Abstract
Loss-of-function mutations in 1-acylglycerol-3-phosphate O-acyltransferase (AGPAT) 2 in humans and mice result in loss of both the white and brown adipose tissues from birth. AGPAT2 generates precursors for the synthesis of glycerophospholipids and triacylglycerols. Loss of adipose tissue, or lipodystrophy, results in hyperinsulinemia, diabetes mellitus, and severe hepatic steatosis. Here, we analyzed biochemical properties of human AGPAT2 and its close homolog, AGPAT1, and we studied their role in liver by transducing their expression via recombinant adenoviruses in Agpat2−/− mice. The in vitro substrate specificities of AGPAT1 and AGPAT2 are quite similar for lysophosphatidic acid and acyl-CoA. Protein homology modeling of both the AGPATs with glycerol-3-phosphate acyltransferase 1 (GPAT1) revealed that they have similar tertiary protein structure, which is consistent with their similar substrate specificities. When co-expressed, both isoforms co-localize to the endoplasmic reticulum. Despite such similarities, restoring AGPAT activity in liver by overexpression of either AGPAT1 or AGPAT2 in Agpat2−/− mice failed to ameliorate the hepatic steatosis. From these studies, we suggest that the role of AGPAT1 or AGPAT2 in liver lipogenesis is minimal and that accumulation of liver fat is primarily a consequence of insulin resistance and loss of adipose tissue in Agpat2−/− mice.
Keywords: Adipose Tissue, Diabetes, Genetic Diseases, Lipodystrophy, Metabolic Diseases, Triacylglycerol, Phospholipid Acyltransferase, Hepatic Steatosis, Phospholipids
Introduction
The 1-acylglycerol-3-phosphate O-acyltransferases (AGPATs)3 are intermediate enzymes in the pathway for the biosynthesis of glycerophospholipids (GPL) and triacylglycerol (TAG) (1, 2). AGPATs esterify the sn-2 position (carbon 2) of 1-acylglycerol-3-phosphate (lysophosphatidic acid (LPA)) to phosphatidic acid (PA) (3–5). Currently, 11 human AGPAT isoforms have been reported, each encoded by a different gene (6–14). Some of these AGPATs also show additional lysophospholipid acyltransferase activities. For example, AGPAT8 and AGPAT9 also possess acyl-CoA: lysocardiolipin acyltransferase 1 (ALCAT1) and acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1) activity, respectively (15–17). AGPAT10 is also reported to have glycerol-3-phosphate acyltransferase (GPAT) activity (18).
Among the AGPAT isoforms, AGPAT1 and AGPAT2 have been studied more extensively than the more recently discovered isoforms 3–11 (19–23). The role of AGPAT2 in cultured cell model systems appears to be providing substrate(s) for synthesis of GPL and TAG. AGPAT2 is expressed most abundantly in adipose tissue, with lower levels in liver and pancreas (19–23). In contrast, AGPAT1 expression is highest in testis followed by pancreas and adipose tissue (19–23).
The role of AGPAT2 in adipocyte biology became apparent when we reported genetic variations in AGPAT2 in patients with congenital generalized lipodystrophy, type 1 (CGL 1) (24). In CGL, lack of adipose tissue from birth results in early onset of insulin resistance, diabetes, hypertriglyceridemia, and hepatic steatosis (25–27). When overexpressed in cultured adipocyte cells, AGPAT2 increases the TAG content but not other GPL (28). The role of AGPAT2 in adipose tissue is further corroborated by the development of Agpat2−/− mice (29). These mice replicate most of the features of human CGL, but some features, such as insulin resistance, seem to be more severe in mice than in humans (29, 30). Conversely, the physiological role of AGPAT1 is less defined.
In the whole animal, the mechanism by which TAG accumulates in livers of Agpat2−/− mice seems to be due to de novo lipogenesis. This TAG accumulation in the livers of Agpat2−/− mice may not be necessarily a direct consequence of hepatic loss of Agpat2 but rather secondary to loss of adipose tissue for TAG storage and/or a result of hyperinsulinemia. Thus, it is unclear what specific role(s) Agpat2 plays in the development of hepatic steatosis. The following is possible: (a) the absence of AGPAT2 in the liver could alter the balance of lipids or signaling molecules that ultimately results in increased TAG formation in the liver, and/or (b) the absence of Agpat2 might prevent normal hydrolysis and oxidation of TAGs and fatty acids, respectively. To determine whether hepatic steatosis was a direct result of loss of AGPAT2 in liver, we expressed AGPAT2 in livers of Agpat2−/− mice using an adenoviral expression system. We also tested whether the closely related isoform, AGPAT1, could compensate for the loss of AGPAT2 expression in livers of these mice. Our reasoning to perform the AGPAT1 compensatory expression was to test the idea that in the absence of a functional AGPAT2 protein in human subjects, the up-regulation of AGPAT1 might alleviate hepatic steatosis.
MATERIALS AND METHODS
Quantitative Real Time PCR in Human Tissue Panel
Quantitative PCR was performed using TaqMan primers and probes designed using Primer Express software and analyzed using ABI Prism 7700 sequence detection system (47). Human cDNA panel was from Clontech. To amplify AGPAT2, 100 pg of cDNA was added to the forward 5′-AACGTGGCGCCTTCCA-3′ and reverse 5′-GAAGTCTTGGTAGGAGGACATGACT-3′ primers along with 6-carboxyfluorescein-labeled probe CTTGCAGTGCAGGCCCAGGTTC and universal mix containing AmpliTaq and appropriate buffers. The PCR was followed for 40 cycles of 94 °C for 15 s and 60 °C for 30 s. Human AGPAT1 was amplified as above but with the following primers: forward 5′-GGTACTCGCAACGACAATGG-3′ reverse 5′-TTGGTGTTGTAGAAGGAGGAGAAG-3′ and 6-carboxyfluorescein-labeled probe CACAGGTGCCCATCGTCCCC. The cDNA was amplified in duplicate along with G3PDH as an internal control. The ΔCt value for each tissue was calculated as ΔCt = (Ct (tissue) − Ct (G3PDH)).
Mouse Liver Tissue Real Time PCR
Total RNA was prepared from mouse livers using RNA STAT-60 (Tel-Test Inc., Friendswood, TX). All RT-PCR were carried out in 96-well plates using the ABI PRISM 7700 sequence detection system (Applied Biosystems) as reported previously (29). Primers used for gene amplification are available upon request.
Generation of Wild Type AGPAT1 and AGPAT2 Recombinant Adenovirus
The recombinant adenovirus was created using the AdEasy adenoviral system (Stratagene, La Jolla, CA) as suggested by the manufacturer and has been described in our recent work (31). Briefly, the coding sequence for human AGPAT1 was amplified from a plasmid carrying the AGPAT1 cDNA using primer pairs (forward 5′-ACGCGTCGACATGGATTTGTGGCCA-3′ and reverse 5′-CCCAAGCTTTCACCCACCGCCC-3′) with restriction sites SalI and HindIII for ease of cloning into pShuttle-CMV vector. The hAGPAT1-pShuttle-CMV was restriction-digested with PmeI and co-transformed with pAdEasy-1 into BJ5183 Escherichia coli (Stratagene, La Jolla, CA) to generate the recombinant plasmid hAGPAT1-pAdEasy-1. This plasmid was then digested with PacI and transfected into AD-293 cells using Lipofectamine-2000. The viral pool that showed the most enzymatic activity was selected for further amplification and purification using the Virabind adenovirus purification kit (Cell Biolabs Inc., San Diego). The AGPAT2 recombinant adenovirus was generated as described for AGPAT1, except that it was amplified using primer pair forward 5′-GGAAGATCTATGGAGCTGTGGCCGTGTCTG-3′ and reverse 5′-CCGCTCGAGCTACTGGGCCGGCTGCACGC-3′. The amplified product was cloned into TA-cloning vector, sequenced, restricted with XhoI, and subcloned into the pShuttle-CMV vector at the same site. The generation of recombinant adenovirus β-galactosidase was generated as above and described earlier (13).
The human AGPAT2 adenovirus was generated using the ViraPower adenoviral expression system (Invitrogen). The coding sequence for AGPAT2 was amplified from the AGPAT2-pshuttle plasmid using the primers forward 5′-CACCATGGAGCTGTGGCCGTG-3′ and reverse 5′-CTACTGGGCCGGCTGCACGC-3′. The PCR product was gel-purified and ligated into pENTR/D-TOPO vector. Insertion was confirmed by restriction digestion and sequencing. Recombination was performed with AGPAT2-pENTR/D-TOPO and pAd/CMV/V5-DEST vectors overnight at room temperature. The recombined AGPAT2-pAD/CMV/V5-DEST plasmid was restriction-digested with PacI and transfected into 293 cells using FuGENE 6 (Roche Diagnostics).
Adenovirus Purification
The amplified adenoviruses were CsCl purified for in vivo use (32).
AGPAT Enzymatic Activity of AGPAT1 and AGPAT2 in Cell Lysate
HEK-293 cells were infected with either AGPAT1 or AGPAT2 recombinant adenovirus at a multiplicity of infection of 150. After 48 h, the infected cells were collected. The cellular viral pellet was resuspended in lysis buffer (100 mm Tris, pH 7.4, 10 mm NaCl) containing protease inhibitor mixture (Roche Diagnostics). The cells were lysed with three freeze/thaw cycles and centrifuged at 3000 × g for 10 min at 4 °C to remove cellular debris. Protein concentrations were determined by a commercially available colorimetric assay (Bio-Rad).
Enzyme activity was determined by measuring the conversion of [3H]LPA to [3H]PA as described previously (9, 33). Briefly, the enzymatic reaction was assembled in 200 μl of 100 mm Tris-HCl buffer, pH 7.4, containing the following: 10 μmol/liter LPA (sn-1-oleoyl-sn-glycerol-3-phosphate, Avanti Polar Lipids, Alabaster, AL), 50 μmol/liter oleoyl-CoA (Sigma), 1 μl of [3H]oleoyl-LPA (specific activity 30–60 Ci/mmol; PerkinElmer Life Sciences), and 1 mg/ml fatty acid-free bovine serum albumin. The reaction was initiated by adding 30 μg of total protein (cell lysate), followed by incubation for 10 min at 37 °C. The reaction was terminated by adding 0.5 ml of 1-butanol containing 1 n HCl to extract phospholipids. The butanol extract was dried under vacuum, and the LPA and PA were resolved by TLC using the solvent system chloroform/methanol/acetic acid/water (85:12.5:12.5:3, v/v). Radioactive spots were identified by co-migration with unlabeled LPA and PA standard and visualized in iodine vapors. The [3H]LPA and [3H]PA spots were scraped and counted for radioactivity (Tri-Carb Liquid Scintillation Counter 3100TR, PerkinElmer Life Sciences).
Additional Lysophospholipid Acyltransferase Activity
To determine whether AGPAT1 and AGPAT2 could acylate any additional lysophospholipids, we used lysophosphatidylcholine, lysophosphatidylethanolamine, lysophosphatidylserine, lysophosphatidylglycerol, and lysophosphatidylinositol, all containing the C18:1 fatty acid at the sn-1 position as substrates. The enzymatic activity was determined as above except that LPA was replaced by the above mentioned lysophospholipids.
Acyl-CoA Specificity
The specificity of esterification of the sn-2 position of [3H]LPA (sn-1-oleoyl-2-hydroxy-sn-glycerol-3-phosphate) was determined by using the following acyl-CoAs: octanoyl (C8:0)-, decanoyl (C10:0)-, lauroyl (C12:0)-, tridecanoyl (C13:0)-, myristoyl (C14:0)-, pentadecanoyl (C15:0)-, palmitoyl (C16:0)-, heptadecanoyl (C17:0)-, stearoyl (C18:0)-, oleoyl (C18:1)-, linoleoyl (C18:2)-, linolenoyl (C18:3; n-3)-, γ-linolenoyl (C18:3; n-6)-, nonadecanoyl (C19:0)-, arachidoyl (C20:0)-, arachidonoyl (C20:4; n-6)-, heneicosanoyl (C21:0)-, behenoyl (C22:0)-, docosahexaenoyl (C22:6; n-3)-, tricosanoyl (C23:0)-, ligneceroyl (C24:0)-, nervonoyl (C24:1; n-9), pentacosanoyl (C25:0)-, and hexacosanoyl (C26:0)-CoA. The assay conditions remained the same as described for the AGPAT activity.
sn-1-acyl-Lysophosphaditic Acid Specificity
To determine the substrate specificity of various LPAs, the following LPA species with various fatty acids at the sn-1 position were used for the assay: myristoyl (C14:0)-, palmitoyl (C16:0)-, arachidonoyl (C20:4)-, arachidoyl (C20:0)-, linoleoyl (C18:2)-, and linolenoyl (C18:3). The enzymatic activity to these LPA species was compared with that of sn-1-oleoyl (C18:1)-2-hydroxy-sn-glycerol-3-phosphate. The enzymatic assay was assembled as before except that unlabeled LPAs and [14C]oleoyl-CoA were used as substrates, and the activity was determined by formation of [14C]PA.
Glycerol-3-phosphate Acyltransferase Assay
The glycerol-3-phosphate acyltransferase activity was determined essentially as described previously (13).
In Silico Modeling of Human AGPAT1 and AGPAT2 Proteins
This is essentially a homology-based modeling of the proteins. The PSIPRED protein structure prediction server was used to predict secondary structure of AGPAT proteins (34), transmembrane domains (35), and protein fold recognition based upon the crystal structure of known proteins (36). Three-dimensional models of human AGPAT1 (accession number: NP_006402.1) and AGPAT2 (accession number: CAH71722.1) were built using the crystal structure of glycerol-3-phosphate acyltransferase (GPAT1) from Cucurbita moscata as a template (Protein Data Bank code 1IUQ) (37). The modeling was performed using AMBER 9 (38) and PyMol (PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC). To facilitate homology modeling based upon GPAT, the GPAT coordinates were modified to exclude the amino-terminal extension (Ala2–His86), the carboxyl-terminal extension (Ile349–Trp368), and the hetero groups (glycerol and sulfate). As such, the AGPAT sequences (AGPAT1 Val58–Asp268; AGPAT2 His52–Thr262) were manually docked to the GPAT sequence using the most conserved residues and structural correspondence. Each of the GPAT residues was then replaced by the corresponding residue of either the human AGPAT1 or AGPAT2 sequences. The length of the conserved regions was kept unchanged. Energy minimizations were then performed using 1000 cycles of the Amber force field (ff99) (39). The resulting structures were inspected visually.
Immunoblot Analysis
Immunoblot analysis was carried out as described previously (33). The cells were lysed in RIPA buffer (150 mm NaCl, 1% Nonidet P-40, 50 mm Tris, pH 8.0) containing protease inhibitors (Roche Diagnostics). Total cell lysate proteins (40 μg) from the AGPAT1, AGPAT2, and LacZ adenovirus-infected cells were resolved on precast 10% SDS-PAGE (Bio-Rad) followed by transfer onto PVDF membranes (Millipore, Billerica, MA). The membranes were blocked with 5% nonfat milk containing 0.2% Tween 20 and then incubated with affinity-purified antibody specific for human AGPAT1 raised against amino acids 208–280. Similarly, blots were probed with affinity-purified antibodies specific for human AGPAT2 (raised against human AGPAT2 amino acids 143–278). Both antibodies were raised in chickens, concentration of 1 μg/μl, and were custom-made by GenWay Biotech (San Diego). Antibodies were used at a dilution of 1:2500 at room temperature for 2 h, followed by incubation with secondary antibody to IgY (goat anti-IgY conjugated to alkaline phosphatase) at a 1:5000 dilution (GenWay Biotech, San Diego) and detected with ECL Plus (GE Healthcare). The same blot was stripped using Restore Western blot stripping buffer (Pierce), according to the manufacturer's protocol, re-probed with GAPDH antibody at 1:5000 dilution (mouse monoclonal, Ambion, Austin, TX), and detected with ECL Plus. In some experiments, liver microsomes were isolated from the livers of mice infected with adenovirus expressing human AGPAT1 and AGPAT2 and used for immunoblotting as above, except that calnexin as an ER marker was used for the microsomal specificity.
Generation of EGFP-tagged Wild Type AGPAT1 and AGPAT2 Expression Vector
The plasmid carrying the human cDNA for AGPAT1 was amplified with primer pair (forward 5′-CCGCTCGAGATGGATTTGTGGCCA-3′ and reverse 5′-CCGGAATTCAACCCACCGCCCC-3′) and cloned in-frame to the expression plasmid pEGFP-N3 (Clontech) in the XhoI and EcoRI sites. Restriction sites are underlined. Similarly, human AGPAT2 was amplified with primer pair (forward 5′-CCCTCGAGATGGAGCTGTGGCCGTGTCTGGC-3′ and reverse 5′-CGGGATCCCTTCTGGGCCGGCTGCACGCCAG-3′) and cloned in the XhoI and BamHI sites. The expression plasmids were sequenced to ascertain the orientation and sequence errors.
Generation of Recombinant mCherry-AGPAT1 Expression Vector
The human AGPAT1 was amplified from the plasmid containing the AGPAT1 open reading frame (pDrive-hAGPAT1) with the primers forward 5′-CCGCTCGAGATGGATTTGTGGCCA-3′ and reverse 5′-CCGGAATTCAACCCACCGCCCC-3′ carrying the restriction sites XhoI and EcoRI, respectively. The amplified product was cloned again in pDrive vector, sequenced for PCR errors, and cloned in the KpnI and XhoI sites of mCherry vector.
Subcellular Fraction of the Cells Expressing AGPAT1-EGFP and AGPAT2-EGFP Fusion Protein
This was achieved by stably expressing AGPAT1-EGFP and AGPAT2-EGFP in the Chinese hamster ovary (CHO) cells. The cells were collected and subjected to homogenization. The homogenate was further subjected to differential centrifugation. The 14,000 and 100,000 × g pellets correspond to mitochondrial and microsomal fractions, respectively. These fractions were washed twice in the same lysis buffer. Protein was determined as described above.
Immunofluorescence Microscopy
Stably expressing AGPAT1-EGFP and AGPAT2-EGFP CHO cells were grown on glass coverslips 1 day before the experiment. Cells were processed as described previously (31). Primary hepatocytes were obtained as described earlier (40) and plated on collagen-coated coverslips. A day after seeding, the cells were transfected with the above plasmid, and 48 h after the transfection, cells were processed as described previously (31, 41).
Infection of Wild Type and Agpat2−/− Mice with Recombinant Adenovirus Expressing Human AGPAT1 and AGPAT2
All mice of mixed genetic background (129.v/C57/6J) between the ages of 20 and 25 weeks were randomly divided into groups of 6–8 mice and under light anesthesia were injected with 1.0 × 1011 viral particles in ∼100 μl of PBS. After 1 week, mice were sacrificed, and their livers were collected, snap-frozen in liquid nitrogen, and stored at −70 °C until needed. Blood was drawn in EDTA tubes, and the plasma was separated immediately and kept at −70 °C until further use. Animals used in this study were approved by the Institutional Use and Care of Animals and BioSafety Committee at the University of Texas Southwestern Medical Center at Dallas.
Quantification of LPA and PA from Mouse Liver Homogenates
Approximately 250 mg of liver was homogenized in 2 ml of 100 mm Tris-HCl, pH 7.5, containing 10 mm NaCl. Cellular debris was removed by centrifugation at 2800 rpm for 5 min at 4 °C, and a small portion of the homogenate was used to extract DNA. DNA was extracted with Easy-DNA kit (Invitrogen) following the manufacturer's protocol. A known volume of the homogenate was used to extract LPA and PA using 1-butanol, 1 n HCl. To determine the extraction efficiency, 0.1 μCi of [3H]oleoyl-LPA (PerkinElmer Life Sciences) was added to the homogenate followed by centrifugation at 2800 rpm for 5 min at 4 °C. The upper aqueous phase was transferred into a new tube. The extraction was repeated twice with water-saturated butanol (1:4), and the upper aqueous phase was combined with the previous extract and dried under vacuum. The dried phospholipid extract was resuspended in a known volume of chloroform. Standard phospholipids 1-oleoyl-2-hydroxy-sn-glycero-3-phosphate (LPA) and 1,2-dioleoyl-sn-glycero-3-phosphate (PA) were from Avanti Polar Lipids (Alabaster, AL). LPA and PA standards were resolved using chloroform/methanol/water/ammonium hydroxide gradient (Buffer A 80:19.5:0:0.5, v/v, and Buffer B 60:34:5:0.5, v/v) on a Supelcosil LC-Diol column (5 μm; 25 cm × 4.6 mm, Sigma) connected to a 1525 HPLC (Waters Corp., Milford, MA) equipped with an evaporative light scattering detector (Waters 2420). The flow rate of the mobile phase was maintained at 1 ml/min, and the separations were performed using a linear gradient (ranging from linear A/B 50:50 to 100% B 0–14 min followed by 14–20 min at 100% B). The HPLC column was maintained at room temperature, and the nebulizer was equilibrated to 35 °C, and the evaporator column of the detector was maintained at 80 °C and flushed with pure N2 gas at 30 p.s.i. Standard curves were generated using various concentrations of LPA and PA and used to determine the concentration of liver LPA and PA. The concentrations of LPA and PA were estimated by determining the area under the peak using Breeze software (version 3.30 SPA) and normalized to DNA.
Biochemical Measurements
Liver AGPAT activity was determined as described above. To determine TAG and cholesterol concentrations, ∼100–150 mg of frozen livers were homogenized in Folch solution (chloroform/methanol 2:1, v/v). The organic phase was separated and collected by adding buffered saline and dried. The extract was reconstituted in the same extraction buffer, and TAG and cholesterol levels were measured using reagents from Infinity Liquid Stable Reagents (Fisher Diagnostics, Middletown, VA). Plasma cholesterol, TAG, and glucose were measured using Dry-slide technology (Vitros 250 Analyzer from Ortho Clinical Diagnostic). All slides were purchased from Cardinal Health, Inc., and their catalog numbers are as follows: SP1707801 for glucose, SP1669829 for cholesterol, and SP1336544 for TAG. All measurements were carried out at the Mouse Metabolic Phenotyping Core at the University of Texas Southwestern Medical Center. Because plasma insulin concentrations were extremely high, they were measured with the kit obtained from Crystal Chem (Downers Grove, IL) in its high range mode, which allows measurements up to 64 ng/ml.
Statistical Analyses
Statistical significance was calculated either by Dunnett's test (for two groups) or by two-way analysis of variance (for multiple groups). p < 0.05 was considered significant.
RESULTS
Expression of AGPAT1 and AGPAT2 in Human Tissue Panel
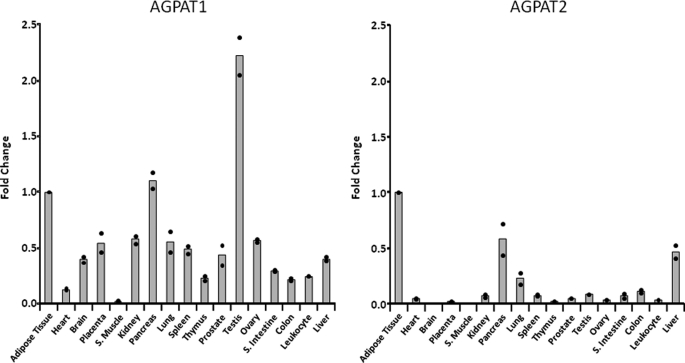
Human AGPAT1 is located within the class III MHC locus on chromosome 6p21.3 and AGPAT2 on chromosome 9q34.3 (23). Inspecting the flanking sequences for both the genes revealed that they have almost identical genes flanking their 5′- and 3′-regions, indicating that the two genes might be a result of partial duplication of the chromosomes. This would indicate a similar, if not identical, tissue expression pattern. However, this is not the case. Quantitative real time PCR revealed that human AGPAT1 is ubiquitously expressed compared with AGPAT2, which is more tissue-restricted (Fig. 1). When compared with omental adipose tissue (AT), AGPAT1 is expressed 2-fold more in the testis compared with all other tissues. Expression of AGPAT1 in pancreas is similar to that in AT. AGPAT1 is undetectable in skeletal muscle, whereas all other tissues express 50% or less compared with AT. In whole brain, the expression of AGPAT1 was detectable, but inspection of the microarray data from multiple tissues showed that within the brain the prefrontal cortex had the highest AGPAT1 expression (42). In contrast, AGPAT2 is most abundantly expressed in AT (this study). In the next two highest expressing tissues, the pancreas and liver, the expression of AGPAT2 was about 50% less than in the AT. In all other tissues, the expression of AGPAT2 is only 20% of AT. A previous study employing a semi-quantitative Northern blot analysis corroborated our finding (21). However, that study did not include AT (21).
FIGURE 1.
Expression of human AGPAT1 and AGPAT2 in tissues. The ΔCt values are shown as quantified by TaqMan real time PCR. Shown as ●, the individual fold changes to adipose tissue (individual ΔCt values were normalized to internal control, G3PDH). The bars represent the mean fold changes as compared with adipose tissue taken as one. Fold increase between various tissues were calculated as 2−ΔΔCt (56). A, AGPAT1; B, AGPAT2. S. muscle, skeletal muscle and S. intestine, small intestine.
Subcellular Localization of the Human AGPAT1 and AGPAT2
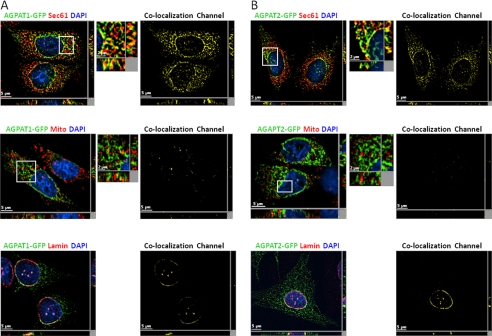
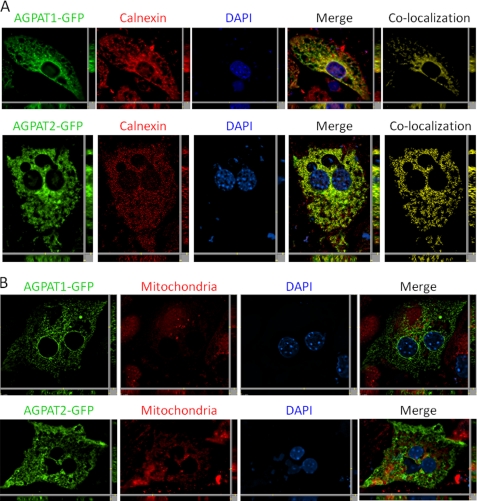
Expression of human AGPAT1-EGFP and AGPAT2-EGFP in CHO cells shows that both the proteins are co-localized with sec61-β, an endoplasmic reticulum (ER)-specific protein (Fig. 2, A and B), corroborating previous studies (20, 21). We also observed that these proteins co-localized with lamin A/C, which is a nuclear lamina protein, indicating the presence of AGPAT1 and AGPAT2 in the nuclear envelope as well. However, the co-localization of AGPAT1 was less than AGPAT2 in the nuclear envelope. We also observed some co-localization of AGPAT1-EGFP in the mitochondrial fraction, but further experiments using either CHO cells expressing only the native AGPAT2 protein or subcellular fractionation of mouse liver revealed this to be due to overexpression in the CHO cells (see below). When AGPAT1-EGFP or AGPAT2-EGFP was expressed in primary mouse hepatocytes, both the proteins localized to the ER only (Fig. 3A) and not to the mitochondria (Fig. 3B).
FIGURE 2.
Localization of AGPAT1-EGFP and AGPAT2-EGFP to endo-membranes in cultured cells. A, CHO cells overexpressing AGPAT1-EGFP were fixed in methanol, incubated with antibody sec61-β (specific for endoplasmic reticulum) and lamin A/C (specific for nuclear lamina), and imaged for green and red fluorescence using fluorescence microscopy. Shown are representative images for AGPAT1 (green fluorescence), sec61-β (red fluorescence), DAPI (blue fluorescence), co-localization channel (yellow fluorescence). The AGPAT1-GFP-expressing cells were incubated with MitoTracker Red dye, fixed in 4% paraformaldehyde, and imaged as before. B, fluorescence images for AGPAT2-EGFP expressed in CHO cells. Cells for sec61-β, lamin A/C and mitochondria were processed as above. Shown for each image is a single z-stack image, whereas the x and y axis shows the z-stacks. Scale bar, 5 μm for larger images and 2 μm for higher magnification images.
FIGURE 3.
Localization of human AGPAT1-EGFP and AGPAT2-EGFP to endoplasmic reticulum in mouse primary hepatocytes. A, mouse primary hepatocytes with exogenously expressed human AGPAT1-EGFP and human AGPAT2-EGFP were fixed in methanol and incubated with antibody calnexin (specific for endoplasmic reticulum) and imaged for green and red fluorescence using fluorescence microscopy. Shown are representative images for AGPAT1 and AGPAT2 (green fluorescence), calnexin (red fluorescence), DAPI (blue fluorescence), co-localization channel (yellow fluorescence), and the merged image. B, AGPAT1-EGFP- and AGPAT2-EGFP-expressing cells were incubated with MitoTracker Red dye, fixed in 4% paraformaldehyde, and imaged as before. Shown for each image is a single z-stack image, and the x and y axis shows the z-stacks. Scale bar, 5 μm.
Determination of AGPAT1-EGFP and AGPAT2-EGFP Fusion Protein Enzymatic Activity in Subcellular Fraction
The CHO cells stably expressing AGPAT1-EGFP and AGPAT2-EGFP fusion protein were subfractionated, and the enzymatic activity was determined in the mitochondrial and ER subcellular fraction. Immunoblots probed with sec61-β antibodies, showed that the mitochondrial fraction was devoid of any ER contamination (data not shown). Both AGPAT isoforms have significant activity in the microsomes. We did detect some AGPAT activity in the mitochondrial fraction obtained from AGPAT1-EGFP-expressing CHO cells but not with AGPAT2-EGFP-expressing cells. This was further confirmed when additional blots were probed with antibodies specific for AGPAT1 and AGPAT2. Although we observed a hint of AGPAT1-EGFP localizing to the mitochondria, computational analysis further revealed that human AGPAT1 had a very low probability of 0.25 to be localized to the mitochondria (43). This indicated that if overexpressed, the AGPAT1 protein might localize to additional subcellular organelles like mitochondria.
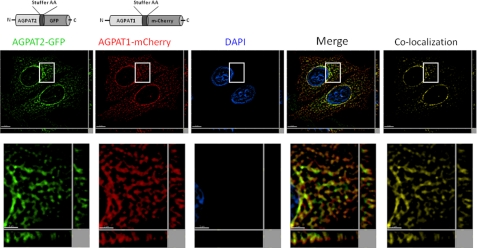
Co-expression of Human AGPAT1-RED and AGPAT2-EGFP in CHO Cells
When individually expressed, both the AGPAT1 and AGPAT2 green fluorescent-tagged proteins targeted primarily to the ER. To test if both the proteins target to different subdomains of the ER or localize to other subcellular compartments of the cell, we co-expressed the AGPAT1 cherry (red) fluorescent protein along with AGPAT2 green fluorescent-tagged protein. As shown in Fig. 4, both proteins localize in the ER in the same ER subdomains. This co-localization was analyzed statistically, and it revealed a high degree of fluorescence signal overlap (Pearson's correlation coefficient r = 0.73; with overlap of 81% in red channel and 74% in green channel) for the whole image.
FIGURE 4.
Localization of human AGPAT1-RED and human AGPAT2-EGFP to endo-membranes in cultured cells. CHO cells overexpressing AGPAT2-EGFP were transfected with AGPAT1-RED and were fixed in methanol, stained with DAPI, and imaged for green and red fluorescence using fluorescence microscopy. Shown are representative images for AGPAT2 (green fluorescence), AGPAT1 (red fluorescence), DAPI (blue fluorescence), co-localization channel (yellow fluorescence), and the merged image. Shown for each image is a single z-stack image, and the x and y axis shows the z-stacks. Scale bar, 5 μm for larger images and 2 μm for higher magnification images.
AGPAT Immunoblots and Activity of Expressed Human AGPAT1 and AGPAT2
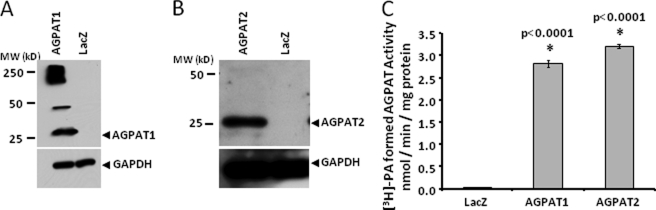
To characterize the recombinant AGPAT1 and AGPAT2 proteins, we expressed the recombinant adenovirus in AD293 cells, and 48 h after infection the cells were collected for immunoblots and in vitro AGPAT activity. As shown in Fig. 5, both the proteins were detectable when probed with specific antibodies to human AGPAT1 and AGPAT2. The expected molecular mass of human AGPAT1 and AGPAT2 protein was around 32 kDa. However, these proteins migrate around 25 kDa on SDS-PAGE. There could be several explanations for this observation. Both proteins have a predicted signal peptide that is most likely cleaved. The predicted cleavage site for human AGPAT1 and AGPAT2 was located between residues 26–27 and 23–24, respectively (prediction using SignalP 3.0 Server). This would result in a mass of around 28 kDa for both the proteins. Alternatively, the protein could undergo post-translational modification such that the protein migrates faster than expected or there could be an additional internal translational start site. Similar observations were made when the proteins were expressed in the insect cells, Sf9, as well (44). In our expression system, human AGPAT1 tends to aggregate forming either a dimer (50 kDa) or a multimer of around 250 kDa. However, when expressed in insect cells, the protein aggregation of human AGPAT1 was not observed (45). The recombinant human AGPAT2 protein showed only one specific protein band. Both isoforms are enzymatically active.
FIGURE 5.
Enzymatic activity and Western blot of wild type human AGPAT1 and AGPAT2 expressed in AD293 cells. A and B, Western blot for the recombinant AGPAT1 and AGPAT2 proteins from whole cell lysate probed with antibody specific for AGPAT1 and AGPAT2 protein. Lysates from cells infected with recombinant β-galactosidase (LacZ) adenovirus were loaded as a negative control. The same blot was stripped and reprobed with full form GAPDH antibody to demonstrate protein loading. C, AGPAT activity in whole cell lysate for AGPAT1 and AGPAT2 as determined by conversion of [3H]LPA to [3H]PA in the presence of oleoyl-CoA and expressed as product [3H]PA formed (nmol per min per mg of protein). The LPA to PA conversion by recombinant LacZ adenovirus was used as a control. Not shown is the conversion of substrate in the absence of enzyme. Each bar represents mean ± S.D. from three independent experiments carried out in triplicate. p value is shown above the bars.
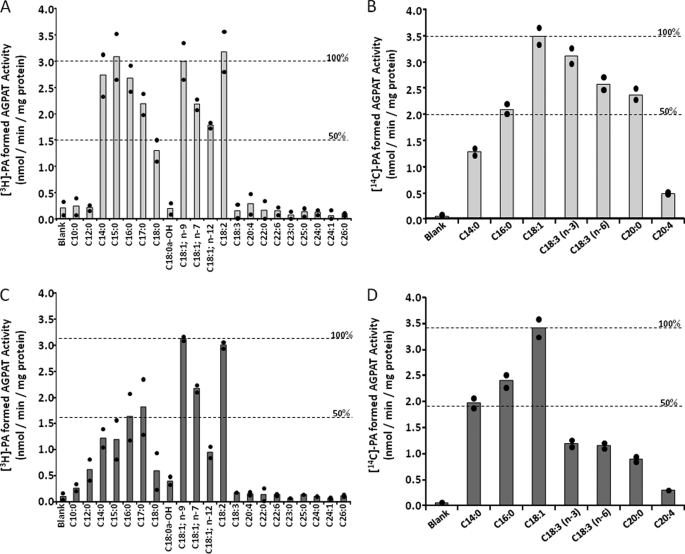
Acyl-CoA Specificity of Human AGPAT1 and AGPAT2
Next we determined the fatty acid specificity of AGPAT1 and AGPAT2 using a panel of commercially available acyl-CoAs consisting of medium chain to very long chain (including odd chain) fatty acids. In our assay, the preferred fatty acid was C18:1 followed by C18:2 for both the isoforms studied (Fig. 6, A and C). Saturated fatty acids (C18:0) were less active. Interestingly, both isoforms also used odd chain fatty acids for LPA acylation. AGPAT1 used C15:0 at approximately the same rate as that of C18:1 or C18:2; however, the AGPAT2 enzymatic activity for C15:0 was ∼50% that of C18:1 or C18:2. The odd chain fatty acid C17:0 was also a substrate for both the enzymes but less active than C15:0. Both isoforms failed to use fatty acids shorter than C14:0 or longer than C20:0, whether saturated or unsaturated. These studies also demonstrated that there is a specific requirement for unsaturation of the fatty acid. Eighteen carbon fatty acids with more than two double bonds were inactive in our assays. Because some prior studies did not use the same LPA used in this study, a direct comparison of the acyl-CoAs specificities was not attempted. For example, using sn-1-C16:0-LPA as acceptor, the rank order of acyl-CoA usage was C14:0 = C18:0 = C20:4 < C16:0 for AGPAT2 enzyme (21). In another study using membrane fractions from insect cells, Sf9, recombinant AGPAT1 protein showed better AGPAT activity with C18:3; n-3- and C16:0-CoAs but reduced activity with C14:0-, C18:1-, C18:0-, and C20:4-CoAs (44). This variation might be due to different host cells (insect cells versus mammalian cells, used in this study) or due to the usage of membrane fractions versus the postnuclear supernatant. In another study using a similar expression system, only C14:0- and C16:0-CoAs were found to be preferred substrates for AGPAT1, whereas C18:1-CoA had very poor activity (45). However, Eberhardt et al. (21) found that C18:1-CoA was the best substrate for AGPAT2. Such results now point to the fact that further studies are needed to resolve these substrate variations, preferably with purified protein.
FIGURE 6.
Acyl-CoA and LPA specificity of recombinant human AGPAT1 and AGPAT2 expressed in AD293 cells. Specificity of human recombinant AGPAT1 (A) and AGPAT2 (C) for acyl-CoA donors was determined using sn-1-oleoyl-lysophosphatidic acid as an acceptor and various short, medium, and long chain fatty acyl-CoA as donors. The enzymatic activity is also compared with C18:1 = 100%, which is shown across the graph as a broken line. Specificity of human recombinant AGPAT1 (B) and AGPAT2 (D) for various species of sn-1-lysophosphatidic acid acceptors was determined using radioactive C18:1 as donor. Activities are expressed as product [3H]PA formed (nmol per min per mg protein). Shown is the 100 and 50% enzymatic activity when compared with C18:1 = 100%. All enzymatic activities were determined in two independent experiments in triplicate. Shown are the means from individual experiments, and each bar represents the mean of two experiments.
LPA Specificity of Human AGPAT1 and AGPAT2
We used various LPAs carrying different fatty acids at the sn-1 position (shown in Fig. 6, B and D) to determine LPA specificity. When C18:1-LPA was used as the acceptor and C18:1-CoA as donor, the maximum rate of conversion of LPA to PA was observed for both the isoforms. Other LPAs were less preferred substrates. Although isoform AGPAT2 appears to show restrictive usage of LPAs, isoform AGPAT1 showed much broader LPA usage. LPAs carrying the C18:3; n-3, C18:3; n-6, and C20:0 were also effective acceptors when incubated with C18:1-CoA as donors in the presence of recombinant AGPAT1 protein. However, with AGPAT2, the conversion was less than 50% that of C18:1-LPA. In our assay, LPA with C20:4 is not an efficient substrate for either enzyme. In one previous study using membrane fractions from insect cells, Sf9, recombinant AGPAT1 protein showed better AGPAT activity with linoleoyl-, palmitoyl-, and myristoyl-LPA but only slightly less with C18:1- and C18:0-LPA (44). This variation might be due to different host cells (insect cells versus mammalian cells) or due to the usage of membrane fractions versus postnuclear supernatant.
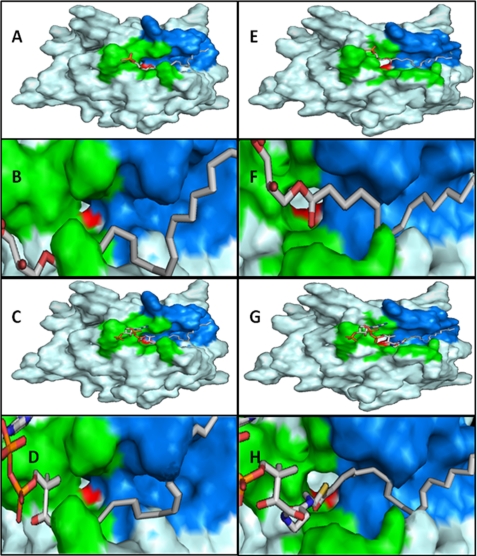
Three-dimensional Models for Human AGPAT1 and AGPAT2 Proteins
Because there is not yet a crystal structure available for AGPAT1 and AGPAT2, the structure of both proteins were modeled based on the available x-ray crystal structure of GPAT (37, 46). Both modeled proteins preserve the basic folding of GPAT. Within the basic structure of AGPAT1 and AGPAT2, it is possible to observe three tunnels also present in GPAT. In the case of AGPAT1 and AGPAT2, one of the tunnels is mostly populated by polar residues (data not shown). The second tunnel is hydrophobic in nature with a length of ∼19 Å (Fig. 7) to accommodate the extended acyl chains of sn-1-C18:0-LPA, which is ∼20 Å. The origin of this tunnel converges with the entrance to the catalytic site H(X)4D and is flanked on the other extreme by positively charged residues that could determine the maximum length of the tunnel by repelling the acyl chain. The tunnel is not closed toward the edge of the protein, which could indicate that this is an area associated with the membrane or in an oligomeric state. It is also interesting to note that ∼20 amino acids that form the hydrophobic tunnel in both human AGPAT1 and AGPAT2 proteins are spread across the protein as shown in Fig. 8. However, a recently crystallized protein, autotaxin, showed a closed tunnel (47). Autotaxin protein was crystallized with LPAs of different lengths. It was observed that the closed environment was capable of accommodating even sn-1-C22:6:0-LPA, which showed a sharp U-turn toward the middle. This indicates that large acyl chains need greater flexibility to fit in the binding pocket. In the case of AGPAT1 and AGPAT2, we observed that acyl-CoAs with a chain of 15 carbons were also preferred. This could be related to the fact that longer acyl chains have limited room to display flexibility. A third tunnel is found on the opposite side of the hydrophobic tunnel just described. Its length is smaller and has no direct access to the active site (data not shown).
FIGURE 7.
Homology modeling of human AGPAT1 and AGPAT2 proteins. Model for human AGPAT1, with LPA 18:1 manually docked (A), close up of LPA 18:1 bound (B), acyl-CoA 18:1 manually docked (C), and close up of acyl-CoA 18:1 bound (D). In the same fashion, the model of AGPAT2 is presented in E–H. The green cluster represents the highly conserved residues that bind the charged regions of the ligands. The blue cluster represents a hydrophobic tunnel that is capable of accommodating acyl chains. The length of the tunnel is ∼19 Å. The red cluster is composed of the conserved catalytic region, H(X)4D. The models clearly show the convergence of the acyl binding pocket and the active site.
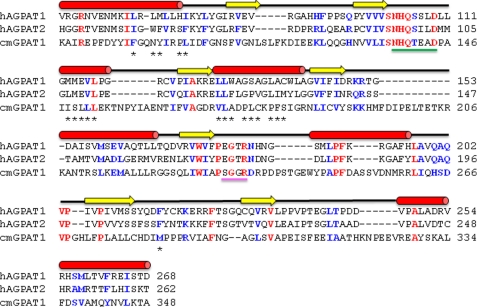
FIGURE 8.
Primary and secondary structure alignment of human AGPAT1 and AGPAT2. Shown are the primary structure alignments of human AGPAT1 (NP_006402.1) and AGPAT2 (CAH71722.1) and squash GPAT (C. moscata BAB17755.1). The secondary structure above the sequences corresponds to that of the AGPAT proteins. α-Helices are colored red; β-sheets are in yellow, and black lines represent coils. The amino acids identified by homology modeling in the hydrophobic tunnel are shown with asterisk. Underlined in green is the catalytic site, with the histidine and aspartate highly conserved amino acids. Underlined in magenta is the EGTR conserved region, with the highly conserved glycine also in the GPAT sequence. These two regions and other highly conserved amino acids were used to establish the homology modeling.
Flanking the other side of the entrance to the active site is a cluster of highly conserved positive residues that have been implicated in GPAT to the binding of the phosphate in glycerol 3-phosphate (37). These residues are highly conserved in both AGPAT1 and AGPAT2. In this region, we could see that the distribution of these positively charged surfaces was different between AGPAT1 and AGPAT2. In AGPAT1, the positive surface is more constricted at the entrance of the hydrophobic tunnel. The role of this difference is unclear, but it could account for ligand affinity differences between AGPAT1 and AGPAT2. It is possible that this charged region guards the entrance of the hydrophobic tunnel, and interactions with the negative portions of LPA or CoA can yield conformational changes that allow the acyl chain to bind inside the tunnel. If we consider that this region could be associated with the membrane, it is also possible to argue that the acyl chain, by binding to the hydrophobic tunnel, creates conformational changes that favor the binding of the charged portions of the LPA or CoA, positioning ligands such that they are at a permissive distance for catalysis.
Additional Acyltransferase Activities
We also determined whether the recombinant proteins AGPAT1 and AGPAT2 also show enzymatic activity for various other lysophospholipids, such as lysophosphatidylcholine, lysophosphatidylethanolamine, lysophosphatidylinositol, lysophosphatidylserine, and lysophosphatidylglycerol. The human AGPAT1 and AGPAT2 proteins were incubated with the above mentioned lysophospholipids carrying C18:1 at their sn-1 position. Both the enzymes have less than 20% of the lysophospholipid acyltransferase activity compared with AGPAT activity (data not shown). These isoforms did not show any GPAT activity (data not shown).
Enzyme Kinetics of Recombinant Human AGPAT1 and AGPAT2 Protein
The preferred substrates for both the enzymes are 1-oleoyl-LPA as acceptor and oleoyl-CoA as fatty acid donor. We then systematically determined the optimum temperature, pH, and length of incubation (data not shown) to further determine the apparent rate constants (Vmax) and the apparent affinity constant (Michaelis-Menten, Km). The apparent Vmax and the apparent Km values for both the enzymes in in vitro assays are similar but not identical (Table 1) when oleoyl-CoA fatty acid was used. Although both the enzymes have similar velocity constants for C18:1 and C18:0, differences were observed with fatty acid C15:0. AGPAT1 has a higher Vmax than AGPAT2. AGPAT1 and AGPAT2 have a higher affinity for C15:0 than C18:1. C18:0 has several fold lower apparent Km values for both the enzymes. This observation is consistent with the substrate preference (for acyl-CoA) determined above. The kinetic parameters for C14:0 are similar to C18:0 for AGPAT1, indicating a strong preference for unsaturated fatty acids. The kinetic parameters for both the enzymes showed very similar enzyme kinetics for the most active LPA (sn-1-oleoyl-LPA). Both enzymes in vitro have very similar velocity and affinity constants.
TABLE 1.
Shown are the mean apparent affinity (Km) and apparent maximum velocity (Vmax) of human AGPAT1 and AGPAT2 (this study) and those of AGPAT2 (44)
Human AGPAT1 and AGPAT2 were expressed in AD293 cells using recombinant adenovirus, whereas human AGPAT2 was expressed in Sf9 cells using recombinant baculovirus. Kinetic parameters for the proteins were determined using unpurified recombinant proteins.
| Enzyme | Substrate | Km | Vmax | Vmax/Km | Ref. |
|---|---|---|---|---|---|
| μm | nmol/min/mg protein | ||||
| AGPAT1 | Acyl-CoA (C15:0) | 3.04 | 86.09 | 28.32 | This study |
| (C18:1) | 39.37 | 77.57 | 1.97 | ||
| (C18:0) | 116.73 | 91.94 | 0.78 | ||
| (C14:0) | 137.97 | 96.21 | 0.70 | ||
| LPA sn-1-C18:1 | 6.0 | 92.0 | 15.33 | ||
| AGPAT2 | Acyl-CoA (C15:0) | 11.05 | 51.61 | 4.67 | |
| (C18:1) | 30.21 | 73.81 | 2.44 | ||
| (C18:0) | 523.97 | 95.55 | 0.18 | ||
| LPA sn-1-C18:1 | 8.29 | 86.05 | 10.38 | ||
| Acyl-CoA (C18:1) | 0.4 | 210 | 525 | 44 | |
| LPA sn-1-C18:1 | 2 | 200 | 100 |
AGPAT Enzymatic Activity in Agpat2−/− and Wild Type Mice
Our in vitro AGPAT activity assay determined the substrate preferences of acyl-CoA and LPA for AGPAT1 and AGPAT2 recombinant proteins. To determine whether this specificity is similar in the livers of mice, which have additional endogenous acyl-CoAs and LPAs present, we also examined the AGPAT activity in the livers of 16-week-old Agpat2−/− and age-matched wild type mice. The liver homogenates of Agpat2−/− mice showed very poor conversion of LPA to PA, which could be attributed to the low expression of AGPAT1 enzyme. The substrate preferences for AGPAT in the wild type mice replicated the results of the in vitro assays (data not shown).
Rescue of Hepatic Steatosis in Agpat2−/− Mice
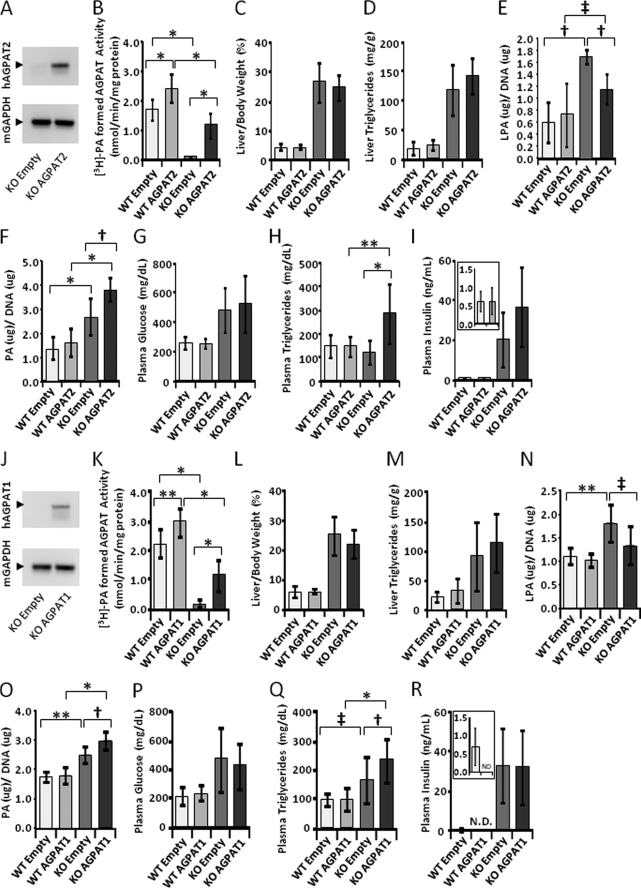
Infection of 20–25-week-old Agpat2−/− mice with the adenovirus expressing AGPAT2 protein did not decrease the hepatic steatosis. The liver weights and TAGs remained the same as those of Agpat2−/− mice that received only empty adenovirus (Fig. 9, C and D). Plasma glucose and insulin concentrations were also the same as mice that received only empty virus (Fig. 9, G and I), although we did observe an increase in plasma TAG rather than a decrease (Fig. 9H). The expression levels of several genes associated with lipid or glucose synthesis were also unchanged by AGPAT2 overexpression in liver (Table 2).This is despite the fact that AGPAT activity was increased by ∼10-fold (Fig. 9, B and K). Similar observations were made with adenoviral AGPAT1 overexpression (Fig. 9, L, M, and P–R).
FIGURE 9.
Liver acyltransferase activity and liver plasma triglyceride, cholesterol, glucose, insulin, lysophosphaditic acid, and phosphatidic acid levels after acute expression of human AGPAT2 and human AGPAT1 in the livers of Agpat2−/−. Groups of 8–15 mice of either wild type or Agpat2−/− genotype were infected with the recombinant adenovirus expressing AGPAT2 and sacrificed 1 week later. A shows a PCR amplification of the human AGPAT2 and mouse GAPDH transcripts. B–F show the liver AGPAT activity, liver weight, triglyceride, lysophosphaditic acid, and phosphatidic acid levels (phospholipids were analyzed in groups of six mice). G–I show plasma glucose, triglyceride, and insulin levels. J–R show PCR amplification and liver and plasma levels after infecting Agpat2−/− mice with recombinant adenovirus expressing AGPAT1. N.D., not determined. The p values are shown above the bars (*, < 0.0001; **, < 0.001; †, < 0.01; ‡, < 0.05).
TABLE 2.
Relative expression of liver mRNAs of Agpat2−/− mice infected with recombinant adenovirus expressing human AGPAT1 and human AGPAT2
Shown are the fold changes of pooled samples from 8 to 15 mice. Values indicate fold change.
| Genotype | Agpat2−/− | Agpat2−/− | Agpat2−/− | Agpat2−/− |
|---|---|---|---|---|
| Sex | Mixed | Mixed | Mixed | Mixed |
| Adenovirus injected | Empty | AGPAT2 | Empty | AGPAT1 |
| Fatty acid and TG synthesis | ||||
| ACL | 1.00 | 0.66 | 1.00 | 0.90 |
| ACC1 | 1.00 | 0.82 | 1.00 | 1.09 |
| FAS | 1.00 | 0.72 | 1.00 | 0.73 |
| Elovl6 | 1.00 | 0.72 | 1.00 | 1.24 |
| SDC-1 | 1.00 | 0.83 | 1.00 | 1.32 |
| SCD-2 | 1.00 | 0.85 | 1.00 | 0.82 |
| Agpat1 | 1.00 | 0.94 | 1.00 | 0.73 |
| Agpat2 | 1.00 | 1.51 | 1.00 | 1.65 |
| Agpat3 | 1.00 | 0.93 | 1.00 | 0.89 |
| Agpat4 | 1.00 | 1.28 | 1.00 | 0.85 |
| Agpat5 | 1.00 | 1.28 | 1.00 | 0.82 |
| Agpat6 | 1.00 | 1.27 | 1.00 | 0.96 |
| Agpat7 | 1.00 | 1.75 | 1.00 | 1.32 |
| Agpat8 | 1.00 | 0.94 | 1.00 | 0.97 |
| Agpat9 | 1.00 | 1.41 | 1.00 | 1.17 |
| Agpat10 | 1.00 | 1.07 | 1.00 | 0.66 |
| Agpat11 | 1.00 | 1.62 | 1.00 | 1.02 |
| Mgat1 | 1.00 | 1.00 | 1.00 | 0.96 |
| Mgat2 | 1.00 | 0.86 | 1.00 | 1.01 |
| Dgat1 | 1.00 | 1.23 | 1.00 | 1.30 |
| Dgat2 | 1.00 | 0.92 | 1.00 | 1.12 |
| Gpat1 | 1.00 | 1.24 | 1.00 | 0.98 |
| PPARG | 1.00 | 0.91 | 1.00 | 1.15 |
| Ppap2a1 | 1.00 | 1.06 | 1.00 | 0.92 |
| Ppap2a2 | 1.00 | 1.07 | 1.00 | 1.01 |
| Ppap2b | 1.00 | 0.81 | 1.00 | 0.99 |
| Ppap2c | 1.00 | 0.82 | 1.00 | 0.82 |
| Lipin 1 | 1.00 | 0.77 | 1.00 | 0.92 |
| Lipin 2 | 1.00 | 1.21 | 1.00 | 0.92 |
| Lipin 3 | 1.00 | 0.82 | 1.00 | 0.61 |
| Fatty acid oxidation | ||||
| PPARA | 1.00 | 1.00 | 1.00 | 1.16 |
| SREBP pathway | ||||
| Srebp-1a | 1.00 | 0.95 | 1.00 | 0.77 |
| Srebp-1c | 1.00 | 1.02 | 1.00 | 0.80 |
| Srebp-2 | 1.00 | 0.76 | 1.00 | 0.86 |
| SCAP | 1.00 | 0.97 | 1.00 | 1.15 |
| Insig-1 | 1.00 | 0.88 | 1.00 | 0.91 |
| Insig-2 | 1.00 | 1.05 | 1.00 | 1.15 |
| Insulin and IGF-1 signaling | ||||
| Insulin receptor | 1.00 | 0.86 | 1.00 | 0.93 |
| IRS-1 | 1.00 | 0.75 | 1.00 | 0.97 |
| IRS-2 | 1.00 | 0.62 | 1.00 | 1.22 |
| IGF-1 | 1.00 | 1.05 | 1.00 | 1.55 |
| IGF-1R | 1.00 | 0.98 | 1.00 | 1.15 |
| IGFbp1 | 1.00 | 0.95 | 1.00 | 0.61 |
| Glucose metabolism | ||||
| ChREBP | 1.00 | 0.76 | 1.00 | 0.94 |
| PEPCK | 1.00 | 0.80 | 1.00 | 0.94 |
| GK | 1.00 | 1.12 | 1.00 | 1.28 |
| Glc-6-P | 1.00 | 0.74 | 1.00 | 0.93 |
| PK | 1.00 | 0.63 | 1.00 | 0.93 |
| NADPH-producing | ||||
| Malic enzyme | 1.00 | 0.77 | 1.00 | 0.67 |
| G6PD | 1.00 | 1.08 | 1.00 | 0.82 |
| 6PGDH | 1.00 | 0.81 | 1.00 | 0.67 |
| Inflammation | ||||
| IL-6 | 1.00 | 1.69 | 1.00 | 1.03 |
| TNF-α | 1.00 | 1.76 | 1.00 | 1.63 |
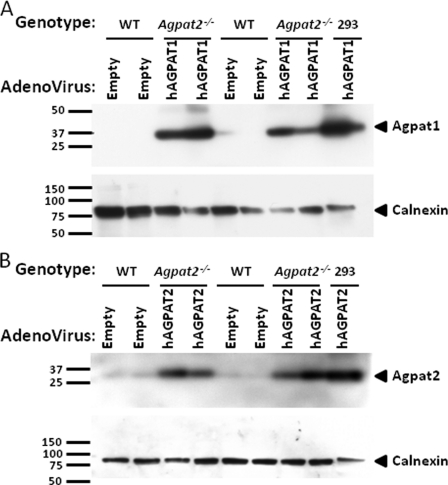
Cell fractionation studies revealed that the exogenously expressed AGPAT1 and AGPAT2 localize to ER, as shown in Fig. 10, A and B. Consistent with this, we observed a modest but statistically significant decrease in the total LPA levels in the livers of mice infected with AGPAT1 and AGPAT2 adenoviruses (Fig. 9, E and N) with concomitant increase in total PA levels (Fig. 9, F and O). This further indicates that the expressed enzymes are functionally active in the livers of Agpat2−/− mice.
FIGURE 10.
Western blot of human AGPAT1 and AGPAT2 in endoplasmic reticulum of mouse livers. A and B, Western blot for the recombinant AGPAT1 and AGPAT2 proteins from ER probed with antibody specific for AGPAT1 and AGPAT2 protein. Lysates from cells infected with recombinant AGPAT1 and AGPAT2 adenovirus expressed in 293 cells were loaded as a positive control. The same blot was stripped and reprobed with calnexin antibody to demonstrate ER specificity.
In some cases of hepatic steatosis, there is a link between expression of proinflammatory cytokine tumor necrosis factor-α (TNF-α) or interleukin-6 (IL-6) (48). We determined the expression of these cytokines in the Agpat2−/− and wild type mice infected with or without recombinant AGPAT1 or AGPAT2 adenovirus. No changes were observed between the genotypes (Table 2). But we did observe a 5–25-fold increase in plasma alanine transaminase and aspartate transaminase between naive mice compared with those that received the adenovirus (data not shown). This is a general response to viral infection.
DISCUSSION
This study revealed that the in vitro substrate specificities of human AGPAT1 and AGPAT2 are very similar but not identical for both LPA and acyl-CoA molecules. The preferred substrates for both the enzymes were C18:1-LPA and C18:1-CoA. Both enzymes will thus produce very similar molecular species of PA in vitro and most likely in vivo as well, suggesting similar function(s). However, we observed that AGPAT1 is also very efficient in utilizing an odd chain fatty acid, C15:0-CoA, as acyl donor. In certain physiological situations, this will produce very different molecular species of PA (1-C18:1–2-C15:0-sn-glycero-3-phosphatidic acid). Usually odd chain fatty acids are below detectable levels in most tissues, and thus their role in synthesis of PA remains unclear. Odd chain fatty acids are synthesized from propionic acid-CoA as the starting substrate instead of acetyl-CoA (49). It is possible that during heavy bacterial colonization of the colon, the level of propionyl-CoA may be elevated and thus increase the odd chain fatty acid and PA containing odd chain fatty acid species. The presence of such, one or more, molecular PA species and their functions in human and mice have yet to be elucidated.
We also determined the Km and Vmax values for some of the substrates. Both AGPAT1 and AGPAT2 isoforms have very similar apparent Km values for the LPA tested in this study. However, variation was seen in the usage of the acyl-CoAs. The rank order of acyl-CoA for AGPAT1 activity was C14:0 = C15:0 > C18:1 > C18:0. Although a similar rank order was observed for AGPAT2, the difference between C15:0 and C18:1 is almost 10- and 2.7-fold for AGPAT1 and AGPAT2, respectively. The reaction velocity did not vary significantly between the various substrates and the enzyme isoforms. Thus, both isoforms have very similar catalytic activity (Vmax/Km) when C18:1-LPA and C15:0-acyl-CoA are used followed by C18:1-LPA and C18:1-CoA. Because odd chain fatty acids are less abundant than even chain fatty acids, it appears that both isoforms will produce very similar PA.
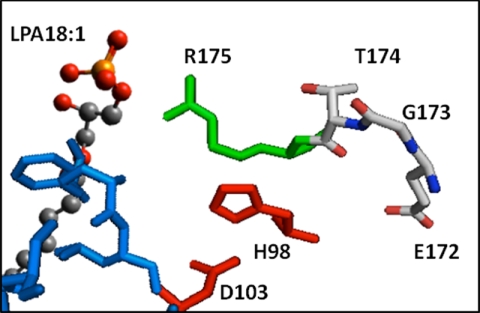
Our in silico modeling of AGPAT1 and AGPAT2 proteins is consistent with the in vitro substrate specificities. The model also sheds light on the role of the highly conserved motif EGTR found in almost all the glycerophospholipid acyltransferases (50). Two motifs are highly conserved among the glycerophospholipid acyltransferases superfamily as follows: an NHX4D in the amino terminus and an EGTR near the middle of the protein. Although the first motif is found to be conserved in all AGPATs (AGPAT1 to AGPAT11), the EGTR motif is not. In AGPAT8, a negatively charged Asp is substituted for positively charged Arg, EGTD (9). Likewise, in human AGPAT10/GPAT3 and AGPAT11, Arg is substituted with Cys, EGTC (13, 14). Both the isoforms still retain AGPAT activity. Thus, the EGTR motif may be conserved among glycerophospholipid acyltransferases, but the arginine is not essential for activity. Fig. 11 displays the relationship between the conserved NHX4D motif and the EGTR motif. The Glu, Gly, Thr, and Arg residues align themselves with the catalytic site His and Asp. By manually docking C18:1-LPA, Arg181 in AGPAT1 and Arg175 in AGPAT2 are most likely related to binding of the charged portions of LPA or CoA. Other conserved residues, Glu, Gly, and Thr are behind the catalytic residues and away from ligand docking. These residues may be involved in the plasticity of the active site during catalysis. Indeed, mutant human AGPAT1 proteins, where Glu is replaced with Asp or Gln, Gly with Leu, and Arg with Ala or Lys, were all enzymatically inactive when expressed in Sf9 cells (45). Only Thr when substituted with Ser retained 50% of the enzymatic activity compared with wild type protein (45). Further site-directed mutagenesis of amino acids identified in the hydrophobic tunnel, which binds the fatty acids, will reveal the role of each of these residues in the substrate specificity of AGPAT1 and AGPAT2 proteins. Similar observations were made with AGPAT1 protein (data not shown).
FIGURE 11.
Folding of the two highly conserved motifs found in glycerophospholipid acyltransferases. This picture of the model of AGPAT2 showing the relationship between a conserved stretch of amino acids (Glu172, Gly173, Thr174, and Arg175) in AGPAT2 that have been proposed to be related to ligand binding. Red is the catalytic site H(X)4D; green is related to the cluster that binds the charged portion of the ligands, and the blue surface is the hydrophobic tunnel that accommodates the acyl chain. By manually docking C18:1-LPA, we observed that Arg175 in AGPAT2 is likely related to binding of the charged portions of LPA. The other conserved amino acids (Glu172, Gly173, and Thr174) in this region are behind the catalytic residues away from ligand docking. The highly conserved Gly173 in AGPAT2 can be involved in the plasticity of the active site during catalysis.
The most intriguing aspect of this study was the fact that acute infection of Agpat2−/− mice with the recombinant adenovirus expressing wild type human AGPAT1 or AGPAT2 protein did not alleviate hepatic steatosis seen in these mice. It is unclear if this is due to the fact that infection was of an acute nature (7 days) versus the chronic hepatic steatosis (20–25 weeks) in the mice or whether AGPAT2 deficiency has no specific role in increasing liver fat. It may very well be that the role of AGPAT2 in the liver is to provide most of the GPL required for the cells to grow and generate cellular membranes or for those required to form the outer monolayer of the lipid droplet formed in liver. Only when these cellular demands are met is the excess PA utilized to synthesize TAG. It should also be mentioned that these mice have an increased rate of fatty acid synthesis (29), which is most likely a consequence of their hyperinsulinemia but is independent of Srebp-1c activation in Agpat2−/− mice (29). Hepatic steatosis in these mice seems not to be due to the activation of proinflammatory response but a simple case of steatosis. Therefore, at this age, the Agpat2−/− mice have not progressed to develop steatohepatitis.
Two other murine models of lipodystrophy have been studied, the A-ZIP/F-1 mouse (51), which selectively expresses a dominant negative protein that heterodimerizes with and inactivates members of the C/EBP and JUN families of b-ZIP transcription factors, and a transgenic aP2-Srebp-1c mouse (52–54). Both mouse models have varying degrees of fat loss. In A-ZIP/F-1 mice, at least a 2-fold increase in serum free fatty acid was observed compared with Agpat2−/− mice, which show no increase in serum free fatty acid in males or a slight decrease in females. Thus, the lack of adipose tissue diverts the free fatty acid and, in conjunction with de novo lipogenesis in the livers of these mice, leads to increased deposition of TAG in the livers of these lipodystrophy mouse models. In fact, this feature of hepatic steatosis is reversible in A-ZIP/F-1 mice upon transplantation of wild type adipose tissue (55). It is to be noted that in these mouse models of lipodystrophy the expression of AGPAT2 remains intact. Because our experiments were performed in an acute setting, i.e. expression of recombinant protein for 1 week, it is possible that the acute expression of AGPAT2 in the liver is not sufficient for the reduction of TAG in the liver. Thus, the role of AGPAT2 in liver lipogenesis may become evident from the studies by generating transgenic mice overexpressing liver-specific AGPAT2 and crossing with the Agpat2−/− mice. Alternatively, the hepatic steatosis develops secondarily as a result of the hyperinsulinemia. Determining which of these mechanisms is responsible for TAG accumulation in livers of Agpat2−/− mice will be the focus of future studies.
Acknowledgments
We thank Akhil Narang (Doris Duke fellow), Diana Varghese, and Ruth Giselle Huet for technical assistance; Kate Luby-Phelps and Abhijit Bugde for their help in cellular imaging; Beverley Adams-Huet for statistical analysis, and Dr. Richard Auchus for suggestions during the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-DK54387, DK081182, and UL1DE019584. This work was also supported by the Southwestern Medical Foundation.
- AGPAT
- 1-acylglycerol-3-phosphate O-acyltransferase
- GPL
- glycerophospholipid
- TAG
- triacylglycerol
- LPA
- lysophosphatidic acid
- PA
- phosphatidic acid
- GPAT
- glycerol-3-phosphate acyltransferase
- ER
- endoplasmic reticulum
- AT
- adipose tissue.
REFERENCES
- 1. McMaster C. R., Jackson T. R. (2004) in Lipid Metabolism and Membrane Biogenesis (Daum G. ed) pp. 5–30, Springer-Verlag, Berlin [Google Scholar]
- 2. Coleman R. A., Lee D. P. (2004) Prog. Lipid Res. 43, 134–176 [DOI] [PubMed] [Google Scholar]
- 3. Leung D. W. (2001) Front. Biosci. 6, D944–D953 [DOI] [PubMed] [Google Scholar]
- 4. Agarwal A. K., Garg A. (2003) Trends Endocrinol. Metab. 14, 214–221 [DOI] [PubMed] [Google Scholar]
- 5. Takeuchi K., Reue K. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E1195–E1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ye G. M., Chen C., Huang S., Han D. D., Guo J. H., Wan B., Yu L. (2005) DNA Seq. 16, 386–390 [DOI] [PubMed] [Google Scholar]
- 7. Vergnes L., Beigneux A. P., Davis R., Watkins S. M., Young S. G., Reue K. (2006) J. Lipid Res. 47, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beigneux A. P., Vergnes L., Qiao X., Quatela S., Davis R., Watkins S. M., Coleman R. A., Walzem R. L., Philips M., Reue K., Young S. G. (2006) J. Lipid Res. 47, 734–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Agarwal A. K., Barnes R. I., Garg A. (2006) Arch. Biochem. Biophys. 449, 64–76 [DOI] [PubMed] [Google Scholar]
- 10. Agarwal A. K., Sukumaran S., Bartz R., Barnes R. I., Garg A. (2007) J. Endocrinol. 193, 445–457 [DOI] [PubMed] [Google Scholar]
- 11. Nagle C. A., Vergnes L., Dejong H., Wang S., Lewin T. M., Reue K., Coleman R. A. (2008) J. Lipid Res. 49, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y. Q., Kuo M. S., Li S., Bui H. H., Peake D. A., Sanders P. E., Thibodeaux S. J., Chu S., Qian Y. W., Zhao Y., Bredt D. S., Moller D. E., Konrad R. J., Beigneux A. P., Young S. G., Cao G. (2008) J. Biol. Chem. 283, 10048–10057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sukumaran S., Barnes R. I., Garg A., Agarwal A. K. (2009) J. Mol. Endocrinol. 42, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agarwal A. K., Garg A. (2010) J. Lipid Res. 51, 2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao J., Liu Y., Lockwood J., Burn P., Shi Y. (2004) J. Biol. Chem. 279, 31727–31734 [DOI] [PubMed] [Google Scholar]
- 16. Chen X., Hyatt B. A., Mucenski M. L., Mason R. J., Shannon J. M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11724–11729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 18. Cao J., Li J. L., Li D., Tobin J. F., Gimeno R. E. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 19695–19700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. West J., Tompkins C. K., Balantac N., Nudelman E., Meengs B., White T., Bursten S., Coleman J., Kumar A., Singer J. W., Leung D. W. (1997) DNA Cell Biol. 16, 691–701 [DOI] [PubMed] [Google Scholar]
- 20. Aguado B., Campbell R. D. (1998) J. Biol. Chem. 273, 4096–4105 [DOI] [PubMed] [Google Scholar]
- 21. Eberhardt C., Gray P. W., Tjoelker L. W. (1997) J. Biol. Chem. 272, 20299–20305 [DOI] [PubMed] [Google Scholar]
- 22. Eberhardt C., Gray P. W., Tjoelker L. W. (1999) Adv. Exp. Med. Biol. 469, 351–356 [DOI] [PubMed] [Google Scholar]
- 23. Eberhardt C., Gray P. W., Tjoelker L. W. (1999) in Eicosanoids and Other Bioactive Lipids in Cancer, Inflammation and Radiation Injury, 4 (Honn K. V., Marnett L. J., Nigam S., Dennis E. A. eds) pp. 351–356, Klumer Academic/Plenum Publishers, NY [Google Scholar]
- 24. Agarwal A. K., Arioglu E., De Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., Garg A. (2002) Nat. Genet. 31, 21–23 [DOI] [PubMed] [Google Scholar]
- 25. Garg A. (2004) N. Engl. J. Med. 350, 1220–1234 [DOI] [PubMed] [Google Scholar]
- 26. Agarwal A. K., Garg A. (2006) Annu. Rev. Med. 57, 297–311 [DOI] [PubMed] [Google Scholar]
- 27. Agarwal A. K., Garg A. (2006) Annu. Rev. Genomics Hum. Genet. 7, 175–199 [DOI] [PubMed] [Google Scholar]
- 28. Gale S. E., Frolov A., Han X., Bickel P. E., Cao L., Bowcock A., Schaffer J. E., Ory D. S. (2006) J. Biol. Chem. 281, 11082–11089 [DOI] [PubMed] [Google Scholar]
- 29. Cortés V. A., Curtis D. E., Sukumaran S., Shao X., Parameswara V., Rashid S., Smith A. R., Ren J., Esser V., Hammer R. E., Agarwal A. K., Horton J. D., Garg A. (2009) Cell Metab. 9, 165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Agarwal A. K., Simha V., Oral E. A., Moran S. A., Gorden P., O'Rahilly S., Zaidi Z., Gurakan F., Arslanian S. A., Klar A., Ricker A., White N. H., Bindl L., Herbst K., Kennel K., Patel S. B., Al-Gazali L., Garg A. (2003) J. Clin. Endocrinol. Metab. 88, 4840–4847 [DOI] [PubMed] [Google Scholar]
- 31. Prasad S. S., Garg A., Agarwal A. K. (2011) J. Lipid Res. 52, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Green M., Wold W. S. (1979) Methods Enzymol. 58, 425–435 [DOI] [PubMed] [Google Scholar]
- 33. Haque W., Garg A., Agarwal A. K. (2005) Biochem. Biophys. Res. Commun. 327, 446–453 [DOI] [PubMed] [Google Scholar]
- 34. Jones D. T. (1999) J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 35. Nugent T., Jones D. T. (2009) BMC Bioinformatics 10, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lobley A., Sadowski M. I., Jones D. T. (2009) Bioinformatics 25, 1761–1767 [DOI] [PubMed] [Google Scholar]
- 37. Tamada T., Feese M. D., Ferri S. R., Kato Y., Yajima R., Toguri T., Kuroki R. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 13–21 [DOI] [PubMed] [Google Scholar]
- 38. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ponder J. W., Case D. A. (2003) Adv. Protein Chem. 66, 27–85 [DOI] [PubMed] [Google Scholar]
- 40. Park S. W., Moon Y. A., Horton J. D. (2004) J. Biol. Chem. 279, 50630–50638 [DOI] [PubMed] [Google Scholar]
- 41. Costes S. V., Daelemans D., Cho E. H., Dobbin Z., Pavlakis G., Lockett S. (2004) Biophys. J. 86, 3993–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S., Hodge C. L., Haase J., Janes J., Huss J. W., 3rd, Su A. I. (2009) Genome Biol. 10, R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Claros M. G., Vincens P. (1996) Eur. J. Biochem. 241, 779–786 [DOI] [PubMed] [Google Scholar]
- 44. Hollenback D., Bonham L., Law L., Rossnagle E., Romero L., Carew H., Tompkins C. K., Leung D. W., Singer J. W., White T. (2006) J. Lipid Res. 47, 593–604 [DOI] [PubMed] [Google Scholar]
- 45. Yamashita A., Nakanishi H., Suzuki H., Kamata R., Tanaka K., Waku K., Sugiura T. (2007) Biochim. Biophys. Acta 1771, 1202–1215 [DOI] [PubMed] [Google Scholar]
- 46. Turnbull A. P., Rafferty J. B., Sedelnikova S. E., Slabas A. R., Schierer T. P., Kroon J. T., Nishida I., Murata N., Simon J. W., Rice D. W. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 451–453 [DOI] [PubMed] [Google Scholar]
- 47. Nishimasu H., Okudaira S., Hama K., Mihara E., Dohmae N., Inoue A., Ishitani R., Takagi J., Aoki J., Nureki O. (2011) Nat. Struct. Mol. Biol. 18, 205–212 [DOI] [PubMed] [Google Scholar]
- 48. Copaci I., Micu L., Voiculescu M. (2006) J. Gastrointestin. Liver Dis. 15, 363–373 [PubMed] [Google Scholar]
- 49. Arora T., Sharma R., Frost G. (2011) Appetite 56, 511–515 [DOI] [PubMed] [Google Scholar]
- 50. Lewin T. M., Wang P., Coleman R. A. (1999) Biochemistry 38, 5764–5771 [DOI] [PubMed] [Google Scholar]
- 51. Moitra J., Mason M. M., Olive M., Krylov D., Gavrilova O., Marcus-Samuels B., Feigenbaum L., Lee E., Aoyama T., Eckhaus M., Reitman M. L., Vinson C. (1998) Genes Dev. 12, 3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shimomura I., Hammer R. E., Richardson J. A., Ikemoto S., Bashmakov Y., Goldstein J. L., Brown M. S. (1998) Genes Dev. 12, 3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shimomura I., Bashmakov Y., Horton J. D. (1999) J. Biol. Chem. 274, 30028–30032 [DOI] [PubMed] [Google Scholar]
- 54. Shimomura I., Hammer R. E., Ikemoto S., Brown M. S., Goldstein J. L. (1999) Nature 401, 73–76 [DOI] [PubMed] [Google Scholar]
- 55. Kim J. K., Gavrilova O., Chen Y., Reitman M. L., Shulman G. I. (2000) J. Biol. Chem. 275, 8456–8460 [DOI] [PubMed] [Google Scholar]
- 56. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]