Background: Regulation of bone remodeling during orthodontic tooth movement.
Results: Tensile stress induces ephrin-B2 expression in PDL fibroblasts, and ephrin-B2-EphB4 interactions induce osteoblastogenesis in osteoblasts of the alveolar bone.
Conclusion: Ephrin-B2-EphB4 signaling between PDLF and osteoblasts of the alveolar bone might contribute to bone remodeling during orthodontic tooth movement.
Significance: Understanding the regulation of bone remodeling during orthodontic tooth movement is a prerequisite for pharmacological interventions.
Keywords: Bone, Fibroblast, Gene Expression, Gene Regulation, Receptor Tyrosine Kinase, Eph Receptors, Ephrin Ligands, Osteogenesis, Tooth Movement
Abstract
During orthodontic tooth movement, the application of adequate orthodontic forces allows teeth to be moved through the alveolar bone. These forces are transmitted through the periodontal ligaments (PDL) to the supporting alveolar bone and lead to deposition or resorption of bone, depending on whether the tissues are exposed to a tensile or compressive mechanical strain. Fibroblasts within the PDL (PDLF) are considered to be mechanoresponsive. The transduction mechanisms from mechanical loading of the PDLF to the initiation of bone remodeling are not clearly understood. Recently, members of the ephrin/Eph family have been shown to be involved in the regulation of bone homeostasis. For the first time, we demonstrate that PDLF exposed to tensile strain induce the expression of ephrin-B2 via a FAK-, Ras-, ERK1/2-, and SP1-dependent pathway. Osteoblasts of the alveolar bone stimulated with ephrin-B2 increased their osteoblastogenic gene expression and showed functional signs of osteoblastic differentiation. In a physiological setting, ephrin-B2-EphB4 signaling between PDLF and osteoblasts of the alveolar bone might contribute to osteogenesis at tension sites during orthodontic tooth movement.
Introduction
Teeth are moved through alveolar bone during orthodontic tooth movement by the application of adequate orthodontic forces. The applied mechanical stress must then be communicated to the alveolar bone via the periodontal ligaments (PDL).2 Mechanosensitive cells of the PDL, the PDL fibroblasts (PDLF), are able to transduce mechanical strain into intracellular signals. This information is then transmitted to other cells like osteoblasts of the alveolar bone. This initiates fundamental processes leading to a profound remodeling of mineralized and nonmineralized periodontal tissues. The sequence of events from the application of mechanical forces to the complex biochemical events commanding the tightly controlled accomplishment of osteogenesis at tension sides and osteoclastogenesis at compressive sides is not completely understood.
Some effectors of physiological bone remodeling have been identified and characterized. Of paramount importance for bone remodeling is the receptor activator of nuclear factor κB/receptor activator of nuclear factor κB ligand/osteoprotegerin signaling pathway (1, 2). For the differentiation and function of osteoblasts, a member of the Runx (Runt-related protein) family of transcription factors, Runx2, is of leading importance. Runx2 binds to the promoter regions of osteoblastic genes and activates, among others, the expression of alkaline phosphatase (ALPL), osteocalcin, bone sialoprotein (BSP-1), and collagen 1A1 (COL1A1), which include hallmarks of osteoblast differentiation.
Ephrins and Ephs are involved in various developmental processes during embryogenesis (3, 4) and have been characterized for their roles in neuronal development, during which they mediate patterning of hindbrain rhombomeres, guidance of migrating neural precursor cell axon guidance (5, 6), and physiological and pathological angiogenesis (7, 8). The Eph receptors include the largest known group of receptor tyrosine kinases. Although they share a conserved domain structure, sequence homology analysis and binding preferences served as criteria for subdividing the 14 mammalian Ephs into two groups: 8 EphA- (EphA1–8) and 6 EphB receptors (EphB1–6). The EphA receptors bind preferentially to the glycosylphosphatidylinositol-linked ephrin-A ligands (ephrins A1–5), althoughthe EphB receptors bind the transmembrane ephrin-B ligands (ephrins B1–3). Ephs, like other receptor tyrosine kinases, initiate signal transduction through autophosphorylation after ligand-receptor engagement, referred to as “forward signaling.” However, in contrast to other soluble ligands for receptor tyrosine kinases, the ephrins possess a unique feature as follows. Ephrins are membrane-bound and capable of receptor-like active signaling (“reverse signaling”), resulting in bi-directional cell-to-cell communication (reviewed in Refs. 6, 9). Major functions of the ephrin/Eph system are within the regulation of cellular motility and alteration in adhesions to the extracellular matrix. Ephrin/Eph interactions frequently modulate cellular repulsion or attraction, indicating a causal relation between ephrins, Ephs, the ECM, and the cytoskeleton. However, the strain that alters the interaction of cells with the ECM and/or influences the organization of the cytoskeleton has an impact on the function and expression of members of the ephrin/Eph family (10, 11).
Recently members of the ephrin/Eph family have been shown to be involved in the regulation of bone homeostasis. Zhao et al. (12) reported an NFATc1-dependent ephrin-B2 expression during the receptor activator of nuclear factor κB ligand-induced differentiation of osteoclasts. Moreover, Zhao et al. (12) could show that osteoblasts expressed the EphB4 receptor, and the bi-directional activation of the ephrin-B2-EphB4 signaling pathway on osteoclasts and osteoblasts led to the suppression of osteoclast differentiation with a concurrent stimulation of osteoblastogenesis and in consequence to bone formation (12, 13).
The mechanism for osteogenesis at tension sites in tooth movement is not well understood. Mechanical forces during orthodontic tooth movement are initially transmitted to the PDL. Fibroblasts within the PDL are the first cellular recipients of mechanical strain. Under cyclic strain, PDL fibroblasts in vitro increased their osteogenic gene expression (14). An in vivo model of tooth movement indicated enhanced expression of Runx2 and the phosphorylation of extracellular signal-regulated kinases 1/2 (pERK1/2) under tension (15). Based on their involvement in bone remodeling, their causal relation between ECM and the cytoskeleton, and also their modulation by mechanical stress, it is tempting to speculate that ephrins and Ephs are modulated by cellular strain in PDLF and/or osteoblasts of the alveolar bone. The anatomical localization of PDLF in proximity to osteoblasts of the alveolar bone would allow cell to cell signaling via ephrin/Eph interactions and might contribute to cellular responses leading to osteogenesis at sites of cellular strain. Besides the interplay between PDL fibroblasts and osteoblasts of the alveolar bone via ephrin/Eph interactions, bi-directional signaling between ephrins and Ephs may also occur within PDLF and osteoblast populations themselves. The communication between ephrin-B2 and EphB4 has recently been shown to be involved in the initiation of osteoblast differentiation within the osteoblast lineage (16). We recently showed at compression sites during orthodontic tooth movement that ephrin-A2 is up-regulated in PDLF and that this up-regulation leads to an attenuated osteogenesis in the alveolar bone (17).
In this study, we provide evidence that ephrin-B2-EphB4 signaling links mechanical strain on PDLF with osteoblastic gene expression in osteoblasts of the alveolar bone. We also investigated the molecular mechanism by which mechanical strain regulates ephrin-B2 gene expression in PDLF. In vitro, ephrin-B2 stimulated osteoblastogenic gene expression, ALP activity, and mineralization in osteoblasts, suggesting that strain-induced ephrin-B2 up-regulation in PDLF contributes to osteogenesis at tension sites of orthodontic tooth movement. Moreover, we have shown that ephrin-B2-EphB4 signaling in PDLF led to the induction of osteoblastogenic gene expression, indicating a dual role for ephrin-B2-EphB4 signaling in osteogenesis at tension sites by stimulating the osteoblastic differentiation of osteoblasts and PDL fibroblasts. Together, these findings establish a role for this ligand/receptor system in osteogenesis during orthodontic tooth movement.
EXPERIMENTAL PROCEDURES
Primary Cell Cultures
Primary PDLF were obtained from juvenile patients (12–20 years) following premolar extraction during orthodontic treatment. Alveolar bone tissue was obtained from patients following osteotomy of third molars. The Ethics Committee (Medical Faculty, University of Heidelberg; Vote S147/2010) has approved the harvest of the tissues, and informed consent was obtained from the patients following explanation of the study.
Small tissue fragments were established as explant cultures in DMEM (Invitrogen) supplemented with 10% fetal calf serum, 2 mm l-glutamine, and antibiotics (penicillin, 100 units/ml, streptomycin 100 μg/ml, kanamycin 50 mg/ml) and antimycotics (amphotericin, 2.5 μg/ml). After reaching 80% confluence, primary PDLF and osteoblasts were used for the experiments. Primary cells were used between passages 3 and 9.
Application of Mechanical Strain
Mechanical strain was carried out according to the method described by Hasegawa et al. (18). Briefly, 3.5 × 103/cm2 PDLF or osteoblast cells were seeded on flexible bottomed dishes (Greiner Bio-One, Frickenhausen, Germany), coated with 20 μg/ml collagen type-I (IBM, Leipzig, Germany) and 10 μg/ml fibronectin (Biomol, Hamburg, Germany), and grown until 80% confluence. The bottom of each dish was strained by induction of a continuous average strain of 2.5% for 2 min to 72 h. The exact durations are given in the figures, representing the individual experiments.
Stimulation with Ephrin-B2-Fc
To test for putative functional consequences of ephrin-B2-dependent EphB4 receptor stimulations, we have used ephrin-B2-Fc chimeras (recombinant ephrin-B2 fused with Fc; R&D Systems, Wiesbaden, Germany). For appropriate signaling, soluble ephrin ligands require preclustering with anti-Fc antibodies (19). Ephrin-B2-Fc was preclustered with anti-human IgG-Fc (1:10 stoichiometry) in cell medium for 30 min at room temperature. Anti-human IgG-Fc alone in cell medium served as a control.
Pharmacological Treatment
U0126 (Merck), a selective inhibitor of MEK1/2, was used to block the MAPK/ERK cascade pathway. Cells were incubated with the concentrations of UO126 indicated in the experiments or with the appropriate amount of DMSO.
siRNA Analyses
Validated siRNAs (FlexiTube Gene Solution) designed to knock down the endogenous expression of FAK and EphB4 and scrambled siRNA as a negative control were purchased from Qiagen (Hilden, Germany). Transfections of siRNA to PDLF or osteoblasts were carried out with HiPerfect reagent (Qiagen). Transfections were performed on 4 × 105 cells with 500 ng of each siRNA and 20 μl of HiPerfect reagent in 60-mm dishes. At 12 h post-transfection, cells were subjected to cellular strain. Knockdown of the mRNA for FAK and EphB4 was monitored using qRT-PCR and for FAK additionally using Western blotting.
Ras Activity Assay
Ras activity (GTP-Ras) was evaluated using a commercially available Ras activation assay (Millipore, Schwalbach, Germany) according to the manufacturer's instructions.
RT-PCR
RT-PCR was used to evaluate the expression of ephrin-A1, ephrin-A2, ephrin-B1, ephrin-B2, ephrin-B3, EphA2, EphA5, EphB2, EphB3, and EphB4 mRNA. Total RNA was isolated from PDLF and osteoblasts using the RNeasy kit (Qiagen; Hilden, Germany) and subjected to reverse transcription using poly(dT) primers. Single-stranded cDNA was used for PCR amplification, according to standard protocols (for annealing temperatures and MgCl2 concentrations, see the primer list) to detect the expression of receptors and ligands. PCR products were separated on 1.5% agarose gels and stained with ethidium bromide. The RT-PCR experiments were performed in triplicate. The primer sequences were as follows: ephrin-A1 (human ephrin-A1), fw, CTG GCCAGG CCC CGC GCT AT, and rv, CAA CCT CAA GCA GCG GTC TT, 55 °C, 1.5 mm; ephrin-A2 (human ephrin-A2), fw, TGC GAC TGA AGG TGT ACG TG, and rv, CGG GCT GCT ACA CGA GTT AT, 55 °C, 1.5 mm; ephrin-B1 (human ephrin-B1), fw, CAG AGC AGG AAA TAC GCT TT, and rv, CAC CGA CAG CCG CGA ACA AT, 55 °C, 1.5 mm; ephrin-B2 (human ephrin-B2), fw, TCC CAA TGC TCA GCG CTT AA, and rv, TAC TTC CTA GTC TAC GGT TC, 55 °C, 1.5 mm; ephrin-B3 (human ephrin-B3), fw, CCG CTC GCA CCA CGA TTA CT, and rv, GCC CGC CGT CTC CGC CAA CA, 55 °C, 1.5 mm; EphA2 (human EphA2), fw, GGG GTG AAG AGC CCC GTA TG, and rv, GTGTGC AAG GCA TCG ACG CT, 55 °C, 1.5 mm; EphA5 (human EphA5), fw, GTA GAG GAA GGC TAT CGT CT, and rv, GAA AAT CTC TGT ATA CCG GC, 55 °C, 2.0 mm; EphB2 (human EphB2), fw, GCC ATT GAG CAG GAC TAT CG, and rv, GAT CAT CTT GTC TAG CGT GT, 66 °C, 1.5 mm; Eph3 (human EphB3), fw, CTC TGC CGC CTC GTT ATG CG, and rv, GCT TCC TGA GGC AGA CGA TA, 55 °C, 1.5 mm; EphB4 (human EphB4), fw, CCT TCC TGC GGC TAA ACG AC, and rv, GTT GAC TAG GAT GTT GCG AG, 66 °C, 1.5 mm.
qRT-PCR Analysis
Total RNA was isolated from cells using the RNeasy kit (Qiagen; Hilden, Germany). RNA integrity was monitored by capillary electrophoresis (Experion System, Bio-Rad) according to the manufacturer's instructions.
Total RNA was subjected to reverse transcription using poly(dT) primers. Single-stranded cDNA was used for qRT-PCR analyses. qRT-PCR was done using the TaqMan chemistry (Applied Biosystems, Darmstadt Germany) or the SYBR Green chemistry (Bio-Rad) on an iCycler Instrument (Bio-Rad). To ensure equal amplification efficiencies, we used predesigned TaqMan gene expression assays (Applied Biosystems) for quantitative detection of EphB4, ephrin-B2, GAPDH, and TATA box-binding protein or RT2 qRT-PCR primer assays (Qiagen, Hilden, Germany) for ALP, Runx2, and GAPDH. For the quantitative detection of FAK and GAPDH, Quantitect primer assays (Qiagen, Hilden, Germany) were used. The assay IDs (TaqMan) were as follows: ephrin-B1, Hs00270004_m1; ephrin-B2, Hs00187950_m1; ephrin-B3, Hs00154861_m1; ephrin-A1, Hs00358886_m1; ephrin-A2, Hs01023290_m1; EphB2, Hs01031827_m1; EphB3, Hs00177903_m1; EphB4, Hs00174752_m1; EphA2, Hs00171656_m1; EphA5, Hs00300724_m1; and GAPDH, Hs03929097_g1 together with TATA box-binding protein, Hs00300806_m1, as the reference/housekeeping gene. The following RT2 qRT-PCR primer assays were used: Runx2, PPH01897B; ALP, PPH01311E, and GAPDH, PPH00150E. The following Quantitect primer assay was used: FAK, QT00057687; GAPDH, QT01192646. The amplification efficiencies were evaluated and found to be comparable and reproducible. Therefore, in stretch and ephrin-B2 stimulation experiments, the relative gene expression, compared with untreated control cells, was determined using the ΔΔCT method. To determine the ephrin-Eph expression in different PDLF and osteoblast populations, the expression was determined using the Qgene software (20).
Immunofluorescence
After strain application, the cells were washed with ice-cold PBS, fixed for 5 min with methanol/acetone (1:1, −20 °C), and air-dried. For immunostaining, membranes of the flexible bottomed dishes were cut in parts. Incubation with primary antibodies (anti-FAK, 1:50, Abcam, Cambridge, UK; anti-p-FAK (Tyr576), 1:50, Santa Cruz Biotechnology, Heidelberg, Germany; anti-α-actinin, 1:200, Sigma) was performed overnight at 4 °C. After washing, membranes were incubated with the appropriate fluorochrome-conjugated antibodies for 1 h at room temperature. Membranes were mounted in Mowiol with DAPI as a counterstain.
Microphotographs were taken using a Leica DMRE microscope equipped with a digital camera (DFC300 FX, Leica, Bensheim, Germany). Image acquisition and processing were done using the Leica application suite software (Leica, Bensheim, Germany).
Immunoprecipitation and Western Blotting
Cell lysates (50 μg/ml) containing protease inhibitor mixture (Roche Diagnostics) were incubated overnight at 4 °C with EphB4-Fc (5 μg/ml). 50 μl of immobilized protein A (Trisacryl GF-2000, Pierce) were added and incubated 2 h at room temperature. Precipitates were washed, lysed, and run on 4–12% NuPAGE gels (Invitrogen). Western-blotted gels were processed as described below.
Protein lysates were obtained by lysis with ice-cold RIPA buffer, supplemented with protease inhibitor mixture (Roche Diagnostics). 25 μg of proteins were separated on 4–12% NuPAGE gels (Invitrogen). The separated proteins were transferred onto PVDF membranes (Invitrogen). Membranes were probed with antibodies against EphB4 (1 μg/ml, H-200, Santa Cruz Biotechnology), anti-phosphotyrosine (1:2000; Cell Signaling/New England Biolabs, Frankfurt, Germany), FAK (1:1000, Abcam, Cambridge, UK), phospho-specific FAK (Tyr576) (1:500, Santa Cruz Biotechnology), ERK1/2, p-ERK1/2 (Thr202/Tyr204) (1:2000 and 1:1000, both Cell Signaling/New England Biolabs, Frankfurt, Germany). The Western Breeze Chromogenic Immunodetection System (Invitrogen) was used for visualization. Blots were scanned, and densitometric analysis was performed (where appropriate) using ImageJ (National Institutes of Health, Bethesda).
Chromatin Immunoprecipitation
After cross-linking the cells with 1% formaldehyde at room temperature for 10 min, cells were harvested, centrifuged, and resuspended in lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris, pH 8.1) supplemented with a protease inhibitor mixture (Roche Diagnostics) and then sonicated (four times for 10 s). Supernatants were diluted 1:10 in dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl, 16.7 mm Tris, pH 8.1) supplemented with protease inhibitor mixture. 20 μl of each sample was reserved as a positive control (“input”). Immunoprecipitation was performed overnight with a specific antibody against Sp1 (Millipore, Schwalbach, Germany) at 4 °C. Then protein A-agarose/salmon sperm DNA (50% slurry) was added and incubated for 1 h at 4 °C. Protein A beads were pelleted by centrifugation and washed sequentially with the following buffers: low salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 150 mm NaCl, and 20 mm Tris, pH 8.1), high salt buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 500 mm NaCl, and 20 mm Tris, pH 8.1), and LiCl wash buffer (0.25 mm LiCl, 1% IGEPAL-CA630, 1% sodium deoxycholate, 1 mm EDTA, and 10 mm Tris, pH 8.1). Bead precipitates were washed twice with Tris-EDTA buffer (1 mm EDTA and 10 mm Tris, pH 8.0) and eluted with 1% SDS, 0.1 m NaHCO3. Eluates were heated for 4 h at 65 °C to reverse the formaldehyde cross-linking. Supernatants were incubated for 1 h at 45 °C with 20 μg/ml proteinase K, and genomic DNA fragments were recovered. PCR was performed with the primers amplifying the part of the ephrin-B2 promoter containing the Sp1-binding site, 5′-ACCCAATGTCGGGAGGGGATG-3′ and 5′-AGCGAGGAGCTGCGCACGCAG-3′ (11).
ALP Activity Assay
Cells were lysed in lysis buffer (0.2% Triton X-100). Lysates were incubated at 37 °C for 30 min with p-nitrophenyl phosphate (alkaline phosphatase substrate, Sigma) in reaction buffer (0.15 m Tris, 0.1 m NaCl, pH 8.5). The reactions were stopped by adding 1 n NaOH. Absorbance at 405 nm was read using a multiwell spectrophotometer (Tecan, Crailsheim, Germany). ALP activity was normalized to the total protein concentration.
Alizarin Red Staining
Cells were fixed with neutral buffered formalin and stained with 2% Alizarin Red, pH 4.3, for 5 min. Cells were rinsed with distilled water and mounted with glycerol gelatin.
Statistics
Results are presented as means ± S.D. Differences between groups were compared using one-way analysis of variance followed by the appropriate post-hoc test. All statistics were performed using SigmaStat software (SPSS Inc., Chicago). Results were considered significant with a p value < 0.05.
RESULTS
Eph Receptors and Ephrin Ligands Are Expressed by PDLF and Osteoblasts of the Alveolar Bone
To study the relevance of ephrins and Ephs in the context of bone remodeling during orthodontic tooth movement, we started our examinations with an expression analysis of ephrin and Eph mRNAs in human primary periodontal cells. PDLF and osteoblasts of the alveolar bone were tested by standard RT-PCR for the presence of five ephrin ligands (ephrin-B1, ephrin-B2, ephrin-B3, ephrin-A1, and ephrin-A2) and five Eph receptors (EphB2, EphB3, EphB4, EphA2, and EphA5) (Fig. 1A). In PDLF and osteoblasts, we detected the mRNA expression of ephrin-B1, ephrin-B2, and ephrin-B3. We could not detect the expression of mRNAs for ephrin-A1 and ephrin-A2. Of the Eph receptors analyzed, EphB2, EphB3, EphB4, EphA5, and EphA2 were detected in osteoblasts of the alveolar bone and in PDLF (Fig. 1A).
FIGURE 1.
Ephrin and Eph receptor expression analysis. A, standard RT-PCR expression analysis of ephrin ligands and Eph receptors in primary human PDLF (upper panel) and primary human osteoblasts (lower panel). B and C, qRT-PCR analysis (TaqMan chemistry) for ephrin ligands and Eph receptors in primary human PDLF (B) and primary human osteoblasts (C). Data represent the summarized results of the analysis of the three PDLF and three osteoblast cell lines used in this study. Data are presented as mean ± S.D., n = 3. All experiments were performed in triplicate.
To gain a more detailed insight into the expression profiles of selected ephrins and Ephs in PDLF and osteoblasts, we have conducted a qPCR analysis using TaqMan chemistry on the three individual PDLF and osteoblast cell lines used in this study. The results are summarized in Fig. 1B for PDLF and Fig. 1C for osteoblasts. The quantitative analysis revealed a specific expression profile of ephrins and Ephs in PDLF and osteoblasts. Both cell types were characterized by high level expressions of ephrin-B1 and EphB2. PDLF displayed higher expressions of ephrin-B2 and ephrin-B3, as compared with osteoblasts. The expression of EphB3 was low in both cell types. EphB4, EphA2, and, unlike the results obtained by standard RT-PCR, EphA5 were present in both PDLF and osteoblasts. We have only detected minimal amounts of ephrin-A2 and ephrin-A5 by means of TaqMan qRT-PCR.
We have found ephrin-B2 expression in PDLF, which has previously not been reported. A possible function has not been investigated yet. We have found ephrin-B2 in osteoblasts; this is line with previous findings (12). The prominent expression of ephrin-B1, which we observed, is in line with its known function during embryonic cell differentiation and bone formation as mutations of ephrin-B1 in humans result in craniofrontonasal syndrome.
Mechanical Strain to PDLF Leads to Spatial Redistribution of FAK and α-Actinin and to Sustained FAK Activation
Mechanical forces applied to the PDLF regulate cellular events via interactions between the ECM and integrin receptors that are located at focal adhesions (FA). A major component of FA is the nonreceptor protein-tyrosine kinase FAK. FAK plays a pivotal role in signaling pathways initiated by integrins. α-Actinin facilitates FA formation and physically links integrin-associated FA complexes with the cytoskeleton (21).
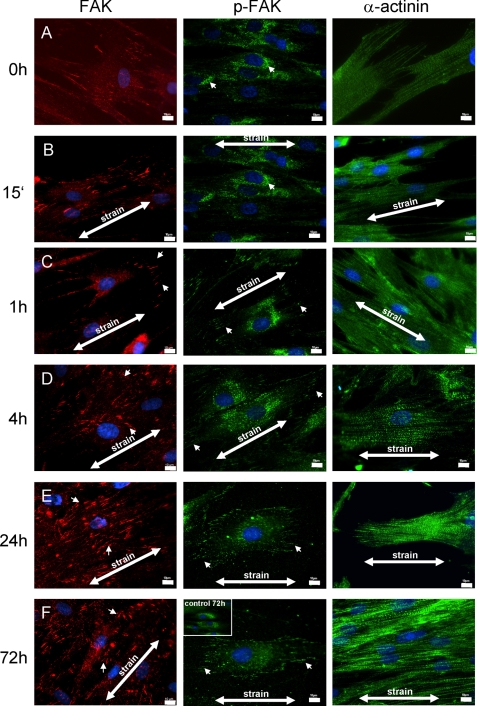
PDLF were subjected to mechanical strain and immunostainings for FAK, and p-FAK and α-actinin were performed to elucidate a possible involvement in mechanotransduction in PDLF. Mechanical loading caused changes in the cellular distribution of FAK and α-actinin in PDLF (Fig. 2). The application of tensile stress induced lamellipodial protrusions at the cell periphery within 15 min without a preferential direction. By 1 h, FAK was recruited to FAs under the lamellipodial protrusion (arrowheads in Fig. 2, C–F; 1–72 h).
FIGURE 2.
Mechanical strain leads to spatial redistribution of FAK and α-actinin and to sustained FAK activation at focal adhesions in PDLF. PDLF were subjected to longitudinal cellular strain (2.5%) for 15 min and 1, 4, 24, and 72 h. The cellular distribution of FAK and α-actinin was assessed by means of immunofluorescent staining, and tyrosine-phosphorylated proteins were stained using an anti-phosphotyrosine-specific antibody. The application of mechanical stress induced lamellipodial protrusions at the cell periphery within 15 min. By 1 h, FAK was recruited to focal adhesions (arrowheads in FAK, C–F, 1–72 h) under the lamellipodial protrusion. In static control cells, α-actinin was primarily localized to regions near the end of actin bundles (α-actinin, A). After exposure to cellular strain, stress fibers formed, and α-actinin was more evenly distributed throughout the cell and along the stress fibers (α-actinin, A–F). p-FAK staining increased in the cellular peripheries (e.g. lamellipodia) with the prolonged application of cellular strain. Unstrained PDLF after 72 h in culture are presented as an inset in the p-FAK 72-h strained microphotograph for comparison. Strain directions are indicated (arrows). Scale bar, 10 μm.
In static control cells, α-actinin was primarily localized to regions near the end of actin bundles (Fig. 2A). After exposure to cellular strain, stress fibers formed, and α-actinin was more evenly distributed throughout the cells and along the stress fibers (Fig. 2, A–F).
We have used a phospho-FAK-specific antibody to localize tyrosine-phosphorylated FAK (Tyr576) during the application of cellular strain. Static control cells and cells stretched for 15 min showed mainly perinuclear staining for p-FAK. During the course of cellular strain, we observed an increase in p-FAK staining in the cellular periphery (e.g. lamellipodia), suggesting a redistribution of phosphorylated/activated FAK.
The expression and activation of FAK were further analyzed using Western blotting with antibodies specific for FAK (total-FAK, t-FAK) and for the Tyr576-phosphorylated and -activated state of FAK (p-FAK) (Fig. 3A). The expression of t-FAK and p-FAK was normalized for β-actin. Densitometric analysis of t-FAK intensities did not reveal a significant impact on the expression of t-FAK (data not shown). Tyr576-phosphorylated FAK, indicating FAK activity, was slightly elevated after 15 min increasing to 18.3% after 30 min. Interestingly, phosphorylation was sustained between 24 and 72 h, peaking at 44.7% elevation (24 h), as compared with static control cells. Nevertheless, FAK phosphorylation did not reach significance during the course of the experiment. These findings suggested a slight but sustained FAK activation throughout the application of mechanical strain in PDLF.
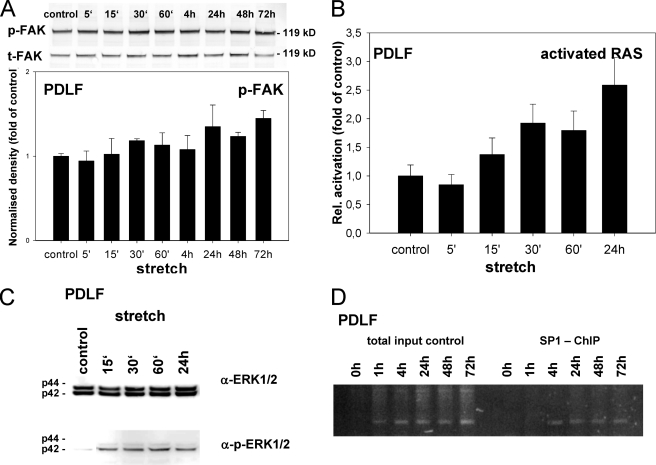
FIGURE 3.
Ephrin-B2 transcription in PDLF is activated via a Ras- and ERK1/2-dependent pathway. Immunostaining for p-FAK suggested an increased phosphorylation status of FAK at sites of FA. Therefore, FAK was further analyzed using immunoblotting against t-FAK and for phosphorylated (Tyr576) FAK (A). t-FAK and p-FAK were normalization for the expression of β-actin (data not shown). Densitometric analysis revealed the p-FAK elevation beginning after 15 min of strain and increasing to 18.3% after 30 min. Phosphorylation was sustained between 24 and 72 h, peaking at 44.7% (24 h) elevation, as compared with static control cells. B, Ras activation assay. PDLF were subjected to mechanical strain. Ras was activated 15 min after the onset of mechanical strain, and a statistically significant activation was found after 24 h. C, Western blotting for total ERK1/2 and phosphorylated ERK1/2 (p-ERK1/2). Induction of ERK1/2 phosphorylation was first detected 15 min after the onset of mechanical stress, being in line with the temporal onset of Ras activation. D, ChIP assays were performed to study the interactions between Sp1 and the ephrin-B2 promoter. Sp1 binding to the ephrin-B2 promoter was detected 4 h after the onset of mechanical strain and sustained until the end of the observation period (72 h). Values are represented as mean ± S.D.
Mechanical Strain on PDLF Activates ERK1/2 Signaling in a Ras-dependent Manner
MAPKs play an important role in the signaling pathway of mechanotransduction in osteoblastic cells and PDLF (22). Downstream signaling events emerged by FAK activation are directly associated with members of the Rho small GTPase family, including RhoA, Cdc42, and Rac (23, 24). FAK is involved in the regulation of cell growth and survival through activation mainly to different pathways, PI3K/PKD1/Akt/PKB and Grb2/SOS/Ras/Raf-1/MEK/ERK. We have focused on the Ras/ERK pathway because of its known involvement in signal transduction events associated with fibroblast/osteoblast differentiation (25, 26).
Therefore, we next sought to study stretch-mediated MAPK activation. First, we investigated whether Ras, an upstream mediator of ERK1/2 activation, is critical for MAPK signaling in mechanically stretched osteoblasts. We performed Ras activation assays based on the affinity of the Ras-binding domain of the Ras effector kinase Raf1 for GTP-Ras, which we used for the affinity purification of GTP-Ras from cell lysates of stretched PDLF (Fig. 3B). Ras was activated 15 min after the onset of mechanical strain. A statistically significant activation was found after 24 h of mechanical strain. We concluded that mechanical strain in PDLF leads to a sustained activation of Ras.
Next, we examined stretch-mediated MAPK activation. The presence of active ERK1/2 and their phosphorylated forms (p-ERK1/2) in PDLF was monitored by Western blotting. Induction of ERK1/2 phosphorylation was first detected 15 min after the onset of mechanical stress, being in line with the temporal onset of Ras activation, and remained detectable during the course of the experiment (Fig. 3C). These findings indicate that in PDLF mechanical strain leads to a Ras-dependent ERK1/2 activation.
Ephrin-B2 Expression Increases in PDLF Exposed to Mechanical Strain
To determine whether ephrin-B2 is modulated by mechanical strain in PDLF, we analyzed the expression of ephrin-B2 and its receptor EphB4. qRT-PCR analysis revealed an increased ephrin-B2 expression in PDLF starting from 1 h after the onset of strain. Significant overexpression was demonstrated after 24 h and was sustained until 72 h, as compared with static control cells (Fig. 4A). To rule out any compensatory activation of the EphB4 receptor in PDLF, we have also checked for the expression of EphB4 mRNA in PDLF. EphB4 was not significantly altered during the course of the experiment (Fig. 4B). As a consequence, we could rule out that possible effects of up-regulated ephrin-B2 on PDLF for the communication with other cell types (e.g. osteoblasts of the alveolar bone) are compensated for by the up-regulation of the EphB4 receptor on PDLF themselves.
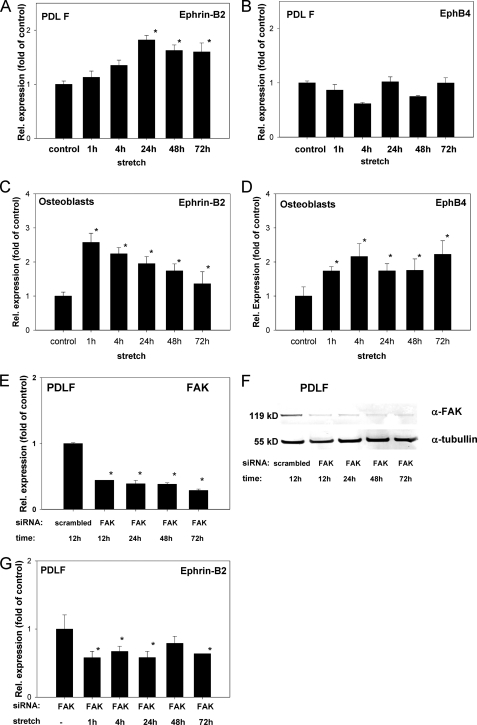
FIGURE 4.
Ephrin-B2 expression increases in PDL fibroblasts and decreases in osteoblasts exposed to mechanical strain. qRT-PCR analyses for ephrin-B2 (A) and EphB4 (B) expression after the application of mechanical strain in PDLF. Mechanical strain was applied for 1–72 h at 2.5% elongation. Significant overexpression for ephrin-B2, as compared with static control cells, was demonstrated after 24 h and sustained until 72 h (A). EphB4 was not significantly altered during the course of the experiment. Ephrin-B2 and EphB4 interactions might be evident within the osteoblast lineage. Therefore, osteoblasts were subjected to mechanical strain and tested for the transcription of ephrin-B2 and EphB4 by means of qRT-PCR (C and D). Ephrin-B2 and EphB4 expression was increased. It is noteworthy that temporal expression pattern of ephrin-B2 in osteoblasts was reciprocal to the temporal ephrin-B2 expression pattern observed in PDLF. Values are represented as mean ± S.D. *, p < 0.05 versus control, n = 3. qRT-PCR experiments were performed in triplicate. FAK might be a prominent mediator of mechano-induced alterations of ephrin-B2 expression in PDLF. siRNA against FAK was used to knock down FAK expression in PDLF. siRNA against FAK and a scrambled control siRNA were transfected to PDLF, and FAK expression was monitored on the mRNA level using qRT-PCR. E, siRNA against FAK attenuated FAK expression in PDLF after 12 h and attenuation remained significant up to 72 h. FAK attenuation mediated by siRNA was also monitored on the protein level using Western blotting against FAK (F), and probing for tubulin served as a loading control. Western blotting confirmed the siRNA-mediated knockdown of FAK in PDLF. PDLF attenuated for FAK using siRNA were subjected to mechanical strain for 1–72 h (G), and ephrin-B2 expression was assessed at discrete time points using qRT-PCR. Unlike WT PDLF, siRNA-transfected PDLF did not show an up-regulation of ephrin-B2 upon the application of mechanical strain.
To link the data on the strain-dependent activation of a FAK-dependent pathway involving Ras and ERK1/2 in PDLF (Fig. 3, A–C) and the strain-dependent up-regulation of ephrin-B2 mRNA expression in PDLF (Fig. 4A), we have used an siRNA approach to knock out FAK in PDLF. Validated siRNAs (Qiagen) against FAK were transfected into PDLF. The effectiveness of the FAK knockdown was monitored using q-PCR (Fig. 4E) and Western blotting against FAK (Fig. 4F) and tubulin, which served as a loading control. FAK expression was significantly reduced on the mRNA level already 12 h post-transfection, and it remained suppressed up to 72 h after transfection. This was of importance as 72 h also represented the maximum duration of strain application on PDLF. Western blotting confirmed the FAK knockdown also on the protein level (Fig. 4F). PDLF with siRNA-attenuated FAK expression were subjected to mechanical strain for 1–72 h, and the ephrin-B2 mRNA expression was monitored at discrete time points using qRT-PCR. The knockdown of FAK in PDLF impeded the strain-dependent up-regulation of ephrin-B2 (Fig. 4G), which had been observed in WT PDLF (Fig. 4A). These results suggest role for FAK in the strain-dependent regulation of ephrin-B2 expression in PDLF.
Ephrin-B2 Expression Increases in Osteoblasts Exposed to Mechanical Strain
Recently, the communication between ephrin-B2 and EphB4 within the osteoblast lineage has been reported, suggesting a paracrine role for ephrin-B2 and EphB4 signaling in osteoblast differentiation (16). It is also known that osteoblasts are responsive to mechanical stimuli. To evaluate possible paracrine relations of ephrin-B2 and EphB4 in osteoblasts of the alveolar bone, we have subjected osteoblasts to mechanical strain and tested for the transcription of ephrin-B2 and EphB4 by means of qRT-PCR (Fig. 4, C and D). We observed an increase in both ephrin-B2 and EphB4 expression. It is noteworthy that the expression of ephrin-B2 in osteoblasts peaked at 1 h and decreased over time. This particular temporal expression pattern was reciprocal to the temporal ephrin-B2 expression pattern observed in PDLF. This suggests a dual role for ephrin-B2 for the onset of osteogenesis in the alveolar bone; paracrine stimulation within the osteoblasts appears to be an early effect, whereas paracrine stimulation between PDLF and osteoblasts may exert a long term effect on osteogenesis.
Transcription Factor Sp1 Is Involved in the Strain-dependent Activation of Ephrin-B2 Transcription in PDLF
ChIP assays were performed to study the interactions between Sp1 and the ephrin-B2 promoter. Chromatin extracts from static or mechanically stressed PDLF were immunoprecipitated with an antibody against Sp1. The immunoprecipitates were subjected to PCR with primers directed against the ephrin-B2 promoter region containing the Sp1 consensus sequence. The ephrin-B2 promoter interacted with Sp1. Sp1 binding to the ephrin-B2 promoter was detected 4 h after the onset of mechanical strain and was sustained during the course of the experiment (Fig. 3D).
The results of the ChIP assays suggested that Sp1 binding to its consensus sequence in the ephrin-B2 promoter is involved in the mechanical stress-dependent activation of ephrin-B2 transcription in PDLF. The induction of Sp1 binding to its consensus sequence in the ephrin-B2 promoter upon the application of mechanical strain to PDLF is likely to be regulated via a pathway involving FAK, Ras, and ERK1/2 (Fig. 3, A–C).
Ephrin-B2-Fc Activates Runx2 and ALP Transcription in Osteoblasts of the Alveolar Bone in a Ras- and ERK1/2-dependent Manner
Based on our studies on PDLF, we sought to further investigate the function of PDLF ephrin-B2 signaling on osteoblasts of the alveolar bone. Osteoblasts express the EphB4 receptor, and bi-directional signaling of ephrin-B2 on PDLF and EphB4 on osteoblasts of the alveolar bone might contribute to the onset of osteogenesis after the application of mechanical strain.
Osteoblasts of the alveolar bone were stimulated with 1 μg/ml ephrin-B2-Fc for 2–10 min. Whole cell lysates were immunoprecipitated with antibodies against EphB4 and probed for EphB4 and for phosphotyrosine. Tyrosine phosphorylation of the EphB4 receptor was detected after 5 min and was sustained through the course of the experiment (Fig. 5A).
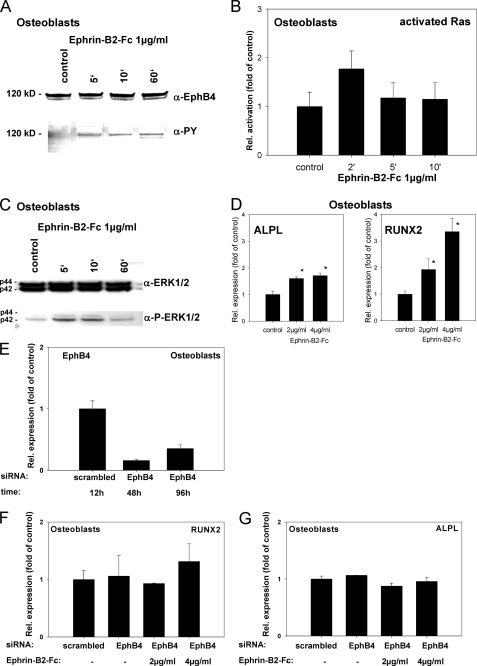
FIGURE 5.
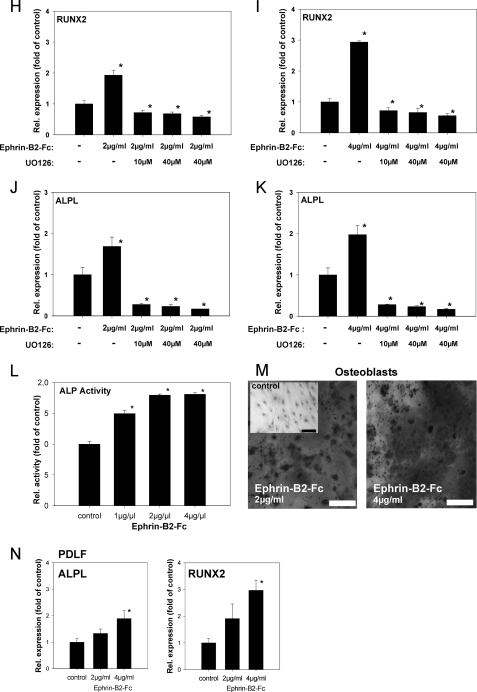
Ephrin-B2 activates Runx2 and ALPL transcription in osteoblasts of the alveolar bone via a Ras- and ERK1/2-dependent pathway. To prove that ephrin-B2-Fc causes EphB4 receptor phosphorylation in osteoblasts of the alveolar bone, osteoblasts were stimulated with 1 μg/ml ephrin-B2-Fc for 5–60 min. Tyrosine phosphorylation of the EphB4 receptor was detected after 5 min and sustained through the course of the experiment (A). To elucidate a putative signaling pathway linking ephrin-B2-Fc stimulation and downstream events, osteoblasts of the alveolar bone were stimulated with 1 μg/ml ephrin-B2-Fc for 2–10 min, and Ras activation was monitored. Ras was transiently activated after ephrin-B2-Fc stimulation (2 min) (B). Ras might transmit downstream signals via ERK1/2. To reveal a possible ERK1/2 dependence of ephrin-B2 downstream signaling, protein lysates of osteoblasts of the alveolar bone stimulated with ephrin-B2-Fc (1 μg/ml) for 5–60 min were probed with antibodies against ERK1/2 and pERK1/2. An increase in phosphorylated ERK1/2 was detected 5 min after stimulation, gradually decreasing to nearly base-line phosphorylation after 60 min (C). To detect further downstream effects of ephrin-B2-dependent signaling in osteoblasts of the alveolar bone, osteoblasts were stimulated with 2 and 4 μg/ml, respectively, of ephrin-B2-Fc for 6 days. qRT-PCR revealed the significant induction of Runx2, as well as ALPL in a dose-dependent manner (D). Although EphB4 is the only EphB receptor exhibiting a binding specificity for a single ligand, ephrin-B2 has binding affinity for other receptors of the EphB-class. To rule out the involvement of other EphB receptors for the ephrin-B2-dependent activation of Runx2 and ALP mRNA expression, we have used siRNA to knock down EphB4 in osteoblasts. siRNA against EphB4 effectively suppressed EphB4 expression in osteoblasts for up to 96 h (E). Stimulation of osteoblasts with 2 and 4 μg/ml ephrin-B2 in the presence of siRNA against EphB4 did not result in an up-regulation of Runx2 (F) or ALP (G) expression as observed in WT osteoblasts (D). The activation of ERK1/2 in osteoblasts upon ephrin-B2 stimulation (C) suggested the involvement of an ERK1/2-dependent pathway in the induction of osteoblastogenic gene expression after ephrin-B2 stimulation. We have used a specific MEK1/2 inhibitor, UO126, to block ERK1/2 activation in osteoblasts. Osteoblasts were stimulated with 2 or 4 μg/ml ephrin-B2 in the absence or presence of different concentrations of UO126. Runx2 (H and I) and ALP (J and K) were monitored by means of qRT-PCR. Already at a concentration of 10 μm, UO126 attenuated the ephrin-B2-dependent activation of Runx2 and its downstream target ALP in osteoblasts. These results suggested an involvement of an ERK1/2-dependent pathway in the ephrin-B2-dependent regulation of Runx2 in osteoblasts. Increased ALP transcription was accompanied by an increase in ALP activity (L). Alizarin Red staining for calcified nodules in osteoblasts stimulated for 6 days with ephrin-B2-Fc (2 and 4 μg/ml) further confirmed the osteoblastogenic effects of ephrin-B2-Fc stimulation. An untreated control is shown as an inset. Scale bar, 20 μm (M). Subpopulations of PDLF might contribute to osteogenesis at tension sites in orthodontic tooth movement. Therefore, we have tested for the expression of Runx2 and ALP in PDLF after stimulation with ephrin-B2-Fc by means of qRT-PCR. Both ALP and Runx2 transcription were up-regulated in a dose-dependent manner in PDLF (N). Up-regulation of ALPL and Runx2 transcription reached significance after stimulation with 4 μg/ml of ephrin-B2-Fc. Values are represented as mean ± S.D. *, p < 0.05 versus control, n = 3.
On whole cell lysates, we performed Ras activation assays based on the Ras-binding domain of the Ras effector kinase Raf1. Ras was transiently activated after ephrin-B2-Fc stimulation (2 min) (Fig. 5B). To obtain more insight in downstream events of EphB4 signaling in osteoblasts of the alveolar bone, we probed whole cell lysates of stimulated osteoblasts of the alveolar bone with antibodies against ERK1/2 and pERK1/2. An increase in phosphorylated ERK1/2 was detected 5 min after stimulation, gradually decreasing to nearly base-line phosphorylation after 60 min (Fig. 5C).
To gain insight in the possible functional consequences of ephrin-B2-dependent stimulation of osteoblasts of the alveolar bone, we analyzed osteoblast-specific gene expression in stimulated osteoblasts. Hallmarks of osteoblast differentiation are the expression of Runx2, the pivotal transcription factor in osteoblast differentiation, and ALP, a downstream target of Runx2.
Osteoblasts of the alveolar bone were stimulated with 2 and 4 μg/ml ephrin-B2-Fc for 6 days. qRT-PCR revealed the significant induction of Runx2 as well as of ALP (Fig. 5D).
Unlike the other promiscuous receptors in the Eph family, the EphB4 receptor exhibits specificity for a single ligand, ephrin-B2, while binding only very weakly to both ephrin-B1 and ephrin-B3. Nevertheless, ephrin-B2 can bind to several receptors within the EphB-receptor class (27). To confirm that the observed activation of Runx2 and ALP in osteoblasts upon stimulation with ephrin-B2 was dependent on EphB4 activation, we have applied an siRNA approach to knock down EphB4 expression in osteoblasts. Validated siRNAs (Qiagen) against EphB4 were transfected into osteoblasts (500 ng/each, 4 × 105 cells, 60-mm dishes). Effectiveness of the siRNA knockdown was monitored using qRT-PCR. Knockdown on the mRNA was evident, and the suppression remained effective for up to 96 h; this was of importance as the stimulation with ephrin-B2 was conducted for 96 h as well. Osteoblasts were transfected with siRNA directed against EphB4 or a scrambled siRNA as a control. Stimulation with 2 or 4 μg/ml ephrin-B2 was conducted for 4 days, and ALP and Runx2 mRNA expression was monitored using qRT-PCR. Unlike WT osteoblasts (Fig. 5D), siRNA-transfected osteoblasts did not reveal an ephrin-B2-dependent up-regulation of ALP and Runx2 (Fig. 5, F and G). Therefore, we could rule out the involvement of an EphB4-independent pathway leading to ALP and Runx2 activation upon ephrin-B2 stimulation.
We have found ERK1/2 activation upon stimulation with ephrin-B2 in osteoblasts. To confirm the involvement of the ERK1/2 pathway in the signaling events leading to the up-regulation of Runx2 and ALP, we have used a specific inhibitor of MEK1/2, UO126. MEK1/2 are the directly upstream-located kinases of ERK1/2 and activate ERK1/2 through phosphorylation of Thr202/Tyr204. Osteoblasts were stimulated with 2 or 4 μg/ml ephrin-B2 in the absence of UO126 and the presence of 10-40 μm UO126. Stimulation with ephrin-B2 in the absence of UO126 confirmed our previous results (Fig. 5, H–K) leading to an induction of Runx2 and ALP expression. UO126 already at 10 μm inhibited the ephrin-B2-dependent activation of Runx2 and ALP expression in osteoblasts, independently of the amount of ephrin-B2 (Fig. 5, H–K). The application of a specific MEK1/2 inhibitor circumvented the ephrin-B2-dependent induction of Runx2 and ALP in osteoblasts, suggesting an involvement of an ERK1/2-dependent pathway in the ephrin-B2-dependent activation of Runx2 and ALP in osteoblasts.
To test for functional consequences of the increased ALP transcription, we performed an ALP activity assay on lysates of stimulated osteoblasts. Stimulation with 1 μg/ml ephrin-B2-Fc already led to a significant increase of ALP activity in osteoblasts of the alveolar bone. Stimulation with 2 μg/ml further increased the ALP activity, whereas 4 μg/ml did not exert an additional effect (Fig. 5L). Alizarin Red staining for calcified nodules in stimulated osteoblasts further confirmed the osteoblastogenic effects of ephrin-B2-Fc stimulation (Fig. 5M).
Together, the results of these experiments suggest that ephrin-B2 leads to a Ras and ERK1/2-dependent activation of the osteoblastogenic transcription factor Runx2 and its downstream target ALP in osteoblasts of the alveolar bone. The activation had functional consequences, as the ALP activity of the cells significantly increased, and the release of calcified matrix was also stimulated. The findings suggest a profound effect of ephrin-B2-Fc via EphB4 receptor activation and Ras/ERK1/2-dependent pathways on the functional properties of osteoblasts of the alveolar bone.
Ephrin-B2-Fc Induces Runx2 and ALPL Transcription in PDLF
Subpopulations of PDLF have been reported to be able to express osteoblast-specific genes (14) and might contribute to osteogenesis at tension sites in orthodontic tooth movement. Here, we showed that mechanical strain in PDLF induces the expression of ephrin-B2. Ephrin-B2-EphB4 signaling in PDLF might have comparable effects, as they have been proposed for the osteoblastic lineage. To find out whether PDLF in our setting might also contribute to osteogenesis, we have tested for the induction of the expression of Runx2 and ALP after stimulation with ephrin-B2-Fc by means of qRT-PCR. ALP and Runx2 transcription were up-regulated in a dose-dependent manner in PDLF after stimulation with ephrin-B2-Fc and reached significance after stimulation with 4 μg/ml ephrin-B2-Fc (Fig. 5N).
These results suggest that a subpopulation of PDLF might contribute to osteogenesis via the induction of osteoblastogenic gene expression. Moreover, this effect might be controlled by the mechanodependent induction of ephrin-B2 in PDLF, which might stimulate osteogenic gene expression in a paracrine manner through ephrin-B2-EphB4 signaling.
DISCUSSION
Teeth are moved through the alveolar bone by the application of orthodontic forces. These forces are then transmitted through the PDL to the supporting alveolar bone leading to deposition or resorption of bone, depending on whether the tissues are exposed to a tensile or compressive mechanical strain.
The first cellular recipients of mechanical strain are mechanoresponsive PDLF. The mechanism for osteogenesis at tension sites is not clearly understood.
The ephrin/Eph system has initially been characterized for its involvement in the regulation of cellular motility and alteration in adhesions to the ECM. Ephrin/Eph interactions frequently modulate cellular repulsion or attraction, indicating a causal relation between ECM, the cytoskeleton, and the ephrin/Eph system. Furthermore, mechanical forces, which alter the interaction of the cells with the ECM and/or influence the organization of the cytoskeleton, have an impact on the function and expression of members of the ephrin/Eph family, at least in endothelial cells (10, 11). These data qualify the ephrin-B2-EphB4 system as a putative regulator of osteogenesis at tension sites in orthodontic tooth movement.
Ephrin-B2-EphB4 signaling at tension sites might play a dual role in promoting osteogenesis. (i) Ephrin-B2-EphB4 signaling might link the initial recipients of the applied mechanical forces, PDLF, to the neighboring osteoblasts of the alveolar bone and might have consequences for the regulation of osteogenesis. (ii) The interplay between ephrin-B2 and EphB4 within the PDLF population itself might have an impact on the contribution to osteogenesis of this cell type.
Therefore, we first set out to study the function of ephrin-B2-EphB4 signaling in an in vitro model for cellular strain on PDLF. Second, we evaluated functional consequences of ephrin-B2-EphB4 interactions in osteoblasts of the alveolar bone.
In orthodontic tooth movement, relatively low static forces are applied during the course of the treatment, and profound effects on tooth positioning might be obvious only after weeks of treatment. The durations used in our experiments were adjusted to represent the effects of a long term treatment on the PDLF and osteoblasts, as well as to elucidate whether initial signaling events are triggered by the applied forces.
Ephrin-B2 is being significantly up-regulated on the transcriptional level in PDLF upon the application of longitudinal cellular strain. In the physiological anatomical setting, PDLF are neighbors of osteoblasts of the alveolar bone, allowing for cell-cell contact between PDLF and osteoblasts of the alveolar bone, a prerequisite for ephrin/Eph interactions.
To test for functional consequences of ephrin-B2-EphB4 interactions on osteoblasts of the alveolar bone, we have stimulated osteoblasts with ephrin-B2-Fc chimeras, pre-clustered with an anti-Fc antibody. Stimulation with ephrin-B2-Fc leads to an increased expression of Runx2, the pivotal transcription factor of osteoblastogenic differentiation. Moreover, ALP, a target gene of Runx2, was found to be up-regulated. In addition, osteoblasts stimulated with ephrin-B2-Fc exposed higher ALP activity together with the deposition of calcified nodules (Alizarin Red staining).
These findings qualify the ephrin-B2-EphB4 interaction between PDLF and osteoblasts of the alveolar bone as an important regulatory mechanism for the onset of osteogenesis at tension sites during orthodontic tooth movement. Furthermore, we revealed a putative mechanism of how mechanical strain might induce the expression of ephrin-B2 in stretched PDLF.
The most widely used initiators of mechanotransduction are integrin receptors (28). Integrins are of paramount importance for mechanotransduction in cells (29, 30). Integrin-mediated signaling includes the downstream activation of adaptor proteins, e.g. FAK and GTPases of the Ras superfamily. Similar to FAK, Ras GTPases are sensitive to mechanical stimuli and play important roles in cellular events in response to these stimuli through interactions with integrins (31).
Therefore, we hypothesized that the induction of ephrin-B2 transcription is dependent on FAK activation. Immunofluorescent staining for FAK and phosphotyrosine revealed a spatial redistribution of FAK during the application of mechanical strain, which was accompanied by a redistribution of phosphorylated FAK at sites of focal interactions. Western blotting revealed a slight increase in Tyr576 phosphorylation of FAK, suggesting the activation of the FAK domain. FAK is a major mechanosensitive kinase linked to integrin signaling, and its role in mechanotransduction has been widely studied using myocytes (32, 33). Given that FAK phosphorylation is correlated with the activation of small GTPases in most of the cell lines studied, it is conclusive that mechanical strain could activate Ras-GTPase pathways in PDLF.
Because mechanical stress leads to cellular messages and outcomes through similar mechanoreceptors and signaling effectors in many types of cells, it is likely that common signaling mechanisms are involved in mechanotransduction pathways. In particular, ERK1/2 is the most prominent kinase activated by mechanical stimuli in most cells examined (34). Many studies have shown that Ras GTPases and FAK are downstream mechanosensors of integrins as well as upstream effectors that induce ERK1/2 phosphorylation in stress-exposed cells (35, 36). These findings suggested a pivotal role for FAK for the induction of the signaling pathway leading to ephrin-B2 regulation upon the induction of mechanical strain in PDLF. To confirm its role for the induction of ephrin-B2 expression, we have used an siRNA approach to knock down FAK expression in PDLF. siRNA attenuated FAK expression on the mRNA and protein level. In contrast to WT PDLF, siRNA-transfected PDLF did not respond to mechanical strain with an induction of ephrin-B2 expression, thereby suggesting an important role of FAK for the mechanodependent regulation of ephrin-B2 in PDLF. Together, these findings demonstrate a FAK-, Ras- and ERK1/2-dependent pathway linking mechanical strain and ephrin-B2 up-regulation in PDLF.
The human ephrin-B2 promoter contains an Sp1 consensus sequence. Sp1 has also been reported to be involved in shear stress-mediated transcriptional up-regulation of genes, including VEGFR-1 and -2 and, via hypoxia-inducible-factor, also ephrin-B2 (37–39) It is also known that ERK1/2 MAPK phosphorylates Sp1 at two threonine residues, Thr453 and Thr739 (40). By using chromatin immunoprecipitation (ChIP), we could reveal that the Sp1 is bound to its consensus site in the ephrin-B2 promoter in response to mechanical stress in PDLF. Therefore, Sp1 might be responsible for the transcriptional activation of the ephrin-B2 gene upon the application of mechanical stress in PDLF. However, the molecular mechanisms by which mechanical stress activates Sp1 remained unclear. Based on our current findings, we propose a mechanotransduction involving FAK, Ras-GTPase, and ERK1/2 leading to the activation of Sp1 in PDLF. The onset of mechanical strain in PDLF leads to the transcriptional activation of ephrin-B2. Ephrin-B2-EphB4 signaling between PDLF and osteoblasts of the alveolar bone might then contribute to osteogenesis during tooth movement.
We could show that osteoblasts of the alveolar bone express the EphB4 receptor, a prerequisite for a bi-directional signaling between ephrin-B2 expressing PDLF and EphB4 expressing osteoblasts of the alveolar bone. In vitro, the stimulation of osteoblasts with preclustered ephrin-B2-Fc has led to the induction of osteoblastogenic gene expression and increased ALP activity along with increased mineralization of osteoblasts. This is in line with the proposed role for the ephrin-B2-EphB4 interaction between osteoblasts and osteoclasts (12) and with the proposed communication within the osteoblastic lineage (16).
The EphB4 receptor is unique in the class of EphB receptors as it exhibits specificity for a single ligand, ephrin-B2 (27, 41, 42). Binding to both ephrin-B1 and ephrin-B3 is only very weak. However, ephrin-B2 can bind to several receptors within the EphB-receptor class. To rule out, the involvement of other receptors of the EphB-class for the observed activation of Runx2 and subsequently ALP in osteoblasts upon stimulation with ephrin-B2, we have applied an siRNA approach to knock down EphB4 expression in osteoblasts. EphB4 was effectively knocked down by siRNA for up to 4 days, the time necessary for the stimulation with ephrin-B2 for the up-regulation of Runx2 and ALP. When siRNA-transfected osteoblasts were stimulated with 2 or 4 μg/ml, an induction of Runx2 and ALP was no longer evident. The knockdown of EphB4 in osteoblasts inhibited the ephrin-B2-dependent activation of Runx2 transcription that was evident in WT osteoblasts. This result confirmed the involvement of ephrin-B2-dependent EphB4 signaling for the activation of Runx2 expression in osteoblasts.
A putative downstream signaling mechanism leading to ephrin-B2-dependent activation of Runx2 expression in osteoblasts might involve ERK1/2, as we have found increased ERK1/2 phosphorylation upon ephrin-B2 stimulation in osteoblasts. To gather further evidence for an ERK1/2-dependent pathway, we have used the specific ERK1/2 inhibitor UO126. The stimulation of osteoblasts with ephrin-B2 in the presence of UO126 did not yield in the activation of Runx2 expression. Runx2 levels remained at or below base-line levels. Concomitantly with the inhibition of Runx2 expression, UO126 also led to the inhibition of ALP expression in UO126-treated ephrin-B2-stimulated osteoblasts. These results indicate a possible involvement of an ERK1/2-dependent pathway in the ephrin-B2-dependent regulation of Runx2 and ALP in osteoblasts.
Stimulation of PDLF with ephrin-B2-Fc led to an increase in the mRNA for Runx2 and ALP, suggesting that at least a subpopulation of the PDLF might contribute directly to osteogenesis at tension sites in tooth movement. This is in line with previous findings indicating an increase of osteoblastic gene expression in PDLF after mechanical stress (14, 15). PDLF and osteoblasts of the alveolar bone might therefore contribute to a significant extent to the induction of osteogenesis at tension sites during orthodontic tooth movement.
In conclusion, the results of our study support an important role for ephrin-B2-EphB4 signaling between PDLF and osteoblasts at tension sites of bone remodeling. These results establish a novel concept in the regulation of bone remodeling during orthodontic tooth movement. Intervention with the function of these molecules may provide novel opportunities for the development of therapeutic strategies aimed at a pharmacological manipulation of tooth movement.
Footnotes
- PDL
- periodontal ligament
- PDLF
- periodontal ligament fibroblasts
- ALPL
- liver/bone/kidney-type alkaline phosphatase
- FA
- focal adhesion
- FA
- focal adhesion
- FAK
- focal adhesion kinase
- fw
- forward primer
- GPI
- glycosylphosphatidylinositol
- p-FAK
- phosphorylated focal adhesion kinase
- qRT-PCR
- quantitative real time PCR
- rv
- reverse primer
- t-FAK
- total focal adhesion kinase
- ECM
- extracellular matrix.
REFERENCES
- 1. Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 2. Boyce B. F., Xing L. (2008) Arch. Biochem. Biophys. 473, 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holder N., Durbin L., Cooke J. (2000) Ernst Schering Res. Found. Workshop, 29, 123–147 [DOI] [PubMed] [Google Scholar]
- 4. Adams R. H., Klein R. (2000) Trends Cardiovasc. Med. 10, 183–188 [DOI] [PubMed] [Google Scholar]
- 5. Wilkinson D. G. (2001) Nat. Rev. Neurosci. 2, 155–164 [DOI] [PubMed] [Google Scholar]
- 6. Kullander K., Klein R. (2002) Nat. Rev. Mol. Cell Biol. 3, 475–486 [DOI] [PubMed] [Google Scholar]
- 7. Pasquale E. B. (2008) Cell 133, 38–52 [DOI] [PubMed] [Google Scholar]
- 8. Erber R., Eichelsbacher U., Powajbo V., Korn T., Djonov V., Lin J., Hammes H. P., Grobholz R., Ullrich A., Vajkoczy P. (2006) EMBO J. 25, 628–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Himanen J. P., Nikolov D. B. (2003) Trends Neurosci. 26, 46–51 [DOI] [PubMed] [Google Scholar]
- 10. Korff T., Braun J., Pfaff D., Augustin H. G., Hecker M. (2008) Blood 112, 73–81 [DOI] [PubMed] [Google Scholar]
- 11. Obi S., Yamamoto K., Shimizu N., Kumagaya S., Masumura T., Sokabe T., Asahara T., Ando J. (2009) J. Appl. Physiol. 106, 203–211 [DOI] [PubMed] [Google Scholar]
- 12. Zhao C., Irie N., Takada Y., Shimoda K., Miyamoto T., Nishiwaki T., Suda T., Matsuo K. (2006) Cell Metab. 4, 111–121 [DOI] [PubMed] [Google Scholar]
- 13. Mundy G. R., Elefteriou F. (2006) Cell 126, 441–443 [DOI] [PubMed] [Google Scholar]
- 14. Wescott D. C., Pinkerton M. N., Gaffey B. J., Beggs K. T., Milne T. J., Meikle M. C. (2007) J. Dent. Res. 86, 1212–1216 [DOI] [PubMed] [Google Scholar]
- 15. Kawarizadeh A., Bourauel C., Götz W., Jäger A. (2005) J. Dent. Res. 84, 902–906 [DOI] [PubMed] [Google Scholar]
- 16. Martin T. J., Allan E. H., Ho P. W., Gooi J. H., Quinn J. M., Gillespie M. T., Krasnoperov V., Sims N. A. (2010) Adv. Exp. Med. Biol. 658, 51–60 [DOI] [PubMed] [Google Scholar]
- 17. Diercke K., Sen S., Kohl A., Lux C. J., Erber R. (2011) J. Dent. Res. 90, 1108–1115 [DOI] [PubMed] [Google Scholar]
- 18. Hasegawa S., Sato S., Saito S., Suzuki Y., Brunette D. M. (1985) Calcif. Tissue Int. 37, 431–436 [DOI] [PubMed] [Google Scholar]
- 19. Egea J., Klein R. (2007) Trends Cell Biol. 17, 230–238 [DOI] [PubMed] [Google Scholar]
- 20. Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. (2002) BioTechniques 32, 1372–1374, 1376,, 1378–1379 [PubMed] [Google Scholar]
- 21. Rajfur Z., Roy P., Otey C., Romer L., Jacobson K. (2002) Nat. Cell Biol. 4, 286–293 [DOI] [PubMed] [Google Scholar]
- 22. Kook S. H., Hwang J. M., Park J. S., Kim E. M., Heo J. S., Jeon Y. M., Lee J. C. (2009) J. Cell. Biochem. 106, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 23. Giancotti F. G., Ruoslahti E. (1999) Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 24. Schoenwaelder S. M., Burridge K. (1999) Curr. Opin. Cell Biol. 11, 274–286 [DOI] [PubMed] [Google Scholar]
- 25. Kono S. J., Oshima Y., Hoshi K., Bonewald L. F., Oda H., Nakamura K., Kawaguchi H., Tanaka S. (2007) Bone 40, 68–74 [DOI] [PubMed] [Google Scholar]
- 26. Boutahar N., Guignandon A., Vico L., Lafage-Proust M. H. (2004) J. Biol. Chem. 279, 30588–30599 [DOI] [PubMed] [Google Scholar]
- 27. Himanen J. P., Rajashankar K. R., Lackmann M., Cowan C. A., Henkemeyer M., Nikolov D. B. (2001) Nature 414, 933–938 [DOI] [PubMed] [Google Scholar]
- 28. Zhang S. J., Truskey G. A., Kraus W. E. (2007) Am. J. Physiol. Cell Physiol 292, C2057–C2069 [DOI] [PubMed] [Google Scholar]
- 29. Katsumi A., Orr A. W., Tzima E., Schwartz M. A. (2004) J. Biol. Chem. 279, 12001–12004 [DOI] [PubMed] [Google Scholar]
- 30. Schwartz M. A., DeSimone D. W. (2008) Curr. Opin. Cell Biol. 20, 551–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bar-Sagi D., Hall A. (2000) Cell 103, 227–238 [DOI] [PubMed] [Google Scholar]
- 32. Dalla Costa A. P., Clemente C. F., Carvalho H. F., Carvalheira J. B., Nadruz W., Jr., Franchini K. G. (2010) Cardiovasc. Res. 86, 421–431 [DOI] [PubMed] [Google Scholar]
- 33. de Oliveira M. V., Marin T. M., Clemente C. F., Costa A. P., Judice C. C., Franchini K. G. (2009) FEBS Lett. 583, 2975–2981 [DOI] [PubMed] [Google Scholar]
- 34. Liedert A., Kaspar D., Claes L., Ignatius A. (2006) Biochem. Biophys. Res. Commun. 342, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 35. Basdra E. K., Papavassiliou A. G., Huber L. A. (1995) Biochim. Biophys. Acta 1268, 209–213 [DOI] [PubMed] [Google Scholar]
- 36. Li B. S., Veeranna, Gu J., Grant P., Pant H. C. (1999) Eur. J. Biochem. 262, 211–217 [DOI] [PubMed] [Google Scholar]
- 37. Abumiya T., Sasaguri T., Taba Y., Miwa Y., Miyagi M. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 907–913 [DOI] [PubMed] [Google Scholar]
- 38. Sohl M., Lanner F., Farnebo F. (2010) Biochem. Biophys. Res. Commun. 391, 24–27 [DOI] [PubMed] [Google Scholar]
- 39. Urbich C., Stein M., Reisinger K., Kaufmann R., Dimmeler S., Gille J. (2003) FEBS Lett. 535, 87–93 [DOI] [PubMed] [Google Scholar]
- 40. Milanini-Mongiat J., Pouysségur J., Pagès G. (2002) J. Biol. Chem. 277, 20631–20639 [DOI] [PubMed] [Google Scholar]
- 41. Gale N. W., Yancopoulos G. D. (1999) Genes Dev. 13, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 42. Myshkin E., Wang B. (2003) J. Chem. Inf. Comput. Sci. 43, 1004–1010 [DOI] [PubMed] [Google Scholar]