Abstract
H2 relaxin is a peptide hormone associated with a number of therapeutically relevant physiological effects, including regulation of collagen metabolism and multiple vascular control pathways. It is currently in phase III clinical trials for the treatment of acute heart failure due to its ability to induce vasodilation and influence renal function. It comprises 53 amino acids and is characterized by two separate polypeptide chains (A-B) that are cross-linked by three disulfide bonds. This size and complex structure represents a considerable challenge for the chemical synthesis of H2 relaxin, a major limiting factor for the exploration of modifications and derivatizations of this peptide, to optimize effect and drug-like characteristics. To address this issue, we describe the solid phase peptide synthesis and structural and functional evaluation of 24 analogues of H2 relaxin with truncations at the termini of its peptide chains. We show that it is possible to significantly truncate both the N and C termini of the B-chain while still retaining potent biological activity. This suggests that these regions are not critical for interactions with the H2 relaxin receptor, RXFP1. In contrast, truncations do reduce the activity of H2 relaxin for the related receptor RXFP2 by improving RXFP1 selectivity. In addition to new mechanistic insights into the function of H2 relaxin, this study identifies a critical active core with 38 amino acids. This minimized core shows similar antifibrotic activity as native H2 relaxin when tested in human BJ3 cells and thus represents an attractive receptor-selective lead for the development of novel relaxin therapeutics.
Keywords: G Protein-coupled Receptors (GPCR), Peptide Chemical Synthesis, Peptide Conformation, Peptide Hormones, Receptors, H2 Relaxin, RXFP1, Structure-Activity
Introduction
Human relaxin-2 or H2 relaxin is a peptide hormone with multiple pleiotropic actions (1). Initially thought to be only a reproductive hormone involved in facilitating delivery of the young, more recent studies have demonstrated that H2 relaxin plays a key role in inflammatory and matrix remodeling processes and possesses potent vasodilatory, angiogenic, and other cardioprotective actions (2). The vasodilatory effects of H2 relaxin are thought to involve promotion of nitric oxide and the gelatinases, matrix metalloproteinase-2 and matrix metalloproteinase-9, in addition to antagonism of the vasoconstricting actions of endothelin-1 and angiotensin II (3). This causes systemic and renal vasodilation, increased arterial compliance, and other vascular changes. These findings have led to current clinical trial evaluation of relaxin as drug for the treatment of patients with acute heart failure (AHF)6 (4, 5). Furthermore, the matrix remodeling actions of H2 relaxin have enhanced its reputation as a rapidly acting but safe antifibrotic agent, which has been further supported by its ability to successfully inhibit and/or reverse fibrosis in every preclinical model of experimental disease evaluated to date (6, 7).
All of the above actions of H2 relaxin are thought to be mediated through its native receptor RXFP1 (originally named LGR7), which is a leucine-rich repeat containing G-protein coupled receptor that is characterized by an unusually large ectodomain (8). H2 relaxin, however, can also bind to and activate the related receptor, RXFP2, which is the native receptor for insulin-like peptide 3 (INSL3) (9), suggesting that potential cross-reactivity may be associated with its diverse actions. Due to its enormous potential as a drug, it would be desirable to produce RXFP1-specific H2 relaxin analogues to minimize any possible side effects elicited by its interaction with RXFP2.
H2 relaxin has an insulin-like core structure containing two chains (A and B) and three disulfide bonds (see Fig. 1) (10, 11). Comparison of the H2 relaxin primary structures with relaxin peptides from different species reveals a remarkable sequence similarity in the mid-region of the B-chain where two arginine residues are located on the same surface of the α-helix (12). This observation led to a study of the role of these amino acids in the function of the hormone. Schwabe et al. (13) first showed that the two arginines (ArgB13 and ArgB17) interact with RXFP1 like a prong and that binding is mediated by both the positive charge and by the hydrogen-bonding network produced by the guanidino group in the side chain of each Arg. A third amino acid in the B-chain, IleB20, has also been reported to be essential for binding to RXFP1 (14), resulting in a triangular contact region referred to as the “RXXXRXXI” binding cassette (14). The residues in this binding cassette have been demonstrated to interact with the large extracellular domain RXFP1 (15, 16). However, it is clear that there is an additional interaction of the peptide with the transmembrane exoloops of the receptor that is predicted to involve the A-chain of the peptide (17). Park et al. (18) recently reported that the A-chain residues ThrA16, LysA17, and PheA23 are involved in interactions either with RXFP1 or RXFP2, although it is unclear whether the described effects are related to a compromised overall structure or disruption of a receptor contact point.
FIGURE 1.
Amino acid sequences of native H2 relaxin and the analogues made for this study. A total of 24 H2 relaxin analogues with B-chain and A-chain truncations as noted were chemically synthesized and characterized.
The N terminus of the A-chain of H2 relaxin has no functional role as reported previously (19, 20). Shortening of the N terminus of the A-chain of porcine (19, 20) or H2 (21) relaxin by up to four residues had no effect on binding or activation of RXFP1 and RXFP2. Further truncation, however, caused both a decrease in affinity and potency and that of structural integrity as assessed by CD and NMR techniques (21). The replacement of these deleted residues with alanines capable of adopting the native α-helical structure without displaying functionality caused significant biological activity and structure to reappear (20, 21). These findings in turn, suggested that no single amino acid in the N-terminal region of the A chain is functionally important, but that the presence of an α-helix is required to maintain the overall fold of the protein which is key to the H2-RXFP1 and H2-RXFP2 interactions (21). In contrast, a similar truncation study on INSL3 showed that it can be truncated at the N terminus of its A-chain by up to nine residues without affecting the binding affinity to its receptor RXFP2, while becoming a high affinity antagonist (22). Thus, H2 relaxin is binding to and activating both RXFP1 and RXFP2 receptors in a similar manner but via a mechanism that is different compared with INSL3 (21).
This structure activity relationship study indicates that the active site of H2 relaxin most likely consists of the mid-region of the B-chain helix and the C-terminal region of the A-chain (18, 23). In this study, we have carried out truncations at other regions within the A- and/or B-chains and evaluated the resulting structure and function of these minimized peptides. We report that B-chain truncations together with the N-terminal A-chain truncations lead to a generation of domain-minimized active H2 relaxin analogues with high selectivity for RXFP1. As H2 relaxin is already in phase III clinical trials for the treatment of AHF (4), the development of smaller, and thus cheaper, RXFP1-specific next generation H2 relaxin analogues will have immediate clinical impact. Additionally, these minimized peptides may also have great potential as treatments for organ fibrosis.
EXPERIMENTAL PROCEDURES
Solid-phase Peptide Synthesis
Native H2 relaxin consists of a 24-residue A-chain and a 29-residue B-chain that are linked by three disulfide bonds (see Fig. 1). All H2 relaxin analogues synthesized in this study contain two chains and three disulfide bonds. Both regioselectively S-protected A- and B-chains were synthesized separately by the continuous flow Fmoc solid-phase method (24) using an automatic PerSeptive Biosystems Pioneer peptide synthesizer (Framingham, MA) or by using microwave-assisted synthesis on a Liberty system (CEM Corp.) (21, 23, 25, 26).
In total, 24 H2 relaxin analogues were synthesized in this study (see Fig. 1). These included the following: four B-chain N-terminal truncated analogues (H2-(B3–29), H2-(B5–29), H2-(B7–29), and H2-(B9–29)), six B-chain C-terminal truncated analogues (H2-(B1–28), H2-(B1–27), H2-(B1–26), H2-(B1–25), H2-(B1–24), and H2-(B1–23)), four B-chain N- and C-terminal truncated analogues (H2-(B7–25), H2-(B7–24), H2-(B8–25), and H2-(B8–24)), and six A- and B-chain truncated analogues (H2-(A2–24)(B7–24), H2-(A3–24)(B7–24), H2-(A4–24)(B7–24), H2-(A5–24)(B7–24), H2-(A7–24)(B7–24), and H2-(A9–24)(7–24)). All of these H2 analogues were amidated at the C termini of both the A- and B-chains. In addition, two free acid analogues were made: H2-(A4–24)(B7–24)-COOH and H2-(A4–24)(B7–24)-COOH. Finally, two <Glu-acylated (pyroglutamic acid) H2 analogues were made: H2-(AZ-5–24)(B7–24) and H2-(AZ-7–24)(B7–24). The solid support used for both A- and B-chains was Fmoc-PAL-PEG-PS for all peptides except for H2-(A4–24)(B7–24)-COOH and H2-(A4–24)(B7–24)-COOH where the first amino acid was pre-loaded on the solid support.
Peptide Characterization
The purity of the synthetic peptides was assessed by analytical RP-HPLC on a Phenomenex C18 column (pore size, 300 Å; particle size, 5 μm; 4.6 × 250 mm) using a gradient of acetonitrile in 0.1% aqueous trifluoroacetic acid. The product was confirmed by MALDI-TOF mass spectrometry using a Bruker Autoflex II instrument (Bremen, Germany) in the linear mode at 19.5 kV. The peptides were quantitated by amino acid analysis of a 24 h acid hydrolysate using a Shimadzu microbore RP-HPLC system.
Ligand Binding Assay
HEK-293T cells stably transfected with RXFP1 and RXFP2 were grown in RPMI 1640 medium (Sigma) supplemented with 10% FCS, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine and plated into 24-well poly-l-lysine-coated plates for whole cell binding assays. Competition binding experiments were performed with either europium-labeled H2 relaxin (26) or Eu3+-labeled INSL3 (27) in the absence or presence of increasing concentrations of unlabeled hormones. Nonspecific binding was determined with an excess of unlabeled peptides (500 nm H2 relaxin or INSL3). All data are presented as the mean ± S.E. of the percentage of the total specific binding of triplicate wells, repeated in at least three separate experiments, and curves were fitted using one-site binding curves in Graphpad Prism 4 (Graphpad Software). Statistical differences in pIC50 values were analyzed using one-way analysis of variance coupled to a Newman Keul's multiple comparison test for multiple group comparisons in Graphpad Prism 4.
Functional cAMP Assay
The influence of the various ligands on cAMP signaling in cells expressing RXFP receptors was assessed using a cAMP reporter gene assay as described previously (28). Briefly, HEK-293T cells in 96-well plates were co-transfected with either RXFP1 or RXFP2 and a pCRE-β-galactosidase reporter plasmid (29). 24 h later, cotransfected cells were treated with increasing concentrations of H2 relaxin analogues or INSL3 in parallel to 10 nm of H2 relaxin or INSL3 for RXFP1- or RXFP2-transfected cells respectively. After 6 h, the cell medium was aspirated, and the cells were frozen at −80 °C overnight. The amount of cAMP-driven β-galactosidase expression in each well was determined as described previously (28). Ligand-induced stimulation of cAMP was expressed as a percentage of the maximum H2 relaxin and INSL3 response for RXFP1 and RXFP2 cells, respectively. Data points were measured in triplicate, and each experiment was repeated at least three times. Statistical differences in pEC50 values were analyzed using one-way analysis of variance coupled to a Newman Keul's multiple comparison test for multiple group comparisons in Graphpad Prism 4.
NMR Structural Analysis
Selected analogues were subjected to solution NMR spectroscopy analysis to investigate the effect of truncations on overall structural features. Samples prepared comprised 1.5, 1.0, 0.7, and 0.3 mg of the peptides H2-(A5–24)(B7–24), H2-(B1–24), H2-(A5–24), and H2-(B7–24), respectively. For each peptide, two-dimensional homonuclear 1H data, including TOCSY and NOESY were recorded at 600 MHz, 298 K, pH ∼ 4. All data were recorded on a Bruker Avance spectrometer and processed using TopSpin (version 2.1) (Bruker). The data were analyzed using the program CARA (30) and assigned using standard two-dimensional sequential assignment strategies (31). The secondary Hα chemical shifts (i.e. deviations of observed chemical shifts from chemical shifts expected for a random coil peptide (32)) were compared with the ones observed in full-length native H2 relaxin and used as indicators of structural changes.
Determination of Collagen Content
The efficacy of the H2-(B7–24) and H2-(A4–24)(B7–24) analogues was compared to the native H2 relaxin peptide via measurement of collagen deposition in BJ3 cells, a human dermal fibroblast cell line (33) which endogenously expresses RXFP1. BJ3 cells were maintained in DMEM containing 17% medium 199, 15% FCS, penicillin (50 units/ml), streptomycin (50 μg/ml), and 1% l-glutamine (DMEM-FCS) and plated at the density of 1 × 106 cells/well (from passages 5–10) in six-well plates for experimental conditions. Cells were then treated for 72 h with varying concentrations of the H2-(B7–24) or H2-(A4–24)(B7–24) analogues alone; with TGF-β1 (2 ng/ml) alone; or with recombinant H2 relaxin (16.8 nm) or the H2-(B7–24) analogue (16.8 nm) or the H2-(A4–24)(B7–24) analogue in TGF-β1 (2 ng/ml)-stimulated cells. The concentration of H2 relaxin used in this study (16.8 nm) was determined previously to have maximal effects in inhibiting TGF-β1-induced collagen synthesis and deposition when applied to other types of fibroblasts (34–36). Untreated cells were used as controls from which basal collagen deposition could be measured. These treatments were carried out three to seven separate times in triplicate.
The amount of collagen deposited by BJ3 cells into the matrix was determined by isolating the cell layers of control and treated samples before subsequently hydrolyzing them with 6 m hydrochloric acid. Hydroxyproline content was then measured from the hydrolyzed samples as described previously (37). Hydroxyproline values were then converted into collagen content by multiplying by a factor of 6.94, based on the finding that hydroxyproline represents ∼14.4% of amino acid composition of collagen in most mammalian tissues (38).
RESULTS
N-terminal Truncations of H2 Relaxin B-chain
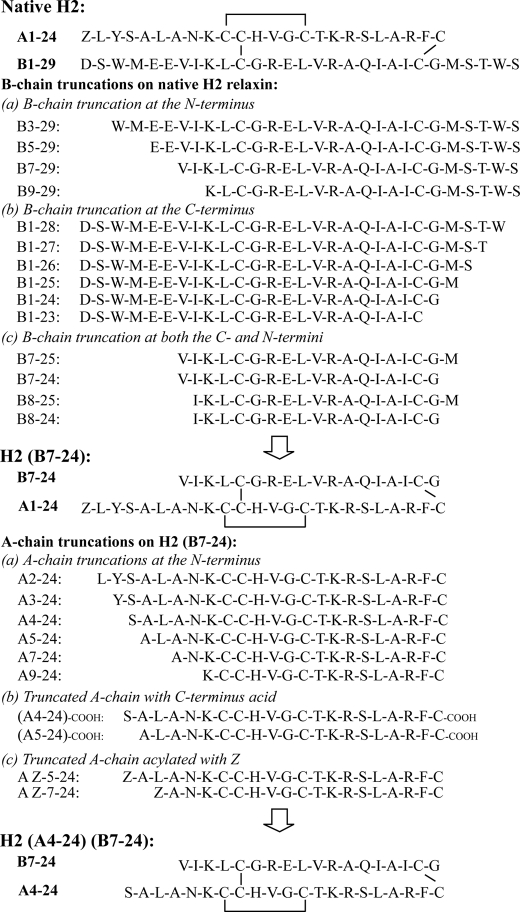
The importance of the N-terminal region of the H2 relaxin B-chain for biological function was investigated by the production and testing of a series of truncated analogues in which up to eight residues were removed (Fig. 1). The synthesis of all analogues was straightforward, and all peptides folded with similar or better yields than native H2 relaxin. The ability of the analogues to bind and activate the RXFP1 receptor was characterized in cell-based assays (Fig. 2 and Table 1, analogues 2–5). Truncations were found to be well tolerated with most analogues retaining close to native affinity and potency at RXFP1. The only peptide showing a statistically significant drop in potency compared with native H2 relaxin was analogue 5, H2-(B9–29). This drop in potency was accompanied by a drop in binding affinity, but this was not significant. Thus, at least residues B1–6 do not significantly contribute to the receptor interaction.
FIGURE 2.
Activity of the B-chain N-terminal-truncated H2 relaxin analogues on the RXFP1 receptor. A, competition binding results of H2 relaxin and the B-chain N-terminal-truncated analogues in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of H2 relaxin and the B-chain N-terminal-truncated analogues in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
TABLE 1.
Competition Binding and Activation of RXFP1 and RXFP2 by H2 relaxin and analogues
| No. | Peptide | Eu3+-relaxin LGR7 pIC50 | LGR7 cAMP pEC50 | Eu3+-INSL3 LGR8 pIC50 | LGR8 cAMP pEC50 |
|---|---|---|---|---|---|
| 1 | H2 (acid) | 9.00 ± 0.06 (8) | 10.35 ± 0.04 (5) | 7.91 ± 0.22 (3) | 9.13 ± 0.06 (3) |
| 2 | H2-(B3–29) | NDa | 10.49 ± 0.33 (3) | ND | ND |
| 3 | H2-(B5–29) | ND | 9.91 ± 0.26 (3) | ND | ND |
| 4 | H2-(B7–29) | 8.76 ± 0.23 (3) | 10.15 ± 0.04 (3) | ND | 9.79 ± 0.47 (3) |
| 5 | H2-(B9–29) | 8.42 ± 0.32 (3) | 8.76 ± 0.19 (3)b | ND | ND |
| 6 | H2-(B1–28) | ND | 10.71 ± 0.23 (4) | ND | ND |
| 7 | H2-(B1–27) | ND | 9.69 ± 0.25 (5) | ND | 8.23 ± 0.34 (4)c |
| 8 | H2-(B1–26) | ND | 10.20 ± 0.10 (4) | ND | ND |
| 9 | H2-(B1–25) | ND | 10.08 ± 0.03 (4) | ND | ND |
| 10 | H2-(B1–24) | 8.56 ± 0.11 (3) | 9.42 ± 0.11 (4)b | ND | ND |
| 11 | H2-(B1–23) | 7.58 ± 0.48 (3)b | 8.29 ± 0.23 (3)b | ND | ND |
| 12 | H2-(B7–25) | ND | 9.32 ± 0.23 (4)d | ND | ND |
| 13 | H2-(B8–25) | ND | 9.04 ± 0.09 (3)b | ND | ND |
| 14 | H2-(B7–24) | 7.44 ± 0.27 (3)b | 9.38 ± 0.32 (3)c | <6 | 7.18 ± 0.15 (4)b |
| 15 | H2-(B8–24) | 7.34 ± 0.15 (3)b | 8.39 ± 0.10 (4)b | ND | ND |
| 16 | H2-(A2–24)(B7–24) | 7.68 ± 0.10 (3)b | 8.55 ± 0.15 (2)b | ND | ND |
| 17 | H2-(A3–24)(B7–24) | 7.21 ± 0.14 (3)b | 8.37 ± 0.12 (3)b | ND | ND |
| 18 | H2-(A4–24)(B7–24) | 6.99 ± 0.06 (3)b | 8.22 ± 0.19 (3)b | <6 | <6 |
| 19 | H2-(A5–24)(B7–24) | 7.39 ± 0.21 (3)b | 8.23 ± 0.11 (4)b | ND | ND |
| 20 | H2-(A7–24)(B7–24) | 6.29 ± 0.30 (3)b | 7.27 ± 0.31 (3)b | ND | ND |
| 21 | H2-(A9–24)(B7–24) | 7.08 ± 0.13 (3)b | 6.78 ± 0.12 (3)b | ND | ND |
| 22 | H2-(A-Z-5–24)(B7–24) | 7.15 ± 0.15 (3)b | 8.39 ± 0.10 (4)b | ND | ND |
| 23 | H2-(A-Z-7–24)(B7–24) | 6.90 ± 0.36 (3)b | 7.89 ± 0.08 (3)b | ND | ND |
| 24 | H2-(A5–24)(B7–24) (acid) | 6.65 ± 0.38 (3)b | 7.61 ± 0.03 (4)b | ND | ND |
| 25 | H2-(A4–24)(B7–24) (acid) | 6.30 ± 0.08 (4)b | 7.51 ± 0.09 (5)b | ND | ND |
a ND, not determined.
b p < 0.001 versus H2 relaxin.
c p < 0.05 versus H2 relaxin.
d p < 0.01 versus H2 relaxin.
C-terminal Truncations of H2 Relaxin B-chain
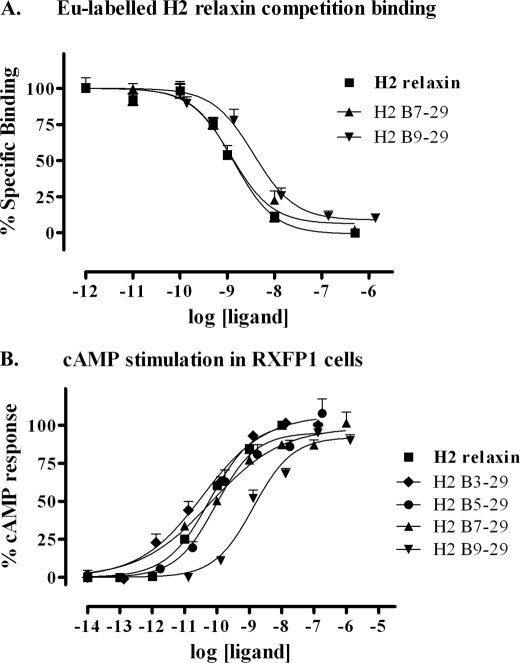
The importance of the C-terminal part of the B-chain for relaxin activity was tested in a similar fashion with stepwise removal of residues up to CysB23 (Fig. 1). Again synthesis was straightforward with all peptides folding in yields similar or improved compared with native H2 relaxin. Analogues with up to five residues removed retained native-like affinity and potency with only analogue 11, H2-(B1–23), showing a statistically significant drop in potency that was accompanied by a significant drop in binding affinity (Fig. 3 and Table 1, analogues 6–11). Thus, residues B25–29 also appear to be dispensable for RXFP1 activation.
FIGURE 3.
Activity of the B-chain C-terminal-truncated H2 relaxin analogues on the RXFP1 receptor. A, competition binding results of H2 relaxin and the B-chain C-terminal-truncated analogues in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of H2 relaxin and the B-chain C-terminal-truncated analogues in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
Combination of N- and C-terminal Truncations of H2 Relaxin B-chain
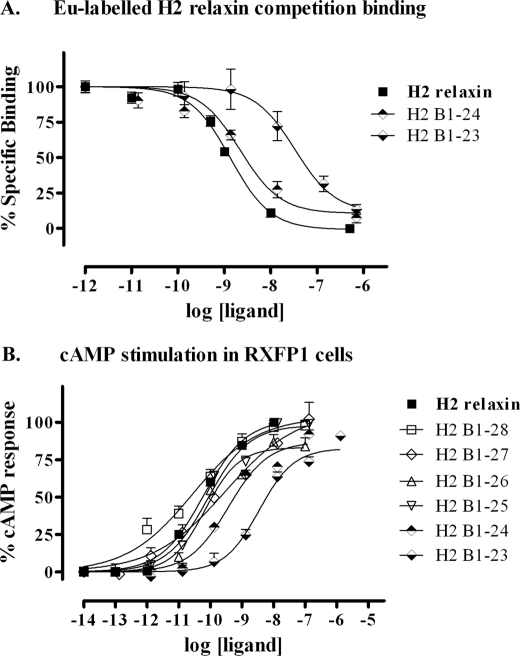
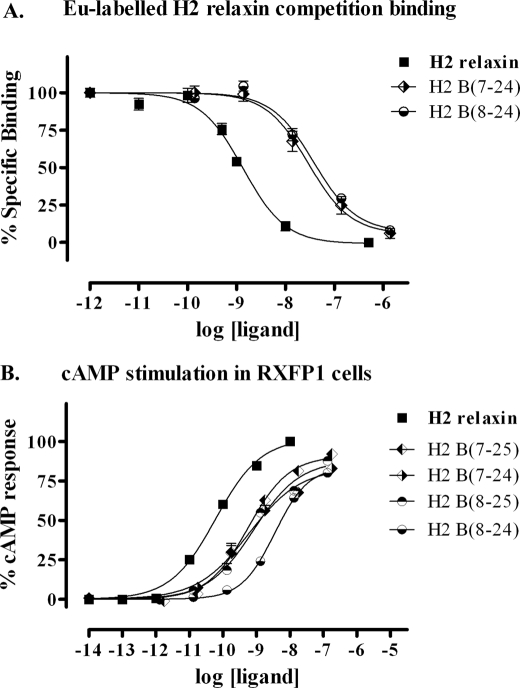
To identify a minimized active B-chain, a set of four peptides with combinations of truncations at both the N and C termini was generated (Fig. 1). Again, no complications were observed throughout the synthetic production with all analogues being produced in high yields and in high purity. All analogues were tested for RXFP1 binding and activity (Fig. 4 and Table 1, analogues 12–15). The four tested peptides showed similar affinity and potency for RXFP1. Thus, the minimal B-chain required for effective activation of RXFP1 was concluded to be the shortest variant B7–24. In comparison with native H2 relaxin, this peptide comprises 20% less amino acids and has an ∼10-fold decrease in activity at the RXFP1 receptor.
FIGURE 4.
Activity of the B-chain N/C-terminal-truncated H2 relaxin analogues on the RXFP1 receptor. A, competition binding results of H2 relaxin and the B-chain N/C-terminal-truncated analogues in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of H2 relaxin and the B-chain N/C-terminal-truncated analogues in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
Combinations of A- and B-chain Truncations of H2 Relaxin
In our previous work, we investigated the role of the H2 relaxin A-chain in the interaction with RXFP1 (18). We showed that analogues with up to five residues truncated retained close to full activity. Thus, to achieve an overall structurally minimized H2 relaxin analogue, we combined the truncated B-chain identified in this work with truncated variants of the H2 relaxin A-chain (Fig. 1). Interestingly, although no drop in activity was previously seen when the A-chain alone was truncated by four to five residues (21), similar truncations in combination with the truncated B-chain did further decrease the potency at RXFP1 by ∼10-fold, with further truncations losing further potency (Fig. 5 and Table 1, analogues 16–21). This likely reflects that the combination of changes leads to a destabilization of the overall structure, which in turn affects both affinity and potency in equal measure. Nonetheless, the H2-(A4–24)(B7–24) peptide retains nanomolar potency at RXFP1 while comprising only 36 residues in comparison with the 53 residues of the native peptide.
FIGURE 5.
Activity of the A- and B-chain-truncated H2 relaxin analogues on the RXFP1 receptor. A, competition binding results of H2 relaxin and the A- and B-chain-truncated analogues in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of H2 relaxin and the A- and B-chain-truncated analogues in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
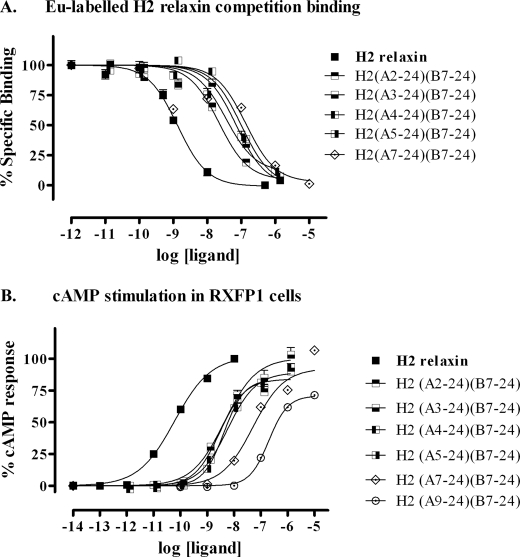
When analyzing the activity of the peptides truncated both in the A- and B-chain, it was somewhat surprising to see that a substantial loss of activity was evident after removal of the first A-chain residue, pyroglutamate (<Glu) (Fig. 5). In native H2 relaxin, the A-chain adopts a helical conformation stretching all the way to the N terminus. Thus, it is possible that the pyroglumate residue is beneficial for the structure, as the positive charge associated with a free N terminus, which would otherwise be present, would be clashing with the helix dipole moment. To investigate this effect, a <Glu residue was added to the N terminus of the A chain-truncated analogues, resulting in analogues H2-(<Glu-A5–24)(B7–24) and H2-(<Glu-A7–24)(B7–24) peptides (Fig. 1). Although increased binding was seen for H2-(<Glu-A7–24)(B7–24) compared with H2-(A7–24)(B7–24), this effect may simply be due to the fact that the peptide chain was extended by one residue, as no significant increase in activity was seen for H2-(<Glu-A5–24)(B7–24) when compared with H2-(A4–24)(B7–24), which comprise the same length A-chains but with the former lacking the free N terminus (Fig. 6, A and B, and Table 1, analogues 22–23). Furthermore, to investigate whether the amidation at the C terminus used for all analogues affected activity, we chemically prepared two peptides with free acid at both chain C termini, H2-(A5–24)(B7–24)-COOH and H2-(A4–24)(B7–24)-COOH, and compared their activity to their respective amidated analogues. The results clearly demonstrated that the amidated truncated peptides are more active than their acid versions (Fig. 6, C and D, and Table 1, analogues 24 and 25). This is not surprising as amidation of the C terminus, like acetylation of the N terminus, would be expected to improve the helicity due to removal of the clash between the negative charge and the dipole moment, thereby stabilizing the structure of the peptides.
FIGURE 6.
Activity of the domain-minimized H2 relaxin analogues on the RXFP1 receptor. A, competition binding results of H2 relaxin and the domain-minimized analogues, H2-(A5–24)(B7–24) and H2-(A7–24)(B7–24), with and without pyroglutamate (<Glu) in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of H2 relaxin and the domain-minimized analogues, H2-(A5–24)(B7–24) and H2-(A7–24)(B7–24), in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate. C, competition binding results of H2 relaxin and the domain minimized analogues, H2-(A4–24)(B7–24) and H2-(A5–24)(B7–24), with and without C-terminal amidation in HEK-293T cells stably expressing the RXFP1 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. D, cAMP activity of H2 relaxin and the domain-minimized analogues, H2-(A4–24)(B7–24) and H2-(A5–24)(B7–24) with/without C-terminal amidation, in HEK-293T cells expressing the RXFP1 receptor using a pCRE-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
Effects of Truncations on INSL3 Receptor RXFP2
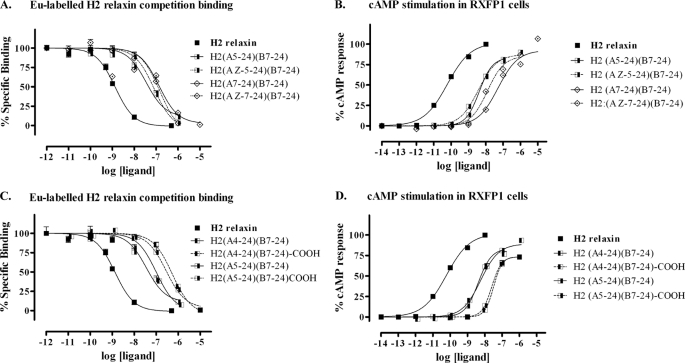
In light of the fact that H2 relaxin is currently in clinical trials for AHF and is showing potential for other clinical uses, it was of interest to investigate the selectivity of the truncated analogues for RXFP1 over RXFP2, as native H2 relaxin is able to activate RXFP2 with high affinity. Truncation of the B-chain C terminus was expected to have a positive effect on the selectivity for RXFP1, as TrpB28, which is conserved in INSL3, is critical for INSL3 binding to RXFP2 (39). This was indeed found to be the case with H2-(B1–24) showing a significant decrease in affinity for, and activation of, RXFP2 (Fig. 7 and Table 1, analogue 10). Truncation of the N terminus of H2 relaxin A-chain was found to further reduce the activation of RXFP2. Thus, the lead molecule H2-(A4–24)(B7–24) is devoid of measureable activity at RXFP2 (Fig. 7 and Table 1, analogue 18).
FIGURE 7.
Activity of the domain-minimized H2 relaxin analogues on the RXFP2 receptor. A, competition binding results of the A- and B-chain-truncated minimized analogues, H2-(B7–24) and H2-(A4–24)(B7–24) compared with INSL3 and H2 relaxin in HEK-293T cells stably expressing the RXFP2 receptor using Eu3+-labeled H2 relaxin as the competitive ligand. Data are expressed as percentage of specific binding and are pooled data from at least three experiments performed in triplicate. B, cAMP activity of the A- and B- chain-truncated minimized analogues, H2-(B7–24) and H2-(A4–24)(B7–24) compared with H2 relaxin in HEK-293T cells expressing the RXFP2 receptor using a pCRE-β-galactosidase reporter gene system. Data are expressed as percentage of maximum H2 relaxin-stimulated cAMP response and are pooled data from at least three experiments performed in triplicate.
Structural Studies of Truncated Analogues
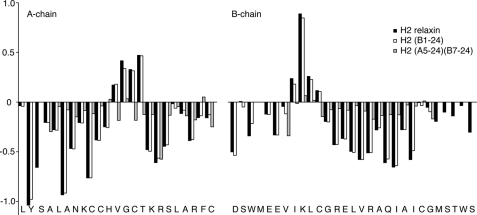
To investigate the effects of the truncations from a structural perspective to see whether there was a link between changes in activity and changes in overall structure, several analogues were subjected to solution NMR spectroscopy analysis and compared with full-length H2 relaxin. Initially, the three different truncations of the A-chain N terminus, the B-chain N terminus, and the B-chain C terminus were studied individually. H2-(B1–24) was found to give high quality data indicative of a well ordered structure. Complete sequential assignments were possible and a comparison of the secondary Hα shifts, which are good indicators of secondary structure, to the ones observed in full-length H2 relaxin showed that all elements of secondary structure were fully retained (Fig. 8). A number of key long range NOEs could also be identified, confirming the overall fold was identical to the one seen in H2 relaxin. In contrast, both H2-(A5–24) and H2-(B7–27) gave spectral data of less quality. This was partially due to the smaller amount of material that was available for these studies, but also overall, the signals were considerably broader, suggesting that the overall structure was destabilized and that conformational rearrangements resulted in significant exchange broadening. As a consequence resonance assignments were not possible for either peptide. A structural analysis was also undertaken on the overall minimized H2-(A5–24)(B7–24) peptide. Although this peptide also suffered from generally broad lines, the effect was less pronounced, and with a higher sample concentration available, near complete backbone assignments was possible. In comparison with full-length H2, the secondary shifts were found to be closer to random coil values, but stretches of negative values are seen throughout the three helical segments, A2–10, A16–24, and B11–22 (Fig. 8), suggesting at least a helical tendency in these regions. However, the overall tightly folded structure appears to have been disrupted.
FIGURE 8.
Secondary Hα chemical shifts of H2 relaxin and analogues. Secondary shifts, i.e. differences between observed chemical shifts and the random coil shifts, are sensitive indicators of secondary structure. Three distinct regions of negative numbers corresponding to the three helical segments and two stretches of positive number corresponding to extended regions are seen in H2 relaxin and H2-(B1–24). Generally smaller values are seen for H2-(A5–24)(B7–24) suggesting a destabilized structure.
Collagen Content Assay
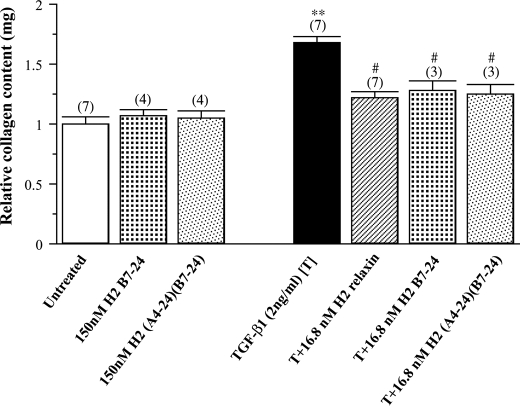
Based on the ability of native H2 relaxin to consistently inhibit TGF-β1-induced aberrant collagen production and deposition when applied to various types of fibroblasts (35, 36, 40–42), the effects of H2 relaxin (16.8 nm) on TGF-β1-stimulated collagen deposition from human BJ3 cells were compared with equivalent concentrations of the H2-(B7–24) and H2-(A4–24)(B7–24) peptides (Fig. 9). TGF-β1 (2 ng/ml) significantly increased collagen deposition in the cell layer by 65–70% (p < 0.01) of that measured in untreated cultures over 72 h, whereas native H2 relaxin (16.8 nm) significantly inhibited this aberrant collagen content by approximately two-thirds (p < 0.05) of that measured in TGF-β1-alone treated cells over the same time period (Fig. 9). Despite having a lower affinity for RXFP1, the H2-(B7–24) and H2-(A4–24)(B7–24) analogues (at an equivalent concentration of 16.8 nm) also significantly decreased TGF-β1-stimulated content by a similar extent to that of the full-length peptide over 72 h (Fig. 9), demonstrating similar efficacy to native H2 relaxin in vitro. Importantly though, these shortened analogues did not markedly influence basal collagen content (up to 150 nm), consistent with previous findings for H2 relaxin (36, 40, 41).
FIGURE 9.
The relative mean ± S.E. collagen content from untreated BJ3 fibroblasts and from cells treated with the H2-B(7–24) or H2-A(4–24), H2-B(7–24) analogues (150 nm) alone; TGF-β1 (2 ng/ml) alone; or with recombinant H2 relaxin (16.8 nm) or the H2-B(7–24) analogue (16.8 nm) or the H2-A(4–24), H2-B(7–24) analogue in TGF-β1 (2 ng/ml)-stimulated cells. All data are expressed as the relative ratio of collagen content measured in the untreated group, which was expressed as 1, whereas the numbers in parentheses represent the number of separate experiments that were conducted for each group in triplicate. **, p < 0.01 versus the untreated group; #, p < 0.05 versus the TGF-β1 alone-treated group.
DISCUSSION
H2 relaxin has been known for >75 years. However, its receptor RXFP1 was only discovered a decade ago (8) which has limited the ability to characterize the structural basis of the mechanism of action of the hormone. The activation of the rather unusual RXFP1 and RXFP2 receptors is complex, requiring several interactions involving the LRR domain (15), the extracellular loops (43), and the N-terminal LDLa module (28), eventually leading to the activation of the receptor (44, 45). Although one key feature, the RXXXRXXI relaxin cassette in the mid-region of the H2 relaxin B-chain and its interaction with the leucine-rich repeats of the ectodomain (14–16), this interaction alone is not sufficient to explain all of the activation events involved. Thus, to search for additional features involved in the activation of RXFP1 and RXFP2, in this study, we chemically generated 24 two-chain variants of H2 relaxin with truncations at different termini. We show that no individual residue within the regions B1–6, B24–28, and A1–4 contributes significantly to the overall affinity of H2 relaxin for RXFP1. However, simultaneous removal of these regions results in a drop in affinity and corresponding potency on RXFP1, presumably due to a compromised overall structure. Nonetheless, our findings demonstrate that it is possible to generate H2 relaxin analogues that remain potent agonists for RXFP1 in vitro but that are only two-thirds the size of native H2 relaxin, thus significantly decreasing the cost and complexity of their chemical synthesis. Importantly, such analogues are fully selective for RXFP1 over RXFP2, in contrast to native H2 relaxin, which is a high affinity ligand for both RXFP1 and RXFP2 (8).
Insights into the Interaction between H2 Relaxin and RXFP1 and RXFP2
Numerous studies have focused on determining the structural features of H2 relaxin that control the interaction with its native receptor RXFP1 and the related INSL3 receptor RXFP2. The identification of the relaxin binding cassette RXXXRXX(I/V) of the H2 relaxin B-chain was made more than a decade ago (14, 46). It was subsequently shown that this mid-region of the B-chain, which is exposed on the face of a B-chain helix is the primary binding site in other relaxins (47, 48). Subsequently, mutations in the LRRs of the ectodomain have identified the corresponding binding site for the cassette in RXFP1. Each of the Arg residues are coordinated by a pair of acidic residues forming a network of hydrogen bonds coordinating the guandinium groups, whereas IleB19 interacts closely with a hydrophobic cluster on the LRR surface (15).
Although the picture of primary binding of H2 relaxin to the LRR domain is well established, very little is known about how the receptor is activated and the signal transferred through the membrane, which requires additional interactions. Studies utilizing the natural cross-reactivity of relaxin peptides for RXFPs and chimeric peptides and receptors have provided important clues. Replacement of RXFP1 extracellular loop 2 with the corresponding region from RXFP2 abolished the binding of H3 relaxin, which effectively activates native RXFP1 but not RXFP2, suggesting that this region is involved in the secondary interaction (43). Furthermore, a chimeric peptide comprising the H3 relaxin B-chain and the A-chain of INSL5 loses the ability of native H3 relaxin to interact with RXFP1 (49). Structural studies on this peptide revealed that this effect is not related to structural changes as the H3 relaxin B-chain adopts an identical conformation in the chimera as when associated with its native A-chain (50). Thus, at least in part, the domain responsible for activation of the RXFP1 and RXFP2 receptors must be located in the A-chain.
Intriguingly, there is substantial evidence that this secondary activation interaction differs significantly between different relaxin peptides and relaxin receptors. Truncation of the N-terminal part of the A-chain in INSL3 results in a high affinity antagonist (19) for the RXFP2 receptor, whereas similar truncations of H2 relaxin results in lower affinity agonists. Thus, the “activation domain” of INSL3 but not H2 relaxin appears to be located in the N-terminal part of the A-chain.
Here, we provide new mechanistic insights firmly ruling out the presence of an activation domain of H2 relaxin in the B-chain N or C termini as both regions can be truncated without loss of activation of the receptor. Instead, analogous to what we reported previously for truncation of the A-chain N terminus, small decreases in affinity and potency of H2 relaxin at the RXFP1 receptor are seen as the termini are being shortened, but importantly, all generated analogues were still able to significantly activate the receptor. This finding points to the regions of H2 relaxin mediating receptor activation as being located in the mid- to C-terminal part of the A-chain. This finding is consistent with studies by Park et al. (18), which suggest that TyrA16 and LysA17 are important for the H2 relaxin interaction with RXFP1. However, the nature of such an interaction is unclear as no drop in affinity for RXFP1 was seen when either of these residues were mutated by Ala but only when replaced by Gly (18), which may suggest a secondary effect related to structural changes. Clearly, further mutational studies targeting the mid- to C-terminal part of H2 relaxin A-chain are required to shed light on its role in receptor activation, in particular for the development of a highly sought after high affinity RXFP1 antagonist.
Effects of Truncations on H2 Relaxin Structural Fold
The finding that, although individual shortening of the A-chain N terminus (21) and the B-chain N and C termini had little effect on the ability of H2 relaxin to interact with RXFP1, combined truncations partially disrupted binding was somewhat surprising. However, NMR structural studies of shortened analogues revealed that in several cases, truncations caused a disruption of the stable overall structure of H2 relaxin, which can explain this feature. We have reported previously that removal of a large part of the H2 relaxin A-chain (residues A1–9) effectively removes any ordered structure of the A-chain, whereas the B-chain retains a helical nature (21). Similar effects are evident for shorter truncations of the A-chain or the B-chain N terminus as evident from the NMR analyses. It is likely that this destabilization is responsible for the reduced affinity of shorter analogues compared with native H2 relaxin but that the active structure can still be induced upon receptor binding.
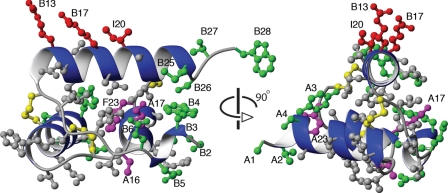
The fold of relaxins is clearly dependent on the integrity of an extensive hydrophobic core (51). Fig. 10 shows the crystal structure of H2 relaxin and illustrates the positioning and interactions of various amino acid side chains. Neither of the removed residues, shown in green, are key residues buried in the hydrophobic core, and as such, their omission would not be expected to be detrimental to the fold. However, residues TyrA3, TrpB4, and GluB6 do form contacts with other parts of the molecules on the fringes of the core; thus, their removal may contribute to the destabilization of overall structure. In contrast, truncations of the B-chain C-terminus had no noticeable effect on the overall structure of H2 relaxin. This is consistent with only limited contacts between residues in this region and other parts of the protein.
FIGURE 10.
Structure of H2 relaxin. Two different views of the crystal structure of H2 relaxin (Protein Data Bank code 6RLX) are shown with side chains of residues removed in the minimized lead peptide H2-(A4–24)(B7–24) shown in green and the conserved disulfide bonds in yellow. Neither of the residues removed are buried in the hydrophobic core however, residues TyrA3, TrpB4, and GluB6 form contacts on the fringes of the core; thus, they may contribute to the decreased stability of the fold of the truncated H2 relaxin.
H2-(A4–24)(B7–24) as Lead for Further Development of RXFP1-specific Therapeutics
H2 relaxin recently completed phase IIb clinical trials (4) and is currently in phase III clinical trials for acute heart failure, due to its positive vasodilatory and renal effects. It is produced for pharmaceutical applications through recombinant expression followed by enzymatic cleavage of the C-peptide to achieve the active two-chain structure. This strategy, although being efficient for native H2 relaxin, is limiting in terms of being able to produce various analogues including chemically modified analogues with non-native amino acids. Such analogues may prove to be valuable as a next generation H2 relaxin agonists and antagonists having improved stability and pharmacokinetic properties. Chemical synthesis, while being more flexible, is on the other hand limited by the size of H2 relaxin and the complexity of joining the two chains by sequential oxidative formation of disulfide bonds. Our result here open the door for development of smaller H2 relaxin variants more readily accessible by synthetic means. Future work will be aimed at identifying weak links in this minimized structure and introducing chemical modifications to improve serum stability and drug-like properties.
As H2 relaxin is currently in clinical trials for AHF and shows potential for the treatment of fibrosis (collagen deposition), selectivity of new analogues for RXFP1 over RXFP2 is a key issue as native H2 relaxin is able to activate RXFP2 with high affinity. Truncation of the B-chain C terminus was expected to have a positive effect on the selectivity for RXFP1, as TrpB28, which is conserved in INSL3, is critical for INSL3 binding to its cognate receptor, RXFP2 (39). This was indeed found to be the case with H2-(B1–27) and H2-(B7–24), both of which lack TrpB28, showing significant decreases in affinity for and activation of RXFP2 (Fig. 8 and Table 1). Combination of a truncated B-chain (B7–24) with N-terminally truncated A-chain (A4–24) was found to further reduce the activation of RXFP2, with the lead molecule H2-(A4–24)(B7–24) being devoid of measureable RXFP2 activity even at micromolar concentration (Fig. 8, Table 1). This result nicely fits with our previous observation that truncations of four to six residues from the N terminus of the A-chain decreased the affinity for and activity of RXFP2 by >2 log units (>100 times) (21).
The receptor (RXFP1) selectivity of our peptide is much more pronounced than the H2 mutant peptide, H2 relaxin FA23A in which the Phe in position 23 of the A-chain is replaced by an Ala which has recently been reported by Park et al. (18). This peptide was claimed to be an RXFP1-specific analogue based on a decrease in affinity primarily for RXFP2. We have recently prepared this mutant and found that it has a pEC50 value of 7.27 for RXFP2,7 whereas the pEC50 value of our peptide, H2-(A4–24)(B7–24) is <6.0.
Importantly, the H2-(A4–24)(B7–24) peptide was then tested on human BJ3 cells that natively express RXFP1 receptors (Fig. 9). Strikingly, despite having a lower affinity for RXFP1, the H2-(A4–24)(B7–24) analogue decreased TGF-β1-stimulated collagen content by a similar extent as that of full-length H2 relaxin. This ability of H2-(A4–24)(B7–24) to strongly reduce collagen content in biologically relevant cells makes it a promising new anti-fibrotic agent.
CONCLUSIONS
In summary, we have identified a critical “active core” of the native H2 sequence, H2-(A4–24)(B7–24). Though it is relatively less potent compared with native peptide, it is still capable of activating RFFP1 receptor at a nanomolar concentration. Importantly, it is also able to mimic the effects of native H2 relaxin on collagen remodeling in fibroblasts. Given that native H2 relaxin has a strong affinity for the INSL3 receptor (RXFP2), future clinical application of H2 relaxin could cause side effects through RXFP2-mediated physiological processes. The H2-(A4–24)(B7–24) analogue will have an advantage over native H2 relaxin in that it is fully selective for RXFP1 over RXFP2, which is thus an ideal starting point for the further development toward the clinic. Finally, the H2-(A4–24)(B7–24) peptide is about one-third smaller in size and thus easier and cheaper to make as a drug compared with H2 relaxin.
Acknowledgments
We are grateful to Tania Ferraro & Sharon Layfield for assistance with biochemical assays and to Feng Lin for amino acid analysis.
This work was supported by National Health and Medical Research Council (NHMRC, Australia) Project Grants 350284 and 508995 (to J. D. W. and R. A. D. B.). Studies at the FNI were supported by the Victorian Government's Operational Infrastructure support program.
M. A. Hossain, K. J. Rosengren, C. S. Samuel, F. Shabanpoor, L. J. Chan, R. A. D. Bathgate, and J. D. Wade, unpublished data.
- AHF
- acute heart failure
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- INSL3
- insulin-like peptide 3
- NHMRC
- National Health and Medical Research Council
- Z
- pyroglutamic acid (also referred to as <Glu).
REFERENCES
- 1. Baccari M. C., Calamai F. (2004) Curr. Protein Pept. Sci. 5, 9–18 [DOI] [PubMed] [Google Scholar]
- 2. Du X. J., Bathgate R. A., Samuel C. S., Dart A. M., Summers R. J. (2010) Nat. Rev. Cardiol. 7, 48–58 [DOI] [PubMed] [Google Scholar]
- 3. Teichman S. L., Unemori E., Teerlink J. R., Cotter G., Metra M. (2010) Curr. Heart Fail. Rep. 7, 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teerlink J. R., Metra M., Felker G. M., Ponikowski P., Voors A. A., Weatherley B. D., Marmor A., Katz A., Grzybowski J., Unemori E., Teichman S. L., Cotter G. (2009) Lancet 373, 1429–1439 [DOI] [PubMed] [Google Scholar]
- 5. Dschietzig T., Richter C., Bartsch C., Laule M., Armbruster F. P., Baumann G., Stangl K. (2001) FASEB J. 15, 2187–2195 [DOI] [PubMed] [Google Scholar]
- 6. Hewitson T. D., Ho W. Y., Samuel C. S. (2010) Endocrinology 151, 4938–4948 [DOI] [PubMed] [Google Scholar]
- 7. Samuel C. S. (2005) Clin. Med. Res. 3, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsu S. Y., Nakabayashi K., Nishi S., Kumagai J., Kudo M., Sherwood O. D., Hsueh A. J. (2002) Science 295, 671–674 [DOI] [PubMed] [Google Scholar]
- 9. Kumagai J., Hsu S. Y., Matsumi H., Roh J. S., Fu P., Wade J. D., Bathgate R. A., Hsueh A. J. (2002) J. Biol. Chem. 277, 31283–31286 [DOI] [PubMed] [Google Scholar]
- 10. Schwabe C., McDonald J. K. (1977) Science 197, 914–915 [DOI] [PubMed] [Google Scholar]
- 11. Chan L. J., Hossain M. A., Samuel C. S., Separovic F., Wade J. D. (2011) Protein Pept. Lett. 18, 220–229 [DOI] [PubMed] [Google Scholar]
- 12. Büllesbach E. E., Schwabe C. (1988) Int. J. Pept. Protein Res. 32, 361–367 [DOI] [PubMed] [Google Scholar]
- 13. Büllesbach E. E., Schwabe C. (2001) J. Pept. Res. 57, 77–83 [DOI] [PubMed] [Google Scholar]
- 14. Büllesbach E. E., Schwabe C. (2000) J. Biol. Chem. 275, 35276–35280 [DOI] [PubMed] [Google Scholar]
- 15. Büllesbach E. E., Schwabe C. (2005) J. Biol. Chem. 280, 14051–14056 [DOI] [PubMed] [Google Scholar]
- 16. Scott D. J., Tregear G. W., Bathgate R. A. (2009) Ann. N.Y. Acad. Sci. 1160, 74–77 [DOI] [PubMed] [Google Scholar]
- 17. Hartley B. J., Scott D. J., Callander G. E., Wilkinson T. N., Ganella D. E., Kong C. K., Layfield S., Ferraro T., Petrie E. J., Bathgate R. A. (2009) Ann. N.Y. Acad. Sci. 1160, 67–73 [DOI] [PubMed] [Google Scholar]
- 18. Park J. I., Semyonov J., Yi W., Chang C. L., Hsu S. Y. (2008) J. Biol. Chem. 283, 32099–32109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Büllesbach E. E., Schwabe C. (1986) Biochemistry 25, 5998–6004 [DOI] [PubMed] [Google Scholar]
- 20. Büllesbach E. E., Schwabe C. (1987) J. Biol. Chem. 262, 12496–12501 [PubMed] [Google Scholar]
- 21. Hossain M. A., Rosengren K. J., Haugaard-Jönsson L. M., Zhang S., Layfield S., Ferraro T., Daly N. L., Tregear G. W., Wade J. D., Bathgate R. A. (2008) J. Biol. Chem. 283, 17287–17297 [DOI] [PubMed] [Google Scholar]
- 22. Büllesbach E. E., Schwabe C. (2005) J. Biol. Chem. 280, 14586–14590 [DOI] [PubMed] [Google Scholar]
- 23. Hossain M. A., Wade J. D. (2010) Curr. Protein Pept. Sci. 11, 719–724 [DOI] [PubMed] [Google Scholar]
- 24. Atherton E., Sheppard R. C. (1998) Solid-Phase Peptide Synthesis: A Practical Approach, IRL Press, Oxford [Google Scholar]
- 25. Hossain M. A., Belgi A., Lin F., Zhang S., Shabanpoor F., Chan L., Belyea C., Truong H. T., Blair A. R., Andrikopoulos S., Tregear G. W., Wade J. D. (2009) Bioconjug. Chem. 20, 1390–1396 [DOI] [PubMed] [Google Scholar]
- 26. Hossain M. A., Rosengren K. J., Zhang S., Bathgate R. A., Tregear G. W., van Lierop B. J., Robinson A. J., Wade J. D. (2009) Org. Biomol. Chem. 7, 1547–1553 [DOI] [PubMed] [Google Scholar]
- 27. Shabanpoor F., Hughes R. A., Bathgate R. A., Zhang S., Scanlon D. B., Lin F., Hossain M. A., Separovic F., Wade J. D. (2008) Bioconjug. Chem. 19, 1456–1463 [DOI] [PubMed] [Google Scholar]
- 28. Scott D. J., Layfield S., Yan Y., Sudo S., Hsueh A. J., Tregear G. W., Bathgate R. A. (2006) J. Biol. Chem. 281, 34942–34954 [DOI] [PubMed] [Google Scholar]
- 29. Chen W., Shields T. S., Stork P. J., Cone R. D. (1995) Anal. Biochem. 226, 349–354 [DOI] [PubMed] [Google Scholar]
- 30. Keller R. L. (2004) The Computer-aided Resonance Assignment Tutorial, CANTINA Verlag, Goldau, Switzerland [Google Scholar]
- 31. Wüthrich K. (1986) NMR of Proteins and Nucleic Acids, Wiley-Interscience, New York [Google Scholar]
- 32. Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
- 33. Hahn W. C., Counter C. M., Lundberg A. S., Beijersbergen R. L., Brooks M. W., Weinberg R. A. (1999) Nature 400, 464–468 [DOI] [PubMed] [Google Scholar]
- 34. Unemori E. N., Pickford L. B., Salles A. L., Piercy C. E., Grove B. H., Erikson M. E., Amento E. P. (1996) J. Clin. Invest. 98, 2739–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Unemori E. N., Amento E. P. (1990) J. Biol. Chem. 265, 10681–10685 [PubMed] [Google Scholar]
- 36. Samuel C. S., Unemori E. N., Mookerjee I., Bathgate R. A., Layfield S. L., Mak J., Tregear G. W., Du X. J. (2004) Endocrinology 145, 4125–4133 [DOI] [PubMed] [Google Scholar]
- 37. Samuel C. S., Butkus A., Coghlan J. P., Bateman J. F. (1996) Endocrinology 137, 3884–3890 [DOI] [PubMed] [Google Scholar]
- 38. Gallop P. M., Paz M. A. (1975) Physiol. Rev. 55, 418–487 [DOI] [PubMed] [Google Scholar]
- 39. Büllesbach E. E., Schwabe C. (1999) Biochemistry 38, 3073–3078 [DOI] [PubMed] [Google Scholar]
- 40. Masterson R., Hewitson T. D., Kelynack K., Martic M., Parry L., Bathgate R., Darby I., Becker G. (2004) Nephrol. Dial. Transplant. 19, 544–552 [DOI] [PubMed] [Google Scholar]
- 41. Tozzi C. A., Poiani G. J., McHugh N. A., Shakarjian M. P., Sharkarjian M. P., Grove B. H., Samuel C. S., Unemori E. N., Riley D. J. (2005) Pulm. Pharmacol. Ther. 18, 346–353 [DOI] [PubMed] [Google Scholar]
- 42. Heeg M. H., Koziolek M. J., Vasko R., Schaefer L., Sharma K., Müller G. A., Strutz F. (2005) Kidney Int. 68, 96–109 [DOI] [PubMed] [Google Scholar]
- 43. Sudo S., Kumagai J., Nishi S., Layfield S., Ferraro T., Bathgate R. A., Hsueh A. J. (2003) J. Biol. Chem. 278, 7855–7862 [DOI] [PubMed] [Google Scholar]
- 44. Svendsen A. M., Vrecl M., Ellis T. M., Heding A., Kristensen J. B., Wade J. D., Bathgate R. A., De Meyts P., Nøhr J. (2008) Endocrinology 149, 1113–1120 [DOI] [PubMed] [Google Scholar]
- 45. Svendsen A. M., Zalesko A., Kønig J., Vrecl M., Heding A., Kristensen J. B., Wade J. D., Bathgate R. A., De Meyts P., Nøhr J. (2008) Mol. Cell Endocrinol. 296, 10–17 [DOI] [PubMed] [Google Scholar]
- 46. Büllesbach E. E., Yang S., Schwabe C. (1992) J. Biol. Chem. 267, 22957–22960 [PubMed] [Google Scholar]
- 47. Rosengren K. J., Zhang S., Lin F., Daly N. L., Scott D. J., Hughes R. A., Bathgate R. A., Craik D. J., Wade J. D. (2006) J. Biol. Chem. 281, 28287–28295 [DOI] [PubMed] [Google Scholar]
- 48. Kuei C., Sutton S., Bonaventure P., Pudiak C., Shelton J., Zhu J., Nepomuceno D., Wu J., Chen J., Kamme F., Seierstad M., Hack M. D., Bathgate R. A., Hossain M. A., Wade J. D., Atack J., Lovenberg T. W., Liu C. (2007) J. Biol. Chem. 282, 25425–25435 [DOI] [PubMed] [Google Scholar]
- 49. Liu C., Chen J., Kuei C., Sutton S., Nepomuceno D., Bonaventure P., Lovenberg T. W. (2005) Mol. Pharmacol. 67, 231–240 [DOI] [PubMed] [Google Scholar]
- 50. Haugaard-Jönsson L. M., Hossain M. A., Daly N. L., Bathgate R. A., Wade J. D., Craik D. J., Rosengren K. J. (2008) J. Biol. Chem. 283, 23811–23818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rosengren K. J., Bathgate R. A., Craik D. J., Daly N. L., Haugaard-Jönsson L. M., Hossain M. A., Wade J. D. (2009) Ann. N.Y. Acad. Sci. 1160, 20–26 [DOI] [PubMed] [Google Scholar]