Background: In Hurler syndrome, heparan sulfate (HS) accumulates and is associated with early childhood mortality.
Results: Accumulated HS is abnormally highly sulfated and positively regulates N-deacetylase/N-sulfotransferase activity during HS biosynthesis.
Conclusion: We have identified a positive feedback loop in HS biosynthesis in Hurler syndrome that exacerbates the disease.
Significance: This will aid the design of therapeutic strategies for Hurler syndrome.
Keywords: Carbohydrate Biosynthesis, Glycosaminoglycan, Heparan Sulfate, Heparin, Lysosomal Storage Disease, Mass Spectrometry (MS), Mucopolysaccharidoses, Mucopolysaccharidosis, N-deacetylase/N-sulfotransferase, Heparan Sulfate
Abstract
Mucopolysaccharide (MPS) diseases are characterized by accumulation of glycosaminoglycans (GAGs) due to deficiencies in lysosomal enzymes responsible for GAG breakdown. Using a murine model of MPSI Hurler (MPSIH), we have quantified the heparan sulfate (HS) accumulation resulting from α-l-iduronidase (Idua) deficiency. HS levels were significantly increased in liver and brain tissue from 12-week-old Idua−/− mice by 87- and 20-fold, respectively. In addition, HS chains were shown to contain significantly increased N-, 2-O-, and 6-O-sulfation. Disaccharide compositional analyses also uncovered an HS disaccharide uniquely enriched in MPSIH, representing the terminal iduronic acid residue capping the non-reducing end of the HS chain, where no further degradation can occur in the absence of Idua. Critically, we identified that excess HS, some of which is colocalized to the Golgi secretory pathway, acts as a positive regulator of HS-sulfation, increasing the N-sulfotransferase activity of HS-modifying N-deacetylase/N-sulfotransferase enzymes. This mechanism may have severe implications during disease progression but, now identified, could help direct improved therapeutic strategies.
Introduction
The mucopolysaccharide (MPS)3 diseases are lysosomal storage disorders characterized by enzyme deficiencies in glycosaminoglycan (GAG) degradation. MPSIH (Hurler syndrome) is caused by the loss of lysosomal α-l-iduronidase (Idua), which hydrolyzes terminal iduronic acid (IdoA) residues from heparan sulfate (HS) and dermatan sulfate (DS) (1). MPSIH manifests early in childhood, presenting with dysostosis multiplex, cardiac and respiratory insufficiency, progressive neurological degeneration, and a severely shortened lifespan (2–4). Currently, diagnosis occurs postsymptomatically via an initial crude test of total GAG secretion in the urine with subsequent confirmation of reduced or absent enzyme activity (4). Few studies have been conducted to determine the degree of accumulation and the amounts and the type of sulfation modifications present in GAGs accumulated in different organs in this disease. This is important, as GAGs not only fulfill structural roles but are also essential for signaling and distribution of multiple growth factors, cytokines, and morphogens (5). The murine model of MPSIH has a targeted disruption in the Idua gene that eliminates activity. Mice show increased urinary GAG levels as well as progressive neurodegeneration and bone and joint disease, making them an excellent model of human disease (2, 6).
During HS biosynthesis, the EXT1-EXT2 polymerase complex adds alternating units of glucuronic acid (GlcA) and N-acetylglucosamine (GlcNAc) to the non-reducing end of the GAG chain. As the chain grows, the first modification enzyme, GlcNAc N-deacetylase/N-sulfotransferase (NDST), removes N-acetyl groups from selected GlcNAc residues and replaces them with sulfate groups, generating short highly sulfated domains (S-domains) separated by unmodified regions (7). N-deacetylation/N-sulfation represents a key regulatory step in HS-sulfation, because NDST action forms a preferential substrate for the subsequent modification reactions, including C5 epimerization of GlcA into IdoA and O-sulfation at various positions (8, 9).
Significantly, the degree of sulfation and sulfation patterning ultimately determines growth factor/morphogen binding. This has been demonstrated by a number of studies using in vitro assays to determine the strength and outcome of binding of protein ligands such as FGF family members (10, 11). Additionally, in vivo analyses have demonstrated that altered sulfation patterning can alter growth factor activity (12, 13). Significantly, mice mutant for various components of the HS biosynthetic and modification pathway, which express abnormal HS patterns, display multiple defects during embryonic development with many not surviving to birth (14). For example, mutation of the Sulf endosulfatase enzymes in mice, which remove 6-O-sulfate groups from HS chains, led to the expression of an oversulfated HS type and early lethality of most embryos, likely because of altered HS-ligand interactions (15, 16).
To improve our understanding of the GAG accumulation in MPSIH, we have quantified the extent of this accumulation in Idua−/− mice and detailed the sulfation patterning of HS from two pathologically relevant tissues, liver and brain. We reveal substantial increases in overall HS sulfation and identify a HS disaccharide uniquely enriched in MPSIH. Studies of HS localization suggest that HS is not confined to lysosomes but instead colocalizes to the Golgi compartment and to other parts of the cell, where it has the potential to interact with multiple factors. Enzyme activity studies suggest that the accumulated HS positively feeds back to the HS biosynthetic pathway, resulting in increased HS sulfation and exacerbating the MPSIH phenotype.
EXPERIMENTAL PROCEDURES
HS Extraction
Three-month-old age- and sex-matched wild-type and Idua−/− mice were sacrificed (n = 3) and the liver and brain dissected. Tissues were individually mechanically disaggregated and Pronase-treated (1 mg/tissue) in 10 ml of PBS for 4 h at 37 °C. Triton X-100 was added to a final concentration of 1% (v/v) for 2 h at room temperature before a further 1 mg of Pronase was added for 4 h at 37 °C. Preparations were then loaded onto a pre-equilibrated 1 cm DEAE-Sephacel column and washed with 50 ml of 0.25 m NaCl/20 mm NaH2PO4.H2O (pH 7) to remove hyaluronan. GAGs were eluted with 5 ml of 1.5 m NaCl/20 mm NaH2PO4.H2O, desalted using a PD10 column (Amersham Biosciences), and freeze-dried.
Human serum samples were obtained from MPSIH children (age range of 4–8 months) at diagnosis or from non-lysosomal disease age matched control patients (n = 3). Blood samples were collected in non-heparinized collection tubes and serum isolated as described previously (17). HS was then purified as described above.
Preparation and Analysis of AMAC-labeled Disaccharides
HS chains were digested using 5 mIU each of heparinase I, II, and III (Seikagaku, Tokyo, Japan) in 100 μl of 0.1 m sodium acetate and 0.1 mm calcium acetate (pH 7). The resulting disaccharides were then freeze-dried with and without prior addition of 0.58 μg Aldurazyme® (laronidase) (Genzyme Corp., Cambridge, MA) as required. Disaccharides were redissolved in 10 μl of 0.1 m 2-aminoacridone (AMAC) in 85% Me2SO/15% acetic acid (v/v) and incubated at room temperature for 20 min. Subsequently, 10 μl of 1 m NaBH3CN was added to each reaction, and the tubes were incubated at room temperature overnight. AMAC-labeled disaccharides were separated by reverse phase high-performance liquid chromatography using a Zorbax Eclipse XDB-C18 RP-HPLC column (3.5 μm, 2.1 mm × 150 mm, Agilent Technologies, Stockport, UK) as described previously (18). Disaccharide types were identified in comparison to AMAC-labeled commercial disaccharide preparations (Iduron, Manchester, UK). A disaccharide labeling efficiency factor was also applied for relative quantification as described previously (18). Duplicate heparinase digestions followed by RP-HPLC were performed for each organ. Preparative RP-HPLC was performed as above, collecting fractions manually by monitoring fluorescence. Small volumes of collected fractions were rerun over the HPLC to check purity. The fractions of interest were then pooled, freeze-dried, and analyzed by mass spectrometry.
Structure Identification by Mass Spectrometry
An LTQTM two-dimensional linear ion trap mass spectrometer equipped with an electrospray ionization source and directly coupled to an HPLC system (Thermo Electron, San Jose, CA) was used for structure identification. The sample enrichment prior to MS analysis was accomplished using an Eclipse XDB-C8 column (3.5 μm, 1.0 mm ID × 150 mm, Agilent Technologies) that was operated for a 12-min linear gradient from 98% solvent A (20 mm NH4OAc) to 100% solvent B acenotrile (ACN) at a flow rate of 75 μl/min. Mass spectra were collected in the negative ion mode. For MSn (n = 1, 2, 3) experiments, the precursor ions were selected with an isolation width of 3.0 Da and an activation energy of 25% normalized collision energy for 30 ms. MS data acquisition and analysis were performed using Xcalibur 2.0 software.
Exact Mass Measurement by FTICR-MS
Exact mass measurements were accomplished using an Apex II FTICR mass spectrometer equipped with an electrospray ionization source and a 7.0-Tesla superconducting magnet (Bruker Daltonics, Billerica, MA). MS analysis was performed after external calibration using beer maltooligosaccharides followed by tuning with disaccharide standards in the negative ion mode at an m/z range of 118–800.
Determination of Enzyme Activities
Brain tissues from wild-type and Idua−/− mice were homogenized in solubilization buffer containing 50 mm Tris/HCl (pH 7.5), 1% Triton X-100, 2 mm EDTA, 2 mm Pefablock, and 10 μg/ml pepstatin A. Protein concentration was determined using the Bio-Rad protein assay. N-deacetylase and N-sulfotransferase activities were essentially analyzed as described previously (19). Briefly, 80 μg of solubilized proteins were incubated with N-deacetylated Escherichia coli capsular K5 polysaccharide substrate (3 μg) and 2 μCi of [35S] 3′-phospho-adenosine-5′-phosphosulfate in 50 mm HEPES (pH 7.4), 10 mm MgCl2, 10 mm MnCl2, 5 mm CaCl2, 3.5 μm NaF, and 1% Triton X-100 in a total volume of 100 μl. After incubation for 30 min at 37 °C, the polysaccharide was precipitated by ethanol overnight. 35S-labeled polysaccharides were separated from excess [35S] 3′-phospho-adenosine-5′-phosphosulfate by centrifugation of the samples through Sephadex G-25 columns (superfine grade, Amersham Biosciences) and quantified by scintillation counting. In a separate experiment, enzyme activity in the extracts was compared before and after removal of endogenous GAGs. After the addition of NaCl to a final concentration of 0.1 m, half of the extracts were passed over DEAE-Sephacel columns equilibrated in solubilization buffer supplemented with 0.1 m NaCl. N-sulfotransferase activity was then measured in both the DEAE non-binding fraction and in the unfractionated extract. O-sulfotransferase activity was performed as described for N-sulfotransferase activity using N/O-desulfated re-N-sulfated heparin as a substrate.
N-deacetylase activity was measured by incubating solubilized extracts (80 μg of protein) and 10,000 cpm of N-[3H]acetyl-labeled K5 polysaccharide for 60 min at 37 °C in 100 μl of 50 mm MES, 10 mm MnCl2, and 1% Triton X-100, pH 6.3. The released [3H]acetate was detected using a biphasic scintillation counting system. Where indicated, N-deacetylase activity was inhibited by the addition of 10 mm N-ethylmaleimide (NEM) to the lysate prior to substrate addition (19).
Real-time PCR
Total RNA was isolated from wild-type and Idua−/− brain tissues using an E.Z.N.A.® Total RNA kit according to the manufacturer's instructions (Omega Bio-Tek, Norcross, GA). Reverse transcription was performed from 1 μg of total RNA with random hexamers and SuperScriptTM II reverse transcriptase (Invitrogen). PCR amplifications were performed using a MiniOpticon real-time PCR detection system (Bio-Rad). The transcript of β-actin was used as a control to normalize expression. Primer sets were as follows: NDST1, 5′-CCACAACTATCACAAAGGCATCG-3′ (forward) and 5′-GAAAGGTGTACTTTAGGGCCAC-3′ (reverse); NDST2, 5′-GTGTGGCAGAATCCCTGTG-3′ (forward) and 5′-GTGCAGGCTCAGGAAGAAGT-3′ (reverse); NDST3, 5′-GGAGCTCTTCTTCACTGTGGTT-3′ (forward) and 5′-TCTGAAGACGCAGGTTGGT-3′ (reverse); NDST4, 5′-GGAGAAAACCTGTGACCATTTAC-3′ (forward) and 5′-CCTTGTGATAGTTGTTGCCATTA-3′(reverse); and β-actin, 5′-AAGAGCTATGAGCTGCCTGA-3′ (forward) and 5′-TACGGATGTCAACGTCACAC-3′ (reverse). PCRs were carried out using SsoFast EvaGreen Supermix (Bio-Rad) using the following conditions: 1 × 95 °C for 10 min, 40 × (95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s). A melting curve (55–95 °C at 1 °C interval) was constructed for each primer pair to ensure the presence of a single gene-specific peak. The relative level of expression of transcripts was calculated as described previously (20).
Immunofluorescent Liver Staining
Livers were excised from 16-week-old wild-type and Idua−/− mice, and the median lobe was trimmed, mounted in O.C.T. and immediately snap-frozen in liquid nitrogen. 30-μm sections were cut using a cryostat, mounted on glass slides, and stored at −80 °C. Sections were fixed for 10 min in acetone, washed, and then processed essentially as described previously (21) using giantin (Abcam, Cambridge, UK) and 10E4 (Seikagaku Corp.) antibodies, followed by goat anti-rat Alexa Fluor 546 and goat anti-mouse IgM Alexa Fluor 488 (Invitrogen). Images were collected using a Nikon C1 confocal on an upright 90i microscope with a ×20/0.5 or ×40/0.75 Plan Apo objective. The confocal settings were as follows: pinhole, 30 μm; scan speed, 400 Hz unidirectional; and format, 1024 × 1024. Images for DAPI, Alexa Fluor 488, and Alexa Fluor 546 were excited with the 405-nm, 488-nm, and 543-nm laser lines, respectively. Only the maximum intensity projections of these three-dimensional stacks are shown in the results.
Statistical Analysis
Statistical analysis was performed using a two-tailed Student's t test assuming equal two-tailed distribution and equal variance. p values of less than or equal to 0.05 were considered significant. The error bars refer to mean ± S.D.
RESULTS
Significant HS Accumulation and Increase in Sulfation in Idua−/− Mice
Recently, a highly sensitive precolumn labeling method has been described that is suitable for limited quantities of biological specimens (18). This technique requires minimal sample processing prior to labeling with AMAC for fluorescent detection. Using this method, we have analyzed HS from age-matched 12-week-old wild-type and Idua−/− mice, an age at which disease pathology is clearly evident in Idua−/− animals (6). GAGs were purified from pathologically relevant liver and brain tissues, and the HS chains were digested into disaccharides using bacterial heparinases. Disaccharides were labeled with AMAC and analyzed using single-step RP-HPLC, revealing six characteristic disaccharides in wild-type tissues (Fig. 1, a and b). These were also present in Idua−/− tissues, although at clearly different proportions and amounts (compare Fig. 1, a and b, y axis). However, Idua−/− samples also contained an unidentified peak (peak U), eluting at approximately 17–18 min, immediately prior to the HexA(2S)-GlcNS disaccharide peak (peak 3) (where HexA is GluA or IdoA), which did not correspond to any known heparinase-generated disaccharide standard (Fig. 1, a and b, U). Interestingly, although clearly present in both Idua−/− organs studied, different relative contributions of this disaccharide were seen between the liver and brain tissues, suggesting that it may represent a disease-specific disaccharide that accumulates at varying amounts in different tissues.
FIGURE 1.
The composition of Idua−/− HS is markedly different from the wild type, with significant increases in all sulfation modifications. RP-HPLC traces of AMAC-labeled heparinase-generated disaccharides from wild-type and Idua−/− liver (a) and brain (b). Peaks are labeled following comparison to known standards: 1, HexA(2S)-GlcNS(6S); 2, HexA-GlcNS(6S); 3, HexA(2S)-GlcNS; 4, HexA-GlcNS; 5, HexA-GlcNAc(6S); 6, HexA-GlcNAc; *, minor free AMAC peak; U, uncharacterized. c and d, relative amount of HS in Idua−/− liver (c) and brain (d) versus the wild type. Disaccharide composition (e and g) and percentage contribution of each modification (f and h) from wild-type and Idua−/− liver (e and f) and brain (g and h). Results are an average from three independent animals with two separate heparinase digests per animal. n = 3. The bars represent mean ± S.D. p values are quoted using a Student's t test and assuming two-tailed distribution and equal variance. NAc, N-acetylated glucosamine; NS, N-sulfated glucosamine; 2S, 2-O-sulfate group; 6S, 6-O-sulfate group.
Integration analysis of disaccharide peak areas enabled relative quantification of HS amounts and disaccharide composition following RP-HPLC (18). Significantly, total levels of HS were approximately 87-fold higher in Idua−/− liver compared with wild-type and approximately 20-fold higher in the brain (Fig. 1, c and d), a remarkable increase at only 12 weeks of age.
Quantification of liver disaccharide composition revealed striking increases in the most sulfated disaccharide HexA(2S)-GlcNS(6S) (2.8-fold) in Idua−/−, mirrored by decreases in all mono-/non-sulfated species (Fig. 1e). This led to an overall 149% increase in 2-O-sulfation and significant increases in both N-(43%) and 6-O-sulfation (56%) (Fig. 1f). The wild-type distribution of disaccharide species was altered in brain samples compared with liver (comparing the wild-type HS composition from Fig. 1, e and f), highlighting HS tissue specificity (22, 23). Critically, similar trends of major increases/decreases were seen in brain Idua−/− samples compared with wild-type samples, as observed in liver (Fig. 1, g and h). Therefore, Idua−/− animals contain not only an excess of non-degraded HS, but the composition of those HS chains is also strikingly altered with significantly increased sulfation.
Excess HS with Increased Sulfation Is Present in Human MPSIH Patient Blood Serum
We have shown previously that excess HS and DS can be detected in the bloodstream of MPSIH patients and acts as a marker of disease (17, 24). Therefore, to establish whether the increase in HS sulfation observed in the MPSIH model mice was a true representation of the human disease, blood serum was collected from MPSIH patients at diagnosis prior to therapy and compared with that of unaffected individuals. The percentage contribution of the most highly sulfated disaccharides in control human serum was substantially lower than that seen in murine liver and brain samples (Fig. 2a compared with Fig. 1, e and g), reflecting the tissue source (22, 23). However, significant total increases in N-, 2-O-, and 6-O-sulfation were apparent in MPSIH serum compared with controls (Fig. 2b), in addition to an approximately 6-fold increase in total HS levels (Fig. 2c), highlighting the alteration in HS structure in affected patients alongside the recognized increase in total HS levels. Peak U was also present in MPSIH serum, although at a lesser proportion than seen in murine liver and brain samples (data not shown).
FIGURE 2.
MPSIH patient blood serum contains significantly increased levels of highly sulfated HS. a, HS disaccharide composition in normal and MPSIH blood serum. b, percentage of HS disaccharides containing each modification. c, relative amount of HS in MPSIH blood serum versus normal. n = 3.
Idua Deficiency Results in an Enrichment of HS Chains with IdoA Residues at the Non-reducing End
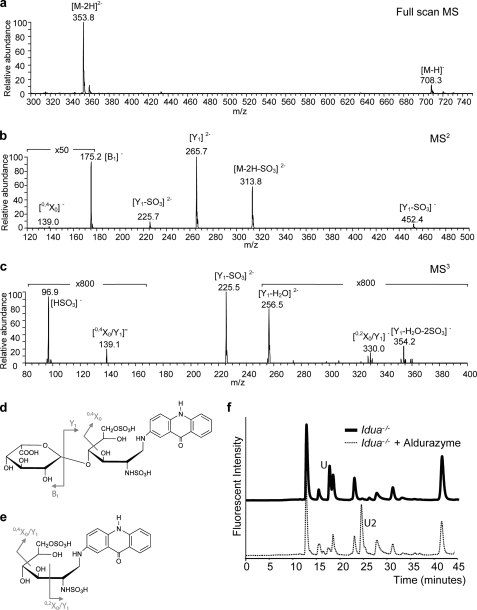
The presence of peak U in all Idua−/− samples (Fig. 1b) suggests that this saccharide may represent the terminal IdoA-containing sequence uniquely enriched by the absence of Idua. To identify its structure, peak U was purified by RP-HPLC and analyzed by mass spectrometry (MS). The full-scan MS of peak U (m/z 353.8, [M-2H]2−) suggested that it was a disaccharide containing two sulfate groups (25) (Fig. 3a). The sulfates were positioned to the reducing end by MS2, which displayed formation of B1, Y1, and 0,4X0 fragments (Fig. 3b). [B1]− (m/z 175.2) and [Y1]2− (m/z 265.7) were glycosidic bond cleavage fragments corresponding to saturated IdoA and a disulfated AMAC-labeled GlcN at the reducing end, respectively. The [0,4X0]− ion (m/z 139.0) was the cross-ring cleavage product identifying the 6-O-sulfation position of GlcN. Confirmation of the 6-O-sulfate group and the positioning of the second sulfation site was elucidated using MS3 (Fig. 3c). The cross-ring cleavages 0,2X0/Y1 (m/z 330.0) and 0,4X0/Y1 (m/z 139.1) identified the N- and 6-O-sulfation sites on GlcN (Fig. 3e).
FIGURE 3.
Identification of a non-reducing end disaccharide unique to Idua−/− animals. a, full-scan mass spectrum of peak U showing the singly ([M-H]−) and doubly ([M-2H]2−) charged precursor ions. b, annotated MS2 of the isolated precursor ion (m/z 353.82−→) showing the product ions of the saturated non-reducing end ([B1]−) and 6-O-sulfate at the reducing end ([0,4X0]−). c, MS3 of peak U (m/z 353.82−→265.72−→) with diagnostic ions for N-sulfate ([0,2X0/Y1]−, m/z 330.0) and 6-O-sulfate ([0,4X0/Y1]−, m/z 139.1) at the reducing end. d, structure of peak U and the cleavage pattern observed in the MS2 spectrum. e, structure of the Y1 fragment and the cleavage pattern observed in the MS3 spectrum. f, RP-HPLC profile of AMAC-labeled heparinase-generated disaccharides from Idua−/− liver before and after Aldurazyme treatment.
Furthermore, exact mass measurement using electrospray ionization-FTICR/MS recorded a mass of m/z 353.5478 ([C25H29N3O17S2]2−) from three sequential MS acquisitions with a resolution of 71,000. The mass error of 2 ppm was calculated from the computed theoretical m/z value of 353.5470. Thus, both methods proposed the structure of peak U as IdoA-GlcNS6S-AMAC (Fig. 3d).
This was further confirmed using Aldurazyme® (laronidase), a polymorphic variant of human Idua used in the clinic. Aldurazyme treatment significantly reduced peak U, indicating successful removal of IdoA. A new fluorescent saccharide (Fig. 3f, U2) was then observed, representing the remaining AMAC-labeled GlcNS6S residue. Heparinase depolymerization of HS results in the introduction of an unsaturated uronate residue at the non-reducing terminal (26, 27). Therefore, saturated IdoA-GlcNS6S-AMAC represents the terminal disaccharide at the non-reducing end of the HS chain (Fig. 3d).
NDST Activity Is Positively Regulated by Excess HS
The lysosomal enzymes act only on the non-reducing terminal residue of GAG chains, sequentially removing saccharides exolytically. Hence, the non-reducing end is an indicator of impaired exoenzyme activity. IdoA residues are much enriched within S-domains, and most HS chains seem to have S-domain sequences at their non-reducing end (28, 29). Thus, it is likely that little breakdown of HS chains will occur in Idua−/− animals, with chains remaining “capped” with terminal IdoA residues. Although heparanase, a HS-specific endo-β-glucuronidase enzyme, is likely to be active in these cells allowing further breakdown of the chain producing new substrates for the lysosomal enzymes, it has previously been shown that only one or two recognition sites are likely to be present per chain (30), limiting its effect. We might therefore expect some increase in the level of sulfation within Idua−/− chains as S-domain digestion is blocked. However it is unlikely that the substantial increase in sulfation observed in these tissues is solely due to this increase in terminal S-domains sequences, instead these results suggest that HS biosynthesis may additionally be altered in Idua−/− tissues.
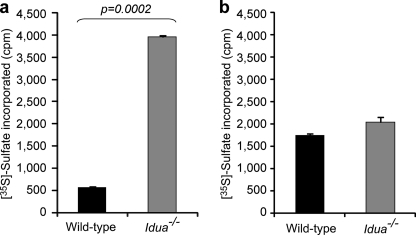
We therefore assessed the activity of NDST enzymes that our previous studies have identified as being key in determining sulfation patterning in HS, with the initial positioning of N sulfate groups ultimately directing further sulfation events (8, 9). NDST enzyme activity was measured in wild-type and Idua−/− brain extracts using N-deacetylated K5 polysaccharide as a substrate. This revealed a substantial increase in N-sulfotransferase activity in Idua−/− extracts compared with the wild type (Fig. 4a). To determine whether this difference reflected a global increase in HS sulfotransferase activity, we also studied HS O-sulfotransferase activity (Fig. 4b). However, only a minor, non-significant increase in activity was observed in Idua−/− tissues. Therefore, these results suggested that there is substantially increased N-sulfotransferase activity in Idua−/− tissues, which will provide a preferential substrate for further sulfate addition, leading to the large increases in overall HS sulfation.
FIGURE 4.
N-sulfotransferase activity is increased in Idua−/− tissues. a, N-sulfotransferase activity in Idua−/− and wild-type brain extracts. b, O-sulfotransferase activity in Idua−/− and wild-type brain extracts.
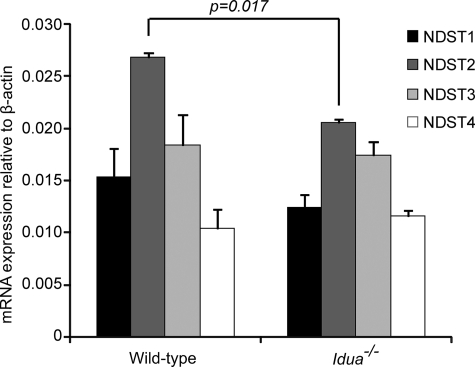
This observed increase in N-sulfotransferase activity may indicate a change in NDST enzyme expression or reflect an enhanced enzymatic activity in Idua−/− cells. Thus, quantitative real-time PCR was used to determine whether this increased activity was due to changes in NDST expression in Idua−/− tissues (Fig. 5). Results revealed equivalent levels of expression of NDST1, NDST3, and NDST4 transcripts together with an actual small but significant decrease in mRNAs encoding NDST2 in Idua−/− brain. Thus, results suggest that increases in NDST enzyme levels are unlikely to affect sulfation, suggesting that enzyme activity may be altered.
FIGURE 5.
Expression of NDST transcripts is similar in Idua−/− and wild-type tissues. Quantitative PCR analysis of the expression of NDSTs in Idua−/− and wild-type brain extracts. mRNA levels were normalized relative to β-actin. The p value for NDST2 is quoted. Other samples were not significant.
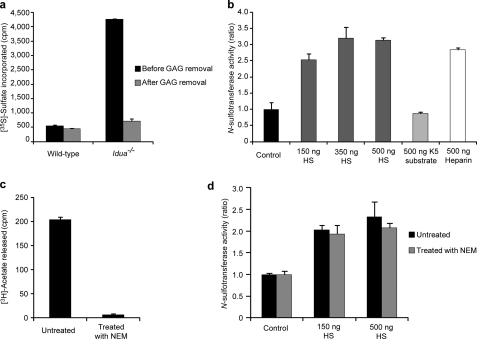
As discussed previously, Idua−/− brain tissue contains substantial levels of accumulated GAGs compared with wild type. Therefore, to investigate whether the accumulation of endogenous GAGs in Idua−/− tissues was enhancing N-sulfotransferase activity, enzyme assays were preformed pre- and post-complete GAG removal. As shown in Fig. 6a, N-sulfotransferase activity was greatly reduced in the Idua−/− extracts after removal of the GAGs, yet activities were still higher than in GAG-depleted wild-type extracts. GAG removal using a DEAE column had no detrimental effect on wild-type N-sulfotransferase activity, with pre- and post-GAG depletion extract producing near identical results.
FIGURE 6.
Excess HS accumulation alters HS biosynthetic enzyme activity, increasing N-sulfotransferase action. a, N-sulfotransferase activity in wild-type and Idua−/− brain extracts in the presence of endogenous GAGs (before GAG removal) or following complete GAG removal. b, N-sulfotransferase activity in wild-type brain extracts in the absence (control) or presence of exogenous polysaccharides (increasing amounts of HS, 500 ng of N-deacetylated K5 substrate, or 500 ng heparin). c, N-deacetylase activity in wild-type brain before or following treatment with NEM. d, N-sulfotransferase activity in wild-type brain before or following treatment with NEM in the presence of exogenous HS. Results shown are the average value obtained for extracts from two animals. The bars represent mean ± S.D.
These results suggested that the presence of excess GAGs in Idua−/− cells may directly alter NDST activity. Thus, using NDST enzyme extracts from wild-type brains, we asked whether the addition of increasing amounts of exogenous HS alongside the N-deacetylated K5 polysaccharide substrate (the substrate used in these N-sulfotransferase assays) could influence NDST activity (Fig. 6b). Results showed that N-sulfotransferase activity was greatly increased with exogenous HS addition. However, when HS was replaced with additional K5 substrate, no effect was seen. Exogenous heparin, which is highly sulfated along its entire length with a similar structure to the S-domain regions of HS, also significantly increased N-sulfotransferase activity. The low concentrations of HS required for this effect compared with available substrate (150 ng of exogenous HS versus 3 μg of K5 substrate) suggests that high levels of accumulating HS have a direct effect on NDST enzyme activity.
To further eliminate the possibility that the exogenous HS is acting as a preferential substrate for the NDSTs instead of the K5 substrate, experiments were repeated in the presence of N-deacetylase inhibitor NEM. Without the N-deactylase activity of the bifunctional NDSTs, native HS is an exceptionally poor substrate for N-sulfotransferase activity, with only a few scattered N-unsubstituted GlcN residues that have the potential for sulfation, in contrast to the N-deacetylated K5 acceptor substrate. Thus, with the addition of NEM, only N-sulfation of the K5 substrate can occur. As expected, N-deacetylase activity was eliminated with NEM addition (Fig. 6c), yet the increase in N-sulfotransferase activity seen with exogenous HS addition remained (d). Thus, our results show that the presence of excess HS can directly increase the N-sulfotransferase activity of NDST enzymes.
Excess HS Localizes to the Golgi and Is Not Restricted to the Lysosome
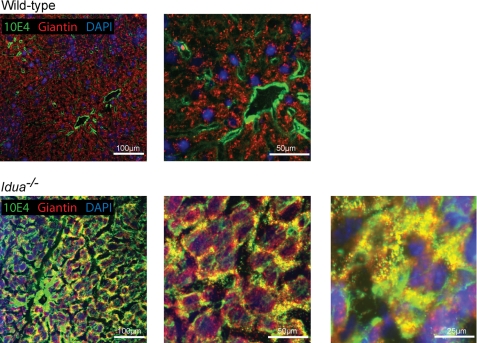
Excess HS in MPS patients is traditionally thought to be restricted to lysosomes. However, the large increases in HS, alterations in HS sulfation, the presence of HS in human serum, and the effect of excess HS on NDST activity suggest that HS may be accumulating outside of the lysosome. To influence biosynthesis, accumulated HS would likely have to be associated with the organelle responsible for HS biosynthesis, the Golgi. To investigate this, tissue sections were co-stained with the anti-HS antibody 10E4, which recognizes a common sulfated HS epitope (31) and the Golgi marker giantin and observed using confocal microscopy. In wild-type liver, HS staining was mainly restricted to the perivascular region of blood vessels, with only very weak staining visible on the plasma membrane of liver cells and within the extracellular matrix (Fig. 7). Significantly increased 10E4 staining was apparent in Idua−/− livers, with strong staining within the cell, at the cell surface, and in the extracellular matrix. In both wild-type and Idua−/− livers, there is extensive giantin staining throughout the cell. However, in the Idua−/− livers, there is apparent colocalization of giantin and 10E4 at the outer edges of cells where 10E4 staining is strongest. This suggests that much of the accumulated HS is not being retained in the lysosome where it might be considered to be sequestered, preventing it from influencing cellular processes and signaling. Instead, it is being redistributed to a variety of cellular and extracellular locations as well as being retained inside the cell within the Golgi apparatus where it can influence HS biosynthesis directly.
FIGURE 7.
Excess HS accumulates in Idua−/− mouse livers and colocalizes with the Golgi. Shown is staining of wild-type and Idua−/− liver with antibodies against HS (10E4, green) and Golgi marker giantin (red). Nuclei (blue) were visualized with DAPI.
DISCUSSION
MPS diseases are characterized as a family of metabolic disorders caused by a deficiency of lysosomal enzymes needed to degrade GAGs. Disease pathology is typically accredited to the accumulation of excess lysosomal GAGs, and available data on GAG structure is conflicting and limited. In addition, there has been little quantification of the extent of GAG accumulation in MPS tissues. Here, we show that in the absence of Idua, HS levels, composition, and localization are significantly altered. The use of highly sensitive analytical techniques enabled the quantification of the changes in HS to be characterized, revealing dramatic increases in HS N-, 6-O-, and 2-O-sulfation. Using 10E4 antibody, which binds the HS chain itself, we were able to show substantial HS staining throughout Idua−/− liver cells and, in particular, colocalization with the Golgi apparatus. We also show that excess HS alters HS biosynthesis, increasing the activity of HS-modifying NDST enzymes. These enzymes carry out the first step in the HS modification pathway, ultimately defining the position of sulfate groups in clusters along the HS chain. Because 2-O- and 6-O-sulfation occur in close proximity to N-sulfate groups, elevated N-sulfation will also result in increased O-sulfation. This leads to the markedly altered HS composition in Idua−/− animals, with likely increases in the size and/or number of S-domain sequences. Thus, this significant accumulation of abnormally highly sulfated HS will ultimately exacerbate the MPSIH phenotype. Critically, elevated HS was also present in the blood serum of MPSIH patients. Compositional analyses of this serum HS indicate that, similar to the Idua−/− mice, HS sulfation is markedly increased in MPSIH patients, demonstrating that the Idua−/− mouse mirrors the human disease well and provides a therapeutically relevant model to study this lysosomal disease.
The current understanding of mechanisms regulating HS biosynthesis is at an early stage. The availability of activated precursors is important, and there is considerable interdependence between the various enzymes, with HS biosynthetic enzymes proposed to functionally interact with one another to form a GAGosome enzyme complex (32, 33). Our identification of a positive feedback signal whereby sulfated HS drives increased NDST activity, which in turn enables subsequent modification via epimerization and O-sulfation, suggests that at least in this disease state, other mechanisms of HS regulation may also need to be taken into account.
The significant accumulation of HS with increased sulfation at non-lysosomal sites has the potential to impact on HS-dependent signaling, dramatically altering cell functions. HS sulfation is often increased in human disease, with specific structural alterations mirroring the sulfation requirements for the growth factor/cytokines involved (34, 35). Increased HS sulfation is known to promote the signaling of some factors while inhibiting the function of others. For example, FGF2 requires a HS sequence containing N- and 2-O-sulfation for FGF2-HS binding; however, at least one 6-O-sulfate group is required for signal transduction, suggesting the involvement of 6-O-sulfation in the formation of an FGF2/HS/FGF-receptor ternary complex (11). On the contrary, high levels of 6-O-sulfation are inhibitory for Wingless signaling, as 6-O-sulfated HS provides a higher affinity binding site for Wnt than its receptor Frizzled, effectively sequestering Wnt protein, preventing signal transduction (36). Thus, it is likely that in Idua−/− animals (and by inference, potentially MPSIH patients) signaling by some factors will be improved because of increased presentation of ligands to their receptors, or inhibited, with excess HS sequestering factors preventing signal transduction. We have recently completed a study that demonstrates that increased sulfated HS in MPSIH patients alters the ability of hematopoietic stem cells to respond to a key factor involved in stem cell migration.4 It is noted that IdoA-containing DS, which also has an important signaling role, is additionally likely to be elevated in Idua−/− animals, compounding disease prognosis.
A previous study detailed the HS structure of cultured human bone marrow-derived multipotent adult progenitor cells isolated from Hurler patients. This identified only a 1.7-fold increase in total GAGs and relatively minor changes in the GAG sulfation patterning, with the most significant finding a 26% decrease in 6-O-sulfation (37), opposite to what we observe in Idua−/− organs. Critically, these cells were adapted to tissue culture prior analysis of HS chain composition, suggesting that this analysis may not accurately reflect GAG structure in vivo (38).
Previous methods identifying diagnostic oligosaccharides in the urine of MPSIH patients required extensive GAG purification followed by MS for visualization (39, 40). Our quick purification, labeling, and RP-HPLC method to analyze small amounts of material from in vivo samples in just a few days, represents a significant advance in the analysis of primary tissues and clinical specimens. The identification of a unique Idua−/−-specific disaccharide residue also acts as a useful diagnostic marker that could be used to monitor disease and therapeutic outcomes. Although it has been suggested that non-reducing end structures of GAG chains will be present in MPS diseases, as the normal substrate for lysosomal enzymes was elucidated many decades ago, to our knowledge this is the first report detailing their visualization. Other MPS types lack different enzymes required for GAG degradation. Thus, it is likely that some or all of these MPS types will also be characterized by the formation of novel unique terminal end structures, providing a new diagnostic tool for the MPS field.
It is also clinically relevant to note that although, as predicted, HS levels were significantly elevated in Idua−/− tissues, results indicated noticeably less storage in the brain compared with the liver, with a simultaneous decrease in the presence of the Idua−/−-specific disaccharide. Brain pathology is notoriously difficult to correct in MPSIH patients because of the blood-brain barrier preventing entry of corrective enzymes except via bone marrow-derived microglia following hematopoietic stem cell transplantation (41, 42). Thus, this difference suggests that brain GAG storage may occur later or more slowly than in other tissues, improving the outlook for therapeutic intervention following early diagnosis. It may also suggest a different pathological threshold for storage in neuronal tissue, suggesting the possibility of highly GAG-sensitive pathological cascades that may, in turn, inform therapeutic targets.
To summarize, we have quantitatively measured a substantial elevation in HS within the liver and brain of Idua−/− mice and revealed significant changes in HS localization and sulfation patterning with significant increases in N-, 6-O-, and 2-O-sulfation. Increased levels of total HS and increased HS sulfation were also observed in blood serum from untreated patients. Disaccharide analysis identified the presence of a non-reducing end disaccharide uniquely elevated in Idua−/− and MPSIH patients. Different non-reducing end structures will unquestionably be present in the other MPS diseases. Tissue extracts from Idua−/− mice show significantly increased NDST enzyme activity, and our additional experiments suggest that the presence of excess HS accumulating in intracellular compartments has a positive stimulatory effect on NDST activity. Thus, we propose a novel hypothesis whereby altered HS biosynthesis contributes to MPSIH pathology. Additionally, the novel peak and structural changes in HS identified in MPSI could permit the development of improved methods for diagnosis and treatment monitoring in this disease.
Acknowledgments
The Bioimaging Facility microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and the University of Manchester Strategic Fund.
This work was supported by funding from The Human Frontier Science Program (to R. J. H. and C. L. R. M.), The Medical Research Council (to C. L. R. M.), The Swedish Research Council (to A. Deligny, A. Dagälv, and L. K.), The Swedish Cancer Society (to A. Deligny, A. Dagälv, and L. K.), Polysackaridforskning AB (to A. Deligny, A. Dagälv, and L. K.), National Institutes of Health Grant NIH GM47356 (to W. W., M. R. N., and J. A. L.), The UK Society for Mucopolysaccharide Diseases (to H. A. W. and B. W. B.), and The National MPS Society (to H. A. W. and B. W. B.).
H. A. Watson and B. Bigger, unpublished data.
- MPS
- mucopolysaccharide
- GAG
- glycosaminoglycan
- IdoA
- iduronic acid
- HS
- heparan sulfate
- DS
- dermatan sulfate
- GlcNAc
- N-acetylglucosamine
- NDST
- N-deacetylase/N-sulfotransferase
- AMAC
- aminoacridone
- NEM
- N-ethylmaleimide
- RP
- reverse-phase
- GlcA
- glucuronic acid
- MPSIH
- Mucopolysaccharide I Hurler
- FTICR-MS
- Fourier transform ion cyclotron resonance mass spectrometry.
REFERENCES
- 1. Fuller M., Brooks D. A., Evangelista M., Hein L. K., Hopwood J. J., Meikle P. J. (2005) Mol. Genet. Metab. 84, 18–24 [DOI] [PubMed] [Google Scholar]
- 2. Braunlin E., Mackey-Bojack S., Panoskaltsis-Mortari A., Berry J. M., McElmurry R. T., Riddle M., Sun L. Y., Clarke L. A., Tolar J., Blazar B. R. (2006) Pediatr. Res. 59, 27–32 [DOI] [PubMed] [Google Scholar]
- 3. Dusing S. C., Rosenberg A., Hiemenz J. R., Piner S., Escolar M. (2005) Pediatr. Phys. Ther. 17, 264–267 [DOI] [PubMed] [Google Scholar]
- 4. Muenzer J., Wraith J. E., Clarke L. A. (2009) Pediatrics 123, 19–29 [DOI] [PubMed] [Google Scholar]
- 5. Bishop J. R., Schuksz M., Esko J. D. (2007) Nature 446, 1030–1037 [DOI] [PubMed] [Google Scholar]
- 6. Clarke L. A., Russell C. S., Pownall S., Warrington C. L., Borowski A., Dimmick J. E., Toone J., Jirik F. R. (1997) Hum. Mol. Genet. 6, 503–511 [DOI] [PubMed] [Google Scholar]
- 7. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 8. Holmborn K., Ledin J., Smeds E., Eriksson I., Kusche-Gullberg M., Kjellén L. (2004) J. Biol. Chem. 279, 42355–42358 [DOI] [PubMed] [Google Scholar]
- 9. Kjellén L. (2003) Biochem. Soc. Trans. 31, 340–342 [DOI] [PubMed] [Google Scholar]
- 10. Kreuger J., Jemth P., Sanders-Lindberg E., Eliahu L., Ron D., Basilico C., Salmivirta M., Lindahl U. (2005) Biochem. J. 389, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pye D. A., Vives R. R., Turnbull J. E., Hyde P., Gallagher J. T. (1998) J. Biol. Chem. 273, 22936–22942 [DOI] [PubMed] [Google Scholar]
- 12. Allen B. L., Filla M. S., Rapraeger A. C. (2001) J. Cell Biol. 155, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allen B. L., Rapraeger A. C. (2003) J. Cell Biol. 163, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forsberg E., Kjellén L. (2001) J. Clin. Invest. 108, 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamanna W. C., Baldwin R. J., Padva M., Kalus I., Ten Dam G., van Kuppevelt T. H., Gallagher J. T., von Figura K., Dierks T., Merry C. L. (2006) Biochem. J. 400, 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamanna W. C., Kalus I., Padva M., Baldwin R. J., Merry C. L., Dierks T. (2007) J. Biotechnol. 129, 290–307 [DOI] [PubMed] [Google Scholar]
- 17. Langford-Smith K. J., Mercer J., Petty J., Tylee K., Church H., Roberts J., Moss G., Jones S., Wynn R., Wraith J. E., Bigger B. W. (2011) J. Inherit. Metab. Dis. 34, 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deakin J. A., Lyon M. (2008) Glycobiology 18, 483–491 [DOI] [PubMed] [Google Scholar]
- 19. Pettersson I., Kusche M., Unger E., Wlad H., Nylund L., Lindahl U., Kjellén L. (1991) J. Biol. Chem. 266, 8044–8049 [PubMed] [Google Scholar]
- 20. Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson C. E., Crawford B. E., Stavridis M., Ten Dam G., Wat A. L., Rushton G., Ward C. M., Wilson V., van Kuppevelt T. H., Esko J. D., Smith A., Gallagher J. T., Merry C. L. (2007) Stem Cells 25, 1913–1923 [DOI] [PubMed] [Google Scholar]
- 22. Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 23. Maccarana M., Sakura Y., Tawada A., Yoshida K., Lindahl U. (1996) J. Biol. Chem. 271, 17804–17810 [DOI] [PubMed] [Google Scholar]
- 24. Langford-Smith K., Arasaradnam M., Wraith J. E., Wynn R., Bigger B. W. (2010) Mol. Genet. Metab. 99, 269–274 [DOI] [PubMed] [Google Scholar]
- 25. Saad O. M., Leary J. A. (2004) J. Am. Soc. Mass Spectrom. 15, 1274–1286 [DOI] [PubMed] [Google Scholar]
- 26. Linhardt R. J., Rice K. G., Kim Y. S., Lohse D. L., Wang H. M., Loganathan D. (1988) Biochem. J. 254, 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Linker A., Hovingh P. (1972) Biochemistry 11, 563–568 [DOI] [PubMed] [Google Scholar]
- 28. Lyon M., Deakin J. A., Gallagher J. T. (1994) J. Biol. Chem. 269, 11208–11215 [PubMed] [Google Scholar]
- 29. Staples G. O., Shi X., Zaia J. (2010) J. Biol. Chem. 285, 18336–18343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fux L., Feibish N., Cohen-Kaplan V., Gingis-Velitski S., Feld S., Geffen C., Vlodavsky I., Ilan N. (2009) Cancer Res. 69, 1758–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. David G., Bai X. M., Van der Schueren B., Cassiman J. J., Van den Berghe H. (1992) J. Cell Biol. 119, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carlsson P., Presto J., Spillmann D., Lindahl U., Kjellén L. (2008) J. Biol. Chem. 283, 20008–20014 [DOI] [PubMed] [Google Scholar]
- 33. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blackhall F. H., Merry C. L., Davies E. J., Jayson G. C. (2001) Br. J. Cancer 85, 1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Esko J. D., Lindahl U. (2001) J. Clin. Invest. 108, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dhoot G. K., Gustafsson M. K., Ai X., Sun W., Standiford D. M., Emerson C. P., Jr. (2001) Science 293, 1663–1666 [DOI] [PubMed] [Google Scholar]
- 37. Pan C., Nelson M. S., Reyes M., Koodie L., Brazil J. J., Stephenson E. J., Zhao R. C., Peters C., Selleck S. B., Stringer S. E., Gupta P. (2005) Blood 106, 1956–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Byers S., Rozaklis T., Brumfield L. K., Ranieri E., Hopwood J. J. (1998) Mol. Genet. Metab. 65, 282–290 [DOI] [PubMed] [Google Scholar]
- 39. Fuller M., Meikle P. J., Hopwood J. J. (2004) Glycobiology 14, 443–450 [DOI] [PubMed] [Google Scholar]
- 40. Fuller M., Rozaklis T., Ramsay S. L., Hopwood J. J., Meikle P. J. (2004) Pediatr. Res. 56, 733–738 [DOI] [PubMed] [Google Scholar]
- 41. Krivit W., Sung J. H., Shapiro E. G., Lockman L. A. (1995) Cell Transplant. 4, 385–392 [DOI] [PubMed] [Google Scholar]
- 42. Wynn R. F., Wraith J. E., Mercer J., O'Meara A., Tylee K., Thornley M., Church H. J., Bigger B. W. (2009) J. Pediatr. 154, 609–611 [DOI] [PubMed] [Google Scholar]