Abstract
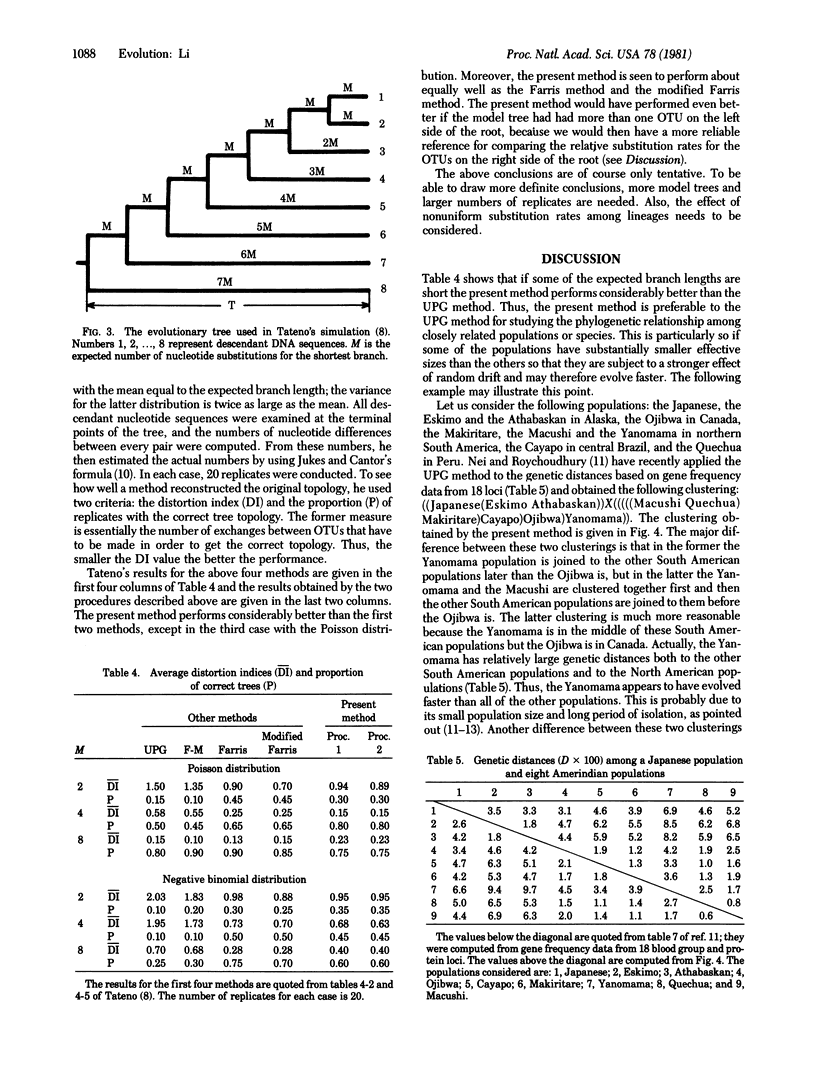
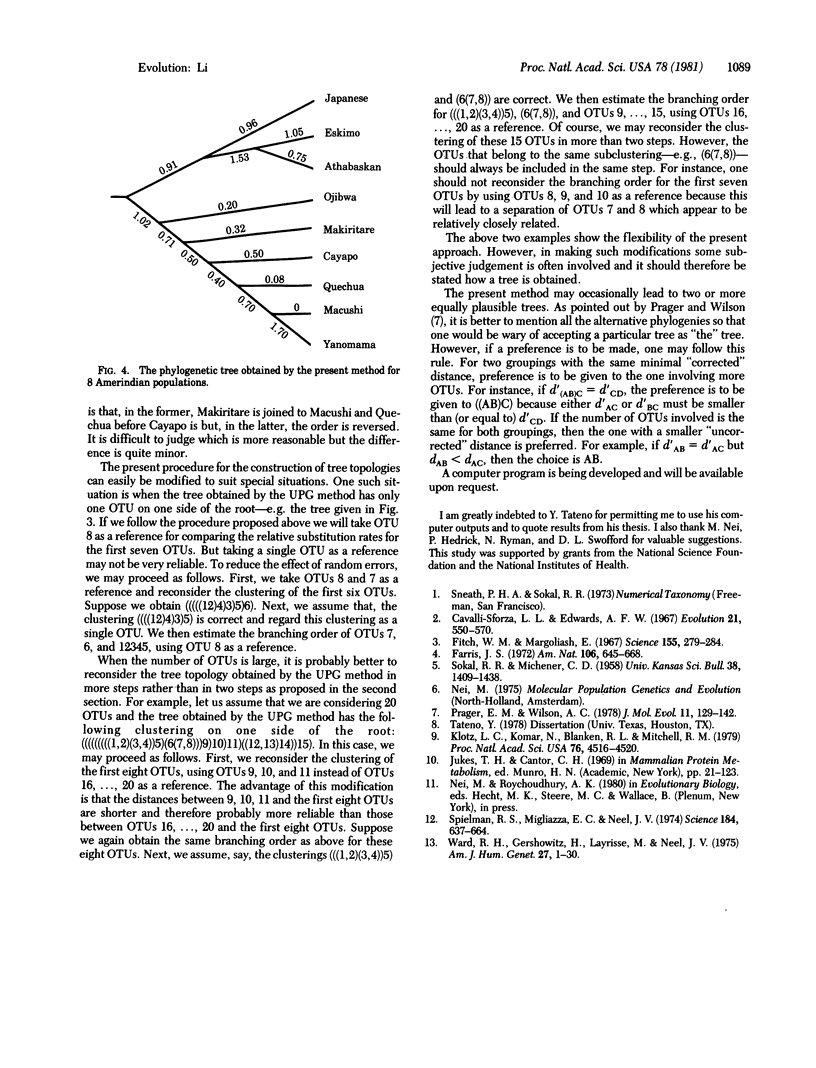
A simple method is proposed for constructing phylogenetic trees from distance matrices. The procedure for constructing tree topologies is similar to that of the unweighted pair-group method (UPG method) but makes corrections for unequal rates of evolution among lineages. The procedure for estimating branch lengths is the same as that of the Fitch and Margoliash method (F-M method) except that it allows no negative branch lengths. The performance of the present procedure for the construction of tree topologies is compared with that of the UPG method, the F-M method, Farris' method, and the modified Farris method by using Tateno's simulation outputs for nucleotide sequence divergence and his results for the performances of the latter four methods [Tateno, Y. (1978) Dissertation (Univ. Texas, Houston, TX). In this limited comparison, the present method performs considerably better than the UPG method and the F-M method and about equally well as the last two methods. The present method appears to be preferable to the UPG method for analysis of data from populations that have not differentiated much. Indeed, an application of the present method to gene frequency data from some Amerindian populations gives a tree topology far more reasonable than that obtained by the UPG method.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Komar N., Blanken R. L., Mitchell R. M. Calculation of evolutionary trees from sequence data. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4516–4520. doi: 10.1073/pnas.76.9.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager E. M., Wilson A. C. Construction of phylogenetic trees for proteins and nucleic acids: empirical evaluation of alternative matrix methods. J Mol Evol. 1978 Jun 20;11(2):129–142. doi: 10.1007/BF01733889. [DOI] [PubMed] [Google Scholar]
- Spielman R. S., Migliazza E. C., Neel J. V. Regional linguistic and genetic differences among Yanomama indians. Science. 1974 May 10;184(4137):637–644. doi: 10.1126/science.184.4137.637. [DOI] [PubMed] [Google Scholar]
- Ward R. H., Gershowitz H., Layrisse M., Neel J. V. The genetic structure of a tribal population, the Yanomama Indians XI. Gene frequencies for 10 blood groups and the ABH-Le secretor traits in the Yanomama and their neighbors; the uniqueness of the tribe. Am J Hum Genet. 1975 Jan;27(1):1–30. [PMC free article] [PubMed] [Google Scholar]



