Abstract
Proteasomal degradation is mediated through modification of target proteins by Lys-48-linked polyubiquitin (polyUb) chain, which interacts with several binding partners in this pathway through hydrophobic surfaces on individual Ub units. However, the previously reported crystal structures of Lys-48-linked diUb exhibit a closed conformation with sequestered hydrophobic surfaces. NMR studies on mutated Lys-48-linked diUb indicated a pH-dependent conformational equilibrium between closed and open states with the predominance of the former under neutral conditions (90% at pH 6.8). To address the question of how Ub-binding proteins can efficiently access the sequestered hydrophobic surfaces of Ub chains, we revisited the conformational dynamics of Lys-48-linked diUb in solution using wild-type diUb and cyclic forms of diUb in which the Ub units are connected through two Lys-48-mediated isopeptide bonds. Our newly determined crystal structure of wild-type diUb showed an open conformation, whereas NMR analyses of cyclic Lys-48-linked diUb in solution revealed that its structure resembled the closed conformation observed in previous crystal structures. Comparison of a chemical shift of wild-type diUb with that of monomeric Ub and cyclic diUb, which mimic the open and closed states, respectively, with regard to the exposure of hydrophobic surfaces to the solvent indicates that wild-type Lys-48-linked diUb in solution predominantly exhibits the open conformation (75% at pH 7.0), which becomes more populated upon lowering pH. The intrinsic properties of Lys-48-linked Ub chains to adopt the open conformation may be advantageous for interacting with Ub-binding proteins.
Keywords: Molecular Dynamics, NMR, Protein Conformation, Ubiquitin, X-ray Crystallography
Introduction
Ubiquitin (Ub)3 is a small protein composed of 76 amino acid residues and plays regulatory roles in various cellular events, including cell cycle progression, DNA repair, transcriptional regulation, and apoptosis (1, 2). These Ub regulatory functions are expressed by its modification of target proteins through the formation of isopeptide linkages at the C terminus. Ubiquitination is catalyzed by the sequential action of the Ub-activating enzyme E1, Ub-conjugating enzyme E2, and Ub-protein ligase E3. The C-terminal group of Ub can also be linked to another Ub (termed the distal and proximal moieties, respectively) through all seven lysine residues at positions 6, 11, 27, 29, 33, 48, and 63 as well as the N terminus, giving rise to various types of polyUb chains (3–7), which mediate diverse signals determining the fate of ubiquitinated proteins. The best characterized case is the Lys-48-linked polyUb chain that serves as a tag for protein degradation by 26 S proteasomes (1).
In the Ub/proteasome-mediated proteolytic pathway, Lys-48-linked polyUb is recognized by several proteins possessing Ub-binding motifs (8, 9). These include the proteasomal subunits S5a/Rpn10 and Rpn13 as well as the Ub receptors hHR23/Rad23, Dsk2/Dph1, and Ddi1/Mud1. NMR studies reported the interactions with the ubiquitin-associated domain of hHR23A and the ubiquitin-interacting motifs of S5a using Lys-48-linked diUb as a minimal model (10, 11). These studies indicated that hydrophobic surfaces, including Val-70 in both Ub units, are involved in the interactions with these Ub-binding motifs.
Conformation of Lys-48-linked diUb has also been characterized in the absence of its binding partners. The crystal structures of Lys-48-linked diUb have been solved for two different crystal forms grown under different crystallization conditions (12, 13). Crystallographic studies showed that Lys-48-linked diUb exhibits a “closed” conformation in which hydrophobic surfaces in both Ub units are packed against each other and shielded from the solvent. Identical Ub-Ub interaction modes have been observed in the crystal structures of an engineered tetraUb (14, 15) in which Lys-48 of the second and fourth Ub units are substituted by thialysine and arginine, respectively, and Gly-76 is deleted in the first Ub, and a cyclic tetraUb (16) in which the C terminus of the first Ub is conjugated to Lys-48 of the fourth Ub.
Conformational characterization of Lys-48-linked diUb in solution has also been performed using NMR spectroscopy (17, 18). Fushman and co-workers reported a series of NMR studies on mutated versions of Lys-48-linked diUb in which the first Ub has a C-terminal extension with Asp-77, and Lys-48 of the second Ub is substituted by cysteine or arginine (19). By inspecting the NMR data based on chemical shift perturbation, residual dipolar coupling (RDC), and relaxation, it has been concluded that Lys-48-linked diUb is at a pH-dependent conformational equilibrium between the closed and open states with respect to the exposure of hydrophobic surfaces and that the closed form is predominant under neutral conditions (90% at pH 6.8) (17, 20, 21). In contrast, Lys-63-linked diUB primarily exhibits an open conformation in solution (18, 22). These data raised the question of how Ub-binding proteins can efficiently access the sequestered hydrophobic surfaces of Lys-48-linked diUb.
To address this issue, we revisited the conformational dynamics of Lys-48-linked diUb in solution using NMR spectroscopy. We modified the protocol for preparing Lys-48-linked diUb using an in vitro enzymatic reaction with E1 and E2–25K, giving rise to wild-type Lys-48-linked diUb and a cyclic form of diUb, in which the Ub units are connected through two Lys-48-mediated isopeptide bonds. We performed structural analyses using these two forms of diUb along with the monomeric Ub to characterize the conformational equilibrium of Lys-48-linked diUb.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Human Ub, Ub E1, and E2–25K were expressed and purified as described previously (23). Yeast ubiquitin hydrase 1 (YUH1) cDNA was inserted into the pGEX vector to encode GST-YUH1. The GST-YUH1 proteins were expressed in Escherichia coli BL21(DE3) CodonPlus and purified using a glutathione-Sepharose column (Amersham Biosciences) in a buffer (50 mm Tris-HCl (pH 8.0) and 0.1 m NaCl). A C-terminally hexahistidine-tagged Ub (Ub-His) protein was expressed from the pET15C plasmid in BL21(DE3) CodonPlus cells and purified using Ni-NTA-agarose (Qiagen). The cells were grown in M9 minimal media containing [15N]NH4Cl (1 g/liter) to produce the isotopically labeled protein. Samples were frozen in liquid nitrogen and stored at −80 °C until use.
Enzymatic Synthesis of Cyclic and Noncyclic Lys-48-linked diUbs
The wild-type and cyclic Lys-48-linked diUbs were prepared by in vitro enzymatic reaction using E2–25K (supplemental Fig. S1A). Ub and Ub-His were mixed at a molar ratio of 2:1 in 50 mm Tris-HCl (pH 8.0) and incubated at 37 °C for 16 h in the presence of 0.2 μm E1, 10 μm E2–25K, 1 mm dithiothreitol, 5 mm MgCl2, 10 mm ATP, 0.6 units/ml creatine phosphokinase, and 1 mm creatine phosphate. Isotopically labeled (unlabeled) Ub was combined with unlabeled (labeled) Ub-His for subunit-specific isotope labeling of wild-type Lys-48-linked diUb. After the reaction, noncyclic Ub chains with the hexahistidine tag were separated from the reaction mixture including cyclic Ub chains by Ni-NTA-agarose (Qiagen). The noncyclic Ub chains with the hexahistidine tag were incubated with YUH1 at 37 °C for 2 h; this resulted in almost complete cleavage of the tag. Finally, noncyclic diUB was separated from longer Ub chains using Resource S (GE Healthcare) cation exchange chromatography. Cyclic Lys-48-linked diUb was isolated from its longer counterparts in a similar manner. Protein purity was verified by SDS-PAGE.
Crystallization and Data Collection
Wild-type Lys-48-linked diUb was crystallized by hanging drop vapor diffusion at 293 K against a well solution containing 20% PEG 3350, 0.2 m Li2SO4 (pH 6.0). Data were collected using synchrotron radiation at BL44XU of SPring-8 (Hyogo, Japan). Diffraction data were processed using iMOSFLM (24) and SCALA (25). The diUb crystal belonged to the monoclinic space group P21 with unit-cell parameters a = 23.8, b = 56.7, c = 46.6 Å; β = 93.3°. Interestingly, this crystal form was the almost identical to that of previously reported tetraUb (PDB code 1TBE, P21, a = 24.1, b = 57.0, c = 47.0 Å; β = 94.1°) (26). This strongly suggests that our crystal structure of wild-type Lys-48-linked diUb is essentially identical with the part of the tetraUb structure found in the asymmetric unit. As expected, the present structure could be solved through utilizing the asymmetric unit coordinate of tetraUb (PDB code 1TBE) with REFMAC5 rigid body refinement (27). On the other hand, we could not obtain the valid molecular replacement solution using monomeric Ub (PDB code 1UBQ) as the search model, possibly due to the multiple crystallinity in the present diffraction data. Further restrained refinement procedures were carried out using REFMAC5 (27). Model fitting to the electron density maps was manually performed using COOT (28). The stereochemical quality of the final model was assessed using PROCHECK (29). The crystallographic parameters and final refinement statistics are summarized in Table 1. Molecular graphics were prepared using PyMOL.
TABLE 1.
Data collection and refinement statistics for wild-type Lys-48-linked diUb
| Data collection | |
| Space group | P21 |
| Unit cell parameters | a = 23.8, b = 56.7, and c = 46.6 Å; α = 90.0, β = 93.3, and γ = 90.0° |
| Wavelength (Å) | 0.9000 |
| Resolution range (Å) | 50.0–2.38 (2.51–2.38)a |
| No. of total reflections | 14,934 |
| No. of unique reflections | 4,854 |
| Completeness (%) | 96.5 (84.2) |
| Redundancy | 3.1 (3.0) |
| Rmerge | 0.149 (0.276) |
| I/σ(I) | 6.2 (4.3) |
| Wilson B (Å2) | 25.6 |
| Refinement | |
| Resolution range (Å) | 20.0–2.39 |
| No. of reflections | 4,607 |
| Rwork/Rfree (%) | 20.3 / 25.2 |
| No. of protein atoms | 1,183 |
| No. of solvent atoms | 73 |
| r.m.s.d. bond length (Å) | 0.012 |
| r.m.s.d. bond angles | 1.19° |
| Ramachandran plot (%) | |
| Most favored regions | 94.6 |
| Additional allowed regions | 4.6 |
| Generously allowed regions | 0.8 |
a Values in parentheses are for the highest resolution shell. A single crystal was used for the data set.
NMR Measurements
NMR samples were prepared at a concentration of 0.5 mm in 90% H2O/10% 2H2O (v/v) and 10 mm sodium phosphate buffer at pH 7.0. All NMR spectra were recorded at 303 K using a JEOL 920 or Bruker DMX500 spectrometer equipped with 5-mm inverse triple-resonance probes with three-axis gradient coils.
The RDC data were extracted from two sets of 1H-15N HSQC and TROSY spectra. One set was recorded from a sample of cyclic Lys-48-linked diUb alone at a concentration of 0.5 mm in 10% D2O and 10 mm sodium phosphate (pH 6.8) containing 0.2 mm NaCl, whereas the other set was recorded with an additional 15 mg/ml of Pf1 phage (ASLA Biotech, Riga) to obtain a weak orientation medium. Spectra were acquired on a JEOL ECA 920 MHz NMR spectrometer. The data were processed using NMRPipe (30) (zero-filled once, square sine apodization in each dimension) and analyzed using Sparky (38).
Grid Search Implementation
We obtained a total of 60 RDCs. The alignment tensor was determined using the software REDCAT (31) and the NMR structure of Ub (PDB code 1D3Z). This model was reoriented to be aligned with one of the tensor axes (REDCAT) for subsequent grid searches. The grid searches were implemented using Xplor-NIH (32). For the first grid search, only the closest Ub pairs without steric clashes in each plane perpendicular to one of the three axes of the principal alignment frame were selected for short energy minimization in which only the following few residues were allowed to move: C-terminal segments (residues 70–76), isopeptide bonds, and a region of eight residues around Lys-48 in each Ub unit (residues 44–51).
Once the correct plane was identified, a second grid search was performed according to the protocol described by Wang et al. (33). Ub dimers with their two subunits too close (<2 Å of interbackbone distance) or too far apart (>4 Å for intermolecularly proximal atomic distance) were removed from the list of models generated by the grid search. The remaining models were finally refined with explicit water, using the same residues as in the first grid search that were allowed to move with addition of the side chains of all residues. The structure models generated were evaluated and ranked by inspecting the criteria on the basis of van der Walls energy, residue-residue pair potentials at an interface (34), and the correlation factor of the experimental RDCs with simulated RDCs (using PALES (35)). We finally isolated a small cluster of possible solutions from these scores.
RESULTS AND DISCUSSION
Preparation of Wild-type and Cyclic Lys-48-linked diUbs
E2–25K catalyzes the formation of cyclic polyUb chains as well as the non-cyclic variety in vitro (36). To prepare wild-type Lys-48-linked diUb by suppressing the formation of the cyclic forms, we used Ub-His. A 2:1 mixture of the wild-type Ub and Ub-His was incubated with E1 and E2–25K in the presence of ATP and Mg2+. After the reaction, noncyclic Ub chains with the hexahistidine tag were separated from the reaction mixture, including cyclic Ub chains, using Ni-NTA-agarose. The hexahistidine tag was cleaved from the noncyclic Ub chains using YUH1. Eventually, wild-type diUb was separated from longer Ub chains by cation exchange chromatography (supplemental Fig. S1B). Similarly, cyclic diUb was isolated from longer cyclic chains.
Crystal Structures of Wild-type Lys-48-linked diUb
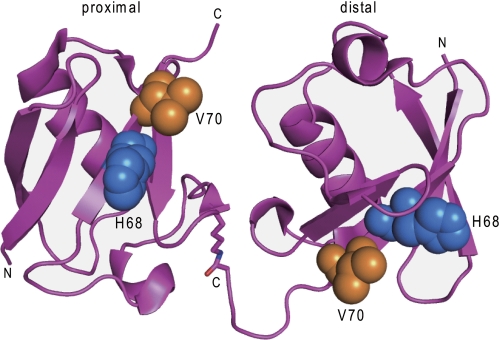
The new crystal form of wild-type Lys-48-linked diUb was determined at a resolution of 2.39 Å with an Rwork of 20.3% and an Rfree of 25.2% (Table 1 and Fig. 1). As expected from its crystallographic parameters, this new form was the same as the asymmetric unit of a previously reported crystal structure of tetraUb (PDB code 1TBE) (26). In the crystal, individual diUb molecules were stabilized in an open conformation without direct contact between hydrophobic surfaces. This was in marked contrast to the previously reported crystal structures of Lys-48-linked diUb, which adopted a closed conformation with sequestered hydrophobic surfaces (supplemental Fig. S2) (12, 13). We suggest that these crystal structures offer snapshot views of the possible conformations of Lys-48-linked diUb in solution.
FIGURE 1.
The crystal structure of wild-type Lys-48-linked diUb. The proximal domain is located on the left, and the distal domain is located on the right. His-68 (blue) and Val-70 (orange) are shown in a space-filling representation. By superimposing the Cα carbons of the distal and proximal Ub units, an r.m.s.d. value of 0.2 Å was obtained for 73 residues, excluding the three C-terminal residues.
Solution Structure of Cyclic Lys-48-linked diUb
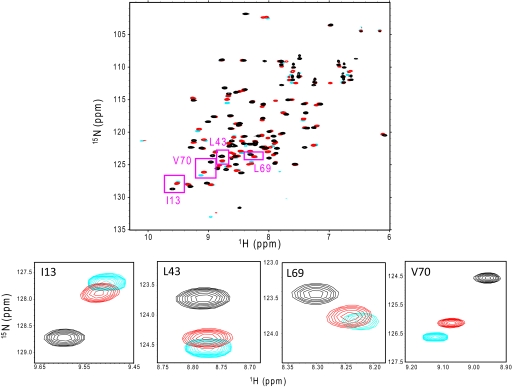
We also determined the structure of the cyclic form of Lys-48-linked diUb in solution by NMR spectroscopy. In its 1H-15N HSQC spectrum, cyclic Lys-48-linked diUb produced a single set of chemical shifts (Fig. 2, supplemental Table S1), indicating that the two Ub units are symmetrically related to each other.
FIGURE 2.
1H-15N HSQC spectra of monomeric Ub (cyan), wild-type Lys-48-linked diUb (red), and cyclic Lys-48-linked diUb (black) at pH 7.0. The close views of spectral regions (boxed), including the peaks from Ile-13, Leu-43, Leu-69, and Val-70, are displayed at the bottom.
To determine the relative orientation and position of the Ub units, we used the RDC-assisted grid search as developed by Wang et al. (33) to construct a three-dimensional model of symmetric protein homo-oligomers. This method is based on the fact that one of the principle axes of the alignment tensor is always parallel to the symmetry axis of the oligomer. For C2-symmetric homodimers, the symmetry axis can be chosen from the three axes of the principal alignment frame, by inspecting RDC data from two independent alignments, as the common axis in both frames. In the case of cyclic Lys-48-linked diUb, however, we found that only one alignment was needed because the two isopeptide bonds linking the two Ub units provided additional structural constraints. Namely, we could successfully define the only plane (perpendicular to one of the three axes of the principal alignment frame) in which the two Ub units could be positioned within the isopeptide bonding distance. An extensive grid search in this plane was performed to determine structures that satisfy the criteria on the basis of van der Waals energy and residue-residue pair potentials at an interface (34) as well as the experimental RDC data (supplemental Fig. S3). The best solution was selected to construct a three-dimensional structure model.
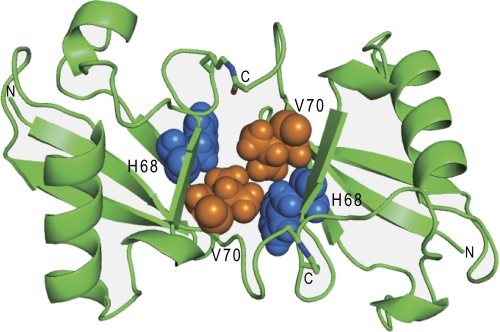
The model revealed that the solution structure of cyclic Lys-48-linked diUb bears a close resemblance to previously reported crystal structures of the noncyclic counterpart (Fig. 3, supplemental Fig. S4) (12, 13). The amino acid residues of cyclic Lys-48-linked diUb showing larger chemical shift differences from those of monomeric Ub were consistently concentrated on the hydrophobic surface(s), suggesting that these regions are involved in the Ub-Ub interaction in cyclic Lys-48-linked diUb (supplemental Fig. S5). This means that cyclic Lys-48-linked diUb exhibits a closed conformation in solution.
FIGURE 3.
Three-dimensional structural model of cyclic Lys-48-linked diUb derived from the NMR data. His-68 (blue) and Val-70 (orange) are shown in a space-filling representation.
NMR Analysis of Conformational Equilibrium of Wild-type Lys-48-linked diUb
We used cyclic Lys-48-linked diUb as an excellent model of the closed state in the following NMR analyses. In addition, monomeric Ub was used because its hydrophobic surface is exposed to the solvent and therefore mimics the hydrophobic surfaces of diUb in an open state. Fig. 2 compares the 1H-15N HSQC spectrum of wild-type Lys-48-linked diUb with that of cyclic Lys-48-linked diUb and monomeric Ub. Wild-type Lys-48-linked diUb exhibited a single set of peaks, except for Phe-45, Ala-46, Gly-47, Lys-48, Gln-49, Tyr-59, Asn-60, Leu-71, Arg-72, Leu-73, Arg-74, Gly-75, and Gly-76, most of which are located around the linkage site, suggesting the structural equivalence of the two Ub units (supplemental Table S1). Intriguingly, each amino acid residue located on the hydrophobic surfaces exhibited an HSQC peak between the peaks originating from the corresponding sites in monomeric Ub and cyclic Lys-48-linked diUb in the same straight line, as exemplified by the peaks originating from Ile-13, Leu-43, Leu-69, and Val-70 (Fig. 2 and supplemental Table S1). This indicates that wild-type Lys-48-linked diUb was in a conformational equilibrium between the open and closed states, which were mimicked by monomeric Ub and cyclic diUb, respectively, with regard to the exposure of the hydrophobic surfaces to the solvent (Fig. 4A).
FIGURE 4.
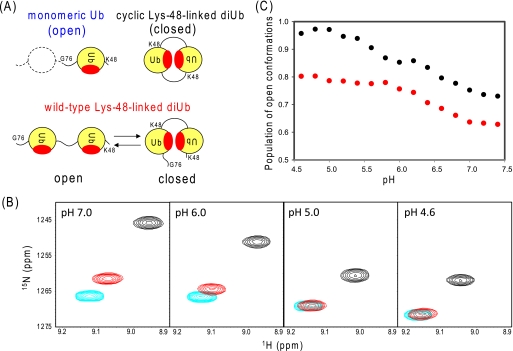
NMR characterization of conformational dynamics of Lys-48-linked diUb in solution. A, cartoon model of the conformational equilibrium of wild-type Lys-48-linked diUb. Cyclic Lys-48-linked diUb was used as a mimic of the closed state, whereas monomeric Ub was used as a mimic of the open state. The hydrophobic surface is colored red. The crystallographic data (supplemental Fig. S2) indicate that the distances between the two Ile-44 Cα atoms are 18.8 and 12.3 Å in the open and closed forms, respectively. B, 1H-15N HSQC peaks originating from Val-70 of monomeric Ub (cyan), wild-type Lys-48-linked diUb (red), and cyclic Lys-48-linked diUb (black) at the following different pH conditions: pH 7.0, 6.0, 5.0, and 4.6, starting from the left. C, pH dependence of the open conformation population of wild-type Lys-48-linked diUb (black) and its His-68→Val mutant (red) estimated on the basis of the dividing ratio of chemical shift differences of Val-70.
Because wild-type Lys-48-linked diUb undergoes a conformational transition between the two states in the fast exchange regime, the population of each conformer can be estimated on the basis of dividing ratios of the chemical shift difference. The dividing ratio was uniform between the monomeric and cyclic forms for all peaks observed, indicating that the population of open and closed conformers was 75 and 25%, respectively, in wild-type Lys-48-linked diUb (Fig. 2 and supplemental Table S1). This result was inconsistent with that of previous reports on mutated Lys-48-linked diUb in which the population of closed conformers was 90% (17, 20, 21). One possible explanation for this discrepancy is that the mutations affected the conformational equilibrium of Lys-48-linked diUb (see below).
pH Titration Experiment
We next examined the pH dependence of the conformational equilibrium of wild-type Lys-48-linked diUb by conducting NMR pH titration experiments. In the three Ub forms, the peaks from amino acid residues located on the hydrophobic surface(s) were all shifted on lowering of pH, but still aligned in the same straight line for each residue and maintained a uniform dividing ratio (Fig. 4B and supplemental Tables S1 and S2 and Fig. S6A). This allowed us to estimate the pH-dependent population shifts in the open and closed conformers of wild-type Lys-48-linked diUb in a quantitative manner. The open conformation became more populated on lowering of pH and eventually almost completely predominated at pH 4.5 (Fig. 4C and supplemental Fig. S6B). The pH-dependent conformational change suggests that protonation of some titratable groups in the hydrophobic surfaces destabilizes the closed conformation. The pH-dependent population shift can be divided into two phases with transition midpoints at pH 6.6 and 5.4, suggesting that at least two titratable groups are involved in this molecular event.
It has been suggested that ionization of the His-68 side chains in both Ub units, which are in spatial proximity to each other in the closed conformation (Fig. 3), is responsible for segregation of the two Ub units due to electrostatic repulsion (21). To examine this hypothesis, we prepared a mutated Ub with valine substitution at His-68 and cyclic and noncyclic Lys-48-linked diUb composed only of this Ub mutant. The effect of this mutation on conformational equilibrium was investigated by observing their NMR spectra. Based on the dividing ratios of the chemical shift difference, it was revealed that, in the H68V mutant of Lys-48-linked di-Ub, the closed conformation is more populated and the pH-dependent domain segregation is less pronounced with the disappearance of the transition at pH 5.4 (Fig. 4C and supplemental Fig. S6B). The pKa value of His-68 in monomeric Ub has been reported to be 5.5 (37); this value coincides with the those of wild-type Lys-48-linked diUb (data not shown). The observations can be explained by the removal of the positive repulsive charges and enhancement of hydrophobicity at the Ub-Ub interface, resulting from valine substitution at His-68. Although these results are consistent with the hypothetical model, the mutated diUb still exhibited a pH-dependent population shift with the midpoint at pH 6.6, which should be attributed to other titratable group(s). Around this pH, small but significant chemical shift changes were observed for the peak originating from Lys-48 and those from C-terminal segments (including Arg-72 and Arg-74), even in the H68V mutant of Lys-48-linked diUb (supplemental Fig. S7). Therefore, the observed chemical shift changes could be attributed to the protonation(s) of Lys-48 in the distal Ub unit and/or the titratable amino acid residue(s) located in the C-terminal segments, including the proximal C terminus itself. Unfortunately, it was not possible to specify the titratable residue(s) responsible for triggering the conformational transition that occurred around pH 6.6 because any mutation of these amino acid residues precludes the formation of the cyclic form. However, the importance of the electrostatic factors of the distal Lys-48 residue and proximal C-terminal segment could be exemplified by the previous NMR studies using the mutations at these sites, i.e. the substitution of Lys-48 and C-terminal extension with Asp-77, which have underscored the predominant population of the closed form (17, 20, 21). The hydrophobic surface of Ub is surrounded by basic amino acid residues such as Lys-6, Arg-42, Lys-48, His-68, and Arg-72. Therefore, the removal of these positive charges is expected to reduce the electrostatic repulsion between the two Ub units in the closed conformation, as demonstrated by the H68V mutation. Similarly, the addition of a negative charge at the Ub-Ub interface would increase the population of the closed form.
In conclusion, we demonstrated that wild-type Lys-48-linked diUb predominantly exhibits an open conformation in solution. Although protonation of His-68 and other unidentified titratable group(s) contribute to further increase in the population of molecules in the open conformation, 75% of the molecules already exist in the open form even under physiological conditions. This intrinsic property of Lys-48-linked Ub chains to adopt the open conformation may be advantageous for interacting with the Ub-binding proteins. The present study not only provides insights into molecular recognition events mediated by Ub chains but also offers new strategies for probing the conformational dynamics of multidomain proteins.
Acknowledgments
cDNA encoding E2–25K was kindly provided by Dr. Keiji Tanaka (The Tokyo Metropolitan Institute, Tokyo, Japan). We acknowledge Dr. Charles Schwieters (National Institutes of Health) for useful advice and for providing the Xplor-NIH code.
This work was supported by Grant-in-aid for Scientific Research on Innovative Areas 20107004, Grant-in-aid for Scientific Research (B) 21370050, the Targeted Proteins Research Program, and the Nanotechnology Network Project from the Ministry of Education, Culture, Sports, Science, and Technology.
The atomic coordinates and structure factors (code 3AUL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S7.
- Ub
- ubiquitin
- RDC
- residual dipolar coupling
- Ni-NTA
- nickel-nitrilotriacetic acid
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- HSQC
- heteronuclear single-quantum correlation
- TROSY
- transverse relaxation-optimized spectroscopy.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M., Eddins M. J. (2004) Biochim. Biophys. Acta 1695, 55–72 [DOI] [PubMed] [Google Scholar]
- 3. Baboshina O. V., Haas A. L. (1996) J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 4. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 5. Kirisako T., Kamei K., Murata S., Kato M., Fukumoto H., Kanie M., Sano S., Tokunaga F., Tanaka K., Iwai K. (2006) EMBO J. 25, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 7. Ikeda F., Dikic I. (2008) EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dikic I., Wakatsuki S., Walters K. J. (2009) Nat. Rev. Mol. Cell Biol. 10, 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Varadan R., Assfalg M., Raasi S., Pickart C., Fushman D. (2005) Mol. Cell 18, 687–698 [DOI] [PubMed] [Google Scholar]
- 11. Zhang N., Wang Q., Ehlinger A., Randles L., Lary J. W., Kang Y., Haririnia A., Storaska A. J., Cole J. L., Fushman D., Walters K. J. (2009) Mol. Cell 35, 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cook W. J., Jeffrey L. C., Carson M., Chen Z., Pickart C. M. (1992) J. Biol. Chem. 267, 16467–16471 [DOI] [PubMed] [Google Scholar]
- 13. Trempe J. F., Brown N. R., Noble M. E., Endicott J. A. (2010) Acta Crystallogr Sect. F Struct. Biol. Cryst Commun. 66, 994–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Phillips C. L., Thrower J., Pickart C. M., Hill C. P. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 341–344 [DOI] [PubMed] [Google Scholar]
- 15. Eddins M. J., Varadan R., Fushman D., Pickart C. M., Wolberger C. (2007) J. Mol. Biol. 367, 204–211 [DOI] [PubMed] [Google Scholar]
- 16. Satoh T., Sakata E., Yamamoto S., Yamaguchi Y., Sumiyoshi A., Wakatsuki S., Kato K. (2010) Biochem. Biophys. Res. Commun. 400, 329–333 [DOI] [PubMed] [Google Scholar]
- 17. Varadan R., Walker O., Pickart C., Fushman D. (2002) J. Mol. Biol. 324, 637–647 [DOI] [PubMed] [Google Scholar]
- 18. Tenno T., Fujiwara K., Tochio H., Iwai K., Morita E. H., Hayashi H., Murata S., Hiroaki H., Sato M., Tanaka K., Shirakawa M. (2004) Genes Cells 9, 865–875 [DOI] [PubMed] [Google Scholar]
- 19. Piotrowski J., Beal R., Hoffman L., Wilkinson K. D., Cohen R. E., Pickart C. M. (1997) J. Biol. Chem. 272, 23712–23721 [DOI] [PubMed] [Google Scholar]
- 20. Ryabov Y., Fushman D. (2006) Proteins 63, 787–796 [DOI] [PubMed] [Google Scholar]
- 21. Ryabov Y. E., Fushman D. (2007) J. Am. Chem. Soc. 129, 3315–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Varadan R., Assfalg M., Haririnia A., Raasi S., Pickart C., Fushman D. (2004) J. Biol. Chem. 279, 7055–7063 [DOI] [PubMed] [Google Scholar]
- 23. Chen Z., Pickart C. M. (1990) J. Biol. Chem. 265, 21835–21842 [PubMed] [Google Scholar]
- 24. Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 25. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 26. Cook W. J., Jeffrey L. C., Kasperek E., Pickart C. M. (1994) J. Mol. Biol. 236, 601–609 [DOI] [PubMed] [Google Scholar]
- 27. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 31. Valafar H., Prestegard J. H. (2004) J. Magn. Reson. 167, 228–241 [DOI] [PubMed] [Google Scholar]
- 32. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) J. Magn. Reson. 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 33. Wang X., Bansal S., Jiang M., Prestegard J. H. (2008) Protein Sci. 17, 899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moont G., Gabb H. A., Sternberg M. J. (1999) Proteins 35, 364–373 [PubMed] [Google Scholar]
- 35. Zweckstetter M. (2008) Nat. Protoc. 3, 679–690 [DOI] [PubMed] [Google Scholar]
- 36. Yao T., Cohen R. E. (2000) J. Biol. Chem. 275, 36862–36868 [DOI] [PubMed] [Google Scholar]
- 37. Fujiwara K., Tenno T., Sugasawa K., Jee J. G., Ohki I., Kojima C., Tochio H., Hiroaki H., Hanaoka F., Shirakawa M. (2004) J. Biol. Chem. 279, 4760–4767 [DOI] [PubMed] [Google Scholar]
- 38. Goddard T. D., Koeller D. G. (1993) Sparky, Version 3.0, University of California, San Francisco, CA [Google Scholar]