Abstract
Quality control of endoplasmic reticulum proteins involves the identification and engagement of misfolded proteins, dislocation of the misfolded protein across the endoplasmic reticulum (ER) membrane, and ubiquitin-mediated targeting to the proteasome for degradation. Ancient ubiquitous protein 1 (AUP1) physically associates with the mammalian HRD1-SEL1L complex, and AUP1 depletion impairs degradation of misfolded ER proteins. One of the functions of AUP1 in ER quality control is to recruit the soluble E2 ubiquitin-conjugating enzyme UBE2G2. We further show that the CUE domain of AUP1 regulates polyubiquitylation and facilitates the interaction of AUP1 with the HRD1 complex and with dislocation substrates. AUP1 localizes both to the ER and to lipid droplets. The AUP1 expression level affects the abundance of cellular lipid droplets and as such represents the first protein with lipid droplet regulatory activity to be linked to ER quality control. These findings indicate a possible connection between ER protein quality control and lipid droplets.
Keywords: E3 Ubiquitin Ligase, Endoplasmic Reticulum (ER), ER Quality Control, Lipid Droplet, Ubiquitin
Introduction
Eukaryotic cells possess an efficient system to detect and remove misfolded proteins from the endoplasmic reticulum (ER)2 (1). The HRD1 complex specializes in the removal of ER proteins with defects in their lumenal domains (2–4). Once a lumenal ER protein has exhausted its folding options, it is directed to the quality control machinery by a set of proteins that includes OS9, XTP3-B, and SEL1L (5–7). The misfolded protein is then dislocated from the ER. Transportation across the ER lipid bilayer is generally believed to proceed through a proteinaceous channel, such as the complexes nucleated by Sec61 (8), Derlin1 (9, 10), or the E3 ligase Hrd1p (11). The dislocated protein is ubiquitylated and deglycosylated prior to its degradation by the proteasome in the cytoplasm. Ubiquitylation is a three-step process. E1 activates ubiquitin in an ATP-dependent reaction, followed by formation of a thioester-linked ubiquitin-E2 complex. An E3 ubiquitin ligase then catalyzes transfer of ubiquitin onto the intended substrate. In the case of the HRD1-SEL1L complex, UBE2G2 (also known as UBC7) serves as the E2 and HRD1 as the E3 (12). UBE2G2 also acts as the E2-conjugating enzyme for gp78, another E3 ubiquitin ligase that specializes in the ubiquitylation of dislocated ER proteins (13). We have also identified Ubc6e as an E2-conjugating enzyme of the HRD1-SEL1L complex (14), suggesting the possibility that protein complexes of overlapping yet distinct composition are involved in ER quality control.
We previously identified ancient ubiquitous protein 1 (AUP1) as a component of the HRD1-SEL1L ER quality control complex and showed that AUP1 is necessary for US11-mediated dislocation of class I MHC heavy chains (14). US11 is a protein encoded by human cytomegalovirus that targets class I MHC heavy chains for destruction as part of its immunoevasive strategy (15). AUP1 has also been proposed to be involved in integrin signaling (16, 17). AUP1 contains a hydrophobic region close to the N terminus that inserts into the membrane such that both termini are found in the cytoplasm (18). AUP1 contains two conserved cytoplasmic domains according to the Ensembl database, an acyltransferase domain and a CUE domain. Acyltransferase domains transfer fatty acids onto phospholipids using a conserved active site histidine and aspartic acid, separated by four amino acids (HX4D) (19). CUE domains are UBA-like domains that bind ubiquitin. Residues on the first and third α-helices of the CUE domain bind to a hydrophobic surface patch of ubiquitin (20, 21). The role of high affinity CUE domains in the monoubiquitylation of an endocytic protein has been elucidated (20, 22, 23), but the role of CUE domains in ER protein quality control has not. We here identify a third region of AUP1 not previously annotated in the domain bioinformatics databases that is necessary for recruitment of UBE2G2. This UBE2G2 binding domain (G2BR) was originally found on the E3 gp78 (24). During manuscript preparation, this G2BR was also identified by another group (18).
At first glance, it is reasonable to hypothesize that in mammalian cells AUP1 merely serves a role similar to that of Cue1p, a component of the yeast Hrd1-Der3p ER protein quality control complex. Both AUP1 and yeast Cue1p are membrane-anchored and recruit its cognate E2 ubiquitin-conjugating enzyme. AUP1 recruits UBE2G2 via a G2 binding region, and yeast Cue1p recruits (25) and enhances (26) the activity of Ubc7p, the yeast UBE2G2 homolog, via a U7 binding region at its C terminus (27). Aside from this similarity, AUP1 and Cue1p are not homologs. Unlike AUP1, yeast Cue1p does not encode a putative acyltransferase domain. The protein domain architecture of AUP1, in which a membrane anchor is followed by an acyltransferase domain and a CUE domain, is conserved in organisms with bilateral symmetry. Also, the CUE domain of yeast Cue1p was reported as dispensable for ER quality control (27), but we identify several important roles for the CUE domain of AUP1. Given these differences, AUP1 may perform additional functions beyond those of yeast Cue1p.
We show that AUP1 is found in both the ER and in lipid droplets, an observation that was suggested by proteomic studies and observed independently by another group (18). Lipid droplets are cytoplasmic organelles that serve as storage depots for cholesteryl esters and triacylglycerols, to be released for membrane biogenesis or as a source of cellular energy via β-oxidation of fatty acids. Lipid droplets are thought to be derived from the ER and are composed of a phospholipid monolayer that surrounds the neutral lipid core (28, 29). The role of lipid droplets also includes a variety of less obvious functions, such as sequestration of histones in embryogenesis (30), involvement in hepatitis C (31) and Chlamydia (32) infections, and proteasomal degradation (33, 34).
Here, we examine the role of AUP1 in ER protein quality control and in lipid droplet accumulation. We expand the role of AUP1 to include general ER quality control of soluble misfolded ER proteins. We find that AUP1 binds UBE2G2 at its C terminus. AUP1-interacting proteins, identified by mass spectrometry, fall into three main categories as follows: ER protein quality control proteins, lipid-modifying enzymes, and subunits of the oligosaccharide transferase complex, further underscoring the connections with the ER. We characterize several functions of the CUE domain in ER protein degradation that have not been described previously, even for Cue1p; the CUE domain mediates the interaction of AUP1 with the ER protein quality control complex, terminally misfolded proteins, and ubiquitylated proteins, and the CUE domain also inhibits ubiquitin chain elongation. Finally, we show that AUP1 localizes to lipid droplets and contributes to their accumulation. These unexpected results suggest that lipid droplets might be important for ER protein quality control. This hypothesis is supported by the observation that dislocation substrates are stabilized in the presence of an inhibitor of lipid-modifying enzymes required for lipid droplet formation and that ubiquitylation can occur in purified lipid droplet fractions.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies have been described: anti-AUP1, anti-UBXD8, and anti-Ubc6e (14); anti-SEL1L (35); and anti-class I MHC HC (HC-70) (9). Other antibodies were purchased as follows: anti-GFP, anti-PDI, anti-GAPDH, and anti-calnexin (Abcam); anti-FLAG and anti-HA-HRP 3F10 (Sigma); immobilized anti-HA 3F10 (Roche Applied Science); anti-OS9 and anti-α1-antitrypsin (Novus); anti-HRD1 (Abgent); anti-p97 (Fitzgerald Industries International); and anti-Myc M2 (Cell Signaling). Anti-ribophorin I antibody was a generous gift of N. Erwin Ivessa (Vienna Biocenter, Vienna, Austria). Anti-STT3B and anti-OST48 antibodies were a generous gift of Reid Gilmore (University of Massachusetts Medical School, Worcester, MA).
Cell Culture, Oleic Acid Treatment, Triacsin C, and Transfection
HeLa cells (ATCC) were cultured in DMEM. For lipid droplet loading, cells were incubated with oleic acid adsorbed to BSA (36), for the concentrations and times indicated. Triacsin C was purchased from Biomol. FuGENE 6 (Roche Applied Science) was used for transfections.
shRNA
shRNA constructs targeting AUP1 were obtained from the RNAi Consortium. shLUC corresponds to clone SHC007, mature sense sequence CGCTGAGTACTTCGAAATGTC, shAUP1-construct A corresponds to clone TRCN0000004269, mature sense sequence TCAGCCAACAGCCCTAACATT, and shAUP1-B corresponds to clone TRCN0000004272, mature sense sequence ACACCTTTCGACCACAACATA. Lentivirus was made, and HeLa cells were infected as described (35). Cells were grown in DMEM supplemented with 1 μg/ml puromycin, and experiments were performed 4 days post-infection.
DNA Constructs
mAT and mCUE mutations were made to AUP1 using site-directed mutagenesis with the following primers (forward primers are given); for mAT (H96A), GGTCCTCATTTCCAACgcTGTGACACCTTTCGACC; for mCUE (E306K, V307A, and L308A), GCTCAGAGAGTCAAGaaggcagcaCCCCATGTGCCATTG; and (Δ333–334), CTTGACTATCACTAATGAGGGGGCCGTAGCTTTC. For the ΔG2BR construct, the following reverse primer was used in PCR: CGTGAATTCTCACTTGGCAAATGTTAGGGCTG. Constructs were cloned into pcDNA3.1+ (Clontech) (untagged and N-terminal HA) or pEGFP-C1/N1 (Clontech). GFP-AUP1 constructs also contain silent mutations rendering them resistant to shAUP1-B. UBE2G2 was obtained from Open Biosystems and cloned into pcDNA3.1+ with an N-terminal Myc epitope tag. HA-ubiquitin was also cloned into pcDNA3.1+.
Immunoprecipitation and Immunoblotting
Cells were lysed on ice in buffer containing 25 mm Tris, pH 7.4, 5 mm MgCl2, 150 mm NaCl, and the detergent indicated, 0.5% Nonidet P-40 or 1% digitonin. Protein concentration of the cytoplasmic fraction was determined and equalized across the samples. Samples were incubated for 3 h at 4 °C with the antibody and protein A-agarose beads or immobilized anti-HA as indicated. The immunoprecipitates were washed, boiled in reducing sample buffer, and run on SDS-PAGE. Proteins were transferred to PVDF membrane, and membranes were blocked in PBS/Tween/milk, incubated with primary and secondary antibodies, washed, and developed with Western Lighting Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
Pulse-Chase Metabolic Labeling
HeLa cells were starved for 45 min in DMEM lacking cysteine and methionine. 250 μCi of 35S-labeled cysteine and methionine (PerkinElmer Life Sciences) was added to each sample during the pulse (10 min for US11 and 15 min for RI332 and NHK). Media containing cold cysteine and methionine were added, and equal numbers of cells were removed at the indicated chase times. Cells were lysed by agitation in a small volume of 1% SDS in PBS and then diluted in Nonidet P-40 lysis buffer. Incorporation of radioactivity was measured by TCA precipitation and liquid scintillation spectrometry to equalize, for each sample, the amount of input radioactivity to that at the zero chase time point. 35S-Labeled proteins were visualized on x-ray film and quantified by phosphorimaging.
Mass Spectrometry
Approximately 90 million HeLa cells were transfected with HA-AUP1 or empty vector (pcDNA3.1+) plasmid and 3F10 beads were used for immunoprecipitation. Proteins were visualized in the SDS-polyacrylamide gel by silver staining. The entire lane was excised from the gel in 4 × 10-mm strips, and each individual gel slice was subjected to trypsinolysis. Disulfide bonds were reduced and alkylated prior to trypsinolysis. Recovered peptides were analyzed by reversed-phase liquid chromatography electrospray ionization mass spectrometry using a Waters NanoAcquity pump coupled to a ThermoFisher LTQ linear in nanoflow configuration. The mass spectrometer was operated in a dependent data acquisition mode where the five most abundant peptides detected in full scan mode were subjected to daughter ion fragmentation. Peptides were identified from the MS data using SEQUEST algorithms that searched a human-specific database generated from the NCBI database. SEQUEST filters used for indication of positive peptide identification were as follows: XCorr versus charge state = 1.5, 2.00, 2.50; Sp − preliminary score = 500. Data interpretation from all bands was aided by the MSRAT program (Protein Forest).
In Vitro Ubiquitylation
Following digitonin lysis and immunoprecipitation, 100 nm E1 (Boston Biochem) and 60 μm FLAG-ubiquitin (Boston Biochem) were added in an ATP-regenerating buffer (50 mm Tris, pH 7.6, 5 mm MgCl2, 5 mm ATP, 10 mm creatine phosphate, 3.5 units/ml creatine kinase) and kept at 37 °C for 1 h with gentle shaking.
Immunofluorescence
Cells were grown on glass coverslips, fixed in 4% paraformaldehyde, and permeabilized in 0.1% Triton X-100. Permeabilized cells were incubated with primary and secondary (Alexa Fluor 568-labeled) antibodies and washed with PBS before being mounted on a slide with Fluoromount-G (Southern Biotech). Imaging was performed at 37 °C on an inverted spinning disk confocal microscope (Nikon TE2000-U) using a Nikon ×100 magnification, 1.4 numerical aperture, differential interference contrast oil lens, and Hamamatsu ORCA camera using Metamorph Imaging software as described (37).
Electron Microscopy
Cells were fixed in 2.5% glutaraldehyde, 3% paraformaldehyde, with 5% sucrose in 0.1 m sodium cacodylate buffer, pH 7.4. Cells were then postfixed in 1% OsO4 in veronal-acetate buffer. The cells were stained in block overnight with 0.5% uranyl acetate in veronal-acetate buffer, pH 6.0, dehydrated, and embedded in Spurr's resin. Sections were cut on a Reichert Ultracut E microtome with a Diatome diamond knife at a thickness setting of 50 nm and stained with 2% uranyl acetate, followed by 0.1% lead citrate. Samples were examined using an FEI Tecnai Spirit TEM at 80 kV and imaged with an AMT camera.
Flow Cytometry
HeLa cells were transduced with shRNA. Three days post-infection, 0.4 mm oleic acid was added to half of the samples for 16 h. Lipid droplets were stained with 10 μg/ml BODIPY 493/503 for 2 h as described (38). Median BODIPY 493/503 intensity was measured on a FACSCalibur (BD Biosciences) using forward scatter measurements to exclude dead cells. Data were analyzed using FlowJo software.
XBP-1 Splicing Assay
Total cellular RNA was isolated from cells using the Qiagen RNeasy kit. cDNA was made using the Superscript II reverse transcriptase from Invitrogen. XBP-1 was amplified using the primers TCCTTCTGGGTAGACCTCTGGGAG (forward) and CAAGGGGAATGAAGTGAGGCCAG (reverse), which flank the splice site.
Isolation of Lipid Droplets
Lipid droplets were isolated from lipid-loaded 293T cells generally as described (36). Briefly, cells were homogenized by 20 strokes in a glass Potter-Elvehjem homogenizer with a loose-fitting Teflon pestle in hypotonic lysis medium (HLM) (20 mm Tris, pH 7.4, 1 mm EDTA, protease inhibitor mixture (Roche Applied Science)). After sedimentation of unbroken cells and nuclei, cell lysate was adjusted to 20% sucrose in HLM and applied to the bottom of a 13.2-ml tube. 5 ml of HLM containing 5% sucrose was layered on top, followed by HLM to the top (about 6 ml). Gradients were centrifuged for 90 min at 28,000 × g in an SW41Ti rotor (Beckman) and allowed to coast to a stop. Fractions were collected from bottom to top using a long needle inserted to the bottom of the tube attached to a peristaltic pump. The lipid droplet fraction was the top-most ∼50 μl. Lipid droplets were solubilized by incubation in a sonicating water bath for 2 h at 37 °C in 5% SDS.
RESULTS
AUP1 Facilitates the Dislocation of Misfolded Proteins from the ER
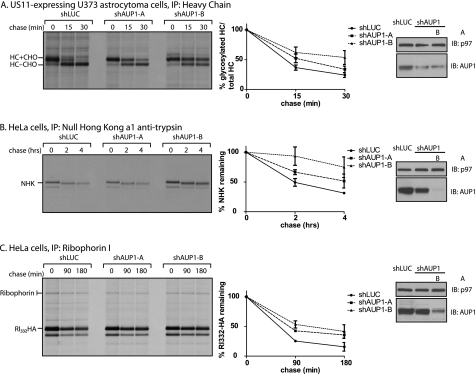
AUP1 has been implicated in the US11-mediated disposal of class I major histocompatibility complex HC based on the observation that GFP-tagged versions of AUP1 act in dominant-interfering fashion and impair the US11-mediated disposal pathway (14). We confirmed this result using shRNA-mediated reduction of AUP1 levels. US11-expressing cells were transduced with a control shRNA specific for luciferase or one of two different shRNAs that target AUP1. These cells were then subjected to pulse-chase analysis and immunoprecipitation of class I heavy chains. The addition of the proteasome inhibitor ZL3VS allowed recovery of both glycosylated (HC + CHO) and deglycosylated, cytoplasmically disposed, heavy chains (HC-CHO). Cells with decreased levels of AUP1 (as determined by Western blot), exhibited slower kinetics of heavy chain removal from the ER (Fig. 1A).
FIGURE 1.
AUP1 is involved in ER quality control. A, US11-expressing astrocytoma cells were transduced with shRNA specific against luciferase (shLUC), as a control, or one of two different constructs targeting AUP1 (A or B). Four days post-transduction, cells were pulse-labeled with 35S-labeled cysteine and methionine. Samples were taken at the indicated chase times, and class I MHC heavy chain was recovered from the lysates. Immunoprecipitates (IP) were separated by SDS-PAGE and imaged by autoradiography. Amount of recovered protein was quantified by phosphorimagery and is shown as a percentage of glycosylated heavy chain compared with total heavy chain. Error bars represent standard deviation of three individual experiments. The level of AUP1 depletion was determined by immunoblotting (IB) with p97 as a loading control. The same experiment as described for A was performed using shRNA-transduced HeLa cells transfected with NHK (B) or RI332-HA (C). NHK was immunoprecipitated using an anti-α1-antitrypsin antibody. RI332-HA was immunoprecipitated using an anti-ribophorin I antibody that recovers both the full-length and the misfolded fragment of ribophorin I. Quantification in B and C shows the percentage of protein remaining compared with the amount recovered at the 0-min chase time.
Two soluble misfolded proteins, ribophorin I fragment (ribophorin I, amino acids 1–332 (RI332)) and the Null Hong Kong variant of α1-antitrypsin (NHK), are known to also use the HRD1 complex for their removal from the ER. NHK and RI332 are ER lumenal proteins harboring mutations that prevent them from folding correctly and thus serve as dislocation substrates (39). Both RI332-HA and NHK are stabilized in AUP1-depleted cells, showing that AUP1 contributes to their dislocation. The magnitude of the effect on dislocation is comparable with those observed for other interventions in ER quality control, such as depletion of members of the Derlin family (40), OS9 (5), and AUP1-GFP dominant negatives. The two AUP1 shRNAs have different efficiencies in HeLa cells; construct A shows ∼50% reduction of AUP1 levels, and construct B shows ∼90% reduction. Furthermore, the level of impairment of dislocation negatively correlated with the level of AUP1, i.e. greater reduction of AUP1 levels leads to slower dislocation rates (Fig. 1, B and C).
AUP1 Binds Proteins Involved in ER Protein Quality Control, Lipid Modification, and Glycosylation
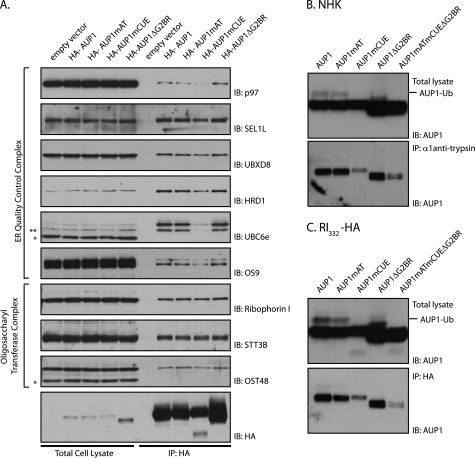
We identified proteins that interact with AUP1 by mass spectrometry. HA-tagged AUP1 was transfected into HeLa cells and recovered from digitonin lysates with anti-HA antibody and resolved by SDS-PAGE (supplemental Fig. 1). We identified the co-precipitating proteins by LC/MS-MS (Table 1). Many of the known components of the HRD1 dislocation complex were recovered: HRD1, SEL1L, OS9, UBXD8, p97, and UBE2G2. We also recovered several components of the oligosaccharyltransferase complex, ribophorin I and II (OST1 and OST2) and OST48. In addition, we identified several proteins involved in lipid modification as follows: long chain fatty acyl-CoA ligase 3 (ACSL3), serine palmitoyltransferase subunit 1, and lysophosphatidylcholine acyltransferase 1 (LPCAT1). (See supplemental material for a complete list of AUP1-interacting proteins.) In those cases where the necessary antibodies were available, we could confirm by immunoprecipitation and immunoblot the interactions established by mass spectrometry (Fig. 3A). None of the reported interacting proteins were present in the bead control pulldown from the cells transfected with empty vector.
TABLE 1.
AUP1-interacting proteins
A partial list of proteins recovered from the HA-AUP1 immunoprecipitates and identified by LC/MS-MS is given. Unique peptides indicate the total number of different peptides recovered from the immunoprecipitate. GenBankTM GI numbers and calculated molecular masses (Da) are also given. Data analysis was performed by MSRAT.
| Protein accession no. | Molecular mass | Unique peptides | Sequence coverage | |
|---|---|---|---|---|
| Da | % | |||
| ER quality control | ||||
| AUP1 | gi|31712030 | 45,758 | 13 | 45 |
| p97 | gi|6005942 | 89,266 | 13 | 20 |
| UBE2G2 | gi|33359701 | 15,603 | 5 | 59 |
| SEL1L | gi|19923669 | 88,699 | 10 | 19 |
| OS9 | gi|63252870 | 73,775 | 6 | 13 |
| UBXD8 | gi|24797106 | 52,591 | 6 | 20 |
| HRD1 | gi|27436927 | 67,641 | 3 | 5 |
| Oligosaccharide transferase complex | ||||
| Ribophorin I | gi|4506675 | 68,527 | 16 | 33 |
| Ribophorin II | gi|209413738 | 67,682 | 10 | 27 |
| Oligosaccharide transferase 48 | gi|20070197 | 50,670 | 5 | 13 |
| Lipid metabolism | ||||
| Acyl-CoA synthetase 3 | gi|42794754 | 80,368 | 5 | 9 |
| Serine palmitoyltransferase subunit 1 | gi|5454084 | 52,711 | 4 | 8 |
| Lysophosphatidylcholine acyltransferase 1 | gi|33946291 | 59,114 | 3 | 8 |
FIGURE 3.
CUE domain of AUP1 mediates interaction with ER quality control proteins and terminally misfolded proteins. A, HeLa cells were transfected with empty vector or one of the HA-AUP1 WT or mutant constructs. HA-AUP1 was recovered with 3F10 (anti-HA) antibody from digitonin lysates. Immunoblotting (IB) with antibodies for the indicated proteins showed the presence of endogenous proteins in the total cell lysates and immunoprecipitates (IP). * indicates cross-reactive proteins and ** indicates degradation product of UBC6e. B, HeLa cells were transfected with the NHK variant of α1-antitrypsin and one of the AUP1 WT or mutant constructs as indicated. These cells were incubated with 5 μm ZL3VS overnight. NHK was recovered from Nonidet P-40 lysates, and the content of AUP1 in total cell lysates and immunoprecipitates was determined by immunoblotting with an anti-AUP1 antibody. Both endogenous and transiently introduced AUP1s are present in the immunoblots. C, same experiment as in B was repeated with RI332-HA instead of NHK.
AUP1 Binds the E2 Ubiquitin Ligase UBE2G2 via a C-terminal G2 Binding Region
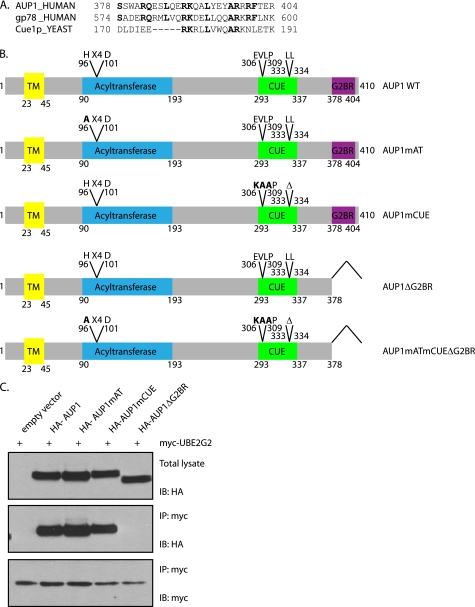
A striking AUP1-interacting protein that we recovered is UBE2G2. This association was also identified in a large scale protein-protein interaction study (41). Five peptides of this 15.6-kDa protein were identified by mass spectrometry, accounting for 59% of its sequence (Table 1). UBE2G2 is a cytosolic E2 ubiquitin ligase, the mammalian homolog of yeast Ubc7p. The mammalian E3 ubiquitin ligase gp78 binds UBE2G2 via a stretch of 27 amino acids found at its C terminus (termed the G2 binding region (G2BR)) (24). An amino acid sequence alignment between the gp78 G2BR and AUP1 identifies a region of high similarity at the C terminus of AUP1 (Fig. 2A). We made constructs of AUP1 lacking its putative G2BR-containing C terminus as well as constructs with mutations in the other two domains of AUP1 described previously (Fig. 2B). Full-length AUP1 and AUP1 with mutations in the acyltransferase or CUE domains interact with UBE2G2, whereas AUP1ΔG2BR does not, as determined by immunoprecipitation for myc-UBE2G2, followed by immunoblotting for HA-AUP1 (Fig. 2C). We thus conclude that AUP1 binds UBE2G2 and that this interaction is dependent on the C terminus of AUP1 that constitutes a G2BR domain.
FIGURE 2.
AUP1 interacts with UBE2G2 via a binding region at its C terminus. A, protein alignment of the G2 binding regions of AUP1 and gp78 shows strong sequence similarity. The U7 region of Cue1p shows weaker similarity. B, AUP1 and the mutant constructs used in this study are represented here. AUP1mAT has an H96A point mutation. AUP1mCUE has residues from 306 to 308 mutated from EVL to KAA and Δ333–334. AUP1ΔG2BR has residues 378–410 deleted. AUP1mATmCUEΔG2BR has H96A, E306K, V307A, L308A, Δ333–334, and Δ378–410. TM, transmembrane. C, HeLa cells were transfected with myc-UBE2G2 and one of the HA-AUP1 WT or mutant constructs. myc-UBE2G2 was recovered from Nonidet P-40 lysates. Immunoblotting (IB) for HA detected associated HA-AUP1. IP, immunoprecipitate.
CUE Domain of AUP1 Mediates Its Binding to ER Quality Control Machinery and Dislocation Substrates
CUE domains possess conserved sequences on their first and third α-helices that bind to hydrophobic patches on the surface of ubiquitin. For AUP1, these areas correspond to a valine-leucine-proline and di-leucine sequence (20). We mutated these regions to compromise the binding of AUP1 to ubiquitin. These mutations affected the association of AUP1 with many components of the ER quality control machinery. HeLa cells were transfected with HA-AUP1 constructs (wild type, mutant acyltransferase domain, mutant CUE domain, or G2BR deletion) or empty vector plasmid as a negative control, and HA-AUP1 was recovered by immunoprecipitation from digitonin lysates. We then immunoblotted for several of the AUP1-interacting proteins identified by mass spectrometry (Table 1). AUP1 with the mutant CUE domain (AUP1mCUE) was less efficient at interacting with most of the proteins involved in dislocation (p97, SEL1L, UBXD8, OS9, UBC6e, and HRD1). Components of the oligosaccharide transferase complex (ribophorin I, OST48, and STT3B) were recruited equally well for all forms of HA-AUP1 (Fig. 3A).
Given the role of AUP1 and the ER quality control complex in processing of misfolded ER proteins, we examined whether AUP1 associates with dislocation substrates. HeLa cells were transfected with AUP1 constructs and one of two terminally misfolded proteins, NHK or RI332-HA. NHK or RI332-HA immunoprecipitates were analyzed for AUP1 content by immunoblotting. AUP1 with CUE domain mutations associated less well with both of the dislocation substrates than did wild type AUP1 (Fig. 3, B and C).
AUP1 Is Modified by Ubiquitin and Binds Ubiquitylated Proteins
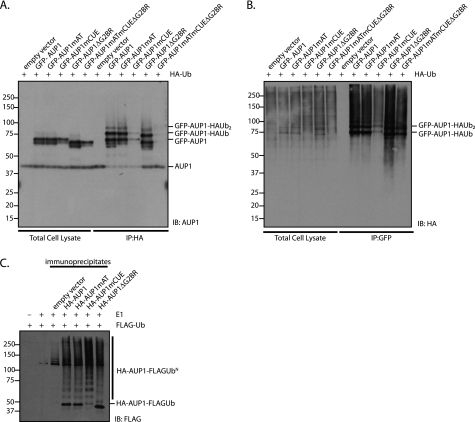
AUP1 is itself modified by ubiquitin. Cells transfected with HA-ubiquitin and GFP-AUP1 were solubilized in mild detergent (digitonin). HA-Ub and HA-Ub-modified proteins were immunoprecipitated from the lysates, followed by immunoblotting for AUP1. Anti-AUP1-reactive material was detected at ∼8 and ∼16-kDa above the expected molecular mass of GFP-AUP1, consistent with mono- and di-ubiquitylation of AUP1 (Fig. 4A). A polypeptide corresponding to the size of unmodified GFP-AUP1 was also detected, suggesting that GFP-AUP1 self-oligomerizes with both ubiquitylated and nonubiquitylated GFP-AUP1 in a digitonin-resistant manner. Very little GFP-AUP1mCUE was recovered, indicating that the CUE domain is essential for interaction with HA-Ub, and GFP-AUP1mCUE is minimally ubiquitylated in vivo. Endogenous AUP1 is also present in the immunoprecipitates but less is recovered in the presence of GFP-AUP1 constructs containing the G2BR, suggesting that the G2BR, and perhaps the associated UBE2G2, competes with endogenous AUP1 for binding with HA-Ub.
FIGURE 4.
AUP1 is both ubiquitylated and binds ubiquitin-modified proteins, and anti-AUP1 immunoprecipitates are able to perform ubiquitin transfer in vitro. A, HeLa cells were transfected with HA-ubiquitin and empty vector or one of the GFP-AUP1 constructs (WT or mutants, as indicated). HA-Ub was recovered with 3F10 (HA-specific) antibody from digitonin lysates supplemented with 2.5 mm N-ethylmaleimide. GFP-AUP1 content in total cell lysates and immunoprecipitates (IP) was determined by immunoblotting (IB) with an anti-AUP1 antibody. B, GFP-AUP1 was recovered with anti-GFP antibody from lysates described in A. HA-Ub content in total cell lysates and immunoprecipitates was determined by immunoblotting with an anti-HA antibody. C, HeLa cells were transfected with empty vector or one of the HA-AUP1 constructs (WT or mutant, as indicated). HA-AUP1 was immunoprecipitated from digitonin lysates. E1 (100 nm), FLAG-ubiquitin (60 μm), and an ATP-regenerating buffer was added to the immunoprecipitates and kept at 37 °C for 60 min. Separate samples containing only FLAG-ubiquitin and buffer or E1, FLAG-ubiquitin, and buffer served as controls. Samples were run on an 8% Tris-Tricine SDS-polyacrylamide gel, and were immunoblotted with a FLAG-specific antibody.
The same experiment was also performed in reverse order; GFP-AUP1 was immunoprecipitated from the lysates followed by immunoblotting for HA-Ub. The amount of HA-reactive material recovered by GFP-AUP1mCUE in digitonin lysates was less than for GFP-AUP1 wild type (Fig. 4B), most likely because the CUE domain of AUP1 mediates the interaction between AUP1 and ubiquitylated proteins. We recovered relatively more HA-reactive material from GFP-AUP1ΔG2BR digitonin lysates than for GFP-AUP1 wild type, consistent with the possibility that the G2BR domain of AUP1 acts as a negative regulator of ubiquitylation or disrupts association of AUP1 with ubiquitylated proteins.
CUE Domains Regulate Ubiquitin Chain Elongation
Given that AUP1 recruits both E2 and E3 enzymes, UBE2G2 and HRD1, respectively, we wondered if the material recovered by immunoprecipitation of HA-AUP1 is sufficient to sustain ubiquitylation in vitro. E1 ubiquitin ligase and FLAG-ubiquitin were added to the HA-AUP1 immunoprecipitates. All HA-AUP1 constructs could be poly-ubiquitylated in vitro, producing the typical ubiquitin “ladder” in anti-FLAG immunoblots (Fig. 4C). There is very little mono-ubiquitylated HA-AUP1mCUE compared with the other constructs, indicating that the CUE domain of AUP1 may negatively regulate polyubiquitylation.
AUP1 Localizes to the ER and Lipid Droplets and Its Overexpression Leads to Accumulation of Lipid Droplets
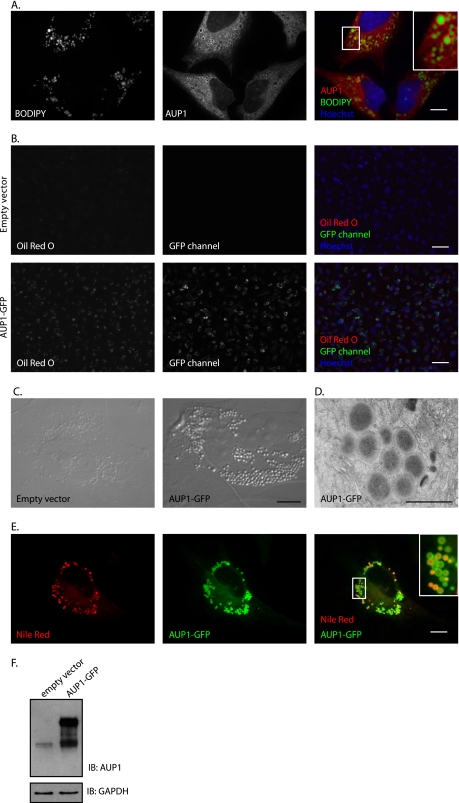
Recent proteomic studies have identified AUP1 as a component of lipid droplets (42–44). We confirmed this result for endogenous AUP1 in HeLa cells by microscopy. We performed anti-AUP1 immunofluorescence microscopy of HeLa cells fed oleic acid to induce lipid droplet formation. Lipid droplets were stained with the lipophilic dye BODIPY 493/503. AUP1 clearly localizes to the periphery of lipid droplets as well as to the ER (Fig. 5A). Similar results were found in A431, Huh7, Madin-Darby canine kidney, and COS7 cells (18).
FIGURE 5.
AUP1 localizes to lipid droplets, and its overexpression results in increased lipid droplets. A, HeLa cells were incubated with 250 μm oleic acid for 16 h. AUP1 was stained with purified anti-AUP1 antibody, and lipid droplets were stained with BODIPY 493/503. Cells were visualized by spinning disk confocal microscopy. Scale bar, 10 μm. B, HeLa cells were transfected with empty vector or AUP1-GFP. Cells were stained with Oil Red O and Hoechst to visualize lipid droplets and nuclei, respectively. Scale bars, 50 μm. C, HeLa cells were stably transduced with empty vector or AUP1-GFP as indicated and imaged by Nomarski imaging. Scale bar, 10 μm. D, US11-expressing astrocytoma cells transduced with AUP1-GFP were imaged by electron microscopy. Scale bar, 1 μm. E, HeLa cells stably transduced with AUP1-GFP were visualized by spinning disk confocal microscopy. Lipid droplets were stained with Nile Red dye. Scale bar, 10 μm. F, lysates of cells used in B were separated by SDS-PAGE and levels of AUP1 and AUP1-GFP were determined by immunoblotting with anti-AUP1 antibody. Immunoblotting (IB) for GAPDH served as a loading control.
Overexpression of several other known protein components of lipid droplets, such as adipose differentiation-related protein, induces the formation of lipid droplets (45). Similarly, cells stably expressing AUP1-GFP under the strong CMV promoter exhibit increased numbers of lipid droplets (46). The magnitude of increased lipid storage due to AUP1-GFP overexpression is readily apparent with Oil Red O stain for lipid droplets (Fig. 5B) and in bright field images (Fig. 5C). Electron microscopy of cells expressing AUP1-GFP produced images of dark electron-dense filled structures characteristic of lipid droplets (Fig. 5D). Confocal microscopy at high magnification with Nile Red co-stain for lipid droplets shows that the overexpressed AUP1-GFP localizes mostly to the lipid droplets as well as to the ER (Fig. 5E). AUP1-GFP expression level is shown in Fig. 5F.
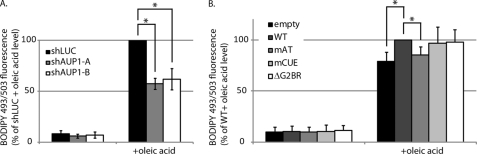
To see if AUP1 plays a role in lipid droplet formation, we looked at the ability of AUP1-depleted cells to form lipid droplets in response to oleic acid treatment. HeLa cells were transduced with the AUP1-targeting shRNA constructs and then either incubated with oleic acid for 16 h or left untreated. Lipid droplets were stained with BODIPY 493/503, and fluorescence intensity levels were determined by flow cytometry. The AUP1-depleted cells accumulated only ∼60% as much lipid droplet staining upon oleic acid treatment as did control cells (Fig. 6A). This effect matches those observed for TIP47-depleted cells (38), a bona fide lipid droplet component. The lipid droplets in oleic acid-fed AUP1-depleted cells were similar in size to those found in control cells (data not shown). Because AUP1 encodes a putative acyltransferase domain, we wondered whether this domain is important for lipid droplet formation. The level of lipid droplet staining of HeLa cells transfected with wild type or mutant acyltransferase AUP1 was determined by FACS (Fig. 6B). Oleic acid-fed cells with the wild type AUP1 construct exhibit higher levels of lipid droplet staining compared with control cells and those transfected with AUP1 containing a point mutation in the acyltransferase domain active site. It is noteworthy that the effect is seen in the presence of the endogenous AUP1 and that the other mutant constructs induce lipid droplet staining levels on par with wild type AUP1.
FIGURE 6.
AUP1 is involved in the accumulation of lipid droplets. A, HeLa cells were transduced with the shRNA constructs used in Fig. 1. Three days post-transduction, cells were left untreated or incubated with 0.4 mm oleic acid for 16 h. Lipid droplets were stained with BODIPY 493/503, and the amount of cellular fluorescence was determined by flow cytometry. Error bars represent standard deviation from three independent experiments. * indicates p < 0.05 by Student's t test. B, HeLa cells were transfected with empty vector or HA-AUP1 (WT or mutants, as indicated). One day post-transfection, cells were left untreated or incubated with 0.4 mm oleic acid for 16 h. Lipid droplets were stained with BODIPY 493/503, and the amount of cellular fluorescence was determined by flow cytometry. Error bars represent standard deviation from three independent experiments. * indicates p < 0.05 by Student's t test.
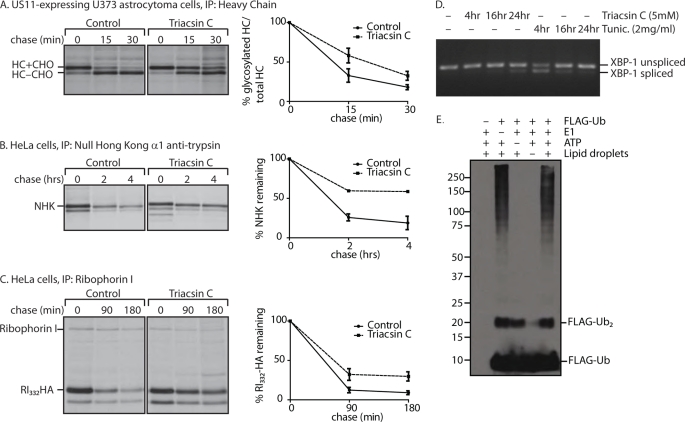
Pharmacological Inhibition of Lipid Droplet Formation Perturbs Dislocation
Upon finding that AUP1 plays a role in both lipid droplet formation and ER protein quality control, we wondered whether the two processes were functionally related. Inhibition of a subset of long chain acyl-CoA synthetases (ACSL1, -3, and -4) by triacsin C impairs lipid droplet formation (47, 48). In cells treated with this inhibitor, dislocation of MHC class I heavy chain in US11-expressing cells was slowed, as was the degradation of two soluble dislocation substrates, NHK and RI332-HA (Fig. 7, A–C). Triacsin C treatment does not disrupt the folding capacity of the ER because XBP-1 splicing, a read-out of unfolded protein response activation, was only slightly triggered after longer incubation times (Fig. 7D). The minor induction of the unfolded protein response may be a result of impaired dislocation and the gradual accumulation of misfolded proteins over time. As a comparison, tunicamycin, an inhibitor of N-linked glycosylation, causes significant XBP-1 splicing at much earlier time points.
FIGURE 7.
Pharmacological inhibition of lipid droplet formation affects dislocation. A,US11-expressing astrocytoma cells were treated with 5 μm triacsin C for 24 h. Cells were pulse-labeled with 35S-labeled cysteine and methionine. Samples were taken at the indicated chase times, and class I MHC heavy chain was recovered from the lysates. Immunoprecipitates (IP) were separated by SDS-PAGE and imaged by autoradiography. Amount of recovered protein was quantified by phosphorimagery and is shown as a percentage of glycosylated heavy chain compared with total heavy chain. Error bars represent standard deviation of three individual experiments. Solid line, control; dashed line, triacsin C-treated cells. B, HeLa cells were transfected with NHK, and the same experiment as in A was performed, using anti-α1-antitrypsin antibody for immunoprecipitation. C, HeLa cells were transfected with RI332-HA, and the same experiment as in A was performed, using anti-ribophorin I antibody for immunoprecipitation. Quantification in B and C shows the percentage of protein remaining compared with the amount recovered at the 0-min chase time. D, HeLa cells were treated with 5 μm triacsin or 2 μg/ml tunicamycin for the indicated times. RNA was purified and reverse-transcribed. XBP-1 cDNA was amplified by PCR, run on a 2% agarose gel, and visualized by UV. E, lipid droplets were isolated from oleic acid-fed 293T cells by flotation through a sucrose gradient. The lipid droplet fraction was added to an in vitro ubiquitylation assay with 60 μm FLAG-Ub, 100 nm E1, and ATP-regenerating mixture as indicated and incubated for an hour at 37 °C. Lipid droplets were solubilized in a 37 °C sonicating water bath for 2 h, run on a 8% Tris-Tricine gel, and immunoblotted with anti-FLAG antibody.
Lipid Droplets Are Ubiquitylation-proficient Organelles
Given that AUP1 is found on lipid droplets and in complex with UBE2G2 (Fig. 5) (18), it seemed plausible that ubiquitylation could occur on lipid droplets. Lipid droplets were isolated from oleic acid-fed 293T cells and added to in vitro ubiquitylation assays. This flotation method of fractionation largely separates lipid droplets from the cytoplasm and ER (data not shown). Ubiquitylation did occur in the lipid droplet fraction and did not require the addition of exogenous E1 enzyme (Fig. 7E). Absent a more stringent method to separate the ER completely from lipid droplets, we cannot exclude the contribution of contaminating ER to this result.
DISCUSSION
AUP1 contributes to the degradation of misfolded ER proteins and affects the intracellular abundance of lipid droplets. AUP1 localizes to both the ER and to the surface of lipid droplets. Reduced levels of AUP1 impair the ability of the cell to efficiently degrade the soluble terminally misfolded proteins RI332-HA and NHK. Cells that lack AUP1 form fewer lipid droplets, and overexpression of AUP1-GFP induces the hyper-accumulation of lipid droplets. AUP1 associates with most of the components of the HRD1 dislocation complex, as well as lipid-modifying proteins and the oligosaccharide transferase complex. The CUE domain and G2BR play important roles in ubiquitylation of dislocated proteins. These domains are provided to HRD1 in trans by AUP1 and to Hrd1p by Cue1p (27), whereas gp78, another E3 ubiquitin ligase, encodes these two domains itself (24).
Lipid Droplet Connection
One of the key differences between AUP1 and yeast Cue1p is the contribution of AUP1 to lipid droplet levels. No such role is apparent for Cue1p, which is not known to localize to lipid droplets. Furthermore, AUP1 interacts with lipid-modifying proteins and encodes a putative acyltransferase domain, which is absent from Cue1p. This putative acyltransferase activity of AUP1 may be directly involved in lipid droplet formation, as is suggested by the FACS data (Fig. 6B), or AUP1 may affect lipid droplet abundance by recruiting other lipid-modifying proteins, such as those identified by mass spectrometry (Table 1), to the site of lipid droplet formation. The dual role of AUP1 may indicate a deeper connection between ER protein quality control and lipid droplet formation. This idea is supported by the fact that pharmacological inhibition of lipid droplet formation affects dislocation. AUP1 may serve as the fulcrum responsible for linking and coordinating these two processes. This proposed connection may be specific to mammalian cells and absent from yeast, as manifest by the differences between AUP1 and Cue1p with regard to lipid droplets. Although our data are consistent with the possibility that the phenomena of lipid droplet formation and dislocation are linked via AUP1, we cannot exclude the alternative possibility, namely that the role of AUP1 in these two processes is mechanistically distinct.
How could lipid droplets be involved in ER protein quality control? In one model, the lipid rearrangements required to form lipid droplets may facilitate the movement of misfolded proteins from the ER to the cytoplasm (49). The incipient lipid droplets could coat the hydrophobic regions of the misfolded proteins, including any transmembrane domains. Alternatively, AUP1 may shuttle the cytoplasmic dislocated proteins from the ER membrane to lipid droplets en route to their proteasomal destruction. Lipid droplets have been proposed to be a storage depot for other aggregation-prone proteins (30). Likewise, misfolded ER proteins could temporarily be parked on lipid droplets when the load of dislocated proteins exceeds the capacity or local availability of proteasomes. Such is the case for a mammalian ER quality control substrate, HMG-CoA reductase, that is also found associated with lipid droplets post-dislocation (50).
Several connections between lipid droplets and the ubiquitin/proteasome system have been suggested in other studies. Components of the proteasome have been identified in lipid droplet preparations by mass spectrometry. The proteasome is responsible for the degradation of two major protein components of lipid droplets, adipose differentiation-related protein (51, 52) and perilipin (53). Cells treated with proteasome inhibitor resulted in the accumulation of ubiquitylated proteins in the lipid droplet-containing fraction (33). As we show here, lipid droplets sustain ubiquitylation in vitro. The E3 enzyme responsible for this ubiquitylation has not been identified, but it may be an E3 recruited to lipid droplets, such as AIP4 (54) or an ER resident E3, because lipid droplets are often found in close proximity to lipid droplets.
CUE Domain
Although the CUE domain was originally identified in Cue1p, the role of the CUE domain in ER protein quality control had not been determined. The function of the CUE domain is best understood for non-ER quality control systems, specifically yeast Vps9p, which binds ubiquitin, is monoubiquitylated, and regulates endocytosis. The CUE domain of Vps9p exhibits higher affinity for ubiquitin than does Cue1p. The CUE domain of AUP1 is predicted to also have a lower affinity for ubiquitin, based on the sequence of the ubiquitin-interaction motif. This raised a number of questions. Do these lower affinity CUE domains have a function in ubiquitylation? If so, what role do CUE domains play in ER protein quality control? The CUE domain of Cue1p was reported to be dispensable in ER protein quality control and even suggested to be vestigial (27). By contrast, we find that the CUE domain of AUP1 facilitates several important protein-protein interactions. The difference between these findings may indicate that Cue1p and AUP1 differ with regard to the importance of the CUE domain, or perhaps the ER quality control-associated CUE domain is responsible for fine-tuning ubiquitylation, possibly in a substrate-specific manner, which could have easily been overlooked in the yeast studies.
The CUE domain of AUP1 is crucial for binding ubiquitylation substrates (Fig. 3, B and C). The lack of association between AUP1mCUE and the terminally misfolded ER proteins may be due to the fact that AUP1mCUE is not efficiently incorporated into the HRD1 complex (Fig. 3A). Alternatively, AUP1 may interact directly with the dislocation substrates, possibly via ubiquitin modifications, which would account for the effect of the CUE domain mutations on the interaction of AUP1 with RI332-HA and NHK. Similarly, the CUE domain of gp78 is important for binding two of its substrates, CFTRΔF508 and Huntingtin (55, 56). Thus CUE domains may fulfill a conserved role of binding ubiquitylation substrates.
AUP1 is ubiquitylated, and the ubiquitin chain length is dependent on its CUE domain. Other proteins that contain a ubiquitin binding domain (such as Vps9p with a CUE domain and Eps15 and Hrs with ubiquitin-interacting motifs) are also ubiquitylated, but in those cases they undergo mono-ubiquitylation exclusively (20, 23, 57, 58). Both CUE domains and ubiquitin-interacting motifs bind ubiquitin in close proximity to its Lys-48 residue, and in this way are thought to block chain extension (21). When the CUE or ubiquitin-interacting motif domains of these proteins are deleted, the proteins are no longer ubiquitylated (22, 58). AUP1, by contrast, is both monoubiquitylated and polyubiquitylated in vivo when the CUE domain is intact but not when the CUE domain is mutated (Fig. 4, A and B). In vitro studies show that AUP1mCUE constructs can be ubiquitylated under optimal conditions and, in contrast to wild type AUP1, do not show a preference for monoubiquitylation (Fig. 4C). Could the affinity of the CUE domain for ubiquitin affect the resulting chain length? The wild type CUE domain of Vps9p exhibits the highest affinity and thus completely blocks the Lys-48 residue resulting in purely monoubiquitylation, although the wild type CUE domain of AUP1 binds ubiquitin less tightly resulting in mono- and polyubiquitylation, and the mutant CUE domain of AUP1 binds ubiquitin even more poorly and thus does not inhibit ubiquitin chain formation in any way resulting in polyubiquitylation. Thus AUP1 CUE domain-mediated binding of ubiquitin may serve a regulatory function to control polyubiquitin chain formation on AUP1 or the dislocated ER protein. Although we do not know the exact role of the ubiquitylation of AUP1, an attractive hypothesis is its possible involvement in forming ubiquitin chains that are then transferred en bloc to substrates via UBE2G2. The idea that a ubiquitin binding domain is involved in chain formation has also been hypothesized for E2–25K (59), an E2-conjugating enzyme that contains a UBA domain (60).
G2BR Domain
The G2BR domain we identify in AUP1 is similar to the U7 domain of Cue1p, and both are essential for recruitment of their cognate E2 (UBE2G2 for AUP1 and Ubc7p for Cue1p). The U7 domain of Cue1p enhances the activity of Ubc7p in vitro (26, 27). The G2BR enhances the affinity of UBE2G2 for the RING domain containing E3 gp78, and it enhances ubiquitylation mediated by gp78 and HRD1 in vitro (61). Our analysis of the G2BR indicates that AUP1 binding of UBE2G2 may actually negatively regulate the ubiquitylation of AUP1 and dislocated substrates, because AUP1 interacts with more ubiquitylated protein when the G2BR is deleted (Fig. 4B).
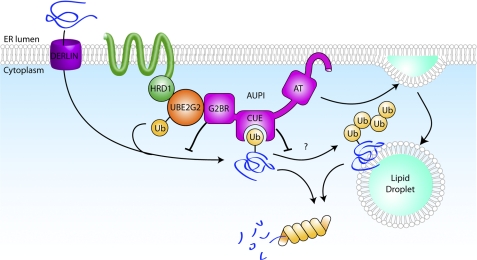
Model
A working model for the role of AUP1 places it in the HRD1-SEL1L complex (Fig. 8). AUP1 may need to either bind ubiquitylated proteins or be ubiquitylated itself before it can be incorporated into the dislocation complex. Both of these functions are dependent on an intact CUE domain. Once the entire complex is assembled, it can proceed to process misfolded ER proteins. Ubiquitin is transferred from E1 to UBE2G2, an E2 that is recruited to the site of dislocation by the G2BR of AUP1. HRD1, an E3, is allosterically activated by the G2BR and catalyzes the transfer of ubiquitin from UBE2G2 to misfolded ER proteins. The G2BR limits the transfer of ubiquitin and thus the interaction of AUP1 with ubiquitylated proteins. This negative regulation may serve as a feedback mechanism to exert control over degradation rates or to serve as an additional checkpoint in quality control. AUP1 then binds the dislocated ER proteins via interactions between its CUE domain and the ubiquitin moiety covalently attached to dislocated ER proteins. This CUE domain-ubiquitin interaction also regulates ubiquitin chain extension. Finally, the dislocated substrates are delivered to the proteasome where they are degraded, but they may first be stored on lipid droplets under certain conditions. Thus, AUP1 is involved in several steps of ER protein quality control that include ubiquitylation and processing.
FIGURE 8.
Model. A possible model for the role of AUP1 in processing of misfolded ER proteins is presented. AUP1 recruits UBE2G2, the E2 that works with HRD1 to ubiquitylate dislocated ER proteins. AUP1 binds the misfolded ER protein and regulates the ubiquitylation and polyubiquitylation of dislocation substrates via its G2BR and CUE domain, respectively. AUP1 is also involved in the formation of lipid droplets, a cytoplasmic organelle that may serve as a temporary storage location for misfolded ER proteins. Further explanation can be found under “Discussion.”
Acknowledgments
We thank Britta Mueller, Sumana Sanyal, Max Popp, Carla Guimaraes, and Annemarthe van der Veen for valuable discussions; Nicki Watson for performing the electron microscopy experiment; Tom DiCesare for help with graphics, and N. Erwin Ivessa and Reid Gilmore for antibodies.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ER
- endoplasmic reticulum
- NHK
- Null Hong Kong variant of α1 anti-trypsin
- CUE
- coupling of ubiquitin conjugation to endoplasmic reticulum degradation
- G2BR
- UBE2G2 binding region
- HC
- heavy chain
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Hebert D. N., Bernasconi R., Molinari M. (2010) Semin. Cell Dev. Biol. 21, 526–532 [DOI] [PubMed] [Google Scholar]
- 2. Carvalho P., Goder V., Rapoport T. A. (2006) Cell 126, 361–373 [DOI] [PubMed] [Google Scholar]
- 3. Denic V., Quan E. M., Weissman J. S. (2006) Cell 126, 349–359 [DOI] [PubMed] [Google Scholar]
- 4. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. (2008) Nat. Cell Biol. 10, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mueller B., Lilley B. N., Ploegh H. L. (2006) J. Cell Biol. 175, 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernasconi R., Pertel T., Luban J., Molinari M. (2008) J. Biol. Chem. 283, 16446–16454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Scott D. C., Schekman R. (2008) J. Cell Biol. 181, 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lilley B. N., Ploegh H. L. (2004) Nature 429, 834–840 [DOI] [PubMed] [Google Scholar]
- 10. Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 11. Carvalho P., Stanley A. M., Rapoport T. A. (2010) Cell 143, 579–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 13. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mueller B., Klemm E. J., Spooner E., Claessen J. H., Ploegh H. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12325–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L. (1996) Cell 84, 769–779 [DOI] [PubMed] [Google Scholar]
- 16. Kato A., Oshimi K. (2009) Platelets 20, 105–110 [DOI] [PubMed] [Google Scholar]
- 17. Kato A., Kawamata N., Tamayose K., Egashira M., Miura R., Fujimura T., Murayama K., Oshimi K. (2002) J. Biol. Chem. 277, 28934–28941 [DOI] [PubMed] [Google Scholar]
- 18. Spandl J., Lohmann D., Kuerschner L., Moessinger C., Thiele C. (2011) J. Biol. Chem. 286, 5599–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heath R. J., Rock C. O. (1998) J. Bacteriol. 180, 1425–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prag G., Misra S., Jones E. A., Ghirlando R., Davies B. A., Horazdovsky B. F., Hurley J. H. (2003) Cell 113, 609–620 [DOI] [PubMed] [Google Scholar]
- 21. Kang R. S., Daniels C. M., Francis S. A., Shih S. C., Salerno W. J., Hicke L., Radhakrishnan I. (2003) Cell 113, 621–630 [DOI] [PubMed] [Google Scholar]
- 22. Davies B. A., Topp J. D., Sfeir A. J., Katzmann D. J., Carney D. S., Tall G. G., Friedberg A. S., Deng L., Chen Z., Horazdovsky B. F. (2003) J. Biol. Chem. 278, 19826–19833 [DOI] [PubMed] [Google Scholar]
- 23. Shih S. C., Prag G., Francis S. A., Sutanto M. A., Hurley J. H., Hicke L. (2003) EMBO J. 22, 1273–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen B., Mariano J., Tsai Y. C., Chan A. H., Cohen M., Weissman A. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Biederer T., Volkwein C., Sommer T. (1997) Science 278, 1806–1809 [DOI] [PubMed] [Google Scholar]
- 26. Bazirgan O. A., Hampton R. Y. (2008) J. Biol. Chem. 283, 12797–12810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kostova Z., Mariano J., Scholz S., Koenig C., Weissman A. M. (2009) J. Cell Sci. 122, 1374–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Farese R. V., Jr., Walther T. C. (2009) Cell 139, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin S., Parton R. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 373–378 [DOI] [PubMed] [Google Scholar]
- 30. Cermelli S., Guo Y., Gross S. P., Welte M. A. (2006) Curr. Biol. 16, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 31. Miyanari Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., Shimotohno K. (2007) Nat. Cell Biol. 9, 1089–1097 [DOI] [PubMed] [Google Scholar]
- 32. Cocchiaro J. L., Kumar Y., Fischer E. R., Hackstadt T., Valdivia R. H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ohsaki Y., Cheng J., Fujita A., Tokumoto T., Fujimoto T. (2006) Mol. Biol. Cell 17, 2674–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo Y., Walther T. C., Rao M., Stuurman N., Goshima G., Terayama K., Wong J. S., Vale R. D., Walter P., Farese R. V. (2008) Nature 453, 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lilley B. N., Ploegh H. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 14296–14301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brasaemle D. L., Wolins N. E. (2006) Curr. Protoc. Cell Biol. Chapter 3, Unit 3.15 [DOI] [PubMed] [Google Scholar]
- 37. Vyas J. M., Kim Y. M., Artavanis-Tsakonas K., Love J. C., Van der Veen A. G., Ploegh H. L. (2007) J. Immunol. 178, 7199–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bulankina A. V., Deggerich A., Wenzel D., Mutenda K., Wittmann J. G., Rudolph M. G., Burger K. N., Höning S. (2009) J. Cell Biol. 185, 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsao Y. S., Ivessa N. E., Adesnik M., Sabatini D. D., Kreibich G. (1992) J. Cell Biol. 116, 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oda Y., Okada T., Yoshida H., Kaufman R. J., Nagata K., Mori K. (2006) J. Cell Biol. 172, 383–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sato S., Fukasawa M., Yamakawa Y., Natsume T., Suzuki T., Shoji I., Aizaki H., Miyamura T., Nishijima M. (2006) J. Biochem. 139, 921–930 [DOI] [PubMed] [Google Scholar]
- 43. Wan H. C., Melo R. C., Jin Z., Dvorak A. M., Weller P. F. (2007) FASEB J. 21, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brasaemle D. L., Dolios G., Shapiro L., Wang R. (2004) J. Biol. Chem. 279, 46835–46842 [DOI] [PubMed] [Google Scholar]
- 45. Imamura M., Inoguchi T., Ikuyama S., Taniguchi S., Kobayashi K., Nakashima N., Nawata H. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E775–E783 [DOI] [PubMed] [Google Scholar]
- 46. Spandl J., White D. J., Peychl J., Thiele C. (2009) Traffic 10, 1579–1584 [DOI] [PubMed] [Google Scholar]
- 47. Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., Takano T. (2007) J. Lipid Res. 48, 1280–1292 [DOI] [PubMed] [Google Scholar]
- 48. Igal R. A., Wang P., Coleman R. A. (1997) Biochem. J. 324, 529–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ploegh H. L. (2007) Nature 448, 435–438 [DOI] [PubMed] [Google Scholar]
- 50. Hartman I. Z., Liu P., Zehmer J. K., Luby-Phelps K., Jo Y., Anderson R. G., DeBose-Boyd R. A. (2010) J. Biol. Chem. 285, 19288–19298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., Sasabe N., Aoki J., Arai H., Takano T. (2006) J. Lipid Res. 47, 87–98 [DOI] [PubMed] [Google Scholar]
- 52. Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., Londos C. (2005) J. Biol. Chem. 280, 42841–42847 [DOI] [PubMed] [Google Scholar]
- 53. Xu G., Sztalryd C., Londos C. (2006) Biochim. Biophys Acta 1761, 83–90 [DOI] [PubMed] [Google Scholar]
- 54. Edwards T. L., Clowes V. E., Tsang H. T., Connell J. W., Sanderson C. M., Luzio J. P., Reid E. (2009) Biochem. J. 423, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang H., Liu C., Zhong Y., Luo S., Monteiro M. J., Fang S. (2010) PloS one 5, e8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Klapisz E., Sorokina I., Lemeer S., Pijnenburg M., Verkleij A. J., van Bergen en Henegouwen P. M. (2002) J. Biol. Chem. 277, 30746–30753 [DOI] [PubMed] [Google Scholar]
- 58. Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 59. Hochstrasser M. (2006) Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 60. Pichler A., Knipscheer P., Oberhofer E., van Dijk W. J., Körner R., Olsen J. V., Jentsch S., Melchior F., Sixma T. K. (2005) Nat. Struct. Mol. Biol. 12, 264–269 [DOI] [PubMed] [Google Scholar]
- 61. Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]