Abstract
The p53 protein is a primary mediator of cellular apoptosis and growth arrest after exposure to DNA-damaging agents. Previous work has shown that the majority of childhood acute lymphoblastic leukemia (ALL) cases express a wild type p53 gene, although the functionality of the p53 pathway has rarely been validated. In the present study, the integrity of the p53 pathway was investigated in a panel of ALL cell lines and xenografts established from direct patient explants in immune-deficient mice. A focused real-time quantitative reverse transcription PCR array of known p53-regulated genes identified p21WAF1 (CDKN1A) as the highest ranked gene to be differentially expressed between B-cell precursor (BCP)-ALL and T-ALL xenografts following exposure to the DNA-damaging drug etoposide. Lack of p21WAF1 induction was observed in six of seven T-ALL xenograft lines, as well as primary T-ALL cells following irradiation exposure, despite an otherwise functional p53 response. Repression of p21WAF1 in T-ALL cells was associated with decreased acetylated H3K9 localized at its promoter compared with BCP-ALL cells, together with increased CpG methylation within the first exon and intron. Although the histone deacetylase inhibitor vorinostat failed to induce p21WAF1 in T-ALL samples, the combination of vorinostat and the demethylating agent decitabine reactivated expression of the silenced p21WAF1 gene in the Molt-4 T-ALL cell line. Considering the known anti-apoptotic function of p21WAF1, our findings have significant implications for the responses of T- versus BCP-ALL cells to chemotherapeutic drugs that induce p21WAF1.
Keywords: DNA Damage, DNA Methylation, Epigenetics, Histone Modification, p53, Acute Lymphoblastic Leukemia, Decitabine, Etoposide, p21WAF1, Vorinostat
Introduction
Combination chemotherapy is highly effective in the treatment of childhood acute lymphoblastic leukemia (ALL)2 (1), and the efficacy of chemotherapeutic drugs is thought to be dependent on their ability to induce apoptosis in leukemia cells (2). The p53 protein and its signaling pathway play a central role in modulating the cellular response to DNA-damaging drug-induced apoptosis (3) or cell cycle arrest (4). Wild type p53 can activate an apoptotic response through a transcription-dependent or -independent mechanism (5). The ability of p53 to induce cell cycle arrest depends on the transcriptional activation of a number of target genes, including p21WAF1 (CDKN1A) (6).
A high proportion of cancers express a mutated or deleted form of the p53 gene, resulting in an impaired response to chemotherapy agents (7). Such p53 gene mutations have rarely been described in primary childhood ALL cells, with the overall incidence at diagnosis reported to be 1–2% (8), increasing to 19% in B-lineage and 33% of T-lineage relapse cases (9). However as an alternative to p53 mutations, it has been suggested that inactivation of wild type p53 in leukemia cells may occur through binding to its principal cellular regulator human homolog of mouse double minute 2 (Hdm-2) (10). Although defective p53 function itself cannot entirely account for the prevalence of faulty responses to chemotherapy, deficiencies in other genes within the p53 pathway may be involved in impaired apoptotic responses leading to the emergence of chemotherapy resistance.
The cyclin-dependent kinase inhibitor, p21WAF1, is a well characterized modulator of p53-induced cell cycle arrest after DNA damage (6) and is recognized as an important tumor suppressor gene. Defective p21WAF1 activation has been implicated in leukemogenesis (11), which appears to conflict with its apparent role in cell death inhibition. Reports have demonstrated that p21WAF1 expression is sufficient to protect cells from apoptotic stimuli (12, 13), whereas suppression of p21WAF1 by extrinsic means enhances sensitivity to drug-induced cell death (14–17). An absence of p21WAF1 expression has been noted in T-ALL cells (18), whereas high levels of p21WAF1 in acute myeloid leukemia cells were associated with a poor prognosis (19). Although inactivation of p21WAF1 mRNA could also be mediated by the miR-106b family (20), other epigenetic processes could likewise play a role in transcriptional inactivation of p21WAF1, as mutations in the p21WAF1 gene are rare in human malignancies (21) .
Epigenetic gene regulation is characterized by methylation of CpG dinucleotides and remodeling of the chromatin structure by post-translational histone modifications. Methylation of CpG islands in promoter regions is strongly associated with gene silencing (22), whereas histone deacetylation leads to transcriptionally inert chromatin (23). DNA hypermethylation of the p21WAF1 gene has been reported in p21WAF1-silenced cells, including at the distal promoter in rhabdomyosarcoma cells (24) and across the Sp1/Sp3 binding sites in ALL primary samples (25), T-ALL cell lines, adult T-cell leukemia/lymphoma cells (26), and lung cancer cells (27). CpG methylation across exon 1/intron 1 of the rat p21WAF1 gene also has been reported (28). Hemizygous CpG methylation has been described in ALL cell lines (29, 30). However in contrast to these reports, CpG methylation was not detectable in a range of neoplastic cells, including ALL patient blasts where p21WAF1 mRNA disruption was evident (29–33).
This report characterizes p53-induced signaling pathways in leukemia cell lines, patient-derived ALL xenografts and primary blasts by assessing their response to the DNA-damaging agents, etoposide and irradiation, and the Hdm-2 small molecule inhibitor, nutlin-3. Although p53 transcriptional activity was intact, we reveal a lineage-dependent epigenetic repression of the p21WAF1 gene in T-ALL, but not B-cell precursor (BCP)-ALL, cells that could be reversed by simultaneous exposure of cells to the histone deacetylase inhibitor vorinostat and the demethylating agent decitabine. Our study provides novel observations on the differential lineage-dependent regulation of p21WAF1 expression in leukemia cells.
EXPERIMENTAL PROCEDURES
Patient Clinical Samples
Leukemia cells were obtained from the Centre for Children's Cancer and Blood Disorders at Sydney Children's Hospital from children presenting with ALL at initial diagnosis or at bone marrow relapse who were enrolled in the Australia and New Zealand Children's Cancer Study Group Study VII (1998–2001) or Study VIII (2002 to present), or from the Royal Victoria Infirmary, Newcastle upon Tyne, UK (from June 2002 to June 2009). The protocol for harvesting and storage of the specimens is detailed in previous work (34). Bone marrow mononuclear cells were isolated by density gradient centrifugation using Lymphoprep (Axis-Shield, Oslo, Norway). Isolated leukemic blasts were cryopreserved in 10% dimethyl sulfoxide (DMSO) (v/v) in FBS (Thermo Trace, Victoria, Australia). Thawed patient blasts were resuspended and seeded at 5 × 105/ml in RPMI 1640 medium containing glutamine (both from Invitrogen) and 10% FBS and exposed to irradiation or MG132 (Sigma-Aldrich) dissolved in DMSO (Sigma-Aldrich). The use of patient-derived leukemia cells was approved by regional ethical committee approval for Australian and UK samples. Minimal residual disease in BCP-ALL patients, 28 days after start of treatment, was measured as described previously (35).
In Vitro Culture and Cytotoxicity Assays of Cell Lines and Xenografted Patient Cells
BCP-ALL (PreB-697, Reh, Nalm-6), BCP lymphoblastoid (TK6), or T-ALL (CCRF-CEM (CEM), Jurkat, Molt-4) cell lines were obtained either from the ATCC (Manassas, VA) or the European Collection of Cell Cultures (Salisbury, UK) and were maintained in RPMI 1640 medium supplemented with 10% FBS and penicillin (100 units/ml), streptomycin (100 μg/ml), and l-glutamine (2 mm) supplied from Invitrogen. Xenografts were previously established from direct patient explants (36) and are representative of BCP-ALL and T-ALL derived from patients with diverse treatment outcomes (see supplemental Table S1). Xenograft cells were retrieved from cryostorage and prepared for in vitro culture as described previously (37).
For in vitro drug exposure experiments, cells were equilibrated in a humidified atmosphere overnight at 37 °C, 5% CO2, prior to the addition of etoposide (Sigma-Aldrich), vorinostat (kindly provided by Merck, Sharp and Dohme Corp. (Rahway, NJ) and the National Cancer Institute (Bethesda, MD)), nutlin-3 or decitabine (Sigma-Aldrich), or an equivalent volume of DMSO. In vitro sensitivity was assessed and viability calculated after 48 h exposure to nutlin-3 using the colorimetric methyl-thiazolyl-tetrazolium (Sigma-Aldrich) assay, as described previously (37).
Analysis of mRNA Expression by Real-time PCR and RT-qPCR Array
RNA extraction and real-time quantitative reverse transcription PCR (RT-qPCR) of gene expression in cells exposed to etoposide or vorinostat was carried out as described previously (37). Primers and probes for p21WAF1 (Hs00355782_m1) were purchased from Applied Biosystems (Foster City, CA). Elongation factor-1α (EF-1α) was used as an internal standard in each reaction (primers EF-1αF, 5′-CTGAACCATCCAGGCCAAAT-3′; EF-1αR, 5′-GCCGTGTGGCAATCCAAT-3′; probe, 5′-VIC-AGCGCCGGCTATGCCCCTG-TAMRA-3′). RNA from irradiated primary ALL cells and cell lines was extracted using the Qiagen RNeasy Mini kit (Qiagen, Crawley, UK), and cDNA synthesis was carried out using the Applied Biosystem High-Capacity cDNA Reverse Transcriptase kit. Primers and probes for p21WAF1 (Hs00355782_m1) and β-actin (VIC-TAMARA, endogenous control, catalog no. 4310881E) were purchased from Applied Biosystems. Triplicate samples were assayed with the ABI 7500 or 7700 Fast Real-time PCR System (Applied Biosystems) using the TaqMan® Universal PCR MasterMix (Applied Biosystems), and RT-qPCR was carried out according to the manufacturer's instructions. The p21WAF1 mRNA levels of irradiated cells were normalized to an endogenous gene and expressed relative to p21WAF1 mRNA expression in a control cell line. Specific details can be found in the figure legends.
For RT-qPCR arrays, total RNA was isolated as described in the manufacturer's protocol for the RT2-qPCR RNA isolation kit (SABiosciences, SuperArray, Frederick, MD) from xenograft cells that had been exposed to etoposide (5 μm for 3 or 12 h). The quality and concentration of extracted RNA was evaluated with a Nanodrop ND-1000 spectrophotometer (Thermo Scientific) and a Bioanalyzer 2100 (Agilent Technologies) according to the manufacturer's instructions. cDNA was synthesized using the RT2 Profiler PCR array first strand kit (SABiosciences) following the manufacturer's recommendations. cDNA samples were loaded into individual wells of a Human p53 Signaling Pathway RT-qPCR array (SABiosciences). Samples were subsequently assayed on an ABI 7700 Fast Real-time PCR System (Applied Biosystems) and analyzed according to the manufacturer's protocol. The acquired data were log-transformed using log10(x), where x is the sample ΔΔCt value. Log-transformed data from individual arrays were analyzed using the GenePattern limma module to assess the degree of differential gene expression comparing BCP-ALL with T-ALL (38). Results were visualized on a heat map and genes are shown in descending significance of induction in BCP-ALL relative to T-ALL.
Analysis of Protein Expression
Whole cell lysates were prepared from xenograft cells and leukemia cell lines as described previously (37). Equal amounts of protein (25 μg) were separated by NuPAGE gel electrophoresis and electro-transferred to PVDF membranes according to the manufacturer's instructions (Invitrogen). Membranes were probed with antibodies for p21WAF1 (BD Transduction Laboratories, San Diego, CA), wild type p53 (Santa Cruz Biotechnology, Santa Cruz, CA), and actin (Sigma-Aldrich), followed by HRP-conjugated donkey anti-rabbit or sheep anti-mouse secondary antibody (GE Healthcare). Bound secondary antibodies were detected by chemiluminescence and visualized by autoradiography detection and phosphorimaging using a VersaDoc 5000 Imaging System (Bio-Rad). Data were analyzed using QuantityOne software (version 4.00; Bio-Rad). For irradiated primary ALL cells and cell lines, lysates were separated on 4–20% Tris-glycine gels (Invitrogen) and electro-transferred onto a PVDF membrane. Immunoblots were probed for p21WAF1 (clone SX118BD or 70; BD Biosciences), p53 (Clone D0–7, Novacastra, Newcastle upon Tyne, UK), tubulin (Sigma-Aldrich), or actin (Clone JLA20, Calbiochem, Nottingham, UK) and detected with HRP-conjugated secondary antibodies (Dako, Ely, UK). The densitometry of scanned immunoblots was carried out using AIDA image analysis software (Straubenhardt, Germany).
ChIP Assays
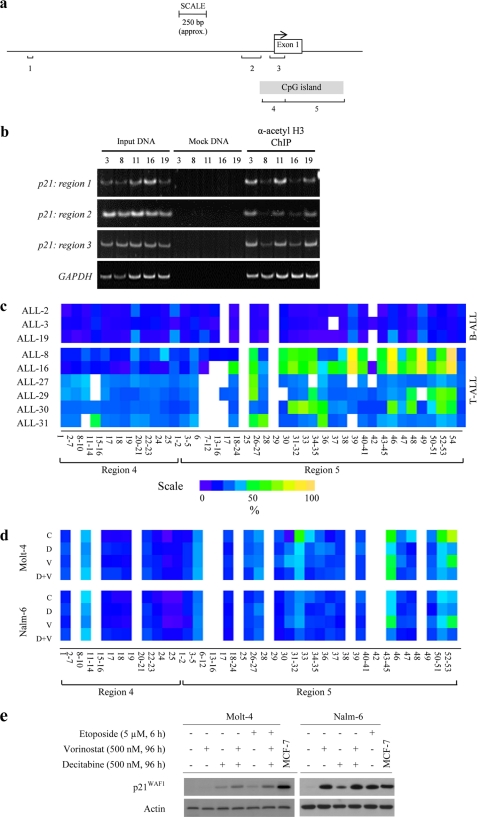
ChIP analysis of histone acetylation was conducted as described previously (39), using immunoglobulin raised against acetyl-histone 3 (Lys-9/14(H3K9)) (Millipore, Billerica, MA), and processed according to the manufacturer's instructions. DNA from protein-associated complexes and corresponding input samples was recovered using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and assayed by SYBR-Green qPCR under standard conditions (supplemental Table S2). The fold enrichment of samples was expressed relative to mock-treated control and normalized to acetylated H3K9 at the GAPDH locus in each PCR sample (40). PCR products were separated by PAGE, gels were stained with SYBR Green and imaged using a VersaDoc 5000 Imaging System (Bio-Rad).
SEQUENOM Cytosine Methylation Analysis
Quantitative high-throughput methylation analysis of CpG sites in the p21WAF1 gene was carried out using the MassARRAY® EpityperTM system (Sequenom, San Diego, CA) as described previously (41). Briefly, genomic DNA was isolated from leukemia cells as described previously (39), and these samples were sent to Australian Genome Research Facility (University of Queensland, Brisbane, Australia) for sample preparation and analysis. Two bisulfite reactions (regions 4 and 5) were designed, which covered all CpGs across the island. The primer sequences are shown in supplemental Table S2. After determining CpG methylation by MALDI-TOF MS, data were quantified by Quantitative Methylation Analysis software (Sequenom), and methylation ratios were plotted from the mean value of three replicate amplicons using Heatmap.2 in R software.
Statistical Comparison and Data Analysis
Unless otherwise stated, all data were compiled and analyzed using GraphPad Prism software (version 4.00). The Mann-Whitney U test and the Student's t test were utilized to determine whether differences between non-normally and normally distributed data, respectively, were significant. The significance level was set at p < 0.05.
RESULTS
Attenuated p21WAF1 Response in T-ALL Cells with Functional p53 Activity
Previous sequence analysis has identified no mutations compared with published wild type sequences of the entire p53 coding region (exons 2 to 11) for ALL-2, -3, -7, -8, -11, -16, and -19 xenografts (34). Cell lines were verified against the p53 coding regions listed on the International Agency for Research in Cancer TP53 database and the COSMIC database. The Jurkat, CEM, and Reh cell lines are reported to contain mutations in their coding region, whereas different sources of the Molt-4 cell line has been identified as mutant and wild type (42, 43). The Nalm-6 and TK6 line were listed as wild type, with no record of the PreB-697 cell line on either databases.
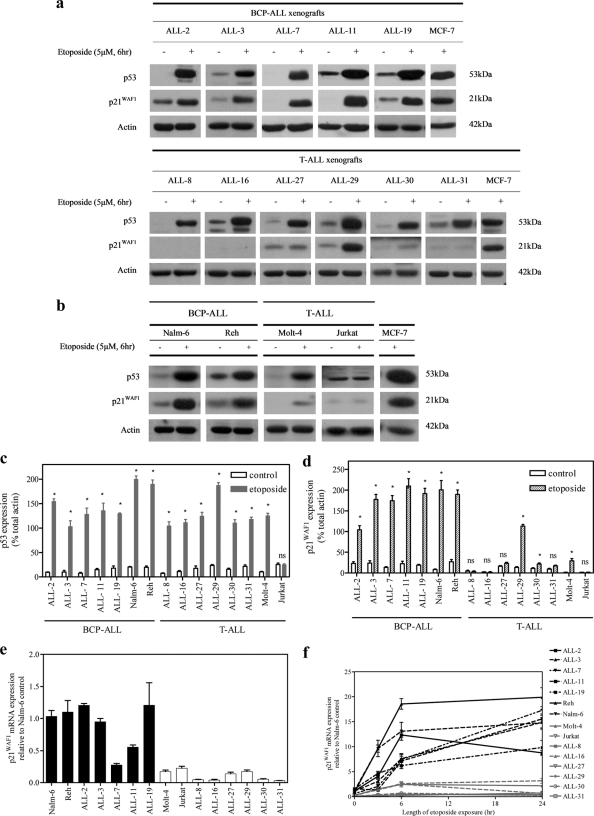
To evaluate the functionality of the p53 pathway in ALL cell lines and in vitro-cultured xenograft cells, p53 and p21WAF1 protein levels were assessed by immunoblotting after exposure to etoposide. Treatment with etoposide (5 μm, 6 h) induced an increase in p53 protein expression in the Molt-4, Reh, and Nalm-6 cell lines, as well as the panel of 11 ALL xenografts (Fig. 1, a–c). In contrast, there was no change in p53 expression in the Jurkat cell line, reflecting an abnormal response to DNA damage. Induction of p21WAF1 protein in response to etoposide was observed in BCP-ALL xenografts and cell lines (Fig. 1, a, upper panel, b, and d), whereas only negligible p21WAF1 levels were detected in the T-ALL Jurkat and Molt-4 cell lines (Fig. 1, b and d). Although induction of p53 occurred in response to etoposide in all T-ALL xenografts (Fig. 1, a, lower panel, and c), notable induction of p21WAF1 occurred in only one of six T-ALL xenograft samples (ALL-29, Fig. 1, a, bottom panel, and d). The basal level of p21WAF1 mRNA was also lower in T-ALL compared with BCP-ALL cells (Fig. 1e). Expression of p21WAF1 mRNA was also measured up to 24 h after etoposide exposure (5 μm). Only a minimal response in p21WAF1 mRNA induction was observed in T-ALL samples (Fig. 1f).
FIGURE 1.
Defective induction of p21WAF1 but not p53 in ALL cell lines and xenografts after etoposide exposure. Cells were exposed to etoposide (5 μm, 6 h), and expression of p53 and p21WAF1 proteins (a–d) or p21WAF1 mRNA (e and f) were assessed by immunoblotting or RT-qPCR, respectively. a, BCP-ALL (upper panel) or T-ALL (lower panel) xenograft cells. b, BCP-ALL and T-ALL cell lines. p53 (c) and p21WAF1 (d) protein levels were expressed relative to actin. Data from individual membranes were normalized to the MCF-7 control lane. e and f, p21WAF1 mRNA expression to the corresponding EF-1α sample and values expressed as fold differences relative to p21WAF1 expression in the Nalm-6 cell line. e, basal levels of expression. f, changes in expression in response to etoposide treatment. Results are expressed as the mean ± S.E. of at least three separate experiments. An asterisk represents values significantly greater than control (p < 0.05) as determined by Mann-Whitney U test; ns, not significant.
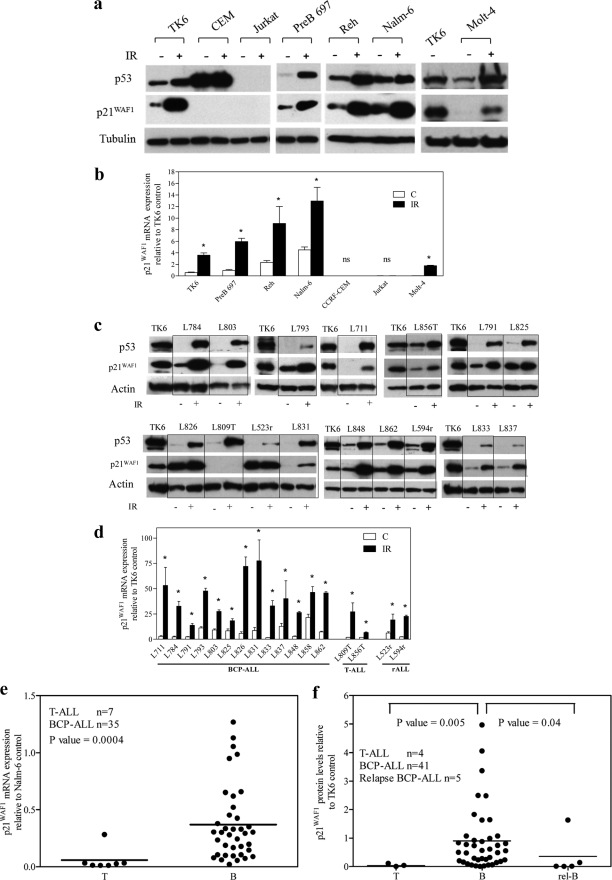
The capacity of the ALL cells to induce p53 and p21 in response to irradiation (6 h, 3.5 cGy) was also assessed. Irradiation-induced up-regulation of p53 and p21WAF1 mRNA and protein reflected those seen with etoposide, with robust induction in the BCP-ALL cell lines Reh and Nalm-6, very low level induction in Molt-4, and no apparent induction in Jurkat (Fig. 2a & b). Consistent with the BCP-ALL panel, an additional BCP-ALL cell line, Pre B-697 and the BCP lymphoblastoid cell line, TK6, showed induction of both p53 and p21WAF1 mRNA and protein to irradiation. In contrast, the p53 mutant T-ALL cell line, CEM, failed to induce p53 or p21WAF1. Irradiation induced p53 protein and p21WAF1 mRNA and protein in BCP-ALL and T-ALL primary patient leukemic blasts, with the exception of one of the two T-ALL samples (L809T) (Fig. 2, c and d). In case L809T p21WAF1 protein was not induced despite p21WAF1 mRNA induction (Fig. 2c and supplemental Fig. S1, a and b). However, concurrent exposure of this T-ALL sample to the proteasomal inhibitor MG132 (5 μm) and 3.5 cGy for 6 h resulted in p21WAF1 protein induction (supplemental Fig. S1c). To confirm that irradiation-induced apoptosis was not the reason for p21WAF1 protein degradation in L809T, the viability of primary cells after irradiation exposure was confirmed by annexin V/ethidium bromide staining (supplemental Table S3). The results demonstrate that primary cells L711, L803, and L856, with similar or even greater levels of apoptosis as L809T 6 h post-irradiation, induced p21WAF1 protein, suggesting that loss of p21WAF1 protein induction in L809T was not due to increased apoptosis in these cells.
FIGURE 2.
Defective induction after irradiation exposure and reduced basal expression of p21 WAF1 protein and mRNA in ALL cell lines and primary cells. ALL cell lines (a and b) or primary cells (c and d) were irradiated (IR) (3.5 cGy), and 6 h later expression of p53 or p21WAF1 proteins (a and c) and mRNA (b and d) were assessed by immunoblot or RT-qPCR, respectively. a and c, a representative blot is shown from at least three independent experiments. c and d, primary T-ALL samples are suffixed with T and bone marrow relapse lysates suffixed with r. b and d, p21WAF1 mRNA expression was normalized to actin and expressed relative to p21WAF1 expression in the untreated TK6 cell line. Results represent the mean ± S.E. of three independent measurements. An asterisk represents a value significantly greater than control (C) (p < 0.05) as determined by paired t test, on log transformed data to equalize variance; ns, not significant. e, expression of p21WAF1 mRNA in primary ALL samples was analyzed by RT-qPCR and normalized to the corresponding EF-1α sample, and values were expressed as fold differences relative to p21WAF1 expression in Nalm-6 cells. f, p21WAF1 protein expression was assessed by immunoblot and normalized to TK6 control cells. In e and f, each symbol represents the mean expression of a primary sample calculated from three individual experiments. Differences between groups were compared using the Mann-Whitney U test. Bars represent the median.
To examine whether low basal levels of p21WAF1 expression in T-ALL xenografts and cell lines observed above was an artifact of xenografting or establishing cell lines, mRNA was analyzed in independent (Australian) primary ALL samples by RT-qPCR. Basal p21WAF1 mRNA levels were significantly lower in T-ALL samples compared with BCP-ALL samples (p = 0.0004; Fig. 2e). Consistent with this finding, measurement of basal levels of p21WAF1 protein by immunoblotting in a separate cohort of ALL patients also showed significantly lower levels in four presentation T-ALL samples compared with 41 presentation and five relapsed BCP-ALL samples (p = 0.005 and p = 0.04 respectively, Fig. 2f and supplemental Fig. S2a). These results indicate that in comparison to BCP-ALL samples, p21WAF1 expression is low in T-ALL primary samples, xenografts, and cell lines. Basal p21WAF1 protein levels in 33 primary BCP-ALL, as assessed by immunoblotting (supplemental Fig. S2a), showed no correlation with presenting white blood cell counts (p = 0.69, supplemental Fig. S2b) or blast count at day 8 of induction therapy (p = 0.94, supplemental Fig. S2c).
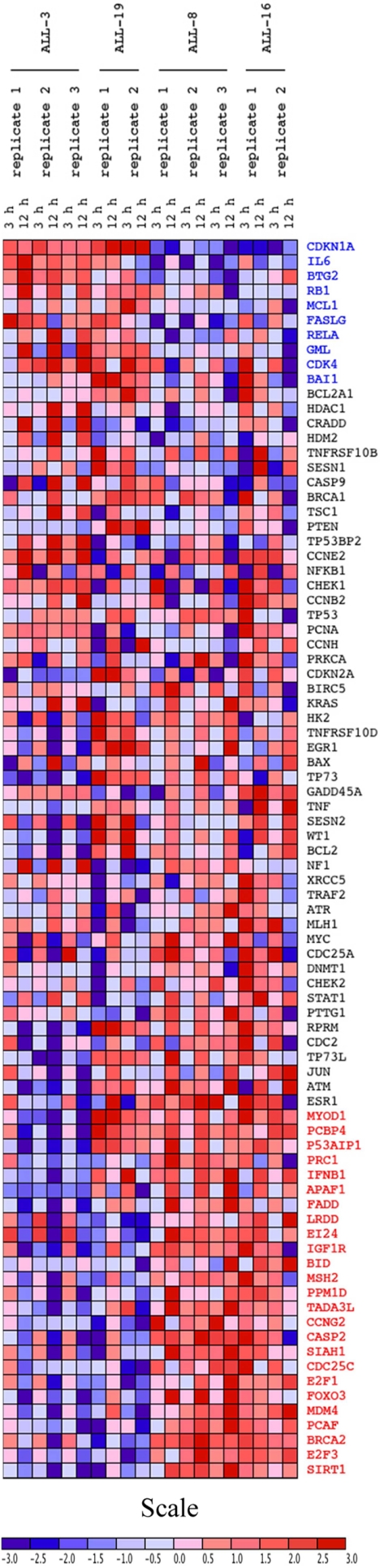
To assess the functionality of the p53 pathway in BCP or T-ALL xenografts derived from patients with diverse treatment outcomes (supplemental Table S1), two BCP-ALL (ALL-3, -19) and two T-ALL (ALL-8, -16) xenografts were exposed in vitro to etoposide and expression of 84 p53-regulated genes assessed relative to control at 3 and 12 h by focused RT-qPCR array. Consistent with an intact p53 response, p53-regulated genes such as Bax, GADD45α, and Hdm-2 were induced for at least one time point for all samples (Fig. 3 and supplemental Table S4). In contrast, the T-ALL samples failed to show p21WAF1 (CDKN1A) induction despite robust induction in ALL-3 and ALL-19. The data were analyzed using the GenePattern limma module in an unbiased fashion to compare differential gene expression in BCP-ALL with T-ALL xenografts (supplemental Table S4). Of the 84 p53-target genes, 35 genes showed significant differential expression between the lineages, with 10 significantly up-regulated genes in BCP-ALL samples and 25 significantly up-regulated genes in the T-ALL samples. The significantly up-regulated genes in the BCP-ALL samples have functional roles in the negative regulation of cell cycle proliferation (n = 3; BTG2, BAI, IL6), positive regulation of cell proliferation (n = 1; CDK4), cell cycle arrest (n = 1; p21WAF1), apoptosis (n = 2; FASLG, GML), and anti-apoptosis (n = 2; MCL1, RELA). Of the genes significantly up-regulated in T-ALL samples, these have a functional role in apoptosis (n = 11; APAF1, BID, CASP2, E124, FADD, FOXO3, LRDD, PCBP4, P53AIP1, SIAH1, SIRT1), cell cycle regulation (n = 9; BRCA2, CCNG2, CDC25C, E2F1, E2F3, IFNB1, MSH2, PRC1, TADA3L), cell proliferation (n = 3; MDM4, PCAF, PPMID), cell growth (n = 1, MYOD1), and also an anti-apoptotic function (n = 1; IGF1R). The p21WAF1 gene demonstrated the most significant difference in expression between the two cell lineages (p = 1.285 × 10−9).
FIGURE 3.
Heat map of p53 target genes from RT-qPCR array in ALL xenografts. BCP-ALL (ALL-3 and ALL-19) and T-ALL (ALL-8 and ALL-16) xenografts were exposed to etoposide (5 μm for 3 h and 12 h) and RNA isolated. An RT-qPCR array was conducted on 84 p53-target genes and log-transformed data from individual replicates were visualized in heat map format. Each column represents an individual replicate for the RT-qPCR array, and each row represents a gene. Genes are listed in order of differential expression of induction between BCP-ALL and T-ALL xenografts. Genes highlighted in blue are significantly up-regulated in BCP-ALL over T-ALL samples, whereas genes highlighted in red are significantly up-regulated in T-ALL over BCP-ALL samples.
To confirm p53 functionality in different ALL lineages, ALL-3, ALL-8, and Jurkat cells were exposed to etoposide (5 μm) or the Hdm-2 inhibitor, nutlin-3 (10 μm), and p53 localization determined by subcellular fractionation and immunoblotting. Increases in p53 protein were observed in the nuclear fractions of ALL-3 and ALL-8 after etoposide and nutlin-3 exposure, with a negligible increase in Jurkat cells (supplemental Fig. S3), whereas cytoplasmic p21WAF1 levels only increased in ALL-3 cells. To further confirm p53 functionality, the sensitivity of xenografts and cell lines to the death-inducing effects of nutlin-3 was tested by methyl-thiazolyl-tetrazolium assay. Although all xenografts and Reh, Nalm-6, and Molt-4 cell lines exhibited IC50 values <10 μm, the IC50 values of the p53 mutant cell lines (CEM and Jurkat) were >100 μm (data not shown).
Vorinostat Fails to Induce p21WAF1 Expression in T-ALL Cell Lines and Xenografts
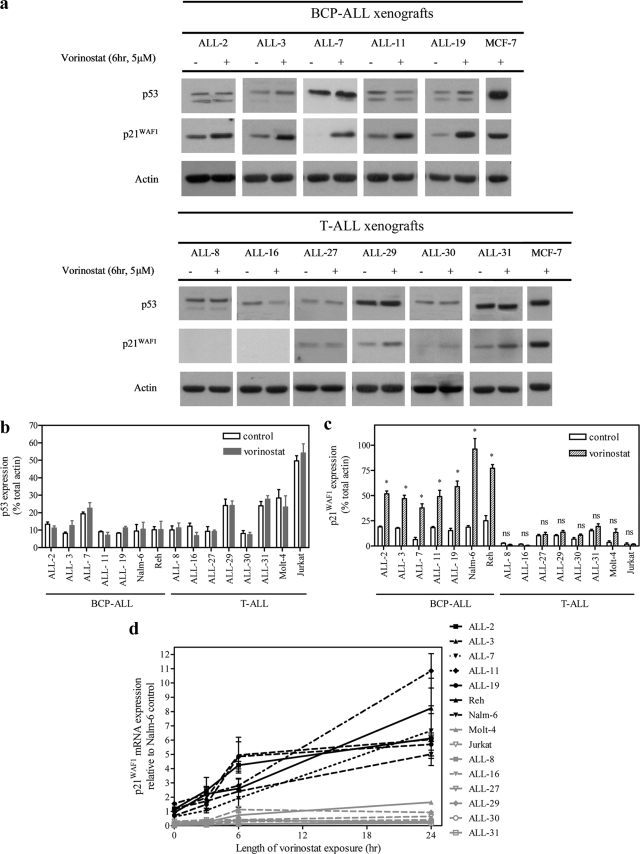
Vorinostat has been shown to induce p21WAF1 transcription independently of p53 function (44). This drug was therefore utilized to determine whether defective p21WAF1 induction in T-ALL cells was specific to p53-inducing agents. Nuclear localization of p53 was used as a marker of p53 transcriptional activity in vorinostat- and etoposide-treated cells. Vorinostat failed to cause increased nuclear localization of p53 in either ALL-3 or ALL-19, which was in contrast to the response of these xenograft cells to etoposide (supplemental Fig. S4, a and b). The ALL xenografts and cell lines were then incubated with vorinostat (5 μm, 6 h), which resulted in a significant induction of p21WAF1, but not p53, mRNA, and protein in BCP-ALL cells (Fig. 4a, upper panel, and b–d), and minimal induction in T-ALL cells (Fig. 4, a, lower panel, and b–d). These results reinforce that T-ALL cells exhibit p53-independent transcriptional repression of the p21WAF1 gene.
FIGURE 4.
a, induction of p21WAF1 protein after exposure of ALL xenograft cells to vorinostat. ALL xenograft cells were exposed to vorinostat (5 μm, 6 h), and expression of p53 or p21WAF1 proteins (a–c) and p21WAF1 mRNA (d) were assessed by immunoblot or RT-qPCR, respectively. a, representative blots of p53 and p21WAF1 proteins in BCP-ALL (upper panel) and T-ALL (lower panel) xenografts. Expression of p53 (b) and p21WAF1 (c) proteins were expressed relative to actin, and data from individual membranes were normalized to the MCF-7 control lane. An asterisk represents values significantly greater than control (p < 0.05) as determined by Mann-Whitney U test; ns, not significant. d, p21WAF1 mRNA expression was normalized to untreated Nalm-6 cells. All quantitative data are expressed as the mean ± S.E. from three separate experiments.
Epigenetic regulation of the p21WAF1 gene locus
To determine whether differential acetylation of histone tails could explain the decreased transcription of p21WAF1 in T-ALL cells, enrichment of acetylated H3K9 (H3K9Ac) was interrogated over three regions of the p21WAF1 promoter in xenografts by ChIP analysis (Fig. 5a). Decreased H3K9Ac enrichment was evident in T-ALL xenografts (ALL-8 and ALL-16) relative to the BCP-ALL xenografts (ALL-3, -11, and -19) over all three regions (Fig. 5b), which was confirmed by ChIP-qPCR (supplemental Fig. 5b). Moreover, exposure of xenograft cells to vorinostat (5 μm, 6 h) resulted in increased H3K9Ac within all three p21WAF1 promoter regions of the BCP-ALL xenografts, with a minimal increase in the T-ALL xenografts (supplemental Fig. 5, a and b). These results indicate a dominant mechanism of p21WAF1 repression in T-ALL cells that is not relieved by inhibition of histone deacetylase activity.
FIGURE 5.
Decreased H3K9Ac and increased CpG methylation at the p21WAF1 locus of T-ALL cells. a, representation of the p21WAF1 genomic locus, illustrating a region spanning −4000 bp from the TSS to the first exon and intron of the p21WAF1 gene. The locations of three regions (1, 2, and 3) amplified by PCR for ChIP analysis are indicated, corresponding to between −2224 bp and +100 bp from the TSS. The TSS is indicated by an arrow at +1. Regions 4 and 5 represent two areas of PCR amplification analyzed for CpG methylation. Region 4 spans from −131 to +97 bp relative to the TSS and contains 25 CpG dinucleotides. Region 5 spans from +73 to +617 bp with 53 CpG nucleotides in this region. b, semi-quantitative ChIP analysis of cells from two T-ALL (ALL-8 and ALL-16) and three BCP-ALL xenografts (ALL-3, ALL-11, and ALL-19) for basal H3K9Ac in regions 1, 2, and 3. Mock samples were treated with IgG only. c and d, CpG methylation at basal levels in nine xenografts (c) or in response to exposure of ALL cell lines to epigenetic modifiers (d) was analyzed by SEQUENOM Epityper assays, which calculates methylation at individual CpG sites across the p21WAF1 locus. Due to the limitations of MALDI-TOF MS to differentiate between CpG sites in close proximity and T-cleavage reactions producing fragments with more than one CpG site, some of the sites could not be resolved individually. Therefore, each colored box shown represents the methylation value for a cluster of CpG sites (e.g. CpG 2–7). Quantified methylation at each site is represented as a heat map ranging from purple (0%) to yellow (100%), with white boxes representing non-detected. In d, Molt-4 and Nalm-6 cells were treated with DMSO control (C), decitabine (D; 500 nm for 96 h), vorinostat (V; 500 nm for 96 h), or a combination of both (D+V). e, cells were treated as in d, and p21WAF1 protein levels were assessed by immunoblotting. Molt-4 cells were also treated with the epigenetic modifying agents and etoposide (5 μm, for the last 6 h of 96 h drug exposure). MCF-7, positive control MCF-7 lysate run concurrently on each gel.
The p21WAF1 locus includes a CpG island spanning the promoter region, transcription start site (TSS), exon 1, and intron 1 (Fig. 5a). Methylation of the p21WAF1 CpG island was analyzed by SEQUENOM MassARRAY Epityper® after amplification of two PCR products, termed region 4 (−131 to +97 bp in relation to the TSS) and region 5 (+73 to +617 bp; Fig. 5a). Low CpG methylation was observed across both regions in three BCP-ALL xenografts (ALL-2, ALL-3, ALL-19), and across region 4 in six T-ALL xenografts (ALL-8, ALL-16, ALL-27, ALL-29, ALL-30, and ALL-31) (Fig. 5c). In contrast, analysis of region 5 revealed a close relationship between the level of CpG methylation and p21WAF1 expression. Methylation levels were low across region 5 in the p21WAF1 functional BCP-ALL xenografts, greatly increased at specific CpG sites in the T-ALL samples that lacked p21WAF1 (ALL-8, ALL-16) and moderately increased at various CpG sites in the xenograft cells that expressed minimal p21WAF1 (ALL-27, ALL-29, ALL-30, ALL-31) (Fig. 5c).
Reversal of p21WAF1 Methylation and Repression by Combined Histone Deacetylase/Methyltransferase Inhibitor Treatment
To test whether the combination of vorinostat and the methyltransferase inhibitor decitabine could reduce CpG methylation in the p21WAF1 gene locus and reconstitute p21WAF1 expression, the p21WAF1-defective cell line Molt-4 and the p21WAF1 functional Nalm-6 cell line were exposed to DMSO (vehicle control), vorinostat, decitabine, or the combination prior to CpG methylation quantification by SEQUENOM MassARRAY analysis (Fig. 5d). Increased basal levels of methylation at specific CpG sites within region 5 were detected in Molt-4 cells (CpG 31–32, 43–45, 50–51, and 52–53) compared with Nalm-6 cells. Decitabine alone caused a reduction in CpG methylation in Molt-4 cells to levels observed in decitabine-treated Nalm-6 cells, particularly at sites between 31 and 53. Vorinostat alone also caused a decrease in CpG methylation at specific sites (e.g. 31–32) in Molt-4 cells, although its effects were more restricted compared with decitabine. The combination of decitabine and vorinostat induced a greater decrease in CpG methylation levels in region 5 than either drug alone, to levels similar to basal CpG methylation in Nalm-6 cells.
Expression of p21WAF1 was evaluated after exposure to the above epigenetic modifiers to determine whether the decrease in CpG methylation corresponded with p21WAF1 induction. Extended exposure of Molt-4 cells to vorinostat (500 nm, 96 h) failed to increase p21WAF1, whereas decitabine alone induced modest p21WAF1 expression (Fig. 5e). Vorinostat and decitabine in combination increased p21WAF1 expression to a greater extent than either drug alone, consistent with their effects on CpG methylation. The addition of etoposide for the last 6 h of drug treatment with vorinostat and decitabine did not further increase p21WAF1 protein levels in Molt-4 cells, demonstrating that induction of p21WAF1 by DNA-damaging agents and epigenetic modifications are not additive. Both single drugs increased p21WAF1 expression in Nalm-6 cells, although the combination was not synergistic.
DISCUSSION
This work has shown that, in contrast to BCP-ALL, p53-independent epigenetic repression of p21WAF1 is prevalent in T-ALL cell lines, xenografts, and primary biopsy specimens, and is associated with reduced H3K9Ac in the p21WAF1 promoter and increased CpG methylation in the first intron/exon of the gene. Impaired p53 function can result in reduced sensitivity of tumor cells to chemotherapeutic drugs (45) and lead to p21WAF1 repression. However, in this analysis, p53 functionality was confirmed by sequencing of the p53 coding region (34), induction and nuclear localization of p53 after exposure to DNA damaging agents, and robust induction of other p53 target genes that were previously used to confirm wild type p53 transcriptional activity after DNA damage, including Bax (5), GADD45α (46), and Hdm-2 (47). Moreover, reduced basal levels of p21WAF1 mRNA and protein in T-ALL versus BCP-ALL were demonstrated in independent cohorts of patients on separate continents, which strengthens the validity of our observations. The down-regulation of p21WAF1 has been previously highlighted in T-ALL cell lines (18, 26, 48) and primary cells (49), although a mechanistic explanation of this phenomenon has yet to be presented.
In this study, the mechanisms and modulation of epigenetic p21WAF1 repression were explored. Vorinostat induced p21WAF1 in BCP-ALL xenografts and cell lines independently of p53 activation, as demonstrated by the lack of p53 induction in whole cell lysates and in nuclear fractions after drug exposure. However, it remains possible that vorinostat acetylates lysine residues on the p53 protein, leading to increased DNA binding affinity without increasing protein expression (50). Vorinostat-mediated induction of p21WAF1 in a variety of cell types has been comprehensively characterized by others. However, the present study of ALL cells shows that p21WAF1 induction following vorinostat treatment was a lineage specific response. Although T-ALL cells showed minimal induction of p21WAF1, BCP-ALL cells exhibited robust induction of p21WAF1 after vorinostat treatment. Together, these results strongly suggest that p21WAF1 is inactivated by epigenetic modification in T-ALL cells. The induction of p21WAF1 in the L809T primary T-ALL sample by the combination of the proteasomal inhibitor MG132 and irradiation highlights the possibility that p21WAF1 may also be inhibited by enhanced degradation pathways.
The epigenetic markers evaluated for the p21WAF1 locus in this study included H3K9Ac and CpG methylation. Histone deacetylation is necessary for stable gene silencing (22). Inability to induce p21WAF1 mRNA after vorinostat exposure corresponded with decreased H3K9Ac within the p21WAF1 promoter region in ALL-8 and ALL-16. Although vorinostat induced an increase in H3K9Ac enrichment across the promoter region in BCP-ALL xenografts, there were minimal changes in T-ALL xenografts, and among these, this did not result in p21WAF1 induction, indicating that histone acetylation alone did not explain the complete lack of induction seen in T-ALL samples.
Methylation of the CpG island in the p21WAF1 promoter region was investigated as another mechanism of gene silencing. There is currently no general consensus on the prevalence and role of CpG methylation in cells with a silenced p21WAF1 gene. The majority of studies to date have been limited to focused regions of the promoter region. To date, there has been only one report on the methylation status of the CpG island that traverses the most proximal end of the promoter region, TSS, exon 1, and 5′ region of intron 1 of the rat p21WAF1 gene. For this reason, only limited conclusions can be drawn on the role of CpG methylation on p21WAF1 silencing in human cells. In this study, quantitative methylation analysis was conducted across this CpG island. CpG hypermethylation was not found in the promoter region immediately upstream of the TSS in any of the ALL samples. However, increased methylation was observed at the 3′ end of exon 1 and 5′ region of intron 1 in T-ALL xenografts that silenced p21WAF1, as well as in the Molt-4 cell line. These results are in accord with a study of silencing in the rat p21WAF1 gene (28). Although many studies suggest that methylation must occur in the promoter regions for a gene to be silenced, studies of the CDKN2B gene have also shown that hypermethylation of a CpG island in the 5′ region, including exons and introns, correlates with gene silencing (51). It was also reported that methylation of the 5′-untranslated exon and first intron, but not with the promoter region per se, correlated with silencing of the ubiquitin 1 gene in the barley plant (52). In a recent comprehensive study of methylation levels across 24,376 genes from genomic DNA derived from the M091 cell line, methylation downstream of TSS, in the region of the first exon, was demonstrated to be more closely correlated with transcriptional silencing than CpG methylation in the promoter region of the gene (53).
Methylation in CpG islands can repress gene expression by providing a site for methyl binding proteins such as MeCP1 and MeCP2 to attach (54) or sterically interfere with the binding of transcription factors. Although this study has demonstrated increased methylation at sites that have not been previously shown as specific binding sites of transcription factors or enhancers, the precise mechanism of transcriptional inactivation of p21WAF1 observed in T-ALL samples remains speculative.
An important finding of the current study is that vorinostat and decitabine acted synergistically to decrease methylation in a cluster of CpG sites across exon 1 and intron 1 of the p21WAF1 gene and to reactivate p21WAF1 in T-ALL cells. Overall, our results indicate divergent and lineage-dependent mechanisms by which the p21WAF1 gene is regulated and repressed in ALL cells. p21WAF1 reactivation in T-ALL cells by combined treatment with histone deacetylase inhibitors and DNA demethylating drugs warrants further investigation as a means to specifically modulate the anti-leukemic effects of chemotherapeutic drugs.
Acknowledgments
We gratefully acknowledge the staff and patients of the Centre for Children's Cancer and Blood Disorders, Sydney Children's Hospital, Australia, and the Royal Victoria Infirmary, Newcastle, UK, for provision of primary ALL samples. Vorinostat was generously provided by Merck, Sharpe and Dohme, Corp. and the National Cancer Institute, National Institutes of Health. We thank Dr. Nicola Brown for assistance with experimental methodology and Dr. Vivek A. Bhadri for assistance with data analysis.
This work was supported by the Children's Cancer Institute Australia for Medical Research, a senior research fellowship (to R. B. L.) from the Australian National Health and Medical Research Council, an Australian postgraduate award (to C. D.), a university postgraduate award (to P. S. B.) and the JGW Patterson Foundation (Newcastle upon Tyne, UK) (to L. A. H.). The Children's Cancer Institute Australia for Medical Research is affiliated with the University of New South Wales and Sydney Children's Hospital.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Methods, Tables S1–S4, and Figs. S1–S5.
- ALL
- acute lymphoblastic leukemia
- BCP
- B-cell precursor
- T-ALL
- T-cell ALL
- DMSO
- dimethyl sulfoxide
- RT-qPCR
- real-time quantitative reverse transcription PCR
- TSS
- transcription start site.
REFERENCES
- 1. Pui C. H., Evans W. E. (2006) N. Engl. J. Med. 354, 166–178 [DOI] [PubMed] [Google Scholar]
- 2. Holleman A., den Boer M. L., Kazemier K. M., Janka-Schaub G. E., Pieters R. (2003) Blood 102, 4541–4546 [DOI] [PubMed] [Google Scholar]
- 3. Brown J. M., Wouters B. G. (1999) Cancer Res. 59, 1391–1399 [PubMed] [Google Scholar]
- 4. Rokudai S., Aikawa Y., Tagata Y., Tsuchida N., Taya Y., Kitabayashi I. (2009) J. Biol. Chem. 284, 237–244 [DOI] [PubMed] [Google Scholar]
- 5. Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 6. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 7. Wattel E., Preudhomme C., Hecquet B., Vanrumbeke M., Quesnel B., Dervite I., Morel P., Fenaux P. (1994) Blood 84, 3148–3157 [PubMed] [Google Scholar]
- 8. Wada M., Bartram C. R., Nakamura H., Hachiya M., Chen D. L., Borenstein J., Miller C. W., Ludwig L., Hansen-Hagge T. E., Ludwig W. D., et al. (1993) Blood 82, 3163–3169 [PubMed] [Google Scholar]
- 9. Blau O., Avigad S., Stark B., Kodman Y., Luria D., Cohen I. J., Zaizov R. (1997) Leuk. Res. 21, 721–729 [DOI] [PubMed] [Google Scholar]
- 10. Marks D. I., Kurz B. W., Link M. P., Ng E., Shuster J. J., Lauer S. J., Brodsky I., Haines D. S. (1996) Blood 87, 1155–1161 [PubMed] [Google Scholar]
- 11. Peterson L. F., Yan M., Zhang D. E. (2007) Blood 109, 4392–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jung C., Motwani M., Kortmansky J., Sirotnak F. M., She Y., Gonen M., Haimovitz-Friedman A., Schwartz G. K. (2003) Clin. Cancer Res. 9, 6052–6061 [PubMed] [Google Scholar]
- 13. Mahyar-Roemer M., Roemer K. (2001) Oncogene 20, 3387–3398 [DOI] [PubMed] [Google Scholar]
- 14. Javelaud D., Besancon F. (2002) J. Biol. Chem. 277, 37949–37954 [DOI] [PubMed] [Google Scholar]
- 15. Tian H., Wittmack E. K., Jorgensen T. J. (2000) Cancer Res. 60, 679–684 [PubMed] [Google Scholar]
- 16. Crescenzi E., Palumbo G., de Boer J., Brady H. J. (2008) Clin. Cancer Res. 14, 1877–1887 [DOI] [PubMed] [Google Scholar]
- 17. Lazzarini R., Moretti S., Orecchia S., Betta P. G., Procopio A., Catalano A. (2008) Clin. Cancer Res. 14, 5099–5107 [DOI] [PubMed] [Google Scholar]
- 18. Scott S. A., Kimura T., Dong W. F., Ichinohasama R., Bergen S., Kerviche A., Sheridan D., DeCoteau J. F. (2004) Leuk. Res. 28, 1293–1301 [DOI] [PubMed] [Google Scholar]
- 19. Zhang W., Kornblau S. M., Kobayashi T., Gambel A., Claxton D., Deisseroth A. B. (1995) Clin. Cancer Res. 1, 1051–1057 [PubMed] [Google Scholar]
- 20. Ivanovska I., Ball A. S., Diaz R. L., Magnus J. F., Kibukawa M., Schelter J. M., Kobayashi S. V., Lim L., Burchard J., Jackson A. L., Linsley P. S., Cleary M. A. (2008) Mol. Cell Biol. 28, 2167–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shiohara M., el-Deiry W. S., Wada M., Nakamaki T., Takeuchi S., Yang R., Chen D. L., Vogelstein B., Koeffler H. P. (1994) Blood 84, 3781–3784 [PubMed] [Google Scholar]
- 22. Jaenisch R., Bird A. (2003) Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 23. Ng H. H., Zhang Y., Hendrich B., Johnson C. A., Turner B. M., Erdjument-Bromage H., Tempst P., Reinberg D., Bird A. (1999) Nat. Genet. 23, 58–61 [DOI] [PubMed] [Google Scholar]
- 24. Chen B., He L., Savell V. H., Jenkins J. J., Parham D. M. (2000) Cancer Res. 60, 3290–3298 [PubMed] [Google Scholar]
- 25. Roman-Gomez J., Castillejo J. A., Jimenez A., Gonzalez M. G., Moreno F., Rodriguez Mdel C., Barrios M., Maldonado J., Torres A. (2002) Blood 99, 2291–2296 [DOI] [PubMed] [Google Scholar]
- 26. Watanabe M., Nakahata S., Hamasaki M., Saito Y., Kawano Y., Hidaka T., Yamashita K., Umeki K., Taki T., Taniwaki M., Okayama A., Morishita K. (2010) J. Virol. 84, 6966–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu W. G., Srinivasan K., Dai Z., Duan W., Druhan L. J., Ding H., Yee L., Villalona-Calero M. A., Plass C., Otterson G. A. (2003) Mol. Cell Biol. 23, 4056–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Allan L. A., Duhig T., Read M., Fried M. (2000) Mol. Cell Biol. 20, 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chim C. S., Wong A. S., Kwong Y. L. (2005) Am. J. Hematol. 80, 282–287 [DOI] [PubMed] [Google Scholar]
- 30. Ying J., Srivastava G., Gao Z., Zhang X., Murray P., Ambinder R., Tao Q. (2004) Blood 103, 743–746 [DOI] [PubMed] [Google Scholar]
- 31. Majid S., Kikuno N., Nelles J., Noonan E., Tanaka Y., Kawamoto K., Hirata H., Li L. C., Zhao H., Okino S. T., Place R. F., Pookot D., Dahiya R. (2008) Cancer Res. 68, 2736–2744 [DOI] [PubMed] [Google Scholar]
- 32. Shen L., Kondo Y., Issa J. P., Garcia-Manero G. (2002) Blood 100, 3432–3433; author reply 3433–3434 [DOI] [PubMed] [Google Scholar]
- 33. Shin J. Y., Kim H. S., Park J., Park J. B., Lee J. Y. (2000) Cancer Res. 60, 262–265 [PubMed] [Google Scholar]
- 34. Lock R. B., Liem N., Farnsworth M. L., Milross C. G., Xue C., Tajbakhsh M., Haber M., Norris M. D., Marshall G. M., Rice A. M. (2002) Blood 99, 4100–4108 [DOI] [PubMed] [Google Scholar]
- 35. Irving J., Jesson J., Virgo P., Case M., Minto L., Eyre L., Noel N., Johansson U., Macey M., Knotts L., Helliwell M., Davies P., Whitby L., Barnett D., Hancock J., Goulden N., Lawson S. (2009) Haematologica 94, 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liem N. L., Papa R. A., Milross C. G., Schmid M. A., Tajbakhsh M., Choi S., Ramirez C. D., Rice A. M., Haber M., Norris M. D., MacKenzie K. L., Lock R. B. (2004) Blood 103, 3905–3914 [DOI] [PubMed] [Google Scholar]
- 37. Bachmann P. S., Gorman R., Papa R. A., Bardell J. E., Ford J., Kees U. R., Marshall G. M., Lock R. B. (2007) Cancer Res. 67, 4482–4490 [DOI] [PubMed] [Google Scholar]
- 38. Smyth G. K. (2004) Stat. Appl. Genet. Mol. Biol. 3, Article 3, DOI: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 39. Bachmann P. S., Piazza R. G., Janes M. E., Wong N. C., Davies C., Mogavero A., Bhadri V. A., Szymanska B., Geninson G., Magistroni V., Cazzaniga G., Biondi A., Miranda-Saavedra D., Göttgens B., Saffery R., Craig J. M., Marshall G. M., Gambacorti-Passerini C., Pimanda J. E., Lock R. B. (2010) Blood 116, 3013–3022 [DOI] [PubMed] [Google Scholar]
- 40. Kimura A. P., Liebhaber S. A., Cooke N. E. (2004) Mol. Endocrinol. 18, 1018–1032 [DOI] [PubMed] [Google Scholar]
- 41. Ehrich M., Nelson M. R., Stanssens P., Zabeau M., Liloglou T., Xinarianos G., Cantor C. R., Field J. K., van den Boom D. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S. V., Hainaut P., Olivier M. (2007) Hum. Mutat. 28, 622–629 [DOI] [PubMed] [Google Scholar]
- 43. Forbes S. A., Bindal N., Bamford S., Cole C., Kok C. Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J. W., Campbell P. J., Stratton M. R., Futreal P. A. (2011) Nucleic Acids Res. 39, D945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Richon V. M., Sandhoff T. W., Rifkind R. A., Marks P. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10014–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. El-Deiry W. S. (2003) Oncogene 22, 7486–7495 [DOI] [PubMed] [Google Scholar]
- 46. Hollander M. C., Alamo I., Jackman J., Wang M. G., McBride O. W., Fornace A. J., Jr. (1993) J. Biol. Chem. 268, 24385–24393 [PubMed] [Google Scholar]
- 47. Barak Y., Juven T., Haffner R., Oren M. (1993) EMBO J. 12, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Min D. J., Moskowitz N. P., Brownstein C., Lee H., Horton T. M., Carroll W. L. (2006) Apoptosis 11, 1977–1986 [DOI] [PubMed] [Google Scholar]
- 49. Chang P. Y., Miyamoto S. (2006) Mol. Cancer Res. 4, 101–112 [DOI] [PubMed] [Google Scholar]
- 50. Gu W., Roeder R. G. (1997) Cell 90, 595–606 [DOI] [PubMed] [Google Scholar]
- 51. Merlo A., Herman J. G., Mao L., Lee D. J., Gabrielson E., Burger P. C., Baylin S. B., Sidransky D. (1995) Nat. Med. 1, 686–692 [DOI] [PubMed] [Google Scholar]
- 52. Meng L., Bregitzer P., Zhang S., Lemaux P. G. (2003) Plant Mol. Biol. 53, 327–340 [DOI] [PubMed] [Google Scholar]
- 53. Brenet F., Moh M., Funk P., Feierstein E., Viale A. J., Socci N. D., Scandura J. M. (2011) PLoS One 6, e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. (1992) Cell 69, 905–914 [DOI] [PubMed] [Google Scholar]