Background: The function of the C. elegans mitochondrial uncoupling protein 4 (ceUCP4) has not been characterized.
Results: Worms deficient in ceUCP4 displayed hypometabolic phenotypes and complex II dysfunction that corresponded with a significant loss of mitochondrial succinate import.
Conclusion: ceUCP4 plays a novel role in the regulation of complex II by controlling succinate transport into mitochondria.
Significance: Understanding how extramitochondrial succinate and ceUCP4 regulate complex II-mediated metabolism is critical for understanding the mechanisms of cellular respiration.
Keywords: Bioenergetics-Electron Transfer Complex, C. elegans, Mitochondrial Metabolism, Mitochondrial Transport, Uncoupling Proteins
Abstract
The novel uncoupling proteins (UCP2–5) are implicated in the mitochondrial control of oxidant production, insulin signaling, and aging. Attempts to understand their functions have been complicated by overlapping expression patterns in most organisms. Caenorhabditis elegans nematodes are unique because they express only one UCP ortholog, ceUCP4 (ucp4). Here, we performed detailed metabolic analyzes in genetically modified nematodes to define the function of the ceUCP4. The knock-out mutant ucp4 (ok195) exhibited sharply decreased mitochondrial succinate-driven (complex II) respiration. However, respiratory coupling and electron transport chain function were normal in ucp4 mitochondria. Surprisingly, isolated ucp4 mitochondria showed markedly decreased succinate uptake. Similarly, ceUCP4 inhibition blocked succinate respiration and import in wild type mitochondria. Genetic and pharmacologic inhibition of complex I function was selectively lethal to ucp4 worms, arguing that ceUCP4-regulated succinate transport is required for optimal complex II function in vivo. Additionally, ceUCP4 deficiency prolonged lifespan in the short-lived mev1 mutant that exhibits complex II-generated oxidant production. These results identify a novel function for ceUCP4 in the regulation of complex II-based metabolism through an unexpected mechanism involving succinate transport.
Introduction
The mitochondrial family of solute carrier proteins (SLC25) is localized to the inner mitochondrial membrane and mediates the transfer of a large variety of metabolic intermediates (e.g. ATP/ADP, carboxylic, and amino acids, ions) between the cytoplasm and mitochondrial matrix. Mitochondrial uncoupling proteins (UCPs)4 comprise the most well characterized subgroup of the SLC25 family and regulate the conductance of protons, among other solutes, across the inner mitochondrial membrane (reviewed in Ref. 2). Commonly referred to as proton leak, the reentry of protons into the mitochondrial matrix creates an energy-dissipating cycle that uncouples the electrochemical proton gradient from ATP synthesis and releases the energy as heat. Brown fat UCP1 (SLC25A7) was identified nearly three decades ago (1), and four UCP1 homologs (UCP2–5) were more recently discovered that exhibit relatively broader tissue distribution compared with UCP1; UCP2 (SLCA25A8; exhibits wide expression, UCP3 (SLC25A9; primarily localized to skeletal muscle, heart, and brown fat), UCP4 (SLC25A27), and UCP5 (SLC25A14, brain-derived mitochondrial carrier 1, BMCP1) are expressed primarily in the central nervous system (2). From a functional perspective, only UCP1 has an established role in metabolic physiology; it generates heat for cold adaptation (3, 4). The diverse tissue distribution of UCP2–5 in most endotherms and the conservation of UCP homologs in a variety of ectotherms support the hypotheses that the novel uncoupling proteins regulate mitochondrial functions apart from, or in addition to, thermogenesis.
In this study we focus on UCP4, the postulated ancestral uncoupling protein from which the others may have diverged (5). UCP4 was identified over a decade ago (6) but to date is not well characterized. A variety of studies have confirmed that UCP2 and UCP3 share significant structural (>50% amino acid) and functional (mitochondrial proton leak) homology with UCP1. By comparison, UCP4 has decreased UCP1 homology (30%), and its role as a classical protonophoric uncoupler is debated. For example, UCP4 is located on a different phylogenetic clade from UCP1–3 and lacks key amino acid residues implicated in proton transport regulation by UCP1–3 (7).
The nematode C. elegans is a powerful model for understanding mechanisms regulating mammalian energy homeostasis because the vast majority of metabolic genes are conserved (8, 9). Unlike most other organisms, C. elegans expresses only a single UCP ortholog, ceUCP4, which shares 46% homology (amino acid) with human UCP4 (5). Thus, the nematode is an attractive model for the characterization of the functions of UCP4 in an animal model. Here, we performed complementary biochemical and genetic studies in both isolated mitochondria and intact nematodes to define the functions of ceUCP4. We show for the first time that ceUCP4 regulates mitochondrial succinate import and complex II-mediated metabolism. These studies provide a new perspective on the potential functions of this ancient UCP1 homolog in the control of mitochondrial oxidative phosphorylation (OXPHOS).
EXPERIMENTAL PROCEDURES
Chemical Reagents
Unless otherwise stated, all chemicals and reagents were purchased from Sigma) with the exception of Nile Red, which was purchased from Invitrogen.
C. elegans Culture and Growth
C. elegans were maintained and cultured as previously described on nematode growth medium plates fed with OP50 Escherichia coli and in liquid cultures (10). N2 (Bristol) wild type, ucp4 (ok195), mev1 (kn-1), and gas1 (fc21) strains were obtained from the Caenorhabditis Genetics Center (CGC). ucp4 (ok195) was generated by the C. elegans Gene Knock-out Consortium and outcrossed to the parental N2 strain >8 times.
RNAi Feeding
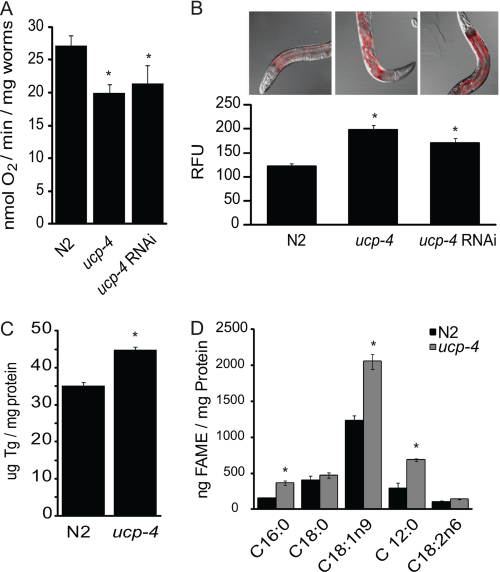
To control for potential residual mutations in the outcrossed RB695 strain as being important for metabolic phenotypes, a 195-bp RNAi targeting construct was amplified from the ceUCP4 coding sequence (5′ to 3′ primers, forward (gca tta gtt gcc gaa acg gtc) and reverse (ccc att cga att cct gtg tag)) and subcloned into the L4440 C. elegans RNAi vector according to established protocols (Addgene). Wild type N2 worms were age-synchronized by bleach-killing gravid adults. Eggs were then hatched and subjected to UCP4 RNAi knockdown by feeding (HT115 transformants). RNAi knockdown efficiency (∼ 80%) was determined by semiquantitative RT-PCR, and phenotype analysis (oxygen consumption) was performed in comparison with ceUCP4 knock-out worms (n = 3, Fig. 1).
FIGURE 1.
ucp4 animals show hypometabolic phenotypes. A, respiration rates (mean ± S.E.) in age synchronized (L1) wild type (N2), ucp4, and N2 worms after ucp4 RNAi knockdown (n = 5) are shown. * indicates p < 0.05. B, shown is lipid granule staining (relative nile red fluorescence units (RFU)) of age synchronized, adult (L4) N2, ucp4, and N2 worms subjected to ucp4 RNAi (n = 4–6). * indicates p < 0.01. C, triglyceride (Tg) quantification (mean ± S.E.) in N2 and ucp4 worm homogenates (n = 3) is shown. * indicates p < 0.01. D, fatty acid methyl ester quantification (mean ± S.E.) in N2 and ucp4 worm homogenates (n = 3) is shown. *, **, and *** indicate significantly different (p < 0.05, 0.01, and 0.001, respectively) from N2 animals.
Mitochondrial Isolation
Mitochondria were isolated from worms as described (10, 11). Briefly, animals were harvested from liquid culture, washed 4×, purged of gut bacteria, and resuspended in 10 ml of mitochondrial isolation buffer (220 mm mannitol, 70 mm sucrose, 5 mm MOPS, 2 mm EDTA). Animals were then slowly homogenized with a Teflon Potter-Elvehjem homogenizer and incubated with the protease subtilisin (5 mg/g worm pellet), and the mitochondria were collected by differential centrifugation. Mitochondrial yield was determined by the bicinchoninic acid protein concentration assay kit according to the manufacturer's instructions (Pierce).
Whole Animal Oxygen Consumption
200 age-matched L1 worms were collected from each strain/treatment in M9 nematode buffer, and rates of oxygen consumption were analyzed with a Clark-type electrode chamber (Instech Laboratories, Plymouth Meeting, PA) in a total volume of 1 ml for 10 min (n = 200 worms/three independent experiments).
Cytochrome c Reduction
Cytochrome c reduction assays in isolated nematode mitochondria were performed in triplicate experiments as described (12). Briefly, 50 μg of isolated mitochondria were added to cytochrome c assay buffer (0.1 m NaPO4, 50 mm EDTA, 25 mm KCN, 1 mm cytochrome c, pH 7.4) supplemented with 500 mm pyruvate, and cytochrome reduction rate was quantified spectrophotometrically.
Mitochondrial Oxygen Consumption
Oxygen consumption analyses were performed (n ≥ 5 times) using a Clark-type electrode as described previously (13, 14) with minor modification. Briefly, respiration of 0.5 mg of mitochondria was assayed by the sequential addition of ADP (1 mm), succinate, malate, durohydroquinone, or N,N,N′,N′-tetramethyl-p-phenylenediamine/ascorbate, ADP (2 mm total concentration), oligomycin (1 μg/ml), and 2,4-dinitrophenol or 2,4-dinitrophenyl (200 μm). 500 μm palmitate was used in the indicated experiments. Guanine nucleotides GDP and GTP had overlapping effects on OXPHOS and were used interchangeably at 1 mm during state 4 respiration. Effects of guanine nucleotides on maximal state 3 respiratory inhibition used a range of GDP doses (0–10 mm, supplemental Fig. S5; 10 mm, Fig. 4, A and C).
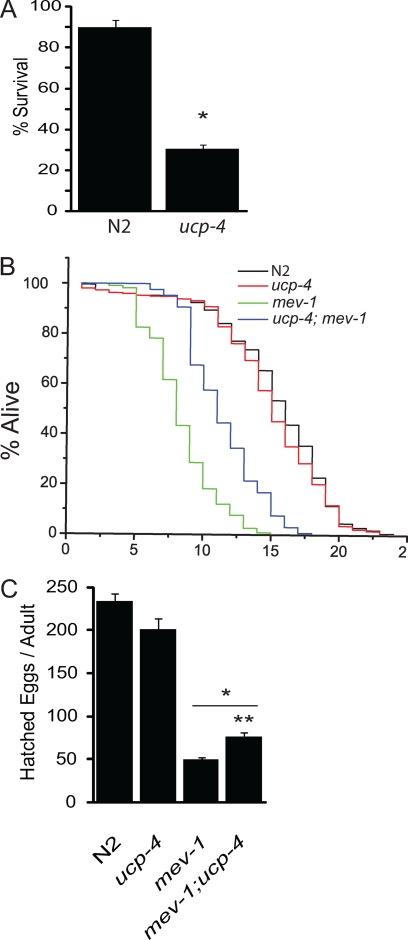
FIGURE 4.
ceUCP4 energizes complex II and shortens mev1 lifespan and fecundity and is required for survival in the absence of complex I function. A, shown is the percent survival (mean ± S.E.) in N2 and ucp4 L2 worms treated with the complex I inhibitor rotenone (48 h) (n = 3). * indicates significantly different (p < 0.001) from N2. B, shown are lifespan analyses in N2, ucp4, mev1 mutants along with ucp4; mev1 double mutant worms. C, no. of hatched eggs per adult in N2, ucp4, and mev1 mutants along with ucp4; mev1 double mutant worms (mean ± S.E., n = 3) is shown. * indicates significantly different from N2, p < 0.001. ** indicates significantly different from mev1, p < 0.01.
Lipid Analysis
C. elegans fat content was visualized/quantified using the lipid-staining dye Nile Red as previously described (15). Fluorescence levels were captured and measured using a Zeiss LSM 510 confocal microscope and LSM510 fluorescence quantification software. Triglycerides were determined in triplicate homogenates of nematodes per strain using a triglyceride determination kit (Wako Diagnostics, Richmond, VA). Fatty acid analysis was performed in triplicate worm cultures per strain as described previously (16). Briefly, adult C. elegans nematodes were washed and gravity-purified 4×. Fatty acid methyl esters were generated from worm pellets using 2.5% methanolic H2SO4 (80 °C for 1 h), extracted with hexane, and analyzed by gas chromatography/mass spectrometry (ThermoFinnigan Trace-MS GC-quadrupole, Waltham, MA).
C. elegans Lifespan and Fecundity Analyses
Approximately 100 age-matched L1 larvae per strain per experiment were plated onto food-replete nematode growth medium plates. Surviving worms were counted and moved to a new plate daily. Death was scored as the absence of a response to slight touch using a thin platinum wire. For fecundity analysis, 12 single L1 larvae from each strain were singly placed on nematode growth medium plates with a lawn of OP50 and embryos counted.
Quantification of Rotenone Survival
Synchronized larval stage 2 N2 and ucp4 worms were transferred to nematode growth medium plates (150/plate) containing a lawn of HT115 and 0.5 mm rotenone. Worms were incubated for 48 h at 20 °C, then counted and scored as either living or dead.
Electron Transport Chain Activity Measurement
Assays were performed on isolated mitochondria as detailed previously (11). In brief, freshly prepared mitochondria were solubilized with cholate to disrupt mitochondrial membranes, and the activities of each complex or multiple complexes were measured spectrophotometrically.
Analysis of Coenzyme Q9 Levels
Lipids were extracted from isolated mitochondria (n = 3) with methanol, and coenzyme Q levels were quantified by HPLC with electrochemical detection as previously described (17). Coenzyme Q9 levels were normalized to that of coenzyme Q6 recovered in individual lipid extracts.
Mitochondrial Substrate Uptake Assays
50 μg of isolated mitochondria were incubated in 1 ml of respiration buffer (120 mm KCl, 5 mm KH2PO4, 3 mm HEPES, 1 mm EGTA, 2 mm MgCl2, 0.3% BSA, pH 7.2) containing 8 μm [14C]succinate or [14C]malate (American Radiolabeled Chemicals, St. Louis, MO) over a time course of 20 min at 37 °C (13). Where indicated, mitochondria were incubated in the presence of either 1 mm GDP, 0.4 μm carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP), or 10 mm n-butylmalonate (18). Mitochondria were collected and washed from extramitochondrial label using a cell harvester (Brandel, Gaithersburg, MD) and quantified for radioactive substrate uptake by scintillation counting. Experiments were repeated 3–6 times.
Statistics
For all quantitative experiments, the data are represented as the mean ± S.E. from at least three independent assays. Differences between independent sample populations were analyzed using an unpaired Student's t test, with significance set a priori as p < 0.05.
RESULTS
ucp4 Worms Exhibit Decreased Metabolic Rate and Increased Lipid Accumulation
We compared respiration rates and markers of adiposity in the wild type parental strain (N2) and the mutant ceUCP4 knock-out strain (ucp4). Age-synchronized L1 larval ucp4 animals exhibited an approximate 20% decrease in overall oxygen consumption compared with wild type, N2 worms (Fig. 1A). Two generations of ucp4 RNAi knockdown (∼80% by RT-PCR, not shown) in N2 worms similarly decreased the metabolic rate, confirming that the respiratory phenotype observed in ucp4 animals was due specifically to the loss of ucp4 (Fig. 1A). Additionally, using the fluorescent lipid indicator nile red in adult (L4) intact worms, we found that both ucp4 deletion and RNAi-based ceUCP4 knockdown led to significantly increased lipid accumulation when compared with N2 (Fig. 1B). Similarly, total triglyceride levels in ucp4 homogenates were 35% increased compared with N2 (Fig. 1C). Gas chromatography/mass spectrometry analyses in adult worm homogenates revealed that ucp4 worms exhibited a significant accumulation of the fatty acids palmitate (C16:0), oleate (C18:1n9), and laurate (C12:0), but not stearate or linoleate, compared with N2 (Fig. 1D). A representative chromatogram for N2 worms is shown in supplemental Fig. S1.
ucp4 Animals Exhibit Decreased Succinate-induced Complex II Respiration
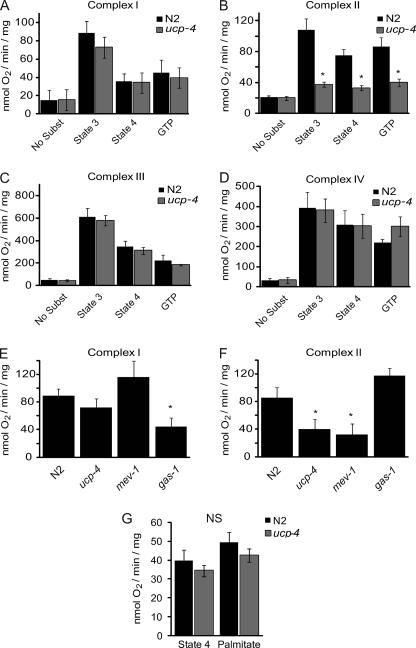
We initially reasoned that the decreased rate of oxygen consumption and increased adiposity of ucp4 animals resulted from decreased respiratory efficiency, rendering the animals prone to positive energy balance and nutrient accumulation. To scrutinize this possibility, detailed analyses of OXPHOS in mitochondria isolated from N2 and ucp4 worms were performed. N2 and ucp4 mitochondria exhibited similar rates of malate-driven, complex I-dependent state 3 and state 4 respiration (Fig. 2A; see a representative respiration assay in supplemental Fig. S2). In contrast, ucp4 mitochondria exhibited a striking, near complete failure to respire on the complex II substrate succinate (Fig. 2B). Moreover, complex III-mediated state 3 and state 4 respiration rates were also similar in N2 and ucp4 mitochondria respiring on the artificial electron donor durohydroquinone (Fig. 2C). Similarly, loss of ceUCP4 had no effect on N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD)/ascorbate-mediated complex IV respiration compared with wild type (Fig. 2D). Abrogation of succinate-driven complex II-mediated state 3 respiration in ucp4 mitochondria was comparable with that observed in mitochondria isolated from the missense mutant mev1 (kn1) that bears a loss of function mutation in cytochrome B of succinate dehydrogenase-complex II (Fig. 2F). In contrast, unlike the complex I mutant gas1 that exhibits a strong reduction in malate respiration (10, 19), no defect in complex I-dependent respiration was observed in mev1 and ucp4 (Fig. 2E).
FIGURE 2.
ucp4 animals have a selective failure of complex II-mediated mitochondrial respiration. A–D, shown are maximal rates of state 3, state 4, and GTP-sensitive respiration (mean ± S.E.) in isolated mitochondria respiring on the following complex-specific substrates: malate for complex I (A), succinate for complex II (B), durohydroquinone for complex III (C), and tetramethylphenylenediamine/ascorbate for complex IV (D) (n = 3–6). * indicates significantly different (p < 0.01) from N2. E and F, comparisons of maximal state 3 respiration rates mediated by complex I (malate) (E) and complex II (succinate) (F) in isolated mitochondria from N2, ucp4, mev1 (complex II mutant), and gas11 (complex I mutant) worms (n = 3–6). * indicates significantly different (p < 0.05) from N2 mitochondria. G, palmitate-stimulated state 4 respiration in isolated mitochondria from N2 and ucp4 worms (n = 3) is shown. No significant (NS) differences were observed between genotypes.
A prototypical feature of mammalian UCP1–3 is that they are activated by fatty acids and inhibited by purine nucleotides (e.g. GDP, GTP). Controversy regarding the regulation of mammalian UCP4–5 by these regulators along with structural divergence from UCP1–3 has led to the notion that UCP4–5 may not function as canonical uncouplers (20, 21). Moreover, similar to mammalian UCP4, ceUCP4 lacks key residues required for UCP1–3 mediated proton transport, but it shares molecular signatures of other anion transport family members (e.g. the dicarboxylate carrier, supplemental Fig. S3) (7). Compared with N2, ucp4 mitochondria failed to show any difference in guanine nucleotide-sensitive state 4 respiration (Fig. 2, A–D), respiratory control ratio (state 3/state 4 respiration, supplemental Fig. S4A), or ADP/O ratio (ATP formed per oxygen consumed, supplemental Fig. S4B) in response to energized complexes I-IV. Moreover, treatment of mitochondria with the fatty acid palmitate failed to increase oligomycin-induced state 4 respiration in N2 mitochondria, and GDP, the prototypical UCP1 inhibitor, also failed to decrease proton leak-dependent, state 4 respiration (Fig. 2G). Taken together, these data indicate that ceUCP4 strongly regulates succinate-mediated complex II activity in nematodes but does not appear to function as a prototypical fatty acid-sensitive uncoupler.
ucp4 Animals Have Normal Electron Transport Chain Function
In nematodes, coenzyme Q9 is the membrane quinone electron carrier for electrons passing from both complexes I and II to III (22). We observed that wild type (N2) and ucp4 mitochondria had comparable levels of coenzyme Q9 (Table 1). We, therefore, hypothesized that a defect in succinate dehydrogenase itself may underlie the complex II dysfunction phenotype in ucp4. Using complex-specific electron donors and inhibitors, we measured the transport-independent activities of electron transport chain enzymes in cholate-solubilized mitochondria using complex-specific substrates. As shown in Table 1, the activities of complexes II, III, and IV were similar in N2 and ucp4 mitochondria, as were the electron flux reactions between complexes I and III and between II and III. Surprisingly, the activity of succinate dehydrogenase was 3-fold increased in ucp4 compared with N2, suggesting that a possible compensation mechanism occurs in vivo to increase complex II function similar to that observed previously in response to knockdown of complex I subunits (23). Importantly, these assays show that the decrease in succinate-induced complex II respiration in intact ucp4 mitochondria cannot be explained by electron transfer defects between succinate and complex II proteins (Fig. 2B). To examine TCA cycle function, we energized N2 and ucp4 mitochondria with the acetyl-CoA generator pyruvate in the presence or absence of complex I or complex II inhibitors and measured rates of cytochrome c reduction. As shown in Table 1, pyruvate-mediated, complex I-independent respiration rates were similar in wild type and ucp4 mitochondria. Combined, the results fail to reveal a defect in electron transport chain function or TCA cycle-generated, complex II-mediated succinate metabolism in response to loss of ceUCP4.
TABLE 1.
UCP4 animals exhibit normal mitochondrial respiratory chain function
Quantification of the levels of coenzyme Q9 in N2 and ucp4 worm homogenates by HPLC (n = 3) is shown. No significant differences were observed between genotypes except for complex II-succinate dehydrogenase, whose activity was induced in ucp4 versus N2 worms. Electron transport assays comparing the activities of the indicated individual or coupled mitochondrial complexes in cholate-solubilized mitochondria from N2 and ucp4 worms are expressed as nmol of substrate/min/mg of protein (n = 4–8). Rates of cytochrome c reduction (per min/mg of protein) are shown in pyruvate-energized isolated mitochondria from N2 and ucp4 worms in the absence (uninhibited) or presence of the complex I inhibitor rotenone or the complex II inhibitor malonate (n = 3). No significant differences were observed.
| Coenzyme Q9 concentration |
||
|---|---|---|
| N2 | ucp4 | |
| pmol of coenzyme Q9/mg of protein | ||
| 3654 ± 379 (n = 6) | 3424.3 ± 266.8 (n = 6) | |
| Enzyme(s) tested | Enzymatic activity |
|
|---|---|---|
| N2 | ucp4 | |
| nmol of substrate/min/mg of protein | ||
| Complexes I–III | 923 ± 50 (n = 3) | 764 ± 139 (n = 5) |
| Complex II | 42 ± 6 (n = 3) | 59 ± 15 (n = 5) |
| Succinate dehydrogenase | 25 ± 2 (n = 3) | 99 ± 32 (n = 5)a |
| Complexes II and III | 124 ± 4 (n = 3) | 147 ± 35 (n = 5) |
| Complex III | 1681 ± 415 (n = 3) | 1673 ± 608 (n = 5) |
| Complex IV | 233,343 ± 31,134 (n = 3) | 248,740 ± 17,634 (n = 3) |
| Cytochrome c reduction |
||
|---|---|---|
| N2 | ucp4 | |
| A550 − A540/min/mg of protein | ||
| Uninhibited | 3.1 ± 0.15 (n = 3) | 2.9 ± 0.05 (n = 3) |
| Rotenone | 2.9 ± 0.14 (n = 3) | 2.6 ± 0.19 (n = 3) |
| Malonate | 1.7 ± 0.12 (n = 3) | 1.9 ± 0.13 (n = 3) |
a Significantly different from N2 (p < 0.05).
ceUCP4 Regulates Mitochondrial Succinate Import
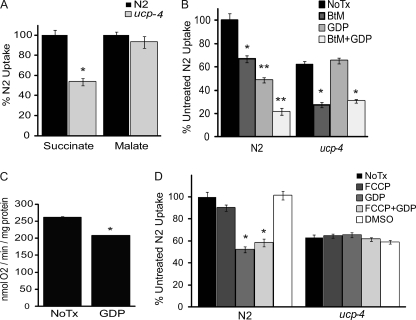
The lack of any apparent defect in succinate-mediated complex II function in permeable ucp4 mitochondria indicated that their failure to respire on succinate may involve a substrate transport mechanism. We quantified the uptake of [14C]succinate and [14C]malate in isolated N2 and ucp4 mitochondria. Compared with N2, malate uptake was normal in ucp4 mitochondria, but succinate uptake was significantly decreased by 43% (Fig. 3A). The ceUCP4-specific effect on succinate uptake was surprising because in a variety of eukaryotes, both succinate and malate transport are believed to be mediated simultaneously by the dicarboxylic acid carrier (DIC) (24, 25).
FIGURE 3.
ceUCP4 regulates mitochondrial succinate uptake. A, shown is [14C]succinate and [14C]malate uptake (mean ± S.E.) in isolated mitochondria from N2 and ucp4 worms (n = 5–7). * indicates significantly different (p < 0.01) from N2. B, [14C]succinate uptake in isolated mitochondria from N2 and ucp4 worms alone (No Tx) or in the presence of the dicarboxylate carrier inhibitor BtM, the UCP inhibitor guanosine nucleotide (GDP), or both (BtM + GDP) is shown. * and ** indicate significantly different (p < 0.01 and p < 0.001, respectively) from the untreated N2 mitochondria. C, shown is the effect of preaddition of GDP on succinate-induced respiration in N2 worms (mean ± S.E., n = 3). D, [14C]succinate uptake in isolated mitochondria from N2 and ucp4 worms (n = 3–4) alone (No Tx) or in the presence of the mitochondrial uncoupler FCCP, the UCP inhibitor GDP, both (FCCP + GDP), or vehicle (DMSO) (n = 3) is shown. * indicates significantly different (p < 0.01) from untreated N2 mitochondria.
We used a pharmacologic approach to further define the specific contributions of ceUCP4 and the DIC to mitochondrial succinate uptake in N2 mitochondria. In the presence of the DIC-specific inhibitor n-butylmalonate (BtM), succinate uptake in N2 mitochondria was decreased by 33%, whereas the UCP inhibitor guanosine diphosphate (GDP) inhibited uptake by 48%. Remarkably, co-treatment of mitochondria with BtM and GDP led to an additive 80% decrease in succinate uptake (Fig. 3B), suggesting that ceUCP4 regulates an alternative succinate import pathway distinct from the canonical dicarboxylate carrier. This notion is further supported by complementary experiments in ucp4 mitochondria. GDP failed to inhibit succinate transport in ucp4 mitochondria, but BtM decreased transport in these mitochondria to a similar extent (∼75%) to that observed in N2 mitochondria co-treated with BtM and GDP (Fig. 3B). Because guanosine nucleotides may also inhibit other mitochondrial carriers in other animals (26), these results imply that the effects of GDP on succinate transport in C. elegans mitochondria are largely due to the specific inhibition of UCP.
The lack of guanosine nucleotide effects on uncoupled, state 4 respiration were defined in experiments where the inhibitors were added to the mitochondria after the exhaustion of ADP (or in the presence of oligomycin) to halt phosphorylating respiration. As shown (Fig. 2), guanine nucleotides failed to affect state 4 respiration in N2 (and ucp4) mitochondria when added after respiratory substrates (e.g. succinate). In contrast, GDP pretreatment before substrate addition significantly inhibited succinate-mediated, maximal state 3 respiration in N2 mitochondria in a dose-dependent manner (Fig. 3C; supplemental Fig. S5B). However, consistent with the normal complex II function in permeable ucp4 mitochondria, GDP failed to inhibit succinate-driven complex II function in permeabilized N2 mitochondria (supplemental Fig. S5A).
A hallmark property of uncoupling proteins is to diminish membrane potential by uncoupling proton transport from ATP synthesis. Although the bulk of our observations imply that ceUCP4 is not a prototypical uncoupler, it remains possible that loss of ceUCP4 may affect succinate transport indirectly by increasing the mitochondrial membrane potential. To examine this possibility, we quantified succinate uptake in isolated mitochondria in the presence of the chemical uncoupler FCCP at a concentration established to decrease mitochondrial membrane potential (18). FCCP failed to rescue succinate import in ucp4 mitochondria and failed to override GDP inhibition of succinate flux in N2 mitochondria (Fig. 3D). Together, these data suggest that ceUCP4-regulated succinate transport is independent of the dicarboxylate carrier and mitochondrial membrane potential.
ceUCP4 Regulates Complex II Function in Vivo
Electrons derived from substrate oxidation primarily enter the electron transport chain through complex I or complex II to drive OXPHOS. Pharmacologic and genetic experiments were performed to scrutinize the in vivo relevancy of ucp4-regulated complex II-driven OXPHOS and succinate transport. N2 and ucp4 L2 worms were treated with the complex I inhibitor rotenone and measured survival after 48 h. Rotenone exposure slightly decreased the survival of N2 animals but led to a striking 70% decrease in the survival of ucp4 animals (Fig. 4A). Similarly, attempts to generate the double mutant ucp4;gas1 failed (data not shown). mev1 worms express a mutation in the cytochrome b subunit of succinate dehydrogenase that results in electron slippage from complex II, leading to the partial reduction of molecular oxygen to form deleterious reactive oxygen species (27). This mutation leads to a dramatically decreased lifespan, ostensibly from the overproduction of complex II-derived mitochondrial oxidants. If ceUCP4 regulates succinate availability for complex II function in vivo, its absence should be protective in the setting of complex II-generated pathology. To test this idea, we generated the double mutant strain mev1:ucp4 and conducted lifespan comparisons with N2, ucp4, and mev1 worms. As previously observed, ucp4 animals had comparable and mev1 had significantly shorter lifespans compared with N2 (28). Notably, a significant rescue of lifespan was observed in the mev1:ucp4 double mutant strain (Fig. 4B). A similar effect was observed when worm fecundity was measured. mev1:ucp4 animals still produced fewer viable offspring than N2 and ucp4 strains but generated 35% more progeny than mev1 worms (Fig. 4C).
DISCUSSION
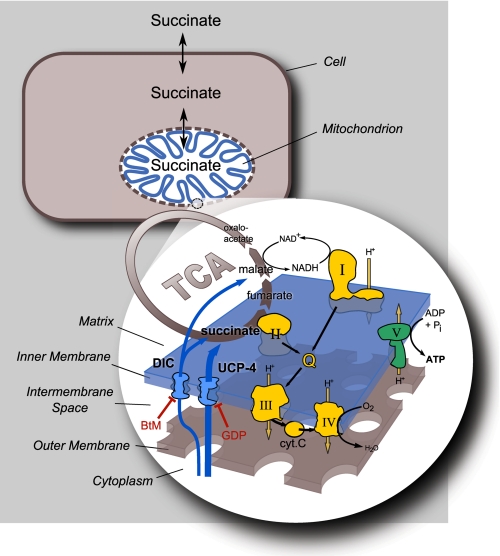
Consistent with their pivotal roles in the regulation of metabolic respiratory efficiency, ucp1–3 are linked in numerous clinical studies with body weight changes and obesity susceptibility. However, in keeping with the hypothesis that ucp4 and ucp5 may not regulate canonical uncoupling, no similar associations have been reported for ucp4 and ucp5 (29). Interestingly, single nucleotide polymorphisms (rs10807344 CC) in ucp4 have been linked to protection against both multiple sclerosis susceptibility and brain leukoaraiosis (30, 31), and others link the same ucp4 mutation to increased schizophrenia incidence (32). We found that mutant ucp4 nematodes exhibited decreased respiration and increased levels of stored triglycerides and several fatty acids species when compared with N2. Surprisingly, the hypometabolic phenotypes of ucp4 animals did not correspond to hyper-coupling of their mitochondria (e.g. decreased state 4 respiration, increased respiratory control). Instead, ucp4 worms displayed defective succinate respiration and fatty acid accumulation phenotypes that closely resembled those observed in the complex II mutant mev1 worm (Fig. 2, supplemental Figs. S6). Observations in isolated mitochondria and intact worms support a model wherein ceUCP4 regulates C. elegans complex II-driven OXPHOS by controlling the anaplerotic mitochondrial import of succinate independently from the canonical dicarboxylate carrier DIC (Fig. 5). To our knowledge, this work identifies for the first time a pathway of mitochondrial succinate import that is controlled by an uncoupling protein family member and that in turn regulates the function of complex II.
FIGURE 5.
Model of UCP4 function. ceUCP4-null mitochondria exhibit a striking decrease in complex II-driven OXPHOS without any other defects in other respiratory chain components, and these defects disappear when mitochondria are permeabilized, suggesting that UCP4 may regulate complex II by regulating mitochondrial succinate import. The main proposed mediator of dicarboxylate (succinate and malate) transport in mitochondria is via the BtM-inhibited DIC. Although malate respiration and transport are not regulated by UCP4 (Figs. 1A and 2A, respectively), maximal rates of succinate respiration and uptake into mitochondria are strongly decreased in the absence of ucp4 (Figs. 1B and 2B, respectively). Moreover, the UCP inhibitor GDP significantly inhibits succinate influx into N2 mitochondria (Fig. 3A). Importantly, intact ucp4 nematodes are far less sensitive compared with wild type to loss of complex I function, which argues that UCP4-mediated succinate uptake from the cytoplasm is required for optimal complex II function in vivo. cyt c, cytochrome c.
Mammalian UCP1–3 and plant PUMP1 regulate fatty acid-activated, guanine nucleotide-inhibited mitochondrial proton leak, a function that is widely considered diagnostic of classical uncoupling protein activity (33). Based on phylogenetic analyses, Hanák and co-workers (5, 34) postulated that UCP4 is a distant, “ancestral” ortholog of UCP1 that may not function in uncoupling because it lacks the signature amino acid residues near the central matrix loop postulated to be necessary for fatty acid-induced UCP1-mediated uncoupling. More recent studies suggest that a novel invertebrate UCP6 ortholog in Anopheles gambiae gave rise to three distinct, functionally divergent UCP clades (UCP4, UCP5, and UCP1–3) early in evolution before the emergence of protostomes and deuterostomes (35). Together, these studies indicate that proton leak-induced thermogenesis may be an evolutionarily recent functional specialization in the UCP1–3 clade and imply that UCP4 and UCP5 may function differently from the more recently evolved orthologs. The hypometabolic phenotypes (decreased respiration, increased adiposity) we observed in ucp4 compared with N2 worms are consistent with the notion that ceUCP4 has functional conservation with mammalian UCP1–3 in the regulation of mitochondrial uncoupling and proton leak (Fig. 1). However, the lack of any discernable differences in state 4 respiration rates between ucp4 and N2 mitochondria and in response to chemical UCP activators and inhibitors in N2 mitochondria supports the notion that prototypical uncoupling may not be the primary mechanism through which ceUCP4 controls worm metabolic physiology (Fig. 2).
Combined with the aforementioned phylogenetic studies, our observations raise questions about whether a physiological proton leak pathway exists in nematode mitochondria and, if so, the identity of the mediator(s) and mechanisms involved. We cannot fully rule out that ceUCP4 may regulate uncoupling in the intact animal under normal living conditions (e.g. soil/low oxygen conditions) or under conditions not used in our studies. UCPregulated proton leak is thought to provide an adaptive mechanism to mitigate the reactive oxidant stress induced by oxygen radicals formed during aerobic respiration (33). It is possible that the relatively low oxygen environment (2–3 versus 21% ambient O2) of soil nematodes failed to provide sufficient selection pressure for the evolution of an uncoupling mechanism in nematodes. Alternatively, most of the more than 30 mitochondrial solute carriers expressed in C. elegans have not been characterized, and one or more of these may mediate uncoupling in the worm in a manner not involving canonical UCP sequence signatures. More work is needed to define the transport and metabolic functions of these nematode mitochondrial transporters.
Cell signaling by tricarboxylic acid cycle intermediates (TCAi) and the regulation of TCAi transport into (anaplerosis) and out of (cataplerosis) the mitochondrial matrix is an important mechanism linking mitochondrial metabolism to cell function and vice versa (36). Numerous functionally diverse mitochondrial solute carrier proteins (∼32 in C. elegans) regulate this complex subcellular compartmental cross-talk by transporting not only TCAi but also a variety of other solutes (phosphate) and small molecules (ATP/ADP) across the inner mitochondrial membrane. TCAi cataplerosis is well established to be critical for a variety of cytoplasmic biosynthetic functions, including gluconeogenesis and the synthesis of fatty acids and nonessential amino acids (36). Cancer cells typically exhibit up-regulated cataplerosis and diminished mitochondrial OXPHOS, a phenotype proposed to increase the mitochondrial export of TCAi substrates necessary to meet the biosynthetic needs (lipid, nucleotide, protein, etc.) of rampant proliferation (37). In concert, TCAi anaplerosis balances the efflux of carbon from the TCA cycle and maintains homeostatic levels of TCA cycle substrates. Although various biochemical aspects of anaplerotic and cataplerotic reactions have been thoroughly described, relatively less is known about the physiological significance of, and mechanisms regulating these processes in eukaryotes.
UCPs have been demonstrated to transport a variety of anions, including diverse fatty acids, pyruvate, and chloride across the inner mitochondrial membrane, but their roles in anaplerosis and cataplerosis have not been defined (38–40). The roughly 45% decrease in succinate uptake in ucp4 mitochondria corresponded to a roughly 70% reduction in succinate-mediated, complex II-driven OXPHOS (Figs. 3 and 4). However, although ucp4 was fatter than N2, consistent with observations from (41), no changes in lifespan or fecundity were observed in ucp4 animals under normal growth conditions. Thus, compensatory metabolic mechanisms may maintain reproductive success and lifespan in ucp4 worms, as implied by experiments in complex I (gas1) and complex II (mev1) mutants showing increased respiration by the unaffected complex (Fig. 2, E and F). Indeed, prevention of compensation through complex I in ucp4 led to marked lethality (Fig. 4). Similar experiments have shown that RNAi knockdown of complex II components led to increased lethality in complex I mutant gas11 worms (42). The defects in succinate import and complex II-mediated OXPHOS observed in ucp4 mitochondria corresponded with decreased metabolic rate and increased fat accumulation in ucp4 worms in vivo. Interestingly, both ucp4 and mev1 strains exhibited a similar defect in succinate-mediated OXPHOS (Fig. 2F) and a strong overlapping pattern of fatty acid accumulation compared with N2 (supplemental Fig. S6). These observations imply that ceUCP4 regulates adiposity in intact worms, presumably through succinate transport-dependent complex II function.
Experiments in cholate-solubilized N2 and ucp4 mitochondria provide strong support for the argument that ceUCP4 controls complex II function through the regulation of mitochondrial succinate import. Unlike observations in intact mitochondria, no effects on succinate-driven complex II activity were observed in solubilized mitochondria (wherein succinate can reach the complex II binding site) in the absence of ceUCP4 (Table 1) or in the presence of GDP in N2 mitochondria (supplemental Fig. S5). Interestingly, total loss of ceUCP4 protein led to roughly similar decreases in mitochondrial succinate uptake (∼45%, Fig. 3) and succinate-induced oxygen consumption (∼ 60%, Fig. 2). However, whereas GDP inhibited mitochondrial succinate uptake in N2 mitochondria to a similar degree as observed in untreated ucp4 mitochondria, GDP only inhibited succinate-induced respiration in wild type worms by ∼ 20%, although the effect was significant (Fig. 3). One interpretation of the relatively decreased efficacy of ceUCP4 inhibition versus ceUCP4 deletion on succinate-induced respiration could be that GDP and succinate may interact with ceUCP4 (or a transport-regulating binding partner) competitively at similar binding sites. Although well beyond the scope of the present study, work using liposomes and reconstituted wild type and mutant ceUCP4 and DIC transporters will be required to understand the precise mechanisms and kinetics underlying the ceUCP4-regulated transport of succinate and to define the impact, if any, of succinate flux on proton transport.
Importantly, the requirement of ceUCP4-regulated succinate import for optimal complex II-mediated metabolism is supported by animal data. UCPs in other organisms are canonically activated by oxidants and in turn increase mitochondrial inner membrane proton leak (2). As mentioned above, the short-lived mev1 worm expresses a loss of function mutation in subunit C of succinate dehydrogenase that leads to the deleterious over-production of succinate-generated, complex II-derived ROS (28, 43, 44). The mev1:ucp4 double mutant exhibited an approximate 50% rescue of the decrease in lifespan induced by the mev1 mutation alone (Fig. 4). This phenotype is opposite to what would be predicted if ceUCP4 regulated classical oxidant-induced, mitochondrial uncoupling in vivo, which is associated with decreased oxidant production and life extension (45). If, on the other hand, ceUCP4 functions predominantly in succinate anaplerosis, one would predict that the succinate-driven decrease in lifespan of the mev1 strain would be diminished in the absence of ceUCP4 (mev1:ucp4) because of decreased mitochondrial succinate availability and consequent oxidant generation. Thus, the toxic and protective responses observed in ucp4 worms in the setting of complex I and II inhibition, respectively, are in keeping with the notion that ceUCP4 plays a vital role in complex II-dependent OXPHOS in the intact animal.
UCPs are increasingly implicated in the control of diverse cellular processes, including synaptic transmission, insulin sensitivity and secretion, and thermoregulation, ostensibly through the regulation of the mitochondrial transport of protons, fatty acids, and reactive oxidant species (2). Interestingly, recent studies in mammalian systems implicate succinate as a second messenger for physiological signal transduction pathways and as a pathophysiologic mediator linking mitochondrial dysfunction to disease. For example, succinate is present in the general circulation at low micromolar concentrations and is the ligand activator for the widely expressed G-protein coupled receptor GPR91, a key mediator of the renal control of blood pressure (46) also involved in retinal angiogenesis (47), hematopoiesis (48), and immunity (49). Succinate also functions as a transcriptional regulator linking mitochondrial function with circadian rhythms in yeast (50) and is a key messenger linking cellular nutrient status with ribosomal RNA production and ribosomal biogenesis (51). In hereditary paragangliomas arising from mutations in the genes encoding subunits of succinate dehydrogenase, cytoplasmic succinate accumulation results from mitochondrial dysfunction, leads to the normoxic stabilization of the cytoplasmic oncogene transcription factor Hif-1α, and promotes tumorigenesis (52). The observations that ceUCP4 optimizes complex II-dependent metabolism in nematodes through its capacity to regulate mitochondrial succinate flux may illuminate a novel UCPdependent mechanism regulating the cross-talk between mitochondrial metabolism and distant cellular functions.
In summary, these studies are the first to provide a detailed characterization of the mitochondrial functions of the sole predicted uncoupling protein in the worm. The significant associations between mammalian UCPs and metabolic disease/aging in experimental and clinical studies are unfortunately met with incomplete understanding of their biochemical functions in mitochondria. Thus, these studies in the simple organism C. elegans lay the foundation for future work aimed at a deeper understanding of the mechanistic underpinnings and physiological outcomes of ceUCP4-mediated succinate transport and complex II regulation and the relevancy of these observations for other UCP homologs.
Acknowledgments
We greatly appreciate the technical assistance of M. Krause, G. Fang, M. Person, M. Falk, K. Smith, and S. Nowinski. We also thank J. E. Sprague, C. L. Hoppel, S. Bratton, C. Wright, K. Hirasaka, J. Basser, T. Terrasso, M. Malladi, and S. Malladi for helpful comments on experiments, data analysis, and manuscript preparation. We thank the Caenorhabditis Genetics Center and the Gene Knock-out Consortium for the nematode strains.
This work was supported, in whole or in part, by National Institutes of Health Grants DK089224 (to E. M. M.), GM58881 and AG026073 (to P. G. M., M. M. S., and E.-B. K.), and GMS 45952 (to C. F. C.), and Training Grant ES007247 (to M. E. P).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- UCP
- uncoupling proteins
- FCCP
- carbonylcyanide-p-trifluoromethoxyphenylhydrazone
- DIC
- dicarboxylic acid carrier
- BtM
- n-butylmalonate
- ceUCP4
- C. elegans mitochondrial uncoupling protein 4
- OXPHOS
- oxidative phosphorylation
- TCAi
- tricarboxylic acid cycle intermediate.
REFERENCES
- 1. Cannon B., Hedin A., Nedergaard J. (1982) FEBS Lett. 150, 129–132 [DOI] [PubMed] [Google Scholar]
- 2. Krauss S., Zhang C. Y., Lowell B. B. (2005) Nat. Rev. Mol. Cell Biol. 6, 248–261 [DOI] [PubMed] [Google Scholar]
- 3. Heaton G. M., Wagenvoord R. J., Kemp A., Jr., Nicholls D. G. (1978) Eur. J. Biochem. 82, 515–521 [DOI] [PubMed] [Google Scholar]
- 4. Enerback S., Jacobsson A., Simpson E. M., Guerra C., Yamashita H., Harper M. E., Kozak L. P. (1997) Nature 387, 90–94 [DOI] [PubMed] [Google Scholar]
- 5. Hanák P., Jezek P. (2001) FEBS Lett. 495, 137–141 [DOI] [PubMed] [Google Scholar]
- 6. Mao W., Yu X. X., Zhong A., Li W., Brush J., Sherwood S. W., Adams S. H., Pan G. (1999) FEBS Lett. 443, 326–330 [DOI] [PubMed] [Google Scholar]
- 7. Robinson A. J., Overy C., Kunji E. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17766–17771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., Liu T., Yip K. Y., Robilotto R., Rechtsteiner A., Ikegami K., Alves P., Chateigner A., Perry M., Morris M., Auerbach R. K., Feng X., Leng J., Vielle A., Niu W., Rhrissorrakrai K., Agarwal A., Alexander R. P., Barber G., Brdlik C. M., Brennan J., Brouillet J. J., Carr A., Cheung M. S., Clawson H., Contrino S., Dannenberg L. O., Dernburg A. F., Desai A., Dick L., Dosé A. C., Du J., Egelhofer T., Ercan S., Euskirchen G., Ewing B., Feingold E. A., Gassmann R., Good P. J., Green P., Gullier F., Gutwein M., Guyer M. S., Habegger L., Han T., Henikoff J. G., Henz S. R., Hinrichs A., Holster H., Hyman T., Iniguez A. L., Janette J., Jensen M., Kato M., Kent W. J., Kephart E., Khivansara V., Khurana E., Kim J. K., Kolasinska-Zwierz P., Lai E. C., Latorre I., Leahey A., Lewis S., Lloyd P., Lochovsky L., Lowdon R. F., Lubling Y., Lyne R., MacCoss M., Mackowiak S. D., Mangone M., McKay S., Mecenas D., Merrihew G., Miller D. M., 3rd, Muroyama A., Murray J. I., Ooi S. L., Pham H., Phippen T., Preston E. A., Rajewsky N., Rätsch G., Rosenbaum H., Rozowsky J., Rutherford K., Ruzanov P., Sarov M., Sasidharan R., Sboner A., Scheid P., Segal E., Shin H., Shou C., Slack F. J., Slightam C., Smith R., Spencer W. C., Stinson E. O., Taing S., Takasaki T., Vafeados D., Voronina K., Wang G., Washington N. L., Whittle C. M., Wu B., Yan K. K., Zeller G., Zha Z., Zhong M., Zhou X., Ahringer J., Strome S., Gunsalus K. C., Micklem G., Liu X. S., Reinke V., Kim S. K., Hillier L. W., Henikoff S., Piano F., Snyder M., Stein L., Lieb J. D., Waterston R. H. (2010) Science 330, 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKay R. M., McKay J. P., Avery L., Graff J. M. (2003) Dev. Cell 4, 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kayser E. B., Morgan P. G., Hoppel C. L., Sedensky M. M. (2001) J. Biol. Chem. 276, 20551–20558 [DOI] [PubMed] [Google Scholar]
- 11. Kayser E. B., Sedensky M. M., Morgan P. G., Hoppel C. L. (2004) J. Biol. Chem. 279, 54479–54486 [DOI] [PubMed] [Google Scholar]
- 12. Lippitt B., McCord J. M., Fridovich I. (1972) J. Biol. Chem. 247, 4688–4690 [PubMed] [Google Scholar]
- 13. Brand M. D., Pakay J. L., Ocloo A., Kokoszka J., Wallace D. C., Brookes P. S., Cornwall E. J. (2005) Biochem. J. 392, 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoppel C., DiMarco J. P., Tandler B. (1979) J. Biol. Chem. 254, 4164–4170 [PubMed] [Google Scholar]
- 15. Mak H. Y., Nelson L. S., Basson M., Johnson C. D., Ruvkun G. (2006) Nat. Genet. 38, 363–368 [DOI] [PubMed] [Google Scholar]
- 16. Brock T. J., Browse J., Watts J. L. (2006) PLoS Genet. 2, e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jonassen T., Marbois B. N., Faull K. F., Clarke C. F., Larsen P. L. (2002) J. Biol. Chem. 277, 45020–45027 [DOI] [PubMed] [Google Scholar]
- 18. Talbot D. A., Lambert A. J., Brand M. D. (2004) FEBS Lett. 556, 111–115 [DOI] [PubMed] [Google Scholar]
- 19. Hartman P. S., Ishii N., Kayser E. B., Morgan P. G., Sedensky M. M. (2001) Mech. Ageing Dev. 122, 1187–1201 [DOI] [PubMed] [Google Scholar]
- 20. Jaburek M., Garlid K. D. (2003) J. Biol. Chem. 278, 25825–25831 [DOI] [PubMed] [Google Scholar]
- 21. Ivanova M. V., Hoang T., McSorley F. R., Krnac G., Smith M. D., Jelokhani-Niaraki M. (2010) Biochemistry 49, 512–521 [DOI] [PubMed] [Google Scholar]
- 22. Jonassen T., Larsen P. L., Clarke C. F. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Falk M. J., Rosenjack J. R., Polyak E., Suthammarak W., Chen Z., Morgan P. G., Sedensky M. M. (2009) PLoS One 4, e6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kaplan R. S., Pedersen P. L. (1985) J. Biol. Chem. 260, 10293–10298 [PubMed] [Google Scholar]
- 25. Palmieri L., Picault N., Arrigoni R., Besin E., Palmieri F., Hodges M. (2008) Biochem. J. 410, 621–629 [DOI] [PubMed] [Google Scholar]
- 26. Parker N., Vidal-Puig A., Brand M. D. (2008) Biosci. Rep. 28, 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishii N., Takahashi K., Tomita S., Keino T., Honda S., Yoshino K., Suzuki K. (1990) Mutat. Res. 237, 165–171 [DOI] [PubMed] [Google Scholar]
- 28. Ishii N., Fujii M., Hartman P. S., Tsuda M., Yasuda K., Senoo-Matsuda N., Yanase S., Ayusawa D., Suzuki K. (1998) Nature 394, 694–697 [DOI] [PubMed] [Google Scholar]
- 29. Lee H. J., Ryu H. J., Shin H. D., Park B. L., Kim J. Y., Cho Y. M., Park K. S., Song J., Oh B. (2008) Clin. Chim. Acta 398, 27–33 [DOI] [PubMed] [Google Scholar]
- 30. Szolnoki Z., Kondacs A., Mandi Y., Bodor A., Somogyvari F. (2009) Neuromolecular Med. 11, 101–105 [DOI] [PubMed] [Google Scholar]
- 31. Szolnoki Z., Kondacs A., Mandi Y., Bodor A., Somogyvari F. (2011) Acta Neurol. Scand. 123, 352–357 [DOI] [PubMed] [Google Scholar]
- 32. Yasuno K., Ando S., Misumi S., Makino S., Kulski J. K., Muratake T., Kaneko N., Amagane H., Someya T., Inoko H., Suga H., Kanemoto K., Tamiya G. (2007) Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 250–253 [DOI] [PubMed] [Google Scholar]
- 33. Jezek P., Jabůrek M., Garlid K. D. (2010) FEBS Lett. 584, 2135–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Urbánková E., Hanák P., Skobisová E., Růzicka M., Jezek P. (2003) Int. J. Biochem. Cell Biol. 35, 212–220 [DOI] [PubMed] [Google Scholar]
- 35. Sokolova I. M., Sokolov E. P. (2005) FEBS Lett. 579, 313–317 [DOI] [PubMed] [Google Scholar]
- 36. Owen O. E., Kalhan S. C., Hanson R. W. (2002) J. Biol. Chem. 277, 30409–30412 [DOI] [PubMed] [Google Scholar]
- 37. DeBerardinis R. J., Lum J. J., Hatzivassiliou G., Thompson C. B. (2008) Cell Metab. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 38. Palmieri F. (2004) Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 39. Klingenberg M., Winkler E., Echtay K. (2001) Biochem. Soc. Trans. 29, 806–811 [DOI] [PubMed] [Google Scholar]
- 40. Smorodchenko A., Rupprecht A., Sarilova I., Ninnemann O., Bräuer A. U., Franke K., Schumacher S., Techritz S., Nitsch R., Schuelke M., Pohl E. E. (2009) Biochim. Biophys. Acta 1788, 2309–2319 [DOI] [PubMed] [Google Scholar]
- 41. Iser W. B., Kim D., Bachman E., Wolkow C. (2005) Mech. Ageing Dev. 126, 1090–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ichimiya H., Huet R. G., Hartman P., Amino H., Kita K., Ishii N. (2002) Mitochondrion 2, 191–198 [DOI] [PubMed] [Google Scholar]
- 43. Ishii N., Ishii T., Hartman P. S. (2006) Exp. Gerontol. 41, 952–956 [DOI] [PubMed] [Google Scholar]
- 44. Senoo-Matsuda N., Yasuda K., Tsuda M., Ohkubo T., Yoshimura S., Nakazawa H., Hartman P. S., Ishii N. (2001) J. Biol. Chem. 276, 41553–41558 [DOI] [PubMed] [Google Scholar]
- 45. Speakman J. R., Talbot D. A., Selman C., Snart S., McLaren J. S., Redman P., Krol E., Jackson D. M., Johnson M. S., Brand M. D. (2004) Aging Cell 3, 87–95 [DOI] [PubMed] [Google Scholar]
- 46. Toma I., Kang J. J., Sipos A., Vargas S., Bansal E., Hanner F., Meer E., Peti-Peterdi J. (2008) J. Clin. Invest. 118, 2526–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sapieha P., Sirinyan M., Hamel D., Zaniolo K., Joyal J. S., Cho J. H., Honoré J. C., Kermorvant-Duchemin E., Varma D. R., Tremblay S., Leduc M., Rihakova L., Hardy P., Klein W. H., Mu X., Mamer O., Lachapelle P., Di Polo A., Beauséjour C., Andelfinger G., Mitchell G., Sennlaub F., Chemtob S. (2008) Nat. Med. 14, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 48. Hakak Y., Lehmann-Bruinsma K., Phillips S., Le T., Liaw C., Connolly D. T., Behan D. P. (2009) J. Leukoc. Biol. 85, 837–843 [DOI] [PubMed] [Google Scholar]
- 49. Rubic T., Lametschwandtner G., Jost S., Hinteregger S., Kund J., Carballido-Perrig N., Schwärzler C., Junt T., Voshol H., Meingassner J. G., Mao X., Werner G., Rot A., Carballido J. M. (2008) Nat. Immunol. 9, 1261–1269 [DOI] [PubMed] [Google Scholar]
- 50. Nakamichi N., Fukushima A., Kusano M., Sakakibara H., Mizuno T., Saito K. (2009) Plant Signal. Behav. 4, 660–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka Y., Okamoto K., Teye K., Umata T., Yamagiwa N., Suto Y., Zhang Y., Tsuneoka M. (2010) EMBO J. 29, 1510–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., Gottlieb E. (2005) Cancer Cell 7, 77–85 [DOI] [PubMed] [Google Scholar]