Background: The molecular basis of GABAA receptor α3 subtype-specific synaptic localization is unknown.
Results: GABAAR α3 interacts with the gephyrin E domain via defined intracellular motifs that partially overlap with glycine receptor binding determinants.
Conclusion: GABAAR subtypes containing α3 are clustered at postsynaptic specializations via direct interactions with gephyrin.
Significance: Distinct binding properties of GABAAR and GlyRs to gephyrin may govern mixed glycinergic/GABAergic transmission.
Keywords: GABA Receptors, Glycine Receptors, Isothermal Titration Calorimetry, Mutagenesis, Synapses, Gephyrin, Synaptic Clustering
Abstract
The multifunctional scaffolding protein gephyrin is a key player in the formation of the postsynaptic scaffold at inhibitory synapses, clustering both inhibitory glycine receptors (GlyRs) and selected GABAA receptor (GABAAR) subtypes. We report a direct interaction between the GABAAR α3 subunit and gephyrin, mapping reciprocal binding sites using mutagenesis, overlay, and yeast two-hybrid assays. This analysis reveals that critical determinants of this interaction are located in the motif FNIVGTTYPI in the GABAAR α3 M3–M4 domain and the motif SMDKAFITVL at the N terminus of the gephyrin E domain. GABAAR α3 gephyrin binding-site mutants were unable to co-localize with endogenous gephyrin in transfected hippocampal neurons, despite being able to traffic to the cell membrane and form functional benzodiazepine-responsive GABAARs in recombinant systems. Interestingly, motifs responsible for interactions with GABAAR α2, GABAAR α3, and collybistin on gephyrin overlap. Curiously, two key residues (Asp-327 and Phe-330) in the GABAAR α2 and α3 binding sites on gephyrin also contribute to GlyR β subunit-E domain interactions. However, isothermal titration calorimetry reveals a 27-fold difference in the interaction strength between GABAAR α3 and GlyR β subunits with gephyrin with dissociation constants of 5.3 μm and 0.2 μm, respectively. Taken together, these observations suggest that clustering of GABAAR α2, α3, and GlyRs by gephyrin is mediated by distinct mechanisms at mixed glycinergic/GABAergic synapses.
Introduction
Inhibitory transmission in the central nervous system is mediated by GABAA and glycine receptors (GlyRs)4 consisting of pentameric combinations of subunits (α1–6, β1–3, γ1–3, δ, ϵ, θ, and π for GABAARs and α1–4 and β for GlyRs), each comprising an extracellular domain, four membrane-spanning helices (M1–M4) and a large intracellular loop between M3 and M4. Cell type-specific gene expression patterns, subunit stoichiometry, and interplay between presynaptic and postsynaptic specializations are thought to underlie the spatial and temporal localization of these receptors. In particular, a postsynaptic matrix of receptor-associated proteins is essential for the dynamic localization of both GABAARs and GlyRs and also recruits components of specific signaling cascades to synapses (1). The multifunctional protein gephyrin (2) is a key player in the clustering of both GlyRs and GABAARs. For example, studies using gephyrin knock-out mice or mRNA knockdown (3–8) have shown a loss of postsynaptic clustering of GlyRs as well as GABAARs containing α2 and γ2 subunits. However, GABAAR α1 and α5 subunit clustering is unaltered in gephyrin knock-out mice (4–7), demonstrating that gephyrin-independent clustering mechanisms also exist in vivo. GABAARs containing the α4, α5, and α6 subunits are located preferentially at extrasynaptic sites (9) and are likely to be localized by other clustering factors, such as the actin-binding protein radixin (10). Hence, although the majority of GlyRs are likely to be clustered by gephyrin, only certain GABAAR subtypes are subject to gephyrin-dependent clustering. Curiously, the subcellular localization of gephyrin is in turn dependent on the presence of certain GABAAR subtypes. For example, targeted deletion of the GABAAR α1, α3, and γ2 subunit genes results in a loss of synaptic gephyrin clusters (11–16), resulting in nonsynaptic dendritic gephyrin aggregates.
The interaction of the GlyR β subunit with the gephyrin E domain has been characterized in detail (17–21). By contrast, a direct interaction of gephyrin with GABAAR subunits has proven elusive, perhaps because of the number of individual GABAAR subunits, splice variants, accessory proteins, and post-translational modifications that could influence these interactions. However, recent studies (22, 23) have demonstrated that the GABAAR α2 subunit interacts directly with both gephyrin and the RhoGEF collybistin (18, 24). Here, we report a detailed characterization of the interaction between the GABAAR α3 subunit and gephyrin in recombinant systems and neuronal cultures, revealing overlapping binding determinants on gephyrin for GABAAR α2, GABAAR α3, and collybistin that are distinct from the E domain GlyR β-subunit binding site.
EXPERIMENTAL PROCEDURES
Expression Constructs and Site-directed Mutagenesis
GABAAR α1, α3, GlyR β subunit, and gephyrin cDNAs were amplified from rat spinal cord or whole brain first-strand cDNA using Pfx DNA polymerase (Invitrogen) and cloned into the yeast two-hybrid vectors pYTH16 or pACT2. Cloning resulted in an in-frame fusion of the GAL4 DNA binding domain (GAL4BD; vector pYTH16) (25) or GAL4 activation domain (GALAD; vector pACT2) to the N termini of all expressed proteins. Mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene), and all constructs were verified by Sanger DNA sequencing. A hemagglutinin (HA) tag (YPYDVPDYA) was inserted between amino acids 32 and 33 of the rat GABAAR α3 subunit (Uniprot P20236; NCBI Entrez Gene ID 24947) using the GeneSOEing (Gene Splicing by Overlap Extension) technique. Because the α3 signal peptide comprises amino acids 1–28, the HA tag is located between amino acids 4 and 5 of the mature polypeptide. There are two published versions of the rat GABAAR subunit α3 intracellular loop in the protein data base Uniprot with regard to amino acid 381, which is either a leucine (nucleotides TTG) or lysine (nucleotides AAG). Because this amino acid is near the critical region for α3 subunit-gephyrin interactions, we compared both constructs in our experiments. Deletions were made in α3L381 using the GeneSOEing technique. GST fusion proteins were constructed by cloning the intracellular loops into an engineered pGEX vector, which provided a C-terminal His6 tag for purification of the fusion protein.
Yeast Two-hybrid Assays
The yeast strain Y190 was co-transformed with pYTH16-GABAAR α1 or α3 or GlyR β subunit intracellular M3–M4 loop bait plasmids together with pACT2-gephyrin prey constructs. pACT2-gephyrin deletions and alanine block mutants were described previously (18, 23). Additional pYTH16-GABAAR α3 deletion mutants and the pYTH16-GABAAR α3inα1 chimera were generated during this study. Transformations were plated on selective dropout media (either −LeuTrpHis +30 mm 3-AT or -LeuTrp). After incubation at 30 °C for 3–6 days, LacZ reporter gene assays were performed as described (18).
Culture and Transfection of Primary Hippocampal Neurons
Hippocampal cultures were made from E18 rats from Charles River as described previously (26). Transfections were made using the Amaxa System with GABAAR α3 and mutant expression constructs in the vector pCI (Promega). Transfected neurons were plated onto poly-l-lysine-coated glass coverslips and maintained in Neurobasal/B27 medium for 18 days. Plasmid DNA used for transfection was prepared with the Endofree maxi kit (Qiagen).
Transfection of HEK293 Cells
HEK293 cells (ATCC CRL-1573) were co-transfected with pCI expression constructs encoding HA-tagged GABAAR α3 and deletion mutants together with the GABAAR β3 subunit. Cells were initially plated on poly-l-lysine-coated glass coverslips in DMEM containing 10% FCS. For transfection we used the TurboFect transfection reagent (Fermentas) and 0.5 μg of pCI GABAAR β3 DNA together with 0.5 μg of pCI GABAAR α3 DNAs (deletions 1–4) according to the manufacturer's protocol using serum-free medium. Three hours after transfection, the culture medium was changed to one also containing 10% FCS. Cells were fixed and stained 48 h after lipofection.
Antibodies
Primary antibodies were anti-HA rabbit polyclonal antibody (dilution 1:100; Santa Cruz Biotechnology), anti-gephyrin mouse monoclonal antibody (dilution 1:100, reconstituted as recommended; Synaptic Systems), and rabbit affinity-purified anti-GABAA receptor α3 antibody (27) (tested for specificity in α3 knock-out brains, at 1 μg/ml). Secondary antibodies were FITC (fluorescein) goat anti-rabbit IgG (H+L) with minimal cross-reactivity (Jackson ImmunoResearch) and Cy3 goat anti-mouse IgG (H+L) with minimal cross-reactivity (Jackson ImmunoResearch).
Immunocytochemistry
Neurons grown on glass coverslips were fixed with 4% paraformaldehyde and 4% sucrose in PBS. HEK293 cells were fixed with 4% paraformaldehyde without sucrose. Nonspecific binding was blocked by incubation with 5% BSA in PBS. Primary and secondary antibodies were diluted in 1% BSA/PBS. Secondary antibodies included FITC and Cy3 anti-rabbit and anti-mouse IgGs. For detecting intracellular epitopes, cells were permeabilized with 0.05% Triton X-100 for 5 min prior to blocking with BSA. After incubation with secondary antibodies, coverslips were washed with PBS and finally with water and mounted using Mowiol mounting solution (Polysciences Inc., Warrington, PA).
Overlay Assays
GST fusion proteins were purified from Escherichia coli BL21 extracts under denaturing conditions on a nickel-nitrilotriacetic acid-Sepharose column as described (Qiagen Expressionist Manual). The protein concentration of purified proteins was quantified using the BCA protein assay (Pierce). Approximately 5 μg of each fusion protein was separated on two 10% polyacrylamide gels (NuPAGE, Invitrogen) using MOPS SDS running buffer (50 mm MOPS, 50 mm Tris-HCl, 0.1% SDS, 1 mm EDTA, pH 7.7). One gel was stained with Coomassie Blue, the other gel was blotted onto a PVDF membrane (Immobilon, Millipore). The membrane was incubated in 7 m guanidinium chloride in renaturation buffer (10 mm HEPES, pH 7.0, 70 mm KCl, 80 mm NaCl, 5 mm EDTA, 1 mm β-mercaptoethanol) for 1 h at 4 °C. This solution was diluted every hour with renaturation buffer to a final concentration of 6, 5, 4, 3, 2, and 1 m guanidinium hydrochloride. Finally the membrane was incubated in renaturation buffer without guanidinium hydrochloride for 1 h, blocked with 5% BSA and 0.03% Triton X-100 in renaturation buffer for 1 h and another 1 h in 1% BSA in detergent-free renaturation buffer. In vitro translation and [35S]methionine labeling of gephyrin was performed using the TNT®T7 Quick Coupled Transcription/Translation kit (Promega) and [35S]methionine (1000 Ci/mmol at 10 mCi/ml). 0.5 μg of the pRK5 gephyrin P1 plasmid was mixed with 2 μl of [35S]methionine, 6 μl of nuclease-free ddH2O, and 40 μl of TNT T7 Master Mix and incubated for 90 min at 30 °C. The blot was incubated overnight at 4 °C in 20 ml of 1% BSA in detergent-free renaturation buffer with 50 μl of in vitro-translated 35S-labeled gephyrin. Subsequently, the blot was washed with 1% BSA in detergent-free renaturation buffer for 1 h and air-dried. The dry membrane was exposed to a PhosphorImager screen for 4 h and analyzed using the Quantity One software. To determine relative gephyrin binding affinities to different GST fusion proteins, the Coomassie-stained gel was analyzed by densitometry, and variations of intensities were used as correction factors when quantifying the overlay assays. The assays were repeated three times under identical conditions. For statistical evaluation, the intensities of bands were compared with the wild-type construct using an unpaired Student's t test with a confidence interval of p < 0.01.
Electrophysiology
Electrophysiological properties of α3β3γ2 GABAARs expressed in Xenopus oocytes were measured using the two-electrode voltage clamp technique. Methods for isolating, culturing, injecting, and defolliculating of oocytes were as described previously (28). In brief, mature female Xenopus laevis (Nasco, Fort Atkinson, WI) were anesthetized in a bath of ice-cold 0.17% Tricain (ethyl-m-aminobenzoate; Sigma) before decapitation and removal of the ovaries. Stage 5–6 oocytes with the follicle cell layer intact were removed from the ovary using a platinum wire loop. Oocytes were stored and incubated at 18 °C in modified Barth's medium (88 mm NaCl, 10 mm HEPES-NaOH, pH 7.4, 2.4 mm NaHCO3, 1 mm KCl, 0.82 mm MgSO4, 0.41 mm CaCl2, 0.34 mm Ca(NO3)2) that was supplemented with 100 units/ml penicillin and 100 μg/ml streptomycin. Oocytes with an intact follicular cell layer were subjected to nuclear injection with a total of 3 ng of cDNA in aqueous solution per oocyte. The subunit ratio was 1:1:5 for α3β3γ2 receptors consisting of wild-type or mutant α3 subunits together with β3 and γ2 subunits. After injection of cDNAs, oocytes were incubated for at least 24 h before the enveloping follicle cell layers were removed. Collagenase treatment (type IA; Sigma), and mechanically defolliculating of the oocytes was performed as described previously (29).
For electrophysiological recordings, oocytes were placed on a nylon grid in a bath of Xenopus Ringer solution (XR; containing 90 mm NaCl, 5 mm HEPES-NaOH, pH 7.4, 1 mm MgCl2, 1 mm KCl, and 1 mm CaCl2). The oocytes were constantly washed by a flow of 6 ml/min XR, which could be switched to XR containing GABA and/or diazepam. Diazepam was diluted into XR from dimethyl sulfoxide solutions resulting in a final concentration of 0.1% dimethyl sulfoxide perfusing the oocytes. Diazepam was preapplied for 30 s before the addition of GABA, which was then co-applied with the diazepam until a peak response was observed. Between two applications, oocytes were washed in XR for up to 15 min to ensure full recovery from desensitization. For current measurements, the oocytes were impaled with two microelectrodes (1–3 megaohms) which were filled with 2 m KCl. Maximum currents measured in cDNA-injected oocytes were in the microampere range for all GABAA receptor subtypes. To test for modulation of GABA-induced currents by diazepam, a concentration of 3 μm GABA was co-applied to the cell with 1 μm diazepam. All recordings were performed at room temperature at a holding potential of −60 mV using a Dagan CA-1B Oocyte Clamp or a Dagan TEV-200A two-electrode voltage clamp (Dagan Corporation, Minneapolis, MN). Data were digitized, recorded, and measured using a Digidata 1322A data acquisition system (Axon Instruments, Union City, CA) and analyzed using GraphPad Prism. Data for GABA-dependent dose-response curve were fitted to the equation Y = Bottom + (Top − Bottom)/(1 + 10(LogEC50−X) * nH), where EC50 is the concentration of the compound that increases the amplitude of the GABA-evoked current by 50%, and nH is the Hill coefficient. Data are given as mean ± S.E. from at least three oocytes and two oocyte batches. Statistical significance was calculated using one-way ANOVA with Bonferroni post hoc test.
Isothermal Titration Calorimetry (ITC) and Native PAGE
Partial GABAAR α3 subunit variants were PCR-amplified and cloned into the NcoI/NotI sites of pETM11. M3–M4 fusion proteins were expressed in the E. coli strain BL21 (Stratagene) as His6-tagged proteins. Cells were grown in LB medium at 30 °C, and protein expression was induced following addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 18 h. Cells were then resuspended in lysis buffer (50 mm Tris-HCl, 500 mm NaCl, pH 8.0), passed through a French pressure cell, and centrifuged (4000 × g). Proteins were initially purified using a 5-ml HisTrap FF crude column according to the instructions of the manufacturer (GE Healthcare). Protein-containing fractions were collected, concentrated, and applied to a 26/60 Superdex 200 size exclusion column (Amersham Biosciences) equilibrated with buffer (10 mm Tris-HCl, 250 mm NaCl, pH 8.0). Pure fractions were pooled, concentrated to 5–100 mg/ml, flash-frozen in 0.5-ml aliquots, and stored at −80 °C. The gephyrin E domain was prepared as described previously (20, 21). Prior to all ITC experiments, gephyrin and GABAAR α3 M3–M4 loop variants were extensively dialyzed against 10 mm Tris-HCl, 250 mm NaCl, 1 mm β-mercaptoethanol, pH 8.0, at 4 °C overnight, followed by filtration and degassing. A 200 μm solution of the GABAAR α3 variants was titrated as the ligand into the sample cell containing 9 μm gephyrin E domain. A volume of 10–15 μl of ligand was added at a time with a total number of 20–30 injections, resulting in a final molar ratio of ligand to protein of 4:1. All experiments were performed using a VP-ITC instrument (MicroCal, Northampton, MA) at 25 °C. Buffer-to-buffer titrations were performed as described above, so that the heat produced by injection, mixing, and dilution could be subtracted prior to curve fitting. The binding enthalpy was measured directly, whereas affinity (KD) and stoichiometry (N) were obtained by data analysis using the ORIGIN software. Native PAGE gels containing 1% agarose and Tris/glycine, pH 8.4, were used to separate the E domain of gephyrin from the E domain-GABAAR α3 complex. 10 μl of the E domain (5 μm) mixed with the respective ligand (80 μm) was loaded in each lane.
RESULTS
Mapping Determinants of GABAAR α3 Subunit Binding to Gephyrin Using Overlay and Yeast Two-hybrid Assays
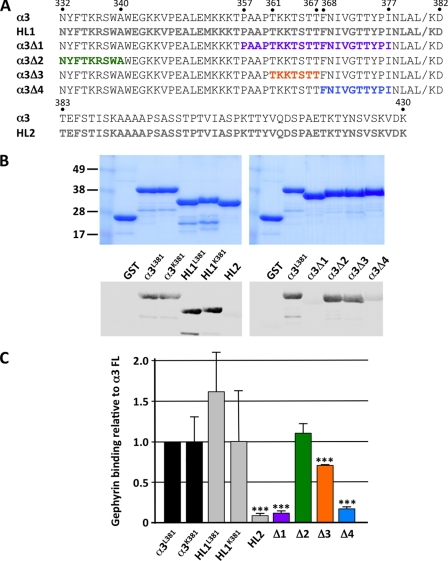
To assess the possible interaction of the GABAAR α3 subunit with gephyrin we performed overlay assays using GST-tagged GABAAR α3 fusion proteins and recombinant 35S-labeled gephyrin, using the abundant P1 isoform that lacks the C3 and C4 cassettes (1). Because the most likely binding site is located in the large intracellular domain between membrane-spanning domains 3 and 4, we split the 99-amino acid domain (constructs HL1 and HL2) and engineered several deletion constructs (Δ1–Δ4; Fig. 1A). The HL1 construct was generated in two versions, due to an apparent polymorphism at amino acid 381 (Leu-381 versus Lys-381). Quantitative analysis (Fig. 1, B and C) revealed that the intact GST-GABAAR α3 M3–M4 domain fusion protein (but not GST alone) showed a robust interaction with recombinant gephyrin (Fig. 1, B and C). This interaction was retained by both versions of the N-terminal portion of the M3–M4 domain (HL1L381 and HL1K381; amino acids Asn-332–Asp-382), but binding of the C-terminal HL2 fragment was negligible (amino acids Thr-383–Lys-430). This suggested that the polymorphism at amino acid 381 does not influence GABAAR α3-gephyrin interactions and, more importantly, that a gephyrin-binding motif is likely to reside in the N-terminal 51 amino acids of the M3–M4 domain. Consistent with this hypothesis, further analysis revealed that α3-gephyrin interactions were most severely reduced by the overlapping deletions α3Δ1 (Pro-357–Ile-377) and α3Δ4 (Phe-368–Ile-377) that lie within the N-terminal HL1 fragment, although deletion Δ3 (Thr-361–Thr-367) also appeared to reduce α3-gephyrin interactions, albeit to a lesser extent than Δ1 and Δ4.
FIGURE 1.
Deletion mapping of gephyrin-interacting residues in the GABAAR α3 subunit intracellular M3–M4 loop. A, sequence of the GABAAR α3 subunit M3–M4 region, showing the position of fusions proteins HL1, HL2 (gray lettering) and deletion mutants α3Δ1 (Pro-357–Ile-377), α3Δ2 (Asn-332–Ala-340), α3Δ3 (Thr-361–Thr-367), α3Δ4 (Phe-368–Ile-377). Residues indicated in color in α3Δ1–4 are missing from these constructs. B, overlay assays using GST-tagged GABAAR α3 fusion proteins and recombinant 35S-labeled gephyrin reveal critical determinants of gephyrin binding. Upper, Coomassie-stained proteins demonstrating equivalent loading of GST fusion proteins. Lower left, overlay assays with full-length GABAAR α3L381 and α3K381 M3–M4 intracellular loop variants and N-terminal fusion proteins HL1L381 and HL1K381 (Asn-332–Asp-382) showed robust α3-gephyrin interactions, but the C-terminal HL2 fragment (Thr-383–Lys-430) showed no detectable gephyrin binding. Lower right, gephyrin interacting with the α3L381 and deletions α3Δ2 and α3Δ3. Interactions were most severely reduced by the overlapping deletions α3Δ1 (Pro-357–Ile-377) and α3Δ4 (Phe-368–Ile-377). C, quantitation of overlay assays revealing significantly reduced GABAAR α3-gephyrin interactions for HL2, α3Δ1, α3Δ3, and α3Δ4. Data represent mean ± S.E. (error bars; n = 3). Significant differences from control values were assessed using an unpaired Student's t test (***, p < 0.001).
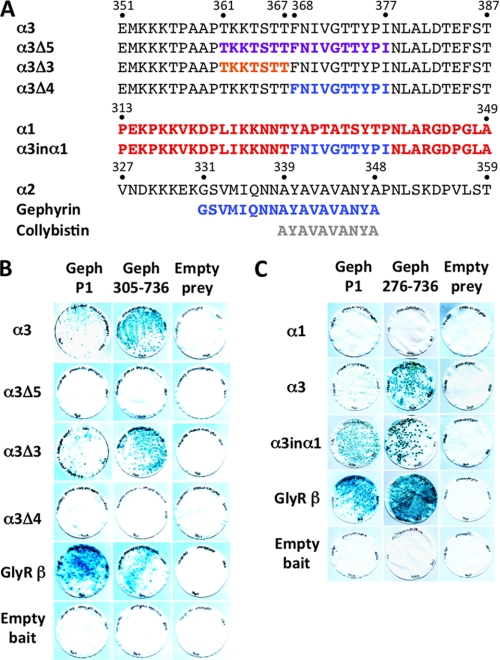
As a second confirmatory assay, we utilized the yeast two-hybrid system to test interactions between the M3–M4 intracellular loops of the GABAAR α1 and α3 subunits and GlyR β (control) with full-length P1 gephyrin and α3 deletion mutants. Although all bait proteins are expressed in yeast (23) the GABAAR α3 and GlyR β baits, but not the GABAAR α1 bait, interact with full-length gephyrin and two N-terminal deletion mutants (Geph276–736 and Geph305–736), as assessed by LacZ freeze-fracture assays (Fig. 2). Using deletion and domain swap mutations (Fig. 2), we found that consistent with overlay assays, deletion of the Thr-361–Ile-377 (α3Δ5) and Phe-368–Ile-377 (α3Δ4) abolished the interaction of the GABAAR α3 subunit with gephyrin (Fig. 2, A and B). By contrast, deletion of Thr-361–Thr-367 (α3Δ3) revealed that this motif was dispensable for GABAAR α3-gephyrin interactions in yeast. Because both overlay and yeast two-hybrid assays suggest that the motif FNIVGTTYPI (Phe-368–Ile-377) contains key determinants of GABAAR α3-gephyrin interactions, we inserted Phe-368–Ile-377 from α3 into a wild-type GABAAR α1 subunit bait, which does not normally interact with gephyrin (23) in yeast (Fig. 2C). This bait (named α3inα1) was able to interact with gephyrin (Fig. 2C), confirming that we have identified key residues involved in the GABAAR α3-gephyrin interaction.
FIGURE 2.
Fine mapping of determinants of GABAAR α3-gephyrin interactions. A, alignment of part of the GABAAR α1, α2, α3 subunit M3–M4 intracellular loop showing the position of artificial GABAAR α3 subunit deletions and the α3inα1 chimera. Binding sites on GABAAR α2 for collybistin and gephyrin are indicated. B and C, both the wild-type GABAAR α3 and GlyR β subunit (control) baits interact with preys for the full-length gephyrin P1 isoform and N-terminal deletions Geph276–736 and Geph305–736. GABAAR α3-gephyrin interactions are abolished by deletion of a 17-amino acid motif in the α3 subunit intracellular loop (α3Δ5; Thr-361–Ile-377). Two further deletions, α3Δ3 (lacking Thr-361–Thr-367) and α3Δ4 (lacking Phe-368–Ile-377) localized crucial binding determinants to the residues Phe-368–Ile-377 (FNIVGTTYPI). Insertion of the Phe-368–Ile-377 motif into the GABAAR α1 subunit (α3inα1) enables the GABAAR α1 subunit bait to interact with the gephyrin prey.
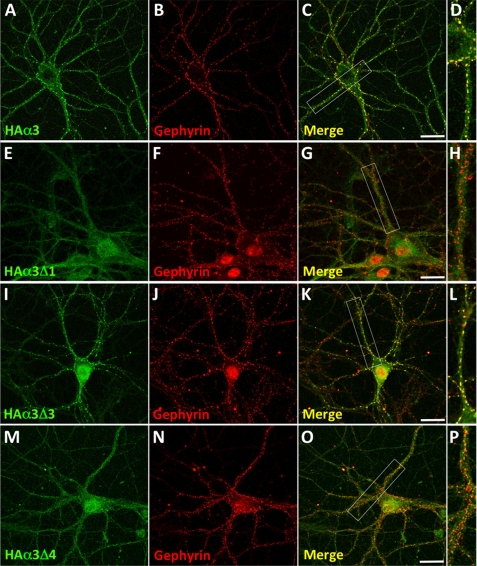
Clustering Properties of HA-tagged GABAAR α3 and Binding-site Mutants in Transfected Hippocampal Neurons
To verify the nature of the identified gephyrin binding motif on GABAAR α3 in a neuronal context, we transfected hippocampal neurons with wild-type HA-tagged GABAAR α3 and selected deletion mutants by nucleofection before plating (Fig. 3). At 17 days in vitro, neurons were immunostained with HA antibodies under nonpermeabilizing conditions and after permeabilization with mAb7a, which recognizes gephyrin. We first examined the subcellular distribution of wild-type HA-α3, which formed clusters on the surfaces of both neuronal processes and the cell body (Fig. 3A) that co-localized with endogenous gephyrin (Fig. 3, B–D). Consistent with our overlay and yeast two-hybrid data, construct HA-α3Δ1 (removing Pro-357–Ile-377) exhibited a diffuse distribution on the plasma membrane and showed little or no co-localization with gephyrin (Fig. 3, E–H). By contrast, HA-α3Δ3 (lacking Thr-361–Thr-367) showed robust co-localization with gephyrin (Fig. 3, I–L), confirming that this motif is dispensable for both gephyrin binding and synaptic targeting. Finally, HA-α3Δ4 (lacking the minimal gephyrin binding motif Phe-368–Ile-377) showed a diffuse cytoplasmic distribution with some small puncta that did not co-localize with endogenous gephyrin (Fig. 3, M–P).
FIGURE 3.
Expression and differential clustering of HA-tagged GABAAR α3 and selected deletion constructs in hippocampal neurons. Transfected 18–21 days in vitro neurons expressing HA-tagged GABAAR α3 (A–D), Δ1 (E–H), Δ3 (I–L), and Δ4 (M–P) were stained with HA antibodies under nonpermeabilizing conditions (without Triton X-100; green) and after permeabilization with 0.05% Triton X-100 with mAb7a against gephyrin (red). Note that wild-type GABAAR α3 and mutant Δ3 display good co-localization with gephyrin, whereas for mutants Δ1 and Δ4 little co-localization is observed. The third panel in each row represents the merged images, and enlargements of the respective dendrites (indicated by white boxes) are displayed in the fourth panel. Scale bars, 25 μm.
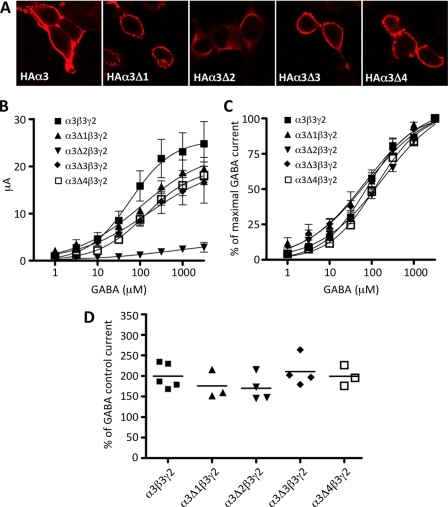
To control for possible alterations in the assembly of the GABAARs containing α3, their capacity to access the plasma membrane and form functional benzodiazepine-responsive GABAARs was assessed using recombinant expression in HEK293 cells (Fig. 4). Using immunofluorescence under permeabilizing and nonpermeabilizing conditions, it was evident that all deletion constructs except α3Δ2 localized to the cell membrane (Fig. 4A). This mutant also showed changes in the maximal response to GABA (Fig. 4B), but normalization of dose-response curves indicated no significant changes in EC50 values (Fig. 4C). Importantly, the deletion mutants affecting the gephyrin binding site (α3Δ1 and α3Δ3; see Fig. 1) had no significant influence on membrane localization or electrophysiological properties of recombinant GABAARs, including benzodiazepine responsiveness (Fig. 4D).
FIGURE 4.
Cell surface expression and functional properties of benzodiazepine-sensitive GABAARs containing α3 deletions. A, HEK293 cells were transfected with HA-tagged GABAAR α3 and deletion constructs (α3Δ1–4) together with the β3 subunit. Two days after lipofection, cells were fixed and stained with anti-HA polyclonal antibody without cell permeabilization. All constructs directed formation of cell surface α3β2 GABAARs with the exception of α3Δ2, which lacks a binding site for the ubiquitin-like protein Plic-1, which presumably causes impaired membrane insertion (35). As the α3Δ2 mutant did not show cell surface expression, cells were permeabilized to allow detection of intracellular antigens. B and C, GABA dose-response curves for α3β3γ2 and α3Δ1–4 deletion constructs together with β3γ2 are shown. C, data are normalized to the maximum GABA current. Data points represent mean ± S.E. (error bars) from at least three oocytes derived from ≥2 batches. EC50 values were compared using one-way ANOVA (with Bonferroni post hoc test) and found to be not significantly different. D, Xenopus oocytes expressing recombinant GABAA receptors containing HA-tagged GABAAR α3 and deletion constructs (α3Δ1–4) together with β3 and γ2 in the presence of 3 μm GABA were challenged with 1 μm diazepam. Stimulation was normalized to the control current at 3 μm GABA. Control current represents 100% stimulation. Data represent means ± S.E. of at least three oocytes. Mean values were determined from recordings of three or four cells.
Mapping a Binding Site for the GABAAR α3 Subunit on Gephyrin
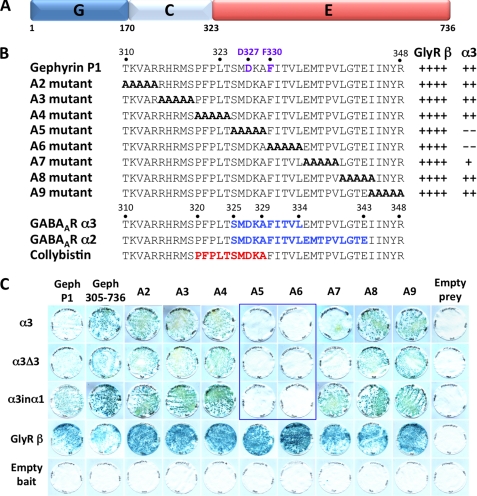
The yeast two-hybrid system was also used to map potential determinants of the GABAAR α3 subunit binding site on the gephyrin G, C, or E domains (Fig. 5A), to determine overlap with previously reported GABAAR or GlyR binding motifs (20, 21, 23). Sequential deletion analysis revealed that the minimal gephyrin prey interacting with the GABAAR α3 bait encompassed amino acids 305–736 (Geph305–736) encoding the E domain and the C-terminal eighteen amino acids of the linker region. Shorter constructs, such as Geph323–736 and Geph305–704, did not mediate this interaction as observed previously for GABAAR α2 and collybistin baits (18, 23). We therefore used alanine-scanning mutagenesis to locate determinants of GABAAR α3 subunit binding to gephyrin (Fig. 5B). Two alanine block mutants (A5 and A6) completely disrupted interactions of the wild-type GABAAR α3 subunit, α3Δ3 and α3inα1 baits with gephyrin, while leaving GlyR β subunit-gephyrin interactions unaffected (Fig. 5, B and C). It is also noteworthy that GABAAR α3 subunit and α3Δ3 interactions with the Ala-7 mutant (335EMPTV339 to 335AAAAA339) were weak compared with interactions with the A2–A4 and A8–A9 mutants, suggesting that robust GABAAR α3 binding might also require one or more amino acids in this sequence. Curiously, however, the GABAAR α3inα1 chimeric bait showed a stronger interaction with this gephyrin mutant. Despite this caveat, the core GABAAR α3 subunit binding motif within the N-terminal part of the gephyrin E domain (SMDKAFITVL; Fig. 1) shows partial overlap with the previously determined collybistin (PFPLTSMDKA) and GABAAR α2 (SMDKAFITVLEMPTVLGTE) binding motifs on gephyrin (18, 23). Interestingly, two residues (Asp-327 and Phe-330) in the minimal GABAAR α2 and α3 binding sites on gephyrin have been previously implicated in GlyR β subunit-E domain interactions (21) (Fig. 5A). However, mutation of Asp-327 and Phe-330 in mutants A5 and A6 respectively, was not sufficient to disrupt GlyR β-E domain interactions in the YTH system (Fig. 5C).
FIGURE 5.
Deletion mapping of the GABAAR α3 binding site on gephyrin. A, schematic of the domain structure of the gephyrin P1 variant shows the positions of the G, C, and E domains. B, protein sequence at the border of the gephyrin C and E domains shows the relative positions of the gephyrin alanine block mutants A2–A9 and interactions with GlyR β and GABAAR α3 (see panel C). The potential GABAAR α3 subunit binding site on gephyrin (amino acids 325–334) is shown together with motifs vital for GABAAR α2 subunit (amino acids 325–343) (23), collybistin (amino acids 320–329) (18), and two residues (Asp-327 and Phe-330) implicated in GlyR β-gephyrin interactions (17) (purple lettering, top sequence). LacZ freeze-fracture assay rankings: ++++, strong interaction; ++, moderate interaction; +, weak interaction; −, no detectable interaction. C, GABAAR α3, α3Δ3, α3inα1, and GlyR β subunit intracellular loop baits were tested for interactions with Geph276–736 and alanine substitution mutants created in this prey (A2–A9). LacZ freeze-fracture assays demonstrate that the GABAAR α3 bait does not interact with mutants A5 and A6 and is weakened in mutant A7, whereas the GlyR β subunit bait interacts with all of these preys.
Comparison of GABAAR α3 and GlyR β Subunit Binding to Gephyrin
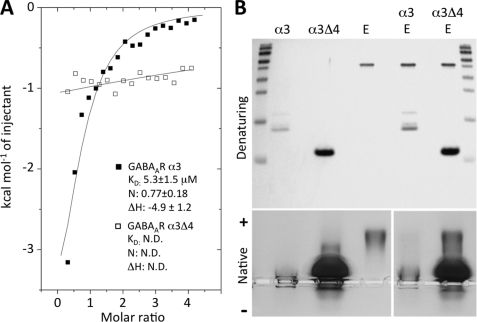
To quantify GABAAR α3 subunit binding to gephyrin and to compare this with GlyR β subunit-gephyrin interactions, we used ITC. GABAAR α3 and the α3Δ4 deletion variant were expressed in E. coli as His6 fusion proteins to determine interactions with a gephyrin E domain fragment (amino acids 318–736) (21). This gephyrin construct has previously been used to characterize GlyR β subunit binding to gephyrin (20, 21). ITC revealed that the intracellular domain of the α3 subunit bound in an exothermic reaction (ΔH = −4.9 ± 1.2 kcal/mol) to the gephyrin E domain with a dissociation constant in the low micromolar range (KD = 5.3 ± 1.5 μm), and a stoichiometry of 0.77 ± 0.18 mol/mol (Fig. 6A). As expected, no binding was observed for α3Δ4 in ITC. The previously analyzed E domain-GlyR β loop interaction was significantly stronger (20, 21) with a KD of 0.2 μm but also displayed a two-site binding behavior with the second lower affinity binding site exhibiting a KD of 11 μm. To further confirm the mapping of the binding site on GABAAR α3 we employed native PAGE to investigate the behaviors of α3 and α3Δ4 with and without the gephyrin E domain. Due to the high isoelectric point of the GABAAR α3 construct (calculated pI of ∼10) it is positively charged at the pH (8.4) at which this experiment was conducted and hence does not enter into the gel. By contrast, the E domain of gephyrin is negatively charged (calculated pI of ∼7) and migrates into the gel. Native PAGE assays confirmed complex formation of the gephyrin E domain with α3, but not α3Δ4 (Fig. 6B).
FIGURE 6.
In vitro analysis of the gephyrin-GABAAR α3 interaction. A, ITC of the gephyrin E domain with GABAAR α3 or α3Δ4. Overlaid binding isotherms of the E domain of gephyrin (318–736, P1 variant) titrated with GABAAR α3 (■) or α3Δ4 (□) intracellular loops are plotted as a function of the molar ratio of GABAAR α3 to gephyrin E domain. Affinity (KD in μm), stoichiometry (N in mol/mol), and enthalpy change (ΔH in kcal/mol) could be determined for GABAAR α3-GephE complex formation, but not for GABAAR α3Δ4. B, upper, SDS-PAGE of GABAAR α3, α3Δ4, and gephyrin E domain constructs and mixtures. Lower, visualization of wild-type GABAAR α3 and α3Δ4 complex formation with the gephyrin E domain (amino acids 318–736, P1 variant) on an agarose gel. At pH 8.4, the E domain hardly enters the gel when bound to the GABAAR α3 intracellular loop (PI ∼10), and GephE is fully complexed with the GABAAR α3 loop. However, no binding is seen for α3Δ4 even when using a 16-fold molar excess of the GABAAR α3 fragment over gephyrin.
DISCUSSION
It is becoming increasingly apparent that GABAAR α subunits not only influence GABAAR physiology, pharmacology, and biological function, but also mediate the synaptic versus extrasynaptic localization of these receptor subtypes via distinct protein-protein interactions. For example, the GABAAR α2 subunit co-localizes with gephyrin in vivo and interacts with both gephyrin and the RhoGEF collybistin via overlapping binding sites (22, 23). For this reason, we considered that other GABAAR α subunits might also be clustered at synapses by these molecules. The α3 subunit was selected for analysis because (i) GABAARs containing this subunit co-localize with gephyrin, e.g. in cerebellar cortex (30), the thalamic reticular nucleus (16), or at perisomatic synapses in the globus pallidus (31) and (ii) gephyrin is mislocalized in GABAAR α3 subunit knock-out mice (15, 16).
Using a combination of deletion mutagenesis, overlay, and yeast two-hybrid assays we have determined that GABAAR α3 specifically interacts with gephyrin via a critical motif (FNIVGTTYPI) that overlaps with sequences that bind gephyrin and collybistin in GABAAR α2. Curiously, very few amino acids are conserved between the equivalent regions of the α2 and α3 subunits, suggesting that either the nature of the amino acids in these motifs is crucial or that conserved amino acids within (Tyr-375) or directly flanking these minimal motifs (e.g. Asn-378) are crucial determinants of gephyrin binding. Certainly, deletion of the minimal gephyrin binding motif prevented synaptic clustering and co-localization of recombinant GABAAR α3 with endogenous gephyrin in cultured neurons.
Using the yeast-two hybrid system, we also mapped crucial determinants of GABAAR α3 binding to gephyrin to the start of the E domain. These residues overlap with previously characterized determinants of GABAAR α2 binding on gephyrin and show partial overlap with key determinants (Asp-327 and Phe-330) of GlyR β binding to gephyrin (21) (Fig. 7) This suggests that binding of GABAARs containing α2, α3, and GlyRs to gephyrin could be mutually exclusive. However, given that mutation of Asp-327 and Phe-330 in mutants A5 and A6 did not appear to disrupt GlyR β-E domain interactions in the YTH system, key differences may exist between GABAAR and GlyR binding to gephyrin. Consistent with this hypothesis, ITC revealed that the GABAAR α3 bound to the gephyrin E-domain in a 1:1 ratio and displayed a lower affinity (KD = 5.3 ± 1.5 μm) than previously observed for GlyR β (two binding sites with KD values of 0.2 μm and 11 μm) (20, 21). Collectively, these observations may explain why GABAAR-gephyrin interactions have been difficult to characterize by immunoprecipitation and are prone to the effects of detergents (22).
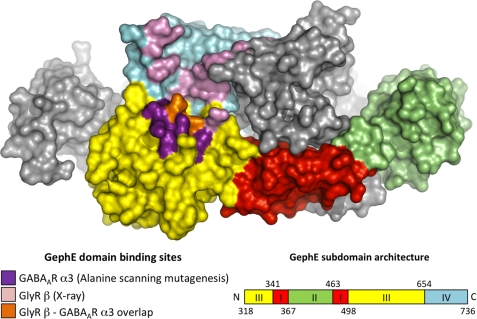
FIGURE 7.
Structural representations of the GlyR β and GABAAR α3 loop binding sites on the gephyrin E domain. Color-coded surface of the gephyrin E domain dimer (21) shows one monomer colored according to the subdomain architecture and the other in gray. Residues involved in GlyR β subunit binding (21) are shown in pink, residues implicated in GABAAR α3 subunit binding by alanine scanning mutagenesis are shown in purple. Amino acids Phe-330 and Asp-327 of gephyrin, which are involved in the binding of both receptor types, are marked in orange.
GABAARs containing the α3 subunit co-localized with gephyrin in both dendritic and perisomatic locations in different brain regions (16, 30, 31). This may reflect different roles for the α3 subunit in mediating dendritic inhibition, affecting the efficacy and plasticity of excitatory synaptic inputs of principal cells, versus perisomatic inhibition, controlling output by synchronizing action potential firing of larger groups of principal cells. Certainly, although GABAA receptors containing the α3 subunit are thought to represent only 10–15% of all GABAA receptors, they are the major GABAA receptor subtype expressed in brain stem monoaminergic nuclei (32, 33). Consistent with these findings, GABAARs containing α3 have been linked to sensorimotor gating and affective and cognitive functions phenotypes (32, 34). A molecular understanding of the basis of synaptic localization of GABAAR α3-containing receptors by gephyrin adds to our knowledge of this interesting receptor subtype.
This work was supported by a Medical Research Council New Investigator Award G0501258 (to K. H.), the EC-FP7 integrated project “Neurocypres,” grant agreement (to W. S.), HEALTH-F4-2008-202088 grant, the Institute of Neurological Disorders and Stroke grants (NS047478, NS48045, NS051195, NS054900, NS056359, and NS065725, to S. J. M.), and the Deutsche Forschungsgemeinschaft Rudolf Virchow Center for Experimental Biomedicine Grants FZ 82 and SFB487 C7 (to H. S.).
- GlyR
- glycine receptor
- GABAAR
- GABAA receptor
- ITC
- isothermal titration calorimetry
- XR
- Xenopus Ringer solution.
REFERENCES
- 1. Fritschy J. M., Harvey R. J., Schwarz G. (2008) Trends Neurosci. 31, 257–264 [DOI] [PubMed] [Google Scholar]
- 2. Prior P., Schmitt B., Grenningloh G., Pribilla I., Multhaup G., Beyreuther K., Maulet Y., Werner P., Langosch D., Kirsch J., Betz H. (1992) Neuron 8, 1161–1170 [DOI] [PubMed] [Google Scholar]
- 3. Feng G., Tintrup H., Kirsch J., Nichol M. C., Kuhse J., Betz H., Sanes J. R. (1998) Science 282, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 4. Kneussel M., Brandstätter J. H., Laube B., Stahl S., Müller U., Betz H. (1999) J. Neurosci. 19, 9289–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fischer F., Kneussel M., Tintrup H., Haverkamp S., Rauen T., Betz H., Wässle H. (2000) J. Comp. Neurol. 427, 634–648 [DOI] [PubMed] [Google Scholar]
- 6. Kneussel M., Brandstätter J. H., Gasnier B., Feng G., Sanes J. R., Betz H. (2001) Mol. Cell. Neurosci. 17, 973–982 [DOI] [PubMed] [Google Scholar]
- 7. Lévi S., Logan S. M., Tovar K. R., Craig A. M. (2004) J. Neurosci. 24, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu W., Jiang M., Miralles C. P., Li R. W., Chen G., de Blas A. L. (2007) Mol. Cell. Neurosci. 36, 484–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belelli D., Harrison N. L., Maguire J., Macdonald R. L., Walker M. C., Cope D. W. (2009) J. Neurosci. 29, 12757–12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Loebrich S., Bähring R., Katsuno T., Tsukita S., Kneussel M. (2006) EMBO J. 25, 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schweizer C., Balsiger S., Bluethmann H., Mansuy I. M., Fritschy J. M., Mohler H., Lüscher B. (2003) Mol. Cell Neurosci. 24, 442–450 [DOI] [PubMed] [Google Scholar]
- 12. Alldred M. J., Mulder-Rosi J., Lingenfelter S. E., Chen G., Lüscher B. (2005) J. Neurosci. 25, 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li R. W., Yu W., Christie S., Miralles C. P., Bai J., Loturco J. J., De Blas A. L. (2005) J. Neurochem. 95, 756–770 [DOI] [PubMed] [Google Scholar]
- 14. Kralic J. E., Sidler C., Parpan F., Homanics G. E., Morrow A. L., Fritschy J. M. (2006) J. Comp. Neurol. 495, 408–421 [DOI] [PubMed] [Google Scholar]
- 15. Studer R., von Boehmer L., Haenggi T., Schweizer C., Benke D., Rudolph U., Fritschy J. M. (2006) Eur. J. Neurosci. 24, 1307–1315 [DOI] [PubMed] [Google Scholar]
- 16. Winsky-Sommerer R., Knapman A., Fedele D. E., Schofield C. M., Vyazovskiy V. V., Rudolph U., Huguenard J. R., Fritschy J. M., Tobler I. (2008) Neuroscience 154, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meyer G., Kirsch J., Betz H., Langosch D. (1995) Neuron 15, 563–572 [DOI] [PubMed] [Google Scholar]
- 18. Harvey K., Duguid I. C., Alldred M. J., Beatty S. E., Ward H., Keep N. H., Lingenfelter S. E., Pearce B. R., Lundgren J., Owen M. J., Smart T. G., Lüscher B., Rees M. I., Harvey R. J. (2004) J. Neurosci. 24, 5816–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sola M., Bavro V. N., Timmins J., Franz T., Ricard-Blum S., Schoehn G., Ruigrok R. W., Paarmann I., Saiyed T., O'Sullivan G. A., Schmitt B., Betz H., Weissenhorn W. (2004) EMBO J. 23, 2510–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schrader N., Kim E. Y., Winking J., Paulukat J., Schindelin H., Schwarz G. (2004) J. Biol. Chem. 279, 18733–18741 [DOI] [PubMed] [Google Scholar]
- 21. Kim E. Y., Schrader N., Smolinsky B., Bedet C., Vannier C., Schwarz G., Schindelin H. (2006) EMBO J. 25, 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tretter V., Jacob T. C., Mukherjee J., Fritschy J. M., Pangalos M. N., Moss S. J. (2008) J. Neurosci. 28, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saiepour L., Fuchs C., Patrizi A., Sassoè-Pognetto M., Harvey R. J., Harvey K. (2010) J. Biol. Chem. 285, 29623–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kins S., Betz H., Kirsch J. (2000) Nat. Neurosci. 3, 22–29 [DOI] [PubMed] [Google Scholar]
- 25. Fuller K. J., Morse M. A., White J. H., Dowell S. J., Sims M. J. (1998) BioTechniques 25, 85–92 [DOI] [PubMed] [Google Scholar]
- 26. Kittler J. T., Thomas P., Tretter V., Bogdanov Y. D., Haucke V., Smart T. G., Moss S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fritschy J. M., Mohler H. (1995) J. Comp. Neurol. 359, 154–194 [DOI] [PubMed] [Google Scholar]
- 28. Sigel E., Baur R., Trube G., Möhler H., Malherbe P. (1990) Neuron 5, 703–711 [DOI] [PubMed] [Google Scholar]
- 29. Li X., Cao H., Zhang C., Furtmueller R., Fuchs K., Huck S., Sieghart W., Deschamps J., Cook J. M. (2003) J. Med. Chem. 46, 5567-5570 [DOI] [PubMed] [Google Scholar]
- 30. Sassoè-Pognetto M., Panzanelli P., Sieghart W., Fritschy J. M. (2000) J. Comp. Neurol. 420, 481–498 [DOI] [PubMed] [Google Scholar]
- 31. Gross A., Sims R. E., Swinny J. D., Sieghart W., Bolam J. P., Stanford I. M. (2011) Eur. J. Neurosci. 33, 868–878 [DOI] [PubMed] [Google Scholar]
- 32. Fiorelli R., Rudolph U., Straub C. J., Feldon J., Yee B. K. (2008) Behav. Pharmacol. 19, 582–596 [DOI] [PubMed] [Google Scholar]
- 33. Corteen N. L., Cole T. M., Sarna A., Sieghart W., Swinny J. D. (2011) Eur. J. Neurosci. 34, 250–262 [DOI] [PubMed] [Google Scholar]
- 34. Yee B. K., Keist R., von Boehmer L., Studer R., Benke D., Hagenbuch N., Dong Y., Malenka R. C., Fritschy J. M., Bluethmann H., Feldon J., Möhler H., Rudolph U. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 17154–17159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bedford F. K., Kittler J. T., Muller E., Thomas P., Uren J. M., Merlo D., Wisden W., Triller A., Smart T. G., Moss S. J. (2001) Nat. Neurosci. 4, 908–916 [DOI] [PubMed] [Google Scholar]