 and MCF-7-KD (with stable knockdown of MLK3) cells. These results also showed faster decay of endogenous β-catenin protein in the MCF-7-KD cells compared with the MCF-7 cells (Fig. 4D).
FIGURE 4.
MLK3 regulates β-catenin expression at a post-translational level. HeLa cells transfected with myc-β-catenin in combination with empty vector (A) or GST-tagged MLK3-WT (B) were treated with 50 μg/ml CHX for the indicated periods of time after recovery in growth medium. The first lane represents the samples harvested without any CHX treatment. Western blot analysis was performed with the antibodies indicated. C, the bar graph shows the ratio of myc-β-catenin and GAPDH obtained from the Western blots of A and B considering the respective CHX untreated values as 100%. D, MCF-7 or MCF-7-KD (with stable MLK3 knockdown) were treated with CHX as in A and analyzed by Western blots with the indicated antibodies.
MLK3 Regulates β-Catenin Protein Stability Independent of GSK3β
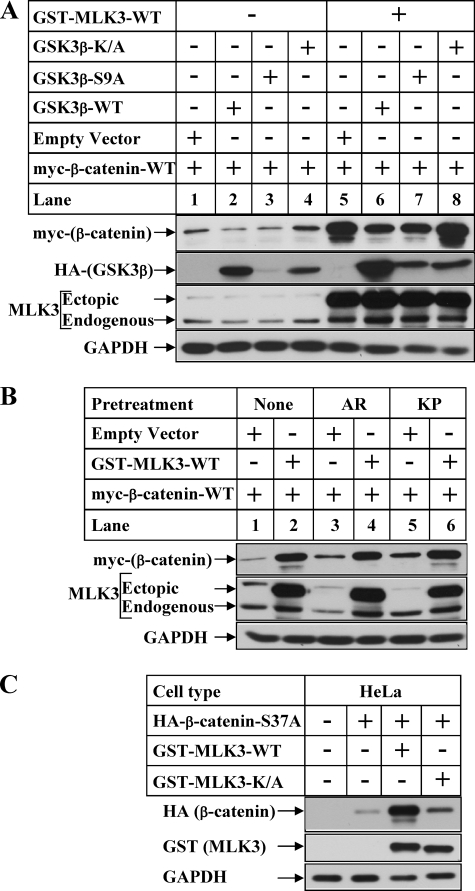
The next sets of studies were designed to understand any participation of GSK3β in MLK3-induced stabilization of β-catenin, as GSK3β is known to regulate β-catenin degradation (32). Overexpression of GSK3β-WT or S9A (phosphorylation-deficient mutant) reduced expression of myc-β-catenin in the absence of MLK3 (Fig. 5A, compare lane 1 with lanes 2 and 3), whereas the kinase inactive form (GSK3βK/A) increased it (lane 4). However, MLK3 overexpression was capable of stabilizing β-catenin irrespective of the presence of GSK3β (lanes 5–8). The slight decrease in β-catenin levels observed in the presence of GSK3β-WT and S9A (compare lane 5 with lanes 6 and 7) are due to the reduction of the basal levels of β-catenin after overexpression of these even in the absence of MLK3 (compare lane 1 with lanes 2 and 3). To confirm these results further, the cells were pretreated with two different pharmacological inhibitors of GSK3β (AR-A014418 and Kenpaullone) as described earlier (33). The goal was to determine whether GSK3β inhibition induced β-catenin stabilization to the same extent as MLK-WT and whether MLK3 is unable to induce β-catenin further when endogenous GSK3β activity is already inhibited. The results showed that inhibition of GSK3β by AR-A014418 or Kenpaullone increased β-catenin expression only minimally (Fig. 5B, compare lanes 1, 3, and 5), which was much less compared with MLK3-induced stabilization (compare lanes 2, 3, and 5). In addition, overexpression of MLK3-WT was capable of inducing β-catenin expression even in the presence of these inhibitors (lanes 3 and 4 and lanes 5 and 6). Furthermore, overexpression of MLK3-WT was capable of inducing the expression β-catenin (S37A) mutant, which is a GSK3β phosphorylation-deficient and degradation-resistant mutant as shown in Fig. 5C. Combined together, these results suggests that MLK3-induced stabilization of β-catenin is independent of GSK3β.
FIGURE 5.
MLK3 increases β-catenin protein stability independent of GSK3β. A, HeLa cells were transiently transfected with myc-β-catenin and HA-GSK3β vectors (WT, S9A, K/A) in the absence (−) or presence (+) of GST-MLK3-WT. Western blot analyses were performed with the cell extracts utilizing the antibodies against myc (for ectopic β-catenin), HA (for ectopic GSK3β), MLK3 (for endogenous and ectopic MLK3), and GAPDH (as control). B, HeLa cells transfected with the indicated plasmids were treated for 20 h with none (lanes 1 and 2) or 10 μm AR-A014418 (AR, lanes 3 and 4) or 5 μm Kenpaullone (KP, lanes 5 and 6) followed by Western blot analysis. C, HeLa cells were transiently transfected with HA-tagged β-catenin-S37A alone (lane 2) or in combination with GST-tagged MLK3-WT (lane 3) or GST-tagged MLK3-K/A (lane 4). Equal amounts of total protein isolated from these cells were subjected to Western blot analyses with antibodies against HA (for ectopic β-catenin) or GST (for ectopic MLK3). The samples blotted with GAPDH antibody served as controls.
MLK3 Inhibits β-Catenin and TCF-mediated Transcriptional Activity
To determine whether MLK3-mediated induction of β-catenin expression also resulted in activation of β-catenin/TCF-dependent transcription (conventional Wnt-signaling pathway), HeLa cells were transfected with a TCF-responsive reporter pGL3OT (with wild-type TCF sites) as described elsewhere (28). Overexpression of β-catenin-WT alone resulted in significant induction of pGL3OT luciferase activity (Fig. 6A, compare lanes 1 and 2), indicative of an activation of β-catenin/TCF-dependent transcription. Surprisingly, overexpression of MLK3-WT resulted in a strong inhibition of this transcriptional activity, an effect that was dependent on MLK3 kinase activity (Fig. 6A, compare lanes 3 and 4). Similarly, overexpression of increasing concentrations of MLK3 showed a concentration-dependent induction of β-catenin expression (supplemental Fig. 4B, myc panel) but a corresponding reduction of TCF-transcriptional activity (supplemental Fig. 4A). In addition, PCR analyses showed an MLK3-mediated reduction of myc and Survivin mRNA expression, the two downstream targets of conventional β-catenin/TCF pathway (Fig. 6C). These results suggested that MLK3 activation leads to an antagonism of the conventional β-catenin/TCF-dependent transcription in cancer cells despite a stabilization of β-catenin protein.
FIGURE 6.
MLK3 inhibits β-catenin and TCF-mediated transcriptional activity. A, HeLa cells were transiently transfected with pGL3OT luciferase (pGL3OT) reporter and β-galactosidase vector (for normalization) along with empty vector (lane 1), β-catenin alone (lane 2), or β-catenin in combination with MLK3-WT (lane 3) or MLK3-K/A (lane 4). Luciferase and β-gal assays were performed as described elsewhere (19) considering the empty vector values as 100%. B, HeLa cells were transfected with pGL3OT and β-gal along with either empty vector (lane 1), β-catenin alone (lane 2), or β-catenin in combination with MLK3-WT (lane 3) or MLK3-WT and KLF4 vectors (lanes 4–6). Luciferase and β-gal assays were performed as in A. Each transfection (A and B) was performed in triplicate, and the data represent the mean ± S.D. of at least two independent experiments. C, shown is a semiquantitative PCR analysis of HeLa cells transfected with empty vector, β-catenin alone, or in combination with MLK3-WT to detect myc or Survivin expression. Actin was used as a control. The bar graphs represent the ratio of myc/β-actin and Survivin/β-actin.
Effect of MLK3 on β-Catenin, TCF, and KLF4 Interaction
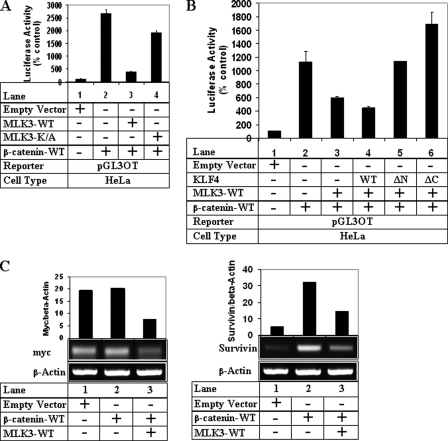
To understand whether MLK3 overexpression antagonized β-catenin/TCF transcriptional activity via modulating their interaction, endogenous TCF4 was immunoprecipitated from HeLa cells following overexpression of GST-tagged MLK3 and myc-tagged β-catenin, and the immunoprecipitates were analyzed by Western blots. These studies indicated increased interaction between TCF4 and β-catenin even in the presence of MLK3-WT (Fig. 7, A, fourth lane, IB: myc, and Fig. 7C, second lane, IB: β-catenin panels), suggesting that MLK3 does not regulate β-catenin/TCF transcriptional activity via reducing the interaction between β-catenin and TCF and indicated the possible recruitment of a repressor to this complex. To understand the involvement of KLF4, which is a known repressor of β-catenin/TCF4 pathway (34), endogenous KLF4 was immunoprecipitated from these cells followed by Western blot analysis to detect β-catenin interaction. Overexpression of MLK3-WT resulted in an increased interaction between KLF4 and β-catenin (Fig. 7, B, fourth lane, IB: myc, and D, second lane, IB: β-catenin panels). To determine whether these proteins also interacted with MLK3, the TCF4 and KLF4 immunoprecipitates were immunoblotted with antibodies against MLK3. These showed that both WT and K/A MLK3 were capable of interacting with TCF4 (Fig. 7, A, fourth and fifth lanes, IB: MLK3) as well as with KLF4 (Fig. 7B, fourth and fifth lanes, IB: MLK3). In addition, immunoprecipitation of endogenous TCF4 and KLF4 showed interaction with endogenous MLK3 (Fig. 7E). These results indicated that MLK3 does not effect β-catenin/TCF interaction and likely antagonizes TCF transcriptional activity via recruiting KLF4 repressor to this complex.
FIGURE 7.
Effect of MLK3 on β-catenin interaction with TCF4 and KLF4. A, HeLa cells were not transfected (first and second lanes) or were transfected with myc-β-catenin alone (third lane) or myc-β-catenin in combination with GST-MLK3-WT (fourth lane) or K/A (fifth lane). Equal amounts of protein were immunoprecipitated (IP) by either a control IgG (first lane) or by an antibody against TCF4 (second through fifth lanes). The immunoprecipitates were then analyzed by Western blots (IB) with antibodies against myc or MLK3. Equal amounts of whole cell extracts (WCE panel) were analyzed by Western blots utilizing TCF4, GST (MLK3), and myc (β-catenin) antibodies to show expression of the respective proteins. B, HeLa cells transfected as in A were immunoprecipitated by either a control IgG (first lane) or by an antibody against KLF4 (second through fifth lanes). The immunoprecipitates were then analyzed by Western blots with antibodies against myc or MLK3. The whole cell extracts panel indicates the expression KLF4 in equal amounts of the cells extracts. C and D, HeLa cells transfected with the indicated vectors were immunoprecipitated with TCF4 (C) or KLF4 (D) as in A. The immunoprecipitates in C were analyzed by Western blots with antibodies against β-catenin and TCF4 and those in D were analyzed with β-catenin antibody. The expression of the respective proteins in the whole cell extract is shown at the bottom. E, equal amounts of total extracts from HeLa cells were immunoprecipitated with either control IgG and TCF4 antibody (upper panel) or control IgG and KLF4 antibody (lower panel). The immunoprecipitates were then analyzed by Western blotting with MLK3 antibody to detect endogenous interaction.
To confirm the participation of KLF4 in these, the effect of MLK3 on β-catenin/TCF responsive luciferase activity was estimated in the presence of KLF4 (WT and ΔN and ΔC mutants). These two mutants of KLF4 in earlier studies were unable to inhibit β-catenin/TCF-mediated transcriptional activity (34), and ΔC served as a dominant negative KLF4 mutant (35). Overexpression of WT-KLF4 synergized with MLK3-WT to inhibit TCF luciferase activity (Fig. 6B, compare lanes 3 and 4). However, the two mutants of KLF4 (ΔN and particularly ΔC) completely abolished MLK3-mediated antagonism of TCF luciferase activity (lanes 5 and 6), suggesting the participation of KLF4 in this.
MLK3 Modulates β-Catenin Pathway in Prostate Cancer Cells
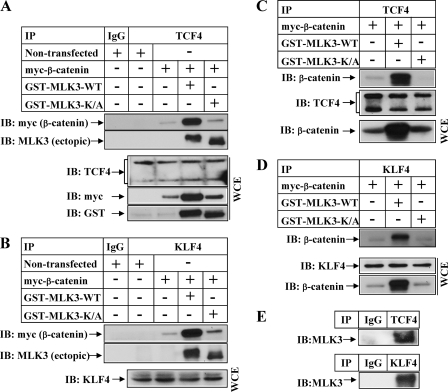
Earlier studies by different groups have shown involvement of β-catenin pathway in prostate cancer (36). To determine whether MLK3 can modulate the β-catenin pathway in prostate cancer cells, MLK3 and β-catenin studies were designed with androgen-sensitive (LNCaP, 22Rv1) and androgen-insensitive (PC3) prostate cancer cells. Overexpression of MLK3-WT and not K/A was capable of inducing the expression of β-catenin in all the cell types tested (Fig. 8, A–C and supplemental Fig. 1B), suggesting that MLK3-induced expression of β-catenin is a generalized event. The fact that both androgen-sensitive (LNCaP) and -insensitive (PC3) cells showed similar effects suggested that this stabilization operates in prostate cancer cells independent of the androgen receptor expression. In addition, MLK3-WT inhibited β-catenin-S37A-induced (Fig. 8E, compare lane 2 with lanes 3 and 4) and β-catenin-WT-induced (Fig. 8D, compare lanes 2 with lanes 3 and 4) TCF-responsive transcriptional activity in a dose-dependent manner. The expression of β-catenin-induced cyclin D1 expression in the 22Rv1 cells was also inhibited by MLK3-WT (Fig. 8F, compare the second lane with the fifth lane, cyclin D1 panel).
FIGURE 8.
MLK3 modulates β-catenin pathway in prostate cancer cells. LNCaP (A), 22Rv1 (B)m and PC3 (C) were transfected with various expression vectors followed by Western blot analysis with the indicated antibodies. D and E, 22Rv1 cells were transiently transfected with pGL3OT luciferase reporter and β-galactosidase vector. In D they were co-transfected with empty vector (lane 1) or β-catenin wild-type alone (lane 2) or with increasing concentration of MLK3-WT (lanes 3 and 4). In E a stabilized mutant form of β-catenin (S37A) was co-transfected either alone (lane 2) or with increasing concentration of MLK3-WT (lanes 3 and 4). Luciferase and β-gal assays were performed as in Fig. 6A. F, 22Rv1 cells were transfected with the indicated plasmids, and after a recovery in growth medium, they were harvested and analyzed by Western blots.
MLK3-WT and β-Catenin Co-expression Regulates Cell Cycle Progression
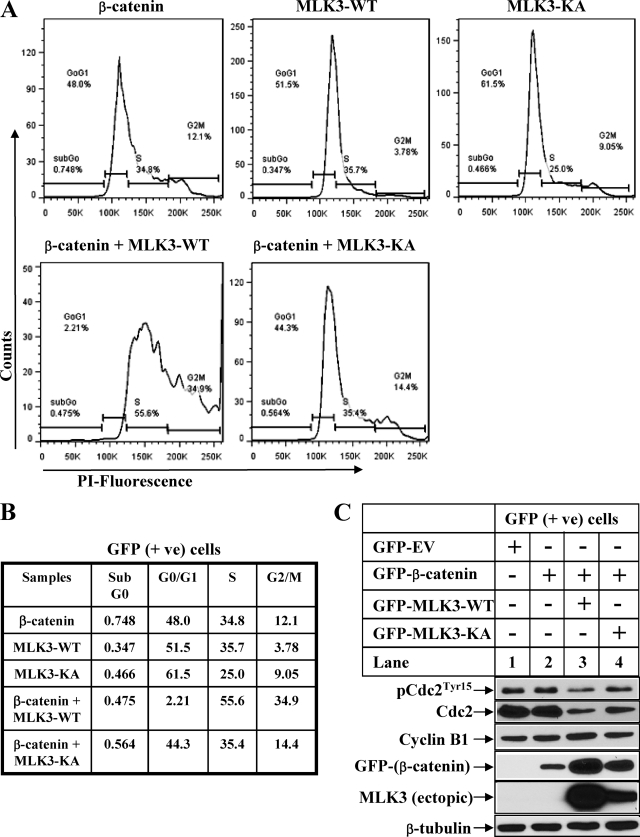
The results described above suggested the involvement of a novel pathway by which MLK3 regulated the β-catenin expression and transcriptional activity. To determine the cellular effects of MLK3 and β-catenin co-expression in cancer cells, GFP-tagged MLK3 and β-catenin were overexpressed in HeLa cells, and cell cycle profile was analyzed in GFP-positive and GFP-negative cells. These results showed a significant increase in G2/M cell population and a corresponding decrease in G0/G1 population after co-expression of MLK3-WT and β-catenin in the GFP-positive cells (Fig. 9, A and B). This cell cycle profile in GFP negative cells (used as negative control) did not show any significant change after overexpression of these two proteins (supplemental Fig. 5, A and B). Western blot analysis of these samples indicated a reduction of total Cdc2 expression associated with a modest increase in cyclin B1 levels specifically in the GFP-positive MLK3-WT and β-catenin-overexpressing cells (Fig. 9C and supplemental Fig. 5C, third lane). Taken together, these suggested that MLK3 can modulate the expression of β-catenin and β-catenin/TCF-mediated transcriptional activity in cancer cells, ultimately leading to cell cycle arrest.
FIGURE 9.
Co-expression of MLK3 and β-catenin inhibits cell cycle progression. A, HeLa cells were transfected with GFP-tagged-β-catenin, -MLK3-WT, and -MLK3-KA either alone or in combination and then subjected to flow cytometry. The percentage of cells in various stages of the cell cycle in GFP-positive population was determined using FACSCanto II, analyzed by FlowJo 7.5.5, and represented by graphs. PI, propidium iodide. B, the table shows the % of GFP cells in various stages of cell cycle as obtained in A. C, HeLa cells were transected as in A, and GFP cells were sorted using FACSAria and analyzed by Western blots with the indicated antibodies.
DISCUSSION
In summary, our results showed that overexpression of MLK3 can induce expression of β-catenin in various cell types including prostate cancer cells. This involves a post-translational stabilization of β-catenin protein. The levels of β-catenin in the normal cells are strictly regulated by a degradation complex that involves GSK3β-adenomatous polyposis coli-axin (8). Because MLK3 is also known to have cross-talk with GSK3β signaling (17), it was conceivable that MLK3-induced stabilization of β-catenin was achieved via targeting members of the β-catenin degradation complex (namely GSK3β). Although the detailed mechanism of how MLK3 stabilizes β-catenin is still unclear, it seems to be independent of GSK3β pathway. This is evident from GSK3β overexpression studies and those with two different GSK3β inhibitors (AR-A014418 and Kenpaullone). Furthermore, MLK3 was able to induce the expression of the GSK3β phosphorylation-deficient β-catenin (S37A) mutant (Fig. 5C). It is unclear at this time whether MLK3 targets members of the other non-conventional pathways of β-catenin degradation for regulating β-catenin expression (28, 37). The effect of MLK3 on β-catenin stabilization required the kinase activity of MLK3, as a kinase dead (K/A) mutant or pretreatment with pan MLK inhibitor (CEP11004) was unable to show this stabilization. MLK3 was, however, capable of interacting with β-catenin independent of its kinase activity and also induced β-catenin phosphorylation in vitro. Identification of the specific MLK3 phosphorylation site(s) on β-catenin in the future will be critical in determining their involvement in β-catenin stabilization, β-catenin/TCF transcriptional activity, and downstream cellular effects.
Our results also showed that despite increasing β-catenin expression, MLK3 inhibited β-catenin/TCF-mediated transcription and antagonized expression of β-catenin target genes. This novel observation suggested that overexpression or stabilization of β-catenin is not sufficient to activate β-catenin/TCF-dependent transcription. This is supported by earlier studies which showed that nuclear localization and interaction of β-catenin with LEF-1 are still not enough to activate TCF-mediated transcription (13). In addition, overexpression of an oncogenic form of β-catenin in the liver was unable to show induction of β-catenin/TCF target genes cyclin D1 and c-myc (38). It is thus likely that once β-catenin is stabilized and complexed with TCF, it needs additional signaling modification to be able to induce optimal TCF-responsive transcription, which can be achieved via recruitment of co-activators and corepressors. In fact, β-catenin/TCF transcriptional activity is induced after recruitment of coactivators such as CBP (CREB-binding protein) (39) and CARM1 (40). Our immunoprecipitation studies with endogenous TCF4 showed no decrease in its interaction with β-catenin in the presence of MLK3, whereas transcriptional activity is repressed. These results suggested that MLK3 does not regulate β-catenin/TCF-mediated transcription via inhibiting β-catenin and TCF interaction but might modulate this complex further downstream. One possible mechanism for this might be the recruitment of a repressor to the β-catenin-TCF complex. Recently, a cross-talk between Kruppel-like factor 4 (KLF4) and β-catenin has been reported (35) where KLF4 inhibits β-catenin/TCF transcriptional activity in a manner that does not interfere with β-catenin/TCF interaction but interferes with coactivator recruitment to β-catenin (34). Interestingly, our immunoprecipitation studies demonstrated increased interaction between KLF4 and β-catenin after MLK3 overexpression and indicate this as a potential mechanism mediating this transcriptional repression. In fact, overexpression of KLF4 ΔC (known to function as dominant negative mutant for the β-catenin/TCF pathway (35)) abolished MLK3-induced repression of β-cat/TCF-responsive transcription. In addition, KLF4 was shown to interact with MLK3 and might undergo additional post-translational modification for its recruitment to β-catenin.
An understanding of the cellular effects that are mediated via MLK3/β-catenin axis is necessary to understand how upstream signaling pathways regulate diverse β-catenin functions. These will also explain why overexpression of β-catenin mediates tumorigenic pathways differentially in various tissues (38, 42, 43). In our studies overexpression of MLK3-WT and β-catenin in combination resulted in attenuation of cell cycle progression at G2/M phase. Although β-catenin overexpression has been linked with a G2 arrest and subsequent apoptosis in earlier studies (44), our results provide the first clue regarding the signaling pathway that targets β-catenin toward this. It will be of interest to determine whether this signaling mechanism is also responsible for other cellular functions mediated by β-catenin (45). The detailed mechanism of how this growth arrest is achieved is still unclear, although our Western data with GFP-positive cells indicate changes in total cdc2 and cyclin B1 expression in the arrested cells. More in-depth studies are needed to confirm their participation.
MLK3 expression has been shown in breast cancer (18), gastric cancer (19), and prostate cancer cells (data not shown), activation of which regulates various pro-oncogenic pathways. β-Catenin is also known to play an important role in prostate cancer as aberrant β-catenin signaling contributes toward prostate tumorigenesis (46). Overexpression of β-catenin induced prostate intraepithelial neoplasia (PIN)-like lesions (47) as well as hyperplasia, leading to high grade PIN (36, 42) in separate studies. β-Catenin mutations have been reported in ∼5% of human prostate cancers (48, 49). In addition, β-catenin can interact with and augment transcriptional function of androgen receptor in a ligand-dependent manner (41, 50). Understanding the upstream signaling mechanism(s), which regulates the β-catenin pathway in prostate cancer cells, is thus critically important. Our results demonstrate for the first time that MLK3 can regulate β-catenin expression and β-catenin/TCF activation in prostate cancer cells. Because this causes an inhibition of β-catenin/TCF transcriptional activation, activating MLK3 might be an effective means of antagonizing aberrant Wnt-signaling in those cancers that have activating β-catenin mutations. This is supported by the fact that in 22Rv1 cells, which has functional Wnt/β-catenin signaling (46), overexpression of MLK3-WT can reduce the expression of β-catenin downstream cyclin D1 (Fig. 8F). In addition, MLK3-induced β-catenin expression seemed to be functional in both androgen-sensitive (LNCaP, 22Rv1) as well as -insensitive (PC3) cells, suggesting that MLK3-based therapy might be effective in the androgen-resistant and more aggressive forms of prostate cancer.
Acknowledgments
We are grateful to Dr. Bert Vogelstein for the pGL3OT luciferase reporter (22), Dr. Wange Lu for the KLF4 vectors (WT, ΔC, and ΔN) (24), Dr. James R Woodgett for the GSK3β expression vectors (WT, K/A, and S9A) (23), Dr. Robert Malenka for the GFP-β-catenin vector (25), and Dr. Stephen Byers for β-catenin vectors (WT and S37A) (26).
This work was supported, in whole or in part, by National Institutes of Health Grants CA121221 (to B. R.), K22 AI078757 and RO1 AI093258 (to E. M. C.), and GM55835 (to A. R.). This work was also supported by Veterans Affairs Merit awards BX000571-01 (to B. R.) and BX000312-01 (to A. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5.
- TCF
- T-cell factor
- DN
- dominant negative
- GSK3β
- glycogen synthase kinase 3 β
- KLF4
- Kruppel-like factor 4
- LEF
- lymphoid enhancer factor
- MLK3
- mixed lineage kinase 3
- NLK
- Nemo-like kinase
- TAK1
- TGFβ-activated kinase 1
- K/A
- kinase dead
- CHX
- cycloheximide
- IP
- immunoprecipitate.
REFERENCES
- 1. Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 2. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 3. Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. (2000) J. Cell Biol. 148, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R., Bradley A. (1999) Nat. Genet. 22, 361–365 [DOI] [PubMed] [Google Scholar]
- 5. Kielman M. F., Rindapää M., Gaspar C., van Poppel N., Breukel C., van Leeuwen S., Taketo M. M., Roberts S., Smits R., Fodde R. (2002) Nat. Genet. 32, 594–605 [DOI] [PubMed] [Google Scholar]
- 6. Miyabayashi T., Teo J. L., Yamamoto M., McMillan M., Nguyen C., Kahn M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5668–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kemler R. (1993) Trends Genet. 9, 317–321 [DOI] [PubMed] [Google Scholar]
- 8. Liu C., Li Y., Semenov M., Han C., Baeg G. H., Tan Y., Zhang Z., Lin X., He X. (2002) Cell 108, 837–847 [DOI] [PubMed] [Google Scholar]
- 9. Behrens J., Jerchow B. A., Würtele M., Grimm J., Asbrand C., Wirtz R., Kühl M., Wedlich D., Birchmeier W. (1998) Science 280, 596–599 [DOI] [PubMed] [Google Scholar]
- 10. Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. (2007) Science 316, 1619–1622 [DOI] [PubMed] [Google Scholar]
- 11. Nelson W. J., Nusse R. (2004) Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Giles R. H., van Es J. H., Clevers H. (2003) Biochim. Biophys. Acta 1653, 1–24 [DOI] [PubMed] [Google Scholar]
- 13. Prieve M. G., Waterman M. L. (1999) Mol. Cell. Biol. 19, 4503–4515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rana A., Gallo K., Godowski P., Hirai S., Ohno S., Zon L., Kyriakis J. M., Avruch J. (1996) J. Biol. Chem. 271, 19025–19028 [DOI] [PubMed] [Google Scholar]
- 15. Gallo K. A., Mark M. R., Scadden D. T., Wang Z., Gu Q., Godowski P. J. (1994) J. Biol. Chem. 269, 15092–15100 [PubMed] [Google Scholar]
- 16. Gallo K. A., Johnson G. L. (2002) Nat. Rev. Mol. Cell Biol. 3, 663–672 [DOI] [PubMed] [Google Scholar]
- 17. Mishra R., Barthwal M. K., Sondarva G., Rana B., Wong L., Chatterjee M., Woodgett J. R., Rana A. (2007) J. Biol. Chem. 282, 30393–30405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rangasamy V., Mishra R., Mehrotra S., Sondarva G., Ray R. S., Rao A., Chatterjee M., Rana B., Rana A. (2010) Cancer Res. 70, 1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mishra P., Senthivinayagam S., Rangasamy V., Sondarva G., Rana B. (2010) Mol. Endocrinol. 24, 598–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ishitani T., Ninomiya-Tsuji J., Nagai S., Nishita M., Meneghini M., Barker N., Waterman M., Bowerman B., Clevers H., Shibuya H., Matsumoto K. (1999) Nature 399, 798–802 [DOI] [PubMed] [Google Scholar]
- 21. Smit L., Baas A., Kuipers J., Korswagen H., van de Wetering M., Clevers H. (2004) J. Biol. Chem. 279, 17232–17240 [DOI] [PubMed] [Google Scholar]
- 22. Morin P. J., Vogelstein B., Kinzler K. W. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7950–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stambolic V., Ruel L., Woodgett J. R. (1996) Curr. Biol. 6, 1664–1668 [DOI] [PubMed] [Google Scholar]
- 24. Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M., Lu W. (2009) Stem Cells 27, 2969–2978 [DOI] [PubMed] [Google Scholar]
- 25. Yu X., Malenka R. C. (2003) Nat. Neurosci. 6, 1169–1177 [DOI] [PubMed] [Google Scholar]
- 26. Orford K., Crockett C., Jensen J. P., Weissman A. M., Byers S. W. (1997) J. Biol. Chem. 272, 24735–24738 [DOI] [PubMed] [Google Scholar]
- 27. Barthwal M. K., Sathyanarayana P., Kundu C. N., Rana B., Pradeep A., Sharma C., Woodgett J. R., Rana A. (2003) J. Biol. Chem. 278, 3897–3902 [DOI] [PubMed] [Google Scholar]
- 28. Sharma C., Pradeep A., Wong L., Rana A., Rana B. (2004) J. Biol. Chem. 279, 35583–35594 [DOI] [PubMed] [Google Scholar]
- 29. Senthivinayagam S., Mishra P., Paramasivam S. K., Yallapragada S., Chatterjee M., Wong L., Rana A., Rana B. (2009) J. Biol. Chem. 284, 13577–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pradeep A., Sharma C., Sathyanarayana P., Albanese C., Fleming J. V., Wang T. C., Wolfe M. M., Baker K. M., Pestell R. G., Rana B. (2004) Oncogene 23, 3689–3699 [DOI] [PubMed] [Google Scholar]
- 31. Sondarva G., Kundu C. N., Mehrotra S., Mishra R., Rangasamy V., Sathyanarayana P., Ray R. S., Rana B., Rana A. (2010) Cell Res. 20, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bienz M., Clevers H. (2000) Cell 103, 311–320 [DOI] [PubMed] [Google Scholar]
- 33. Mishra P., Senthivinayagam S., Rana A., Rana B. (2010) J. Mol. Signal. 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P. M., Chen X., Zhang W., Liu C. (2010) Mol. Cell. Biol. 30, 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang W., Chen X., Kato Y., Evans P. M., Yuan S., Yang J., Rychahou P. G., Yang V. W., He X., Evers B. M., Liu C. (2006) Mol. Cell. Biol. 26, 2055–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu X., Wang Y., Jiang M., Bierie B., Roy-Burman P., Shen M. M., Taketo M. M., Wills M., Matusik R. J. (2009) Prostate 69, 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu J., Stevens J., Rote C. A., Yost H. J., Hu Y., Neufeld K. L., White R. L., Matsunami N. (2001) Mol. Cell 7, 927–936 [DOI] [PubMed] [Google Scholar]
- 38. Cadoret A., Ovejero C., Saadi-Kheddouci S., Souil E., Fabre M., Romagnolo B., Kahn A., Perret C. (2001) Cancer Res. 61, 3245–3249 [PubMed] [Google Scholar]
- 39. Takemaru K. I., Moon R. T. (2000) J. Cell Biol. 149, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Koh S. S., Li H., Lee Y. H., Widelitz R. B., Chuong C. M., Stallcup M. R. (2002) J. Biol. Chem. 277, 26031–26035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Truica C. I., Byers S., Gelmann E. P. (2000) Cancer Res. 60, 4709–4713 [PubMed] [Google Scholar]
- 42. Bierie B., Nozawa M., Renou J. P., Shillingford J. M., Morgan F., Oka T., Taketo M. M., Cardiff R. D., Miyoshi K., Wagner K. U., Robinson G. W., Hennighausen L. (2003) Oncogene 22, 3875–3887 [DOI] [PubMed] [Google Scholar]
- 43. Harada N., Tamai Y., Ishikawa T., Sauer B., Takaku K., Oshima M., Taketo M. M. (1999) EMBO J. 18, 5931–5942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olmeda D., Castel S., Vilaró S., Cano A. (2003) Mol Biol. Cell 14, 2844–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bahmanyar S., Kaplan D. D., Deluca J. G., Giddings T. H., Jr., O'Toole E. T., Winey M., Salmon E. D., Casey P. J., Nelson W. J., Barth A. I. (2008) Genes Dev. 22, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chesire D. R., Ewing C. M., Gage W. R., Isaacs W. B. (2002) Oncogene 21, 2679–2694 [DOI] [PubMed] [Google Scholar]
- 47. Gounari F., Signoretti S., Bronson R., Klein L., Sellers W. R., Kum J., Siermann A., Taketo M. M., von Boehmer H., Khazaie K. (2002) Oncogene 21, 4099–4107 [DOI] [PubMed] [Google Scholar]
- 48. Chesire D. R., Ewing C. M., Sauvageot J., Bova G. S., Isaacs W. B. (2000) Prostate 45, 323–334 [DOI] [PubMed] [Google Scholar]
- 49. Voeller H. J., Truica C. I., Gelmann E. P. (1998) Cancer Res. 58, 2520–2523 [PubMed] [Google Scholar]
- 50. Yang F., Li X., Sharma M., Sasaki C. Y., Longo D. L., Lim B., Sun Z. (2002) J. Biol. Chem. 277, 11336–11344 [DOI] [PubMed] [Google Scholar]