Background: Cohen syndrome, characterized mainly by mental retardation, is caused by loss-of-function mutations in the gene COH1.
Results: COH1 encodes a Golgi matrix protein important for Golgi integrity.
Conclusion: Altered Golgi integrity and function probably underlie Cohen syndrome.
Significance: Our study highlights the importance of regular Golgi function during brain development and maintenance.
Keywords: Cell Biology, Genetic Diseases, Golgi, Human Genetics, Scaffold Proteins, COH1, Cohen Syndrome
Abstract
Loss-of-function mutations in the gene COH1, also known as VPS13B, lead to autosomal recessive Cohen syndrome. However, the cellular distribution and function of the encoded protein COH1 (3997 amino acids), which lacks functional homologies to other mammalian proteins, have remained enigmatic. We show here that COH1 is a peripheral Golgi membrane protein that strongly co-localizes with the cis-Golgi matrix protein GM130. Consistent with its subcellular localization, COH1 depletion using RNAi causes fragmentation of the Golgi ribbon into ministacks. Disruption of Golgi organization observed in fibroblasts from Cohen syndrome patients suggests that Golgi dysfunction contributes to Cohen syndrome pathology. In conclusion, our findings establish COH1 as a Golgi-associated matrix protein required for Golgi integrity.
Introduction
Autosomal recessive Cohen syndrome is characterized by a broad phenotypic spectrum. Obligatory symptoms include moderate to severe mental retardation, progressive postnatal microcephaly, typical facial dysmorphisms with downward-slanting palpebral fissures and a short philtrum, and ophthalmologic problems such as progressive retinal dystrophy and/or myopia. Frequent facultative symptoms comprise intermittent neutropenia, obesity, and short stature (1). The disease locus was mapped to chromosome 8q22 (2, 3), and mutations in the gene COH1 (also known as VPS13B) were found to cause Cohen syndrome (4–7). An expression analysis of murine Coh1 identified highest levels in neurons of adult brain cortical layers II–VI, proposing a role of COH1 in late brain development (6). Accordingly, normal prenatal but disturbed postnatal brain development (8) suggests defects in the terminal differentiation of neurons as contributing to the pathology of Cohen syndrome. However, the molecular pathomechanism of Cohen syndrome and the function of the protein COH1 have remained elusive.
COH1 is a protein of 3997 amino acids (aa)2 without known homologies to other mammalian proteins. It harbors two short regions homologous to yeast vacuolar protein sorting-associated protein 13 (Vps13p), which led to its classification as one of four mammalian VPS13 family members (4, 9). Moreover, based on the partial Vps13p homologous regions it has been speculated that COH1 functions in intracellular membrane traffic. Vps13p, the presumed yeast homolog of COH1, is a peripheral membrane protein that plays a role in the cycling of transmembrane proteins between the trans-Golgi network (TGN) and the prevacuolar compartment as demonstrated via interactions with the yeast endoproteinase Kex2p, the yeast dipeptidyl aminopeptidase Ste13p, and the yeast carboxypeptidase Y vacuolar protein receptor Vps10p (10, 11). Vps13p has been further implicated in spindle pole organization by interacting with yeast centrin Cdc31p (12). Moreover, a function in endocytosis and/or actin function was demonstrated by Vps13p mutations, which were found to increase cytotoxicity of an expanded poly(Q) domain in Rnq1p, a yeast prion-like protein (13). Whether mammalian COH1 carries out similar or additional functions is unknown.
This study provides the first molecular and functional characterization of the protein COH1. We identify COH1 as a peripheral membrane protein localized to the Golgi complex, where it overlaps with the cis-Golgi matrix protein GM130. We demonstrate by RNAi that COH1 is required for maintenance of the Golgi morphology. Consistent with this, human adult skin fibroblasts (HAFs) from Cohen syndrome patients carrying frameshift or nonsense mutations display a similar fragmentation of the Golgi complex. This, together with the observation that COH1 localization to the Golgi is mediated by a C-terminal fragment of 315 aa, specifies an important role of the COH1 in Golgi maintenance. In summary, our results establish COH1 as novel Golgi matrix protein and link Golgi dysfunction to developmental abnormalities in Cohen syndrome.
EXPERIMENTAL PROCEDURES
Materials and Antibodies
All materials were purchased from Sigma unless otherwise stated. Three polyclonal rabbit anti-COH1 antibodies were raised by repeated immunization with the following keyhole limpet hemocyanin-coupled peptides (Eurogentec): STAESTKSSIKPRRMQ (COH1, no. 441), GEEDFVGNDPASTMHQ (COH1, no. 442), and EHYNRQEEWRRQLPE (COH1, no. 755). Anti-COH1 antibodies were purified from serum by peptide affinity chromatography. Antibodies 441, 442, and 755 were used for immunofluorescence experiments; 442 and 755 for Western blot analysis. The following commercial antibodies were used in this study: mouse anti-BAP31 (Alexis), mouse anti-EEA1 (BD Bioscience), mouse anti-ERGIC53 (Alexis), rabbit anti-FLAG (Invitrogen), mouse anti-FLAG M2 (Invitrogen), mouse anti-GAPDH (Ambion), rabbit anti-giantin (Covance), mouse anti-GM130 (BD Bioscience), mouse anti-GMAP210 (BD Bioscience), mouse anti-GS27 (BD Bioscience), mouse anti-LAMP-1 (Developmental Studies Hybridoma Bank), mouse anti-LAMP-2 (Developmental Studies Hybridoma Bank), sheep anti-TGN46 (Serotech), rabbit anti-RAB6 (Santa Cruz Biotechnology), and mouse anti-β-tubulin (Developmental Studies Hybridoma Bank). Monoclonal mouse anti-RAB6 antibody was a generous gift from Angelika Barnekow (University of Muenster). Secondary antibodies used for Western blot analysis were anti-mouse IgG-HRP or anti-rabbit IgG-HRP (both Cell Signaling) and for immunofluorescence analysis anti-rabbit IgG-Alexa Fluor 488, anti-rabbit IgG-Alexa Fluor 555, anti-mouse IgG-Alexa Fluor 488, anti-mouse IgG-Alexa Fluor 555, anti-sheep IgG-Alexa Fluor 488, and anti-sheep IgG-Alexa Fluor 555 (all Invitrogen). 4′,6-Diamidino-2-phenylindole (DAPI) (Invitrogen) was used for nuclear staining. Restriction enzymes were purchased from NEB.
Human COH1 Constructs
For transient expression experiments of COH1, full-length COH1 (according to NM_152564.3, NP_689777.3) and different truncated constructs were cloned as follows. PCR products were amplified using primer pairs with appropriate restriction sites and cDNA from a human cell line (HeLa). Obtained amplicons were subsequently digested and ligated into an expression vector. C-terminally truncated human COH1 constructs were as follows: for COH1_1–504aa, coding nucleotides 1–1512, cloned into EcoRI and NotI sites of pFLAG-CMV5 (Sigma); for COH1_1–1104aa, coding nucleotides 1–3313, into NotI and KpnI sites of pFLAG-CMV5 and pFLAG-CMV6 (Sigma); for COH1_1–2347aa, coding nucleotides 1–7042, into NotI and SalI sites of pFLAG-CMV5; and for COH1_1–3682aa, coding 3314–11048, into the KpnI site of the pFLAG-CMV5_COH1_1–1104aa construct. N-terminally truncated human COH1 constructs were as follows: for COH1_2307–3997aa, coding nucleotides 6922–11991, cloned into NotI and SalI sites of pFLAG-CMV5 together with an N-terminal HA epitope tag; and for EGFP-COH1_3683–3997aa, coding nucleotides 11049–11991 into the KpnI site of pEGFP-C1 (BD Clontech). Full-length human COH1 constructs coding nucleotides 9828–11991 from pFLAG-CMV5_COH1_2307–3997aa were cloned into pFLAG-CMV5_COH1_1–3682aa by digesting both vectors with BspEI and AgeI and subcloning the proper fragments in-frame with the FLAG tag. Full-length untagged COH1_1–3997aa was subsequently cloned into TOPO-TA sites of pcDNA3.1 (Invitrogen) by primer pairs recognizing the start codon and introducing a stop codon. All constructs were confirmed by direct sequencing with BigDyeTM Terminator v3.1 Cycle Sequencing kit (Applied Biosystems) and analysis on an automated DNA analyzer (3730 Applied Biosystems).
Cell Culture and Transient Transfection
HeLa, MCF-7, A549, and LLC-PK1 cells were cultured at 37 °C, 5% CO2 in DMEM supplemented with 5% fetal calf serum (FCS) and 2 mm ultraglutamine. HEK293 cells were cultured at 37 °C, 5% CO2 in α-MEM supplemented with 5% FCS and 2 mm ultraglutamine. Primary HAFs were grown at 37 °C, 5% CO2 in α-MEM supplemented with 10% FCS, 2 mm ultraglutamine, 100 μg/ml penicillin G, and 100 μg/ml streptomycin. Transfection of plasmid DNA was performed using jetPEI (Polyplus transfection) according to the manufacturer's manual. Briefly, 3 μg of plasmid DNA was diluted in 100 μl of sterile 0.9% (w/v) NaCl; this solution was then mixed with an equal volume of a 6% (v/v) jetPEI dilution in sterile 0.9% (w/v) NaCl. After incubation for 20 min at room temperature the transfection solution was added dropwise into the cell culture dish and left for 24 h until subsequent analysis. All cell lines used in this study were purchased from the ATCC. HAFs were obtained from patients and unaffected controls after informed consent.
Drug Treatment
Brefeldin A (BFA, 5 μg/ml), nocodazole (5 μm), or paclitaxel (10 μm) was added directly to the culture medium and incubated for the indicated length of time.
RNA Interference
All small interference RNAs (siRNA) specific for COH1, MAPK1, GAPDH, and negative control (scramble) were purchased from Ambion or Invitrogen. All siRNA target sequences are available on request. siRNAs were resuspended to 50 μm according to the manufacturer's instructions and stored at −80 °C. For siRNA transfection, HeLa cells were grown to 15–30% confluence in 6-well plates and transfected with a 200 nm concentration of each siRNA using INTERFERin (Polyplus transfection) and OptiMEM (Invitrogen) according to the manufacturer's instructions. Briefly, cells were incubated in 1 ml od cell culture medium, siRNAs were diluted to 200 nm in a 100-μl final volume with OptiMEM, and subsequently 2 μl of INTERFERin was added. After a 10-min incubation at room temperature the transfection mix was added dropwise to each well. Transfection was repeated after 12 h, medium was changed 12 h later, and cells were cultured for another 60 h. Finally, cells were prepared for subsequent analysis.
Quantitative PCR (qPCR)
Total RNA was isolated from cell cultures using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. 1 μg of isolated RNA was reverse transcribed using a RevertAid H Minus First Strand cDNA Synthesis kit and random hexamer primers (Fermentas). Primer pairs for cDNA amplification of ACTB (the gene for β-actin) (NM_001101.3), COH1, MAPK1 (NM_002745.4), and GAPDH (NM_002046.3) were designed. All cDNA primer sequences are available on request. mRNA levels were determined by qPCR using cDNA from HAF cultures or siRNA-treated HeLa cells. Each sample was analyzed as triplicate and amplified on an ABI PRISM7500 instrument (Applied Biosystems). Relative mRNA levels were quantified using the comparative Ct method (14). The different mRNA values were normalized against the ACTB or GAPDH mRNA level.
Immunofluorescence and Image Analysis
For staining of overexpressed and endogenous protein, cells were grown on glass coverslips (12 mm; Marienfeld). Cells were fixed with 4% (w/v) paraformaldehyde in PBS at 4 °C or 100% methanol at −20 °C, permeabilized in 1% (v/v) Triton X-100 or 0.1% (w/v) saponin in 3% (w/v) bovine serum albumin (BSA) in PBS, and blocked with 3% (w/v) BSA in PBS. Primary antibodies were applied in 3% BSA in PBS for 5 h at 4 °C, coverslips were washed in PBS, and secondary antibodies were applied in 3% BSA in PBS for 1 h at 4 °C. Coverslips were mounted on slides using Fluoromount-G (SouthernBiotech). Images were taken with a confocal microscope (LSM510; Zeiss). Images for subsequent evaluation were acquired under identical exposure conditions. Image analysis was performed with macros in ImageJ or AxioVision (Zeiss) under identical threshold conditions. Statistical significance was calculated with Student's t test (two-sided, unpaired, homogeneous variation).
Ultrastructural Analysis
Cultured cells were fixed for at least 2 h at 4 °C in 3% glutaraldehyde solution in 0.1 m cacodylate buffer, pH 7.4. Scraped cells were washed in buffer, postfixed for 1 h at 4 °C in 1% osmium tetroxide, rinsed in water, and dehydrated through graded ethanol solutions. After transfer into propylene oxide and embedding in epoxy resin (glycidether 100), ultrathin sections were cut with an ultramicrotome (Reichert Ultracut E) and treated with uranyl acetate as well as lead citrate. Pictures were obtained with an electron microscope (Philips EM 400).
Western Blot Analysis
For Western blot analysis, all samples were diluted in 1× SDS loading buffer and resolved by gel electrophoresis in Tris acetate SDS 3–8% polyacrylamide gradient gels (Invitrogen). Protein concentrations were determined using the BCA protein assay kit (Pierce). After transfer on nitrocellulose membranes by tank blotting and blocking in 5% block milk, 0.2% Nonidet P-40 in 1× TBS, blots were incubated with the appropriate primary and secondary antibodies in 5% blocking milk and finally detected using the ECL reaction (Amersham Biosciences) and visualized on Hyperfilm ECL (Amersham Biosciences).
Isolation of Membrane and Cytosolic Fractions
Transiently transfected HEK293 cells were scraped off culturing dishes and resuspended in HPLC-H2O supplemented with 1× complete proteinase inhibitor mixture (Roche Applied Science). Following cell lysis by a freeze-thaw step, nuclei and cell debris were removed by centrifugation at 5,000 × g for 5 min at 4 °C. Clarified postnuclear cell lysates were centrifuged at 100,000 × g for 30 min at 4 °C in a Ti-50 rotor, and obtained supernatants were stored for further investigation. Membrane pellets were washed once by resuspending in 1× TBS supplemented with 1× complete proteinase inhibitor mixture, 50 mm NaF, 30 mm NaPPi, and 5 mm EDTA (native membrane pellet lysis buffer) and again pelleted by high speed centrifugation as described above. Membrane pellets were resuspended in native membrane pellet lysis buffer and stored at −80 °C for further investigation.
Stripping of Lipid Membranes
Native membrane pellets were used for membrane stripping in 1 m KCl, 0.2 m Na2CO3, and 6 m urea. Briefly, washed membrane pellets were resuspended in the appropriate stripping solution and incubated under regular shaking at 4 °C for 30 min. The separation of stripped membranes (pellet) and peripheral membrane proteins (supernatant) was achieved by centrifugation at 100,000 × g for 30 min at 4 °C in a Ti-50 rotor. Both fractions were stored at −80 °C until further investigation.
Triton X-114 Phase Separation
Triton X-114 phase separation was done as described previously (15). Briefly, transiently transfected HEK293 cells were scraped off culturing dishes and resuspended in 1× PBS supplemented with 1× complete proteinase inhibitor mixture. Following cell lysis by freeze-thawing, nuclei and cell debris were removed by centrifugation at 5,000 × g for 5 min at 4 °C. Cleared postnuclear cell lysates were precondensed with prewashed Triton X-114 solution, 4% (v/v) final concentration, for 10 min on ice. Phase separation occurred by incubation for 3 min at 30 °C and subsequent centrifugation at 1,700 × g for 5 min at 30 °C. Both lower detergent and upper aqueous phase were washed three times by adding 1× PBS or Triton X-114 solution, respectively. For this, condensation for 10 min on ice, incubation for 3 min at 30 °C, and centrifugation at 1,700 × g for 5 min at 30 °C were repeated. Both fractions were stored at −80 °C until further investigation.
Computational Analyses
Primary amino acid sequences of COH1 (NP_689777.3), giantin (NP_004478.3), GM130 (NP_004477.3), and SCYL1BP1 (NP_689494.2) were used for predictions of coiled-coil structures by COILs and of disorder propensities by GlobPlot.
RESULTS
COH1 Localizes to the Golgi Apparatus
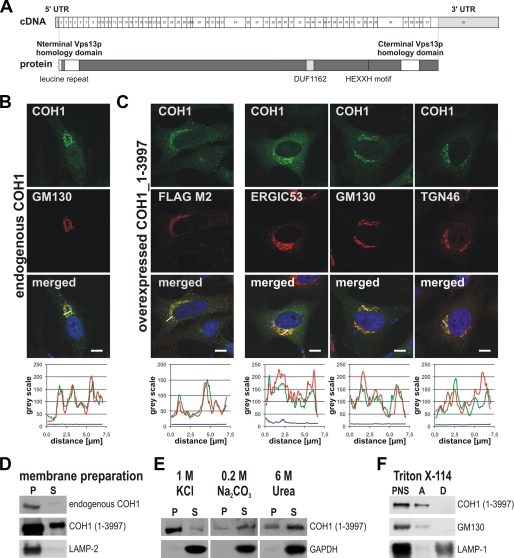
COH1 was predicted to encode a protein of 3,997 aa lacking functional sequence homologies to other mammalian proteins, except for partial homology to yeast Vps13p at its N terminus and within the C-terminal part (Fig. 1A) (4, 9, 11). Because of its relevance for the Cohen syndrome pathology, we decided to study the cell biological role of COH1. Therefore, we cloned the ubiquitously expressed COH1 transcript (pFLAG-CMV5_COH1_1–3997; NM_152564.3) (5, 9) into mammalian expression vectors and generated COH1-specific polyclonal anti-peptide antibodies. First, we analyzed the subcellular localization of endogenous COH1 in HeLa cells. COH1 is enriched in the perinuclear area, where it strongly co-localizes with the cis-Golgi marker GM130 (Fig. 1B). To corroborate these data we expressed epitope-tagged COH1 in HeLa cells and analyzed its distribution by confocal imaging. Both anti-FLAG and anti-COH1 antibodies showed an identical perinuclear distribution of COH1-FLAG at the Golgi complex, suggesting that our antibodies indeed specifically recognize COH1 (Fig. 1C). Untagged COH1 was also found at the Golgi complex in LLC-PK1, A549, and MCF-7 cells, suggesting that COH1 localizes to the Golgi in a broad spectrum of cells and tissues (supplemental Fig. S1A). Exogenously expressed COH1 again strongly co-localized with GM130. Significant co-localization was also detected for the endoplasmic reticulum (ER)-to-Golgi intermediate compartment marker ERGIC53 and for the cycling TGN/recycling endosomal protein TGN46, indicating a more widespread distribution of COH1 in pre- and post-Golgi compartments (Fig. 1C). Immunostaining with B cell receptor-association protein BAP31, early endosome antigen EEA1, or lysosome-associated membrane protein LAMP-2 showed no significant enrichment of COH1 to the ER or the endolysosomal system (supplemental Fig. S1B).
FIGURE 1.
COH1 localizes mainly to the cis-Golgi apparatus. A, schematic representation of the COH1 transcript (NM_152564.3) (upper) and the predicted protein COH1 (lower, gray). White boxes, located at the N terminus and in the C-terminal part of COH1, indicate homolog regions to yeast Vps13p. Light gray box indicates Pfam domain DUF1162 representing also a conserved region within several hypothetical eukaryotic vacuolar protein sorting-related proteins. Further conserved patterns predicted by literature and data base searches (e.g. Prosite) implicate a leucine repeat at the N terminus and a HEXXH motif in the C-terminal part of COH1. B, immunofluorescence staining of endogenous COH1, detected with a specific antipeptide-antibody to residues 326–377 (GEEDFVGNDPASTMHQ), identifying COH1 (green) as perinuclear protein. Co-localization with GM130 (red) demonstrated COH1 as Golgi protein. C, overexpressed full-length COH1 (pFLAG-CMV5_COH1_1–3997) detected with a COH1-specific antibody, recognizing residues 111–126 (STAESTKSSIKPRRMQ) (green), as well as a FLAG tag-specific antibody (red). Both antibodies detected COH1 at the Golgi apparatus, and their co-localization confirmed antibody specificity. COH1 (green) shows considerable overlap with the ER to Golgi intermediate compartment marker ERGIC53 (red), cis-Golgi marker GM130 (red), and trans-Golgi marker TGN46 (red). Images were taken by confocal microscopy. Scale bars, 10 μm. Fluorescence intensity profiles were achieved by ImageJ of the lines depicted in the merged images of B; graph colors match color code in B. D, COH1, a peripheral membrane protein as shown by membrane preparations of post-nuclear protein lysates from control and C-terminal FLAG-tagged COH1_1–3997aa-overexpressing HEK293 cells. Western blot analysis was performed using antibodies specific for COH1 and LAMP-2. COH1 is highly enriched in the pellet (P) fraction but also detectable in the supernatant fraction (S). E, subsequent membrane stripping with 1 m KCl, 0.2 m Na2CO3 (pH 11), or 6 m urea revealing strong association of COH1 with lipid membranes. Separated supernatant and pellet fractions were analyzed by Western blotting using antibodies against COH1 and GAPDH. F, Triton X-114 phase separation confirming COH1 as nonintegral membrane protein. Triton X-114 phase separation was performed on postnuclear protein lysates (PNS) from COH1_1–3997aa-overexpressing HEK293 cells. Western blot analysis using specific antibodies identified COH1 and the peripheral membrane protein GM130 in the aqueous (A) phase whereas the integral membrane protein LAMP-2 was separated into the detergent fraction (D).
COH1 Is a Peripheral Membrane Protein
Yeast Vps13p was described as a peripheral membrane protein of 358 kDa (11). In contrast to this, COH1 was predicted to be embedded into the membrane via 10 transmembrane helices (4). To study the association of COH1 with membranes, we analyzed subcellular fractions from cells expressing endo- and/or exogenous COH1 following high speed centrifugation. Endogenous and full-length COH1-FLAG were partitioned between the membrane and soluble cytosolic fractions (Fig. 1D). The association of COH1 with membranes was further analyzed by producing different N- and C-terminal truncation mutants of COH1. We found all fragments in both membrane as well as cytosolic fractions (supplemental Fig. S2A). By chemical stripping of crude membrane fractions we tested the strength of COH1 membrane association. Treatment of the membrane pellet with 6 m urea or 0.2 m Na2CO3 (pH 11) led to the recovery of substantial fractions into the supernatant, whereas high salt by 1 m KCl washes only partially solubilized full-length or truncated COH1 (Fig. 1E). Phase separation with Triton X-114, which distinguishes integral and peripheral membrane proteins into a detergent and aqueous phase, respectively, accumulated COH1 completely into the aqueous phase, suggesting that it is not integrated into the phospholipid bilayer (Fig. 1F and supplemental Fig. S2B). Together, these results identify COH1 as a peripheral membrane protein of the Golgi apparatus.
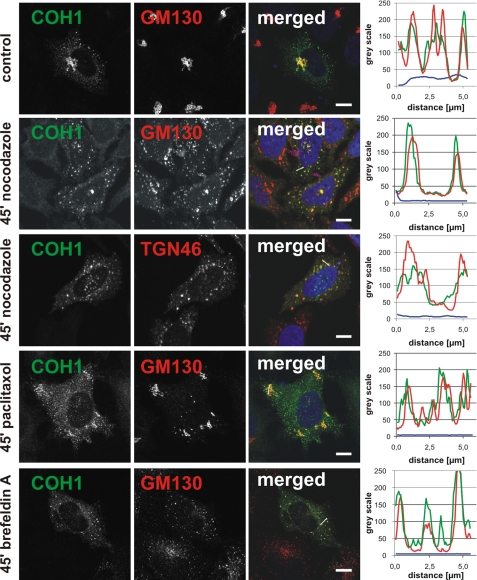
COH1 Associates with Golgi Structures upon Chemical Disruption of the Golgi Apparatus
To investigate further the mode by which COH1 associates with the Golgi complex we made use of the microtubule-depolymerizing agent nocodazole, which induces accumulation of Golgi ministacks adjacent to ER exit sites (16). Following nocodazole treatment, COH1 maintained its association with the resulting GM130- or TGN46-positive Golgi fragments (Fig. 2), indicating that the Golgi localization of COH1 does not depend on Golgi integrity. In line with this, COH1 Golgi association was also not disturbed by the microtubule-stabilizing agent paclitaxel (Fig. 2). As a further tool, we made use of BFA, a drug that interferes with membrane traffic at the Golgi and between the ER and the Golgi complex. BFA inhibits guanine nucleotide exchange on ARF small GTPases, thereby inducing the redistribution of luminal Golgi proteins into the ER and the accumulation of Golgi matrix proteins at ER exit sites (17, 18). Similar to nocodazole, BFA treatment did not affect the extensive co-localization of COH1 with the Golgi matrix protein GM130 (Fig. 2). These findings suggest that COH1 might act as a scaffolding protein by association with the cytoplasmic leaflet of the Golgi complex.
FIGURE 2.
COH1 Golgi localization is not disturbed by microtubule-disrupting agents or interference with retrograde Golgi transport. COH1-overexpressing HeLa cells were treated with nocodazole, paclitaxel, or BFA for the indicated lengths of time. Indirect immunofluorescence analysis was performed with a COH1-specific antibody (green) and GM130 or TGN46 antibodies (both red). Images were taken by confocal microscopy. Scale bars, 10 μm. Fluorescence intensity profiles of the lines are depicted in the merged images; graph colors match color code of merged images. Analysis was performed with ImageJ.
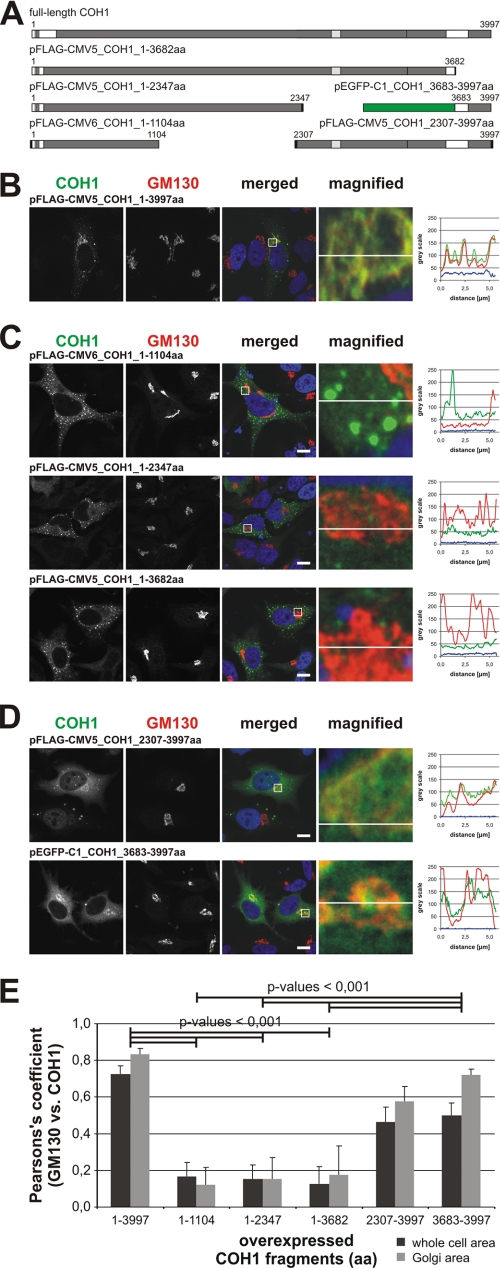
COH1 Golgi Localization Is Mediated by Its C Terminus
To characterize the subcellular targeting of COH1 in more detail, we expressed a series of truncated recombinant variants of COH1 in HeLa cells (Fig. 3A). Although full-length COH1 is targeted to the perinuclear Golgi region (Fig. 3, B and E), none of the C-terminally truncated fragments COH1_1–1104aa, COH1_1–2347aa, and COH1_1–3682aa localized to the Golgi apparatus (Fig. 3, C and E). Further subcellular analysis of the COH1_1–2347aa fragment did not identify the origin of punctate cytoplasmic structures as they did not co-localize with markers of the endolysosomal system such as EEA1 or LAMP-1 (supplemental Fig. S2C). These structures may, thus, potentially represent cytoplasmic aggregates. As the N terminus was insufficient for Golgi targeting, we next tested whether the Golgi targeting information resides within the COH1 C terminus. Indeed a C-terminal fragment, COH1_2307–3997aa, showed partial association with the Golgi apparatus. Further truncation studies narrowed the Golgi targeting determinant down to the C-terminal 315 residues of COH1. Indeed, transplanting this 315-aa C-terminal fragment of COH1 onto EGFP was sufficient to direct the fusion protein EGFP-COH1_3683–3997aa to the Golgi area (Fig. 3, D and E). Because sequence analysis of this C-terminal fragment did not identify significant sequence homologies to other proteins apart from VPS13 family members, these findings reveal a hitherto uncharacterized Golgi targeting domain within the COH1 C terminus.
FIGURE 3.
The C terminus of COH1 mediates its Golgi localization. A, schematic representation of truncated COH1 constructs used in this experiment. Black boxes indicate FLAG tag or HA tag; green box indicates EGFP. B–D, immunofluorescence analysis of paraformaldehyde-fixed and 0.1% saponin-permeabilized HeLa cells. Overexpression of full-length COH1 (B) and N-terminal COH1 fragments (C) was detected with an antibody recognizing residues 326–377 (GEEDFVGNDPASTMHQ), and C-terminal COH1 fragments were stained with an antibody to residues 3706–3720 (EHYNRQEEWRRQLPE) (D). Golgi localization was analyzed by counterstaining of COH1 (green) with GM130 (red). Nuclei were stained with DAPI (blue). Images were taken by confocal microscopy. Scale bars, 10 μm. Fluorescence intensity profiles of the lines are depicted in the merged and magnified images of B–D; graph colors match color code in B–D. Analysis was performed with ImageJ. E, for quantitative analysis of co-localization, Pearson's correlation coefficient calculated using a JACoP plugin in ImageJ software. Average Pearson's coefficients for COH1 and GM130 were estimated for overexpressed full-length COH1 (COH1_1–3997aa), N-terminal (COH1_1–1104aa, COH1_1–2347aa, and COH1_1–3682aa), and C-terminal (COH1_2307–3997aa and COH1_3683–3997aa) fragments. Error bars show S.D. (n = 3 independent experiments with 10 images/condition). Statistical significance was calculated by t test.
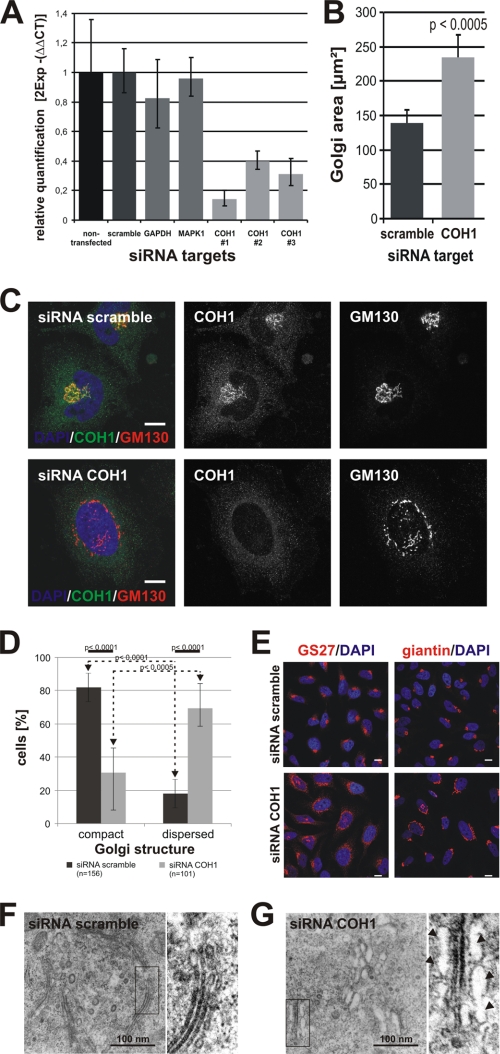
Loss of COH1 Disrupts Golgi Structure
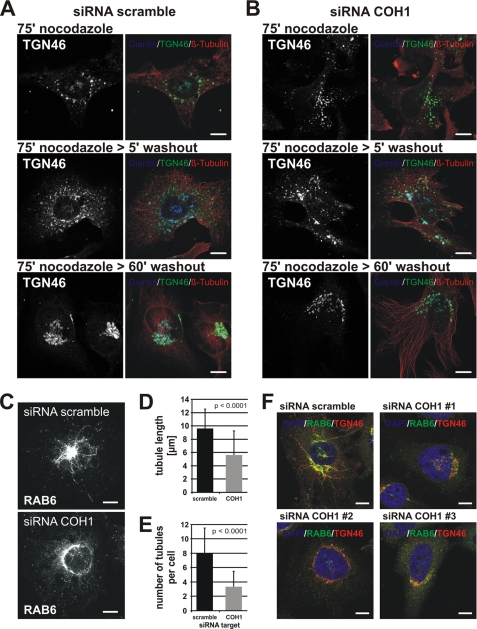
Golgi matrix proteins such as GM130 or p115 play an important role in Golgi structure and biogenesis by interconnecting individual Golgi cisternae to form typical Golgi ribbons (19). Based on our observation that COH1 is a Golgi-localized peripheral membrane protein we analyzed its potential function in Golgi organization. Therefore, we depleted COH1 from HeLa cells by three different siRNAs. Transient transfection of COH1-specific siRNAs effectively diminished COH1 mRNA expression as demonstrated by qPCR analysis (Fig. 4A). This corresponded to the disappearance of perinuclear COH1 staining in immunofluorescence microscopy, confirming down-regulation of COH1 on a protein level (Fig. 4C). By confocal analysis of COH1-depleted cells for different Golgi marker proteins, we observed a severe fragmentation of the Golgi apparatus as evident from the dispersion of the GM130-positive Golgi ribbon into ministacks (Fig. 4C). Quantitative analysis revealed Golgi dispersion in ∼70% of COH1-depleted cells compared with only ∼18% of control cells (Fig. 4, D and E). The degree of Golgi dispersion corresponds to the efficiency of COH1 knockdown measured by qPCR (Fig. 4A). The extent of Golgi fragmentation was further analyzed by determining the Golgi occupied area. These measurements revealed a significant increase of the Golgi area by ∼70% (Fig. 4B). To analyze the Golgi ultrastructure in HeLa cells we turned to electron microscopy. Electron microscopic images showed a normal Golgi morphology with laterally linked, elongated, and flat cisternae in control cells (Fig. 4F). By contrast, COH1-deficient cells contained fragmented Golgi ribbons dispersed into ministacks, confirming the results from light microscopic imaging. Moreover, the lumen of these Golgi ministacks occasionally appeared swollen (Fig. 4G). The morphological disturbance of the Golgi apparatus in the absence of COH1 could either be a consequence of the altered steady-state distribution of Golgi proteins or may reflect the inability of Golgi fragments to reassemble after disassembly. To test the latter possibility directly, we induced Golgi disruption by applying nocodazole and followed Golgi restoration upon nocodazole washout (Fig. 5). Nocodazole treatment induced a comparable dispersion of the Golgi complex into ministacks in scrambled and COH1 siRNA-treated HeLa cells. However, whereas control cells were able to reassemble compact Golgi structures within 60 min after washout of the drug (Fig. 5A), in COH1-depleted cells Golgi fragmentation persisted (Fig. 5B). Thus, COH1 plays a critical role in Golgi (re)assembly.
FIGURE 4.
COH1-deficient HeLa cells display severe Golgi fragmentation. A, efficient siRNA treatment with three independent COH1 siRNAs was confirmed by relative qPCR. COH1 mRNA expression is decreased to ∼16% (siRNA COH1 #1) leftover compared with non- and/or scrambled-transfected HeLa cells. ΔCT values were normalized to ACTB. B, control (transfected with scrambled siRNA) and COH1-deficient HeLa cells were processed for immunofluorescence analysis using different subcellular markers. Confocal images were analyzed using AxioVision. In COH1-deficient cells the Golgi-occupied area (visualized by giantin) is significantly increased. C, endogenous COH1 showed perinuclear Golgi enrichment as demonstrated by co-localization with GM130 in HeLa cells. Efficient RNAi-mediated loss is confirmed by absent COH1 staining; and COH1 depletion induced Golgi dispersion into ministacks as illustrated by GM130 staining. Images of siRNA-treated HeLa cells stained with antibodies specific to COH1 (green) and GM130 (red) as well as DAPI (blue) were taken by confocal microscopy. Scale bars, 10 μm. D, HeLa cells were transiently transfected with scrambled or COH1 siRNA, processed for immunofluorescence, and analyzed by confocal microscopy. For morphological analysis diverse Golgi marker proteins were stained and quantified. Cells with small and more roundly Golgi structures were counted as “compact,” whereas an elongated and/or disconnected staining was counted as “dispersed.” Error bars show S.D., and statistical significance was calculated by t test. E, representative images for quantitative analysis of Golgi morphology quantified in D show fragmented and disconnected Golgi apparatus after COH1 siRNA depletion. Immunofluorescence staining occurred with Golgi marker GS27 and giantin. Scale bars, 10 μm. F and G, representative electron microscopic images from siRNA-treated glutaraldehyde fixed HeLa cells are shown. Control cells display long, flat, and laterally connected Golgi-stacks (F). COH1-deficient cells display disconnected Golgi ministacks. Arrows indicate swollen cisternae (G). Contrast was enhanced simultaneously in magnified electron microscopic images.
FIGURE 5.
COH1 is important for Golgi reassembly and tubulation activities. A and B, HeLa cells transfected with scramble or COH1 siRNA were treated with nocodazole for 75 min at 37 °C. Subsequently, nocodazole was washed out, and cells were incubated at 37 °C for 5 min and 60 min to allow Golgi reassembly. Cells were fixed and stained for giantin (blue), TGN46 (green), and β-tubulin (red) by immunofluorescence and analyzed by confocal microscopy. HeLa cells transfected with scrambled siRNA display a discernible reassembled compact Golgi structure after 60 min of nocodazole washout (A) whereas in COH1-deficients cells the Golgi remained fragmented (B). C–E, siRNA-transfected HeLa cells were treated with 5 μg/ml BFA for 5 min and processed for immunofluorescence analysis using an antibody against the small GTPase RAB6. D and E, quantifications of mean RAB6 tubule length (D) and RAB6 tubule number/cell (E) were performed using AxioVision. At least 30 cells were analyzed per condition and experiment (n = 3). F, representative confocal images showed reduced tubulation activity in COH1-deficient cells upon 5 μg/ml BFA treatment for 5 min not only for RAB6 (green) but also for TGN46 (red). Nuclei were stained with DAPI (blue). Scale bars, 10 μm. Error bars show S.D. Statistical significance was calculated by t test.
Finally, we assessed the role of COH1 in Golgi polarization. To this aim, control or COH1-deficient cells stained for various markers of distinct Golgi subcompartments were analyzed by confocal imaging. The relative distribution of the ER-to-Golgi intermediate compartment marker ERGIC53 versus the medial-Golgi marker giantin was unaffected in HeLa cells lacking COH1 (supplemental Fig. S3A). Moreover, cis-trans-Golgi orientation as demonstrated by co-staining for GMAP210 and TGN46 also appeared normal following COH1 depletion (supplemental Fig. S3B). We conclude that COH1 is not required for Golgi polarization.
COH1-deficient Cells Display Reduced Tubulation Activity upon BFA Treatment
Membrane tubulations emanate from Golgi stacks and are thought to be involved in membrane trafficking and Golgi maintenance (20). BFA was found to enhance the formation of Golgi-derived membrane tubulation (21). To assess whether COH1 is implicated in Golgi tubulation, we used a short BFA treatment (5 μg/ml, 5 min) to visualize the formation of tubular Golgi structures. Incubation of HeLa cells for 5 min with BFA and subsequent staining for RAB6 (Fig. 5C), a central regulator of Golgi tubule formation and vesiculo-tubular transport along microtubules (22), revealed significantly reduced formation of RAB6-positive tubules (3.34 tubules/cell) in COH1-deficient cells compared with control cells (7.94 tubules/cell) (Fig. 5D). The decrease of tubule numbers/cell was accompanied by a significant reduction of the mean tubule length from 9.56 μm in control cells to 5.62 μm in COH1-deficient cells (Fig. 5E). Similar observations were made for the TGN as visualized by TGN46 staining (Fig. 5F). These results establish COH1 as a positive regulator of Golgi-derived tubule formation.
Cohen Syndrome Mutations Disturb Golgi Integrity
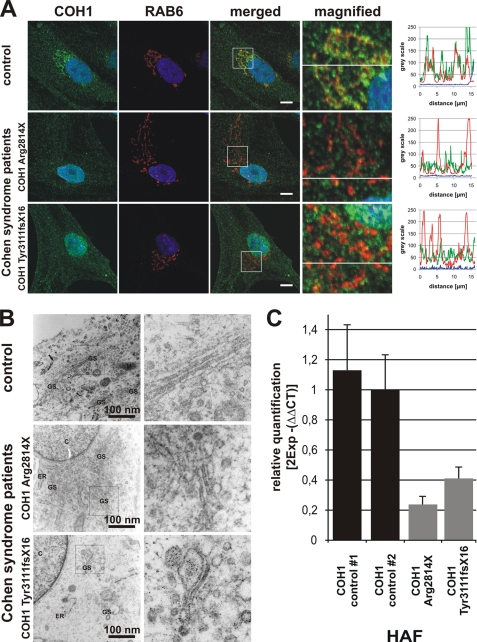
The majority of COH1 mutations is predicted to result in a premature translational stop, suggesting that a loss-of-function underlies Cohen syndrome (5). Such loss-of-function might then cause changes in the Golgi structure similar to those we observed following depletion of COH1 in HeLa cells. To assess the role of COH1 mutations in Golgi integrity we analyzed HAF cultures derived from Cohen syndrome patients carrying the homozygous mutations p.Arg2814X or p.Tyr3111fsX16. COH1 mRNA expression as determined by qPCR was reduced by more than 60% in patient HAFs compared with controls (Fig. 6C), possibly due to nonsense-mediated mRNA decay. Confocal imaging demonstrated the absence of perinuclear COH1 staining, further confirming that COH1 HAF cells are COH1-deficient. Furthermore, COH1 HAF cells showed a disrupted Golgi morphology (Fig. 6A) and thus recapitulate the cellular phenotype upon COH1 knockdown, supporting the idea that COH1 mutations result in COH1 loss-of-function. To analyze these alterations further, HAFs from Cohen syndrome patients and appropriate controls were processed for electron microscopy. Ultrastructural analysis confirmed that COH1 mutations induce fragmentation of the normally laterally linked Golgi ribbon into mini-stacks with swollen cisternae (Fig. 6B). We conclude that COH1 is a peripheral protein required for Golgi maintenance and postulate that loss of Golgi integrity contributes to the pathology of Cohen syndrome.
FIGURE 6.
Golgi fragmentation in HAFs from Cohen syndrome patients. A, immunofluorescence pictures of HAF cultures stained for COH1 (green), RAB6 (red), and DAPI (blue) were taken by confocal microscopy. Note absent COH1 staining at Golgi fragments in HAFs from Cohen syndrome patients with homozygous COH1 mutations p.Arg2814X or p.Tyr3111fsX16. Fluorescence intensity profiles are of the lines depicted in the merged and magnified images of A; graph colors match color code in A. Analysis was performed with ImageJ. Scale bars, 10 μm. B, electron microscopic images were obtained from glutaraldehyde-fixed HAF cultures. HAFs from Cohen syndrome patients display disrupted Golgi ministacks with swollen cisternae. C, qPCR analysis revealed reduced COH1 mRNA level in HAFs derived from Cohen syndrome patients indicating nonsense-mediated mRNA decay. ΔCT values were normalized to GAPDH. Error bars show S.D.
DISCUSSION
Mutations in COH1 are well established to cause autosomal recessive Cohen syndrome (4, 5, 23–25); however, no study has addressed the biochemical characteristics or cellular localization and function of the encoded protein COH1 so far. We provide here by multiple lines of evidence that COH1 is a Golgi-associated protein that co-localizes with the cis-Golgi marker protein GM130. The strong Golgi association of COH1 is preserved even upon disruption of the Golgi architecture by nocodazole, paclitaxel, or following BFA treatment. Biochemical fractionation and partitioning experiments further show that COH1 is a peripheral membrane protein, similar to Vps13p in yeast (4, 9). Vps13p has been found to regulate anterograde and retrograde vesicular transport of transmembrane proteins between the prevacuolar compartment and the TGN (10, 11). In agreement with these data we find COH1 to be required for the maintenance of Golgi stacks and for the reassembly of Golgi cisternae from ministacks. Furthermore, COH1 regulates the formation of Golgi-derived membrane tubules, consistent with its possible function in intracellular membrane traffic. COH1 therefore functionally resembles golgins, Golgi membrane-associated coiled-coil proteins required for the assembly of Golgi stacks (19, 26) via integrating the activity of small GTPases and cytoskeletal elements (27–31). Membrane targeting of golgins occurs via GRAB or GRIP domains as found in GMAP210 or p230, respectively (32–35). COH1, however, contains no predictable GRAB or GRIP domains, and its close association with the Golgi apparatus is mediated at least by its C-terminal 315 residues, which comprise a hitherto uncharacterized Golgi targeting domain. The molecular details by which this domain targets COH1 to the Golgi complex remain to be determined.
Unlike other golgins, such as GM130, COH1 does not contain typical coiled-coil domains (supplemental Fig. S4, A–D) but rather comprises arrays of ordered globular domains of unknown structure (supplemental Fig. S4, E and F). We hypothesize that COH1 via these domains facilitates the assembly of Golgi stacks or fragments. This idea is supported by the severe fragmentation of the Golgi in COH1-depleted cells, whereas the cis-trans polarization of these fragments remained intact.
The altered architecture and dynamics of the Golgi complex in COH1-depleted cells are also reflected by the significant reduced formation of Golgi-derived RAB6-positive membrane tubules. Such membrane tubules have recently been identified to mediate Golgi reassembly (36), a process defective in COH1-deficient cells. Together, these results clearly establish COH1 as a crucial factor in Golgi maintenance and function.
Consistent with our findings in COH1-depleted HeLa cells, fibroblasts from two previously reported Cohen syndrome patients (5, 7) display fragmented Golgi cisternae, suggesting that Golgi maintenance is directly linked to Cohen syndrome pathology. The vast majority of Cohen syndrome-associated mutations presumably result in premature termination of protein translation because of nonsense or frameshift changes. Therefore, we expect that HAF cultures obtained from two Cohen syndrome patients and analyzed here are representative of most COH1 mutations. COH1 HAF cultures display severe nonsense-mediated mRNA decay, an absence of COH1 at the Golgi, and fragmented Golgi structures. These results are consistent with observations from RNAi experiments in HeLa cells and render it highly likely that Cohen syndrome is caused by loss-of-function of the COH1 protein. However, further work still has to consider that persisting COH1 protein fragments, such as truncations of the C-terminal Golgi targeting determinant, might have an impact on Cohen syndrome pathology. Although no phenotype-genotype correlation or mutational hotspot has been identified so far, the impact of rarely detected COH1 missense and in-frame deletion mutations on COH1 protein level and function might in the future be helpful to unravel the pathomechanism of Cohen syndrome.
In summary, our study identifies COH1 as Golgi-associated peripheral membrane protein that is required for maintaining Golgi integrity and function. Our results may form the basis for a more detailed dissection of the molecular function of COH1 in Golgi membrane traffic and are essential to understand better its role in brain development, neuronal function, and in Cohen syndrome pathology.
Acknowledgments
We thank the patients and their families for participating in this study. We thank Muriel Holder-Espinasse for providing one patient fibroblast culture, Petra Schrade and Sebastian Bachmann of the Charité electron microscopy facility for electron microscopic imaging, and Angelika Barnekow for monoclonal RAB6 antibody. We thank Karl Sperling, Peter Nürnberg, and Stefan Mundlos for continuous support.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (to T. M., V. H., D. H., and H. C. H.) and a studentship from the Charité Universitätsmedizin Berlin (to W. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- aa
- amino acid(s)
- BFA
- brefeldin A
- EGFP
- enhanced GFP
- ER
- endoplasmic reticulum
- HAF
- human adult skin fibroblast
- qPCR
- quantitative PCR
- TGN
- trans-Golgi network.
REFERENCES
- 1. Cohen M. M., Jr., Hall B. D., Smith D. W., Graham C. B., Lampert K. J. (1973) J. Pediatr. 83, 280–284 [DOI] [PubMed] [Google Scholar]
- 2. Horn D., Krebsová A., Kunze J., Reis A. (2000) Am. J. Med. Genet. 92, 285–292 [PubMed] [Google Scholar]
- 3. Kolehmainen J., Norio R., Kivitie-Kallio S., Tahvanainen E., de la Chapelle A., Lehesjoki A. E. (1997) Eur. J. Hum. Genet. 5, 206–213 [PubMed] [Google Scholar]
- 4. Kolehmainen J., Black G. C., Saarinen A., Chandler K., Clayton-Smith J., Träskelin A. L., Perveen R., Kivitie-Kallio S., Norio R., Warburg M., Fryns J. P., de la Chapelle A., Lehesjoki A. E. (2003) Am. J. Hum. Genet. 72, 1359–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seifert W., Holder-Espinasse M., Kühnisch J., Kahrizi K., Tzschach A., Garshasbi M., Najmabadi H., Walter Kuss A., Kress W., Laureys G., Loeys B., Brilstra E., Mancini G. M., Dollfus H., Dahan K., Apse K., Hennies H. C., Horn D. (2009) Hum. Mutat. 30, E404–420 [DOI] [PubMed] [Google Scholar]
- 6. Mochida G. H., Rajab A., Eyaid W., Lu A., Al-Nouri D., Kosaki K., Noruzinia M., Sarda P., Ishihara J., Bodell A., Apse K., Walsh C. A. (2004) J. Med. Genet. 41, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hennies H. C., Rauch A., Seifert W., Schumi C., Moser E., Al-Taji E., Tariverdian G., Chrzanowska K. H., Krajewska-Walasek M., Rajab A., Giugliani R., Neumann T. E., Eckl K. M., Karbasiyan M., Reis A., Horn D. (2004) Am. J. Hum. Genet. 75, 138–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kivitie-Kallio S., Autti T., Salonen O., Norio R. (1998) Neuropediatrics 29, 298–301 [DOI] [PubMed] [Google Scholar]
- 9. Velayos-Baeza A., Vettori A., Copley R. R., Dobson-Stone C., Monaco A. P. (2004) Genomics 84, 536–549 [DOI] [PubMed] [Google Scholar]
- 10. Redding K., Brickner J. H., Marschall L. G., Nichols J. W., Fuller R. S. (1996) Mol. Cell. Biol. 16, 6208–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brickner J. H., Fuller R. S. (1997) J. Cell Biol. 139, 23–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kilmartin J. V. (2003) J. Cell Biol. 162, 1211–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meriin A. B., Zhang X., Miliaras N. B., Kazantsev A., Chernoff Y. O., McCaffery J. M., Wendland B., Sherman M. Y. (2003) Mol. Cell. Biol. 23, 7554–7565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 15. Bordier C. (1981) J. Biol. Chem. 256, 1604–1607 [PubMed] [Google Scholar]
- 16. Cole N. B., Sciaky N., Marotta A., Song J., Lippincott-Schwartz J. (1996) Mol. Biol. Cell 7, 631–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lippincott-Schwartz J., Donaldson J. G., Schweizer A., Berger E. G., Hauri H. P., Yuan L. C., Klausner R. D. (1990) Cell 60, 821–836 [DOI] [PubMed] [Google Scholar]
- 18. Klausner R. D., Donaldson J. G., Lippincott-Schwartz J. (1992) J. Cell Biol. 116, 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puthenveedu M. A., Bachert C., Puri S., Lanni F., Linstedt A. D. (2006) Nat. Cell Biol. 8, 238–248 [DOI] [PubMed] [Google Scholar]
- 20. de Figueiredo P., Polizotto R. S., Drecktrah D., Brown W. J. (1999) Mol. Biol. Cell 10, 1763–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lippincott-Schwartz J., Yuan L., Tipper C., Amherdt M., Orci L., Klausner R. D. (1991) Cell 67, 601–616 [DOI] [PubMed] [Google Scholar]
- 22. Valente C., Polishchuk R., De Matteis M. A. (2010) Nat. Cell Biol. 12, 635–638 [DOI] [PubMed] [Google Scholar]
- 23. Parri V., Katzaki E., Uliana V., Scionti F., Tita R., Artuso R., Longo I., Boschloo R., Vijzelaar R., Selicorni A., Brancati F., Dallapiccola B., Zelante L., Hamel C. P., Sarda P., Lalani S. R., Grasso R., Buoni S., Hayek J., Servais L., de Vries B. B., Georgoudi N., Nakou S., Petersen M. B., Mari F., Renieri A., Ariani F. (2010) Eur. J. Hum. Genet. 18, 1133–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Chehadeh-Djebbar S., Faivre L., Moncla A., Aral B., Missirian C., Popovici C., Rump P., Van Essen A., Frances A. M., Gigot N., Cusin V., Masurel-Paulet A., Gueneau L., Payet M., Ragon C., Marle N., Mosca-Boidron A. L., Huet F., Balikova I., Teyssier J. R., Mugneret F., Thauvin-Robinet C., Callier P. (2011) J. Med. Genet., in press [DOI] [PubMed] [Google Scholar]
- 25. El Chehadeh S., Aral B., Gigot N., Thauvin-Robinet C., Donzel A., Delrue M. A., Lacombe D., David A., Burglen L., Philip N., Moncla A., Cormier-Daire V., Rio M., Edery P., Verloes A., Bonneau D., Afenjar A., Jacquette A., Heron D., Sarda P., Pinson L., Doray B., Vigneron J., Leheup B., Frances-Guidet A. M., Dienne G., Holder M., Masurel-Paulet A., Huet F., Teyssier J. R., Faivre L. (2010) J. Med. Genet. 47, 549–553 [DOI] [PubMed] [Google Scholar]
- 26. Diao A., Rahman D., Pappin D. J., Lucocq J., Lowe M. (2003) J. Cell Biol. 160, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goud B., Gleeson P. A. (2010) Trends Cell Biol. 20, 329–336 [DOI] [PubMed] [Google Scholar]
- 28. Donaldson J. G., Lippincott-Schwartz J. (2000) Cell 101, 693–696 [DOI] [PubMed] [Google Scholar]
- 29. Rios R. M., Bornens M. (2003) Curr. Opin. Cell Biol. 15, 60–66 [DOI] [PubMed] [Google Scholar]
- 30. Short B., Barr F. A. (2003) Curr. Biol. 13, R311–313 [PubMed] [Google Scholar]
- 31. Allan V. J., Thompson H. M., McNiven M. A. (2002) Nat. Cell Biol. 4, E236–242 [DOI] [PubMed] [Google Scholar]
- 32. Brown D. L., Heimann K., Lock J., Kjer-Nielsen L., van Vliet C., Stow J. L., Gleeson P. A. (2001) Traffic 2, 336–344 [DOI] [PubMed] [Google Scholar]
- 33. Kjer-Nielsen L., Teasdale R. D., van Vliet C., Gleeson P. A. (1999) Curr. Biol. 9, 385–388 [DOI] [PubMed] [Google Scholar]
- 34. Munro S., Nichols B. J. (1999) Curr. Biol. 9, 377–380 [DOI] [PubMed] [Google Scholar]
- 35. Gillingham A. K., Tong A. H., Boone C., Munro S. (2004) J. Cell Biol. 167, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feinstein T. N., Linstedt A. D. (2008) Mol. Biol. Cell 19, 2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]