Abstract
The Zic transcription factors play critical roles during embryonic development. Mutations in the ZIC2 gene are associated with human holoprosencephaly, but the etiology is still unclear. Here, we report a novel function for ZIC2 as a regulator of β-catenin·TCF4-mediated transcription. We show that ZIC2 can bind directly to the DNA-binding high mobility group box of TCF4 via its zinc finger domain and inhibit the transcriptional activity of the β-catenin·TCF4 complex. However, the binding of TCF4 to DNA was not affected by ZIC2. Zic2 RNA injection completely inhibited β-catenin-induced axis duplication in Xenopus embryos and strongly blocked the ability of β-catenin to induce expression of known Wnt targets in animal caps. Moreover, Zic2 knockdown in transgenic Xenopus Wnt reporter embryos led to ectopic Wnt signaling activity mainly at the midbrain-hindbrain boundary. Together, our results demonstrate a previously unknown role for ZIC2 as a transcriptional regulator of the β-catenin·TCF4 complex.
Keywords: Development, Neurodifferentiation, T-cell Factor (TCF), Wnt Pathway, Xenopus, Holoprosencephaly, Zic2
Introduction
The Zic genes (Zic1–5) encode transcription factors that contain highly conserved C2H2 class zinc finger motifs. They are implicated in development of the dorsal neural tube and neural crest as well as somites and the cerebellum. Although Zic (zinc finger protein of cerebellum) proteins play analogous roles in early neural development, mutations in individual Zic genes result in diverse phenotypes such as cerebellar malformations in ZIC1 mutants (1), HPE2 in ZIC2 mutants (2), and left-right asymmetry in ZIC3 mutants (3). Among members of the Zic family, Zic2 is unique in that it is the only maternally expressed Zic gene, and it is the only one of the Zic genes known to be associated with both major forms of HPE: classic HPE and midline interhemispheric HPE (4). The mechanisms by which Zic2 defects affect brain development are largely unknown.
In Xenopus, Zic2 represses the expression of several Xnr (Xenopus Nodal-related) genes and decreases overall Nodal signaling (5). Moreover, a number of dorsally expressed wnt genes are induced by Zic2. For example, wnt1 is induced at the midbrain-hindbrain boundary (6), whereas wnt4 and wnt8b are induced at the forebrain-midbrain boundary and midbrain (7, 8). It has been suggested that Zic genes function as activators of Wnt signaling by acting directly on the expression of the Wnt ligands (9). However, the role of Zic2 in transcriptional activity of the β-catenin/Tcf complex, has not been studied in detail. Several studies have indicated a crucial role for Zic2 in neuroectodermal differentiation (10–13), the process that is also known to be regulated by Wnt antagonists Dkk1 (Dickkopf-1) and Sfrp2 (14, 15). Expression of Zic2 and inhibition of Wnt signaling have been shown to be required for the specification of anterior neural fates within the neural plate (13, 16–19). When Wnt/β-catenin signals are antagonized, neural progenitors are massively induced (10). These findings prompted us to explore the interaction of ZIC2 and Wnt signaling using in vitro and in vivo models. The results demonstrate a direct interaction of ZIC2 with TCF4 and identify ZIC2 as a negative regulator of canonical Wnt signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
The human embryonic kidney 293T cell line and colon cancer cell lines Caco-2, SW480, HCT116, and DLD-1 were obtained from American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
Plasmids
Human full-length ZIC2, a kind gift from Dr. Y. Yang (20), and ZIC2 deletions were subcloned into pcDNA3.1 (Invitrogen). pFLAG-CMV4-TCF4 and its deletion mutants were kindly provided by Dr. M. Idogawa (21). pFLAG-TCF4-DN101 was generated by PCR. The pCS2-LEFΔN-β-cateninΔN53 construct containing β-catenin residues 53–781 fused to LEF1 with a deletion of residues 7–264 was a kind gift from Dr. P. Vogt (22). The TOPflash reporter, which contains three optimal TCF-binding sites upstream of a minimal c-fos promoter that drives expression of the luciferase gene, and the FOPflash reporter, which contains critical nucleotide replacements within the binding elements, were obtained from Upstate. The human pcDNA3-TCF4E expression vector was a kind gift from Dr. Osamu Tetsu. Myc-pCS2+Zic2 was generated by subcloning the Xenopus Zic2 ORF (a generous gift from Dr. D. W. Houston) into the Myc-pCS2+ vector. The pCS2-Myc-Zic2-ΔTcf plasmid, which lacks the Tcf-binding site (amino acids 339–501), was generated by PCR. Myc-β-catenin in the pCS2+ plasmid was described previously (23). The sequences of all plasmids were verified by sequencing.
Western Blotting
Cells were lysed in lysis buffer (50 mm Tris HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100) supplemented with proteinase inhibitor mixture (Roche Applied Science) and phosphatase inhibitor mixture (Sigma). Protein samples were run on a 10% BisTris gel (Invitrogen) and electroblotted on Hybond-C nitrocellulose membranes (Amersham Biosciences). After incubation with antibody and washing steps, the blots were developed using ECL Western blotting detection reagent (Amersham Biosciences). The following antibodies were used: rabbit anti-ZIC2 polyclonal antibody (Sigma), mouse anti-TCF4 monoclonal antibody clone 6H5-3 (Millipore), anti-FLAG monoclonal antibody M2 and anti-Myc monoclonal antibody (Sigma), anti-β-catenin antibody (BD Transduction Laboratories), anti-GAPDH antibody (Abcam), and anti-cyclin D1 polyclonal antibody (Santa Cruz Biotechnology). For detection of Myc-tagged proteins in embryos, animal caps were dissected at stages 11.5–12 and lysed in radioimmune precipitation assay buffer supplemented with Phospho Stay solution (Novagen), PMSF (Bio-Rad), and protease inhibitor mixture.
ChIP
ChIP experiments were performed as described by Mahmoudi et al. (24). 293T cells were transfected with ZIC2 and TCF4 and, after 48 h, were cross-linked according to the protocol. A total of 5 μg of the indicated anti-TCF4 antibody (Santa Cruz Biotechnology) and ZIC2 (Millipore, Santa Cruz Biotechnology, and anti-ZIC2 antibody generated by Dr. S. Brown) were incubated with the sheared cross-linked chromatin and BSA-blocked protein G beads. Input and immunoprecipitated DNAs were subjected to quantitative PCR using primers designed against the Tcf-bound or unbound region as a negative control as described by Mahmoudi et al. (24). The ZIC2-bound region at the ApoE promoter was described previously (25).
Real-time Semiquantitative PCR Assay
RNA (1 μg) was reverse-transcribed into cDNA using Superscript transcriptase II (Invitrogen). The relative amount of gene transcripts was determined by real-time RT-PCR using the ABI 7500 system (Applied Biosystems). The PCR protocol was carried out as recommended by Applied Biosystems. Standard curves for targets and the housekeeping control gene were based upon the Ct (threshold cycle) values, and the relative concentrations of the standards and the relative concentrations for samples were calculated from the detected Ct values and the equation of the curves. Values obtained for targets were divided by the values of housekeeping genes to normalize for differences in reverse transcription. Genomic contamination of the samples was checked by no-amplification control samples, which did not contain reverse transcriptase enzyme during the cDNA preparation.3
Luciferase Reporter Assays
293T cells were transiently transfected in duplicates or triplicates with a combination of plasmids using FuGENE 6 (Roche Applied Science). The total DNA concentration for each transfection was matched with the empty vector. β-Galactosidase-expressing plasmid (20 ng/well) was cotransfected to normalize the transfection efficiency. 36 h after transfection, cells were washed with cold PBS and lysed in lysis buffer from Promega. Luciferase reporter enzyme activity (Promega) and β-galactosidase (Applied Biosystems) assays were performed according to the manufacturers' protocol. The results are the mean of at least three experiments.
Xenopus Embryos and Microinjections
Xenopus embryos were obtained from adult frogs by hormone-induced egg laying and in vitro fertilization using standard methods. Synthesis of capped RNA was performed with an mMESSAGE mMACHINE kit (Ambion, Austin, TX), and injection was carried out as described previously (26).
Immunoprecipitation
293T cells were transfected with pcDNA3.1-FLAG-ZIC2 and its truncation mutants together with pFLAG-CMV4-TCF4. After 48 h, cells were collected and lysed in lysis buffer containing proteinase inhibitor mixture and Phosphatase Inhibitor Mixture 1 (Sigma). After sonication, the mixture was centrifuged at 12,000 for 30 min. The cell lysates were precleared on protein G-Sepharose beads (Amersham Biosciences) at 4 °C for 3 h on a rotating wheel. Meanwhile, antibody against FLAG, ZIC2 (Sigma), or TCF4 (Upstate) was coupled to protein G-Sepharose beads during incubation on a rotating wheel at 4 °C for 3 h. The precleared cell lysates were divided over the antibody-coupled protein G-Sepharose beads and an equal amount of protein G-Sepharose beads coupled with IgG as a control. After several washing steps in TBS/Tween, the bound protein complexes were solubilized in sample buffer and analyzed by Western blotting using the appropriate antibody.
In Vitro Transcription/Translation
Different deletion peptides of ZIC2 and TCF4 were made using the TnT T7 quick coupled transcription/translation system (Promega). The synthesized peptide was used for pulldown experiments.
EMSA
EMSAs were performed using the LightShift chemiluminescent EMSA kit (Pierce) with slight modifications. Briefly, 5 μg of nuclear extract was preincubated with 50 ng/μl poly(dI-dC), 10 mm Tris-HCl (pH 7.5), 50 mm KCl, 1 mm DTT, 5 mm MgCl2, and 0.05% Nonidet P-40 for 10 min at room temperature. After preincubation, the samples were either directly used in the binding assay or incubated for 1 h at 4 °C with anti-TCF4 antibody. 30 fmol of biotin-labeled probe (Eurogentec) was added in a total volume of 20 μl, and the reaction mixture was incubated for 20 min at room temperature. Subsequently, the DNA·protein complexes were separated from the free probes by electrophoresis on a 6% nondenaturing polyacrylamide gel in 0.5× Tris borate/EDTA buffer at 4 °C. After transferring to a Hybond-C membrane (GE Healthcare), the blot was cross-linked at 120 mJ/cm2 using a Stratalinker cross-linker (Stratagene). The membrane was developed using a streptavidin-conjugated horseradish peroxidase system according to the manufacturer's instructions. The sequences of the upper strands of the oligonucleotides used were CGGGCTTTGATCTTTGCTTAA (TCF4-Wild-for) and CGGGCTTTGGCCTTTGCTTAA (TCF4-Mut-for).
DNA Affinity Precipitation
293T cells were transfected with indicated plasmids. 48 h after transfection, cells were lysed in lysis buffer containing proteinase inhibitors. After centrifugation at 10,000 × g for 20 min at 4 °C, the supernatant was precleared at 4 °C for 2 h with Strep-Tactin beads (IBA) and then incubated at 4 °C for 16 h with 300 ng of biotinylated double-stranded probe corresponding to the TCF4-binding sequence with wild-type sequence 5′-CGGGCTTTGATCTTTGCTTAA-3′ or mutant sequence 5′-CGGGCTTTGGCCTTTGCTTAA-3′. After several washing steps, precipitated DNA·protein complexes were analyzed by Western blotting.
RESULTS
ZIC2 Represses β-Catenin·TCF-mediated Transcriptional Activation of Reporter Constructs
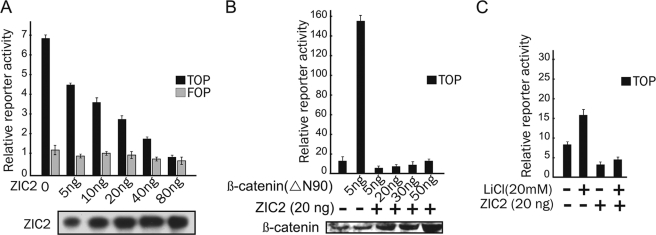
To determine whether ZIC2 interferes with β-catenin·TCF transcriptional activity, we studied the effect of ZIC2 on a luciferase reporter construct containing the wild-type consensus LEF/TCF-binding sites (TOPflash) or the same construct with mutant LEF/TCF-binding sites (FOPflash). ZIC2 overexpression in 293T cells, as well as in other cell lines such as Caco-2, SW480, HCT116, and DLD-1, consistently decreased the TOPflash reporter activity in a dose-dependent manner, whereas FOPflash reporter activity did not change (Fig. 1A and data not shown). ZIC2 strongly inhibited reporter activity even in the presence of excess β-catenin. In 293T cells, overexpression of a constitutively active form of β-catenin (ΔN90-β-catenin) induced TOPflash activity by 15-fold. This activation was significantly (95%) inhibited by coexpression of ZIC2 (Fig. 1B). Increasing amounts of ΔN90-β-catenin failed to abrogate the inhibition of reporter activity by ZIC2 (Fig. 1B). In addition to β-catenin activation by overexpression, ZIC2 significantly inhibited the TOPflash activity induced by LiCl treatment, which is known to stabilize endogenous β-catenin by inhibiting GSK3β activity (Fig. 1C). Together, these data show that ZIC2 overexpression is able to strongly abrogate β-catenin·TCF transcriptional activation of responsive promoters.
FIGURE 1.
ZIC2 negatively regulates the transcriptional activity of the β-catenin·TCF4 complex in 293T cells. A, activity of the TOPflash or FOPflash reporter in 293T cells transfected with ZIC2 expression vector in different amounts. B, TOPflash reporter activity of 293T cells transfected with ZIC2-expressing plasmid (20 ng) and increasing amounts of ΔN90-β-catenin-expressing plasmid (5–50 ng). The Western blot shows the levels of overexpressed proteins under each condition. C, TOPflash reporter activity of 293T cells treated with 20 mm LiCl for 12 h after transfection with the indicated plasmids.
ZIC2 Binds TCF4
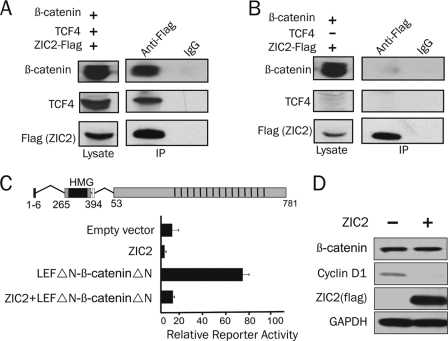
To investigate whether the observed regulatory effect of ZIC2 on the transcriptional activity of the β-catenin·TCF complex occurs via the interaction of ZIC2 with TCF4 and/or β-catenin, we performed immunoprecipitation using 293T cells transfected with expression vectors for ZIC2, TCF4, and β-catenin. ZIC2 was able to immunoprecipitate TCF4 (Fig. 2A). However, β-catenin was co-immunoprecipitated with ZIC2 only in the presence of ectopic TCF4 (Fig. 2B), suggesting that ZIC2 binds to TCF4 but not directly to β-catenin. Next, we sought to analyze if ZIC2-mediated repression of β-catenin·TCF transcriptional activity occurs through inhibition of the binding between β-catenin and TCF4. Therefore, we analyzed the effects of ZIC2 on a chimeric protein that directly fuses the N-terminally truncated form of LEF1, which lacks the β-catenin-binding domain and the two transactivation domains, to β-cateninΔN53, which lacks the N-terminal 53 amino acids, i.e. the destabilizing region of β-catenin (22). If ZIC2 inhibits β-catenin signaling by altering β-catenin binding to LEF/TCF, it should not inhibit the LEFΔN-β-cateninΔN53 fusion protein. The TOPflash reporter was activated by up to 6-fold after transfection with the fusion protein compared with the control, and activation of the TOPflash reporter was reduced to basal levels after ZIC2 cotransfection (Fig. 2C), suggesting that repression by ZIC2 does not rely on competition with β-catenin for LEF/TCF binding. Furthermore, to rule out any effect of ZIC2 on β-catenin protein levels, we analyzed expression of the protein in cells overexpressing ZIC2. As shown in Fig. 2D, ZIC2 overexpression decreased the level of cyclin D1 protein, a well known Wnt target, but had no effect on β-catenin protein levels, indicating that ZIC2 can inhibit a Wnt target gene and that this effect is not due to β-catenin down-regulation or degradation. In addition, ZIC2 did not change the nuclear translocation of β-catenin (data not shown).
FIGURE 2.
ZIC2 binding to TCF4. Cell lysates of 293T cells transfected with ZIC2-FLAG- and β-catenin-expressing plasmids together with TCF4-expressing vector (A) or without TCF4 (B) were subjected to immunoprecipitation (IP) using anti-FLAG antibody. C, schematic representation of LEFΔN-β-cateninΔN53 fusion protein. The graph shows the TOPflash reporter activity of 293T cells after transfection with the expression plasmids as indicated. D, Western blot showing the levels of the indicated proteins after FLAG-tagged ZIC2 overexpression in 293T cells.
Characterization of ZIC2·TCF4-binding Domains
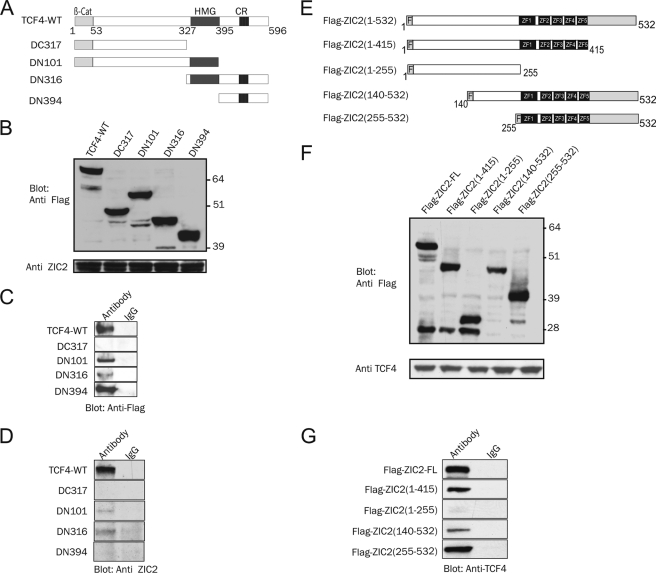
To map further the ZIC2·TCF4 interaction domains, we transfected 293T cells with a ZIC2 expression vector together with truncated forms of FLAG-tagged TCF4. Proteins were immunoprecipitated with anti-ZIC2 antibody (Fig. 3, A–C). In this experiment, full-length TCF4 and constructs carrying the HMG domain (DN101 and DN316), as well as a deletion mutant carrying only the C-terminal domain (DN394), were found to bind ZIC2, whereas the construct lacking both the HMG domain and the C-terminal domain (DC317) failed to co-immunoprecipitate ZIC2 (Fig. 3C). We next tried to confirm these results in a cell-free system by performing an in vitro protein binding assay. In vitro translated ZIC2 was incubated with FLAG-tagged TCF4 and its truncated forms purified with anti-FLAG beads. Only proteins carrying the HMG domain (full-length TCF4, DN101, and DN316) were found to bind ZIC2 (Fig. 3D), suggesting that the HMG domain of TCF4 is essential for direct interaction. We could not confirm the interaction between the C-terminal domain of TCF4 distal to the HMG domain (DN394) and ZIC2 in vitro. This suggests that the interaction of ZIC2 with the C terminus of TCF4 relies on another protein or a post-translational modification that is present in 293T cells but not in reticulocyte lysate. Next, we generated different ZIC2 deletion constructs (Fig. 3, E–G) and evaluated the ability of the truncated ZIC2 proteins to bind TCF4. Full-length ZIC2 and truncation mutants harboring the zinc finger domain of ZIC2 were able to bind to TCF4, whereas mutants lacking the zinc finger domain failed to interact (Fig. 3G), indicating that the zinc finger domain of ZIC2 is essential for TCF4 binding. Together, these data demonstrate that ZIC2 can bind to TCF4 and that the zinc finger domain of ZIC2 and the HMG domain of TCF4 are important for this interaction.
FIGURE 3.
Mapping the ZIC2·TCF4 interaction domains. A, schematic representation of the different TCF4 mutants. The HMG domain and C-terminal region (CR) are represented as dark gray boxes. The β-catenin-interacting domain (β-Cat) is represented as a light gray box. B, Western blot showing transient coexpression of FLAG-tagged TCF4, deletion mutants, and ZIC2 using the indicated antibodies. C, immunoprecipitated proteins using anti-ZIC2 antibody (or IgG as a mock) were subjected to Western blot analysis for FLAG detection. D, immunoblot of in vitro protein binding assay. FLAG-tagged TCF4 and its truncated forms were purified with anti-FLAG beads (with IgG serving as a mock) and incubated with in vitro translated ZIC2, and the precipitated complex was subsequently subjected to immunoblotting using antibody against ZIC2. E, schematic representation of ZIC2 and its deletion constructs. Zinc finger domains are represented as dark gray boxes. F indicates FLAG. F, Western blot showing the protein levels of overexpressed full-length ZIC2, ZIC2 deletions, and TCF4 using the indicated antibodies. G, proteins immunoprecipitated from the lysates in F using anti-FLAG antibody (or IgG as a mock) were analyzed by Western blotting using anti-TCF4 antibody.
ZIC2 Does Not Interfere with the Ability of TCF4 to Bind DNA
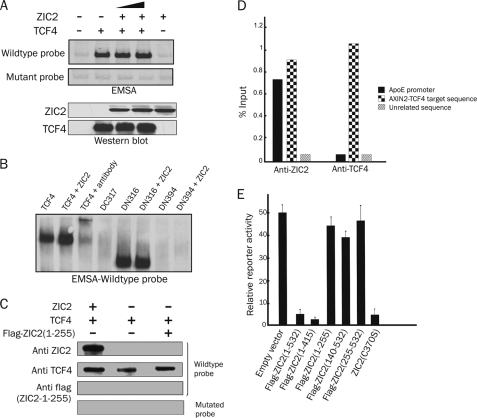
Because of the DNA-binding ability of the zinc finger domain of ZIC2 and the HMG domain of TCF4, we sought to determine whether ZIC2 binds to the TCF4 target sequence. To test this possibility, EMSA was performed using nuclear extracts from 293T cells transfected with ZIC2, TCF4, or TCF4 deletion mutants and incubated with a wild-type (ATCTTT) or mutant (GCCTTT) HMG consensus target sequence. ZIC2 neither bound nor changed the TCF4 binding affinity for the target sequence (Fig. 4A), suggesting that the effect of ZIC2 on the TCF4 complex is not due to inhibition of DNA binding. Only TCF4 and its mutants still harboring the HMG domain were found to specifically bind the probe (Fig. 4B). To confirm that ZIC2 was still bound to TCF4 in the presence of DNA, DNA precipitation assay was performed using DNA probes containing only consensus TCF4-binding sequences. As shown in Fig. 4C, full-length ZIC2 and TCF4 could be pulled down by the DNA probe, suggesting, first, that TCF4 can bind to the probe in the presence of ZIC2 and, second, that ZIC2 can be pulled down by the DNA via TCF4. To address whether TCF4 and ZIC2 are bound to TCF4-binding sites in vivo, we performed ChIP experiments. Using the TCF target sequence in the AXIN2 promoter as a readout in 293T cells, we could show binding of both ZIC2 and TCF4 to this region (Fig. 4D). The known binding sequence of ZIC2 for the ApoE promoter and the known binding sequence of TCF4 for the AXIN2 promoter were used as positive controls, and as a negative control, we used an unbound region of the AXIN2 promoter. ZIC2 enrichment was specific to the TCF4-bound region of AXIN2 and not the unbound downstream control region. These results together suggest that ZIC2 is likely a cofactor of the TCF4 transcriptional repression complex rather than being involved in DNA binding competition.
FIGURE 4.
Complex of ZIC2·TCF4 bound on DNA. A, EMSA was done by incubating a cell extract of 293T cells transfected with different expression plasmids as indicated with biotin-labeled probes containing wild-type or mutant TCF4-binding sequence. The lower panels represent Western blots of the corresponding samples as indicated. B, EMSA using 293T cells cotransfected with full-length TCF4 and its deletions together with ZIC2-expressing plasmid. Where indicated, the samples were preincubated with antibody against TCF4. C, DNA affinity precipitation of 293T cells transfected with the indicated expression vectors was performed as described under “Experimental Procedures.” The lysates were incubated with wild-type or mutant biotinylated probes corresponding to the TCF4-binding sequence, and the precipitated DNA·protein complexes were analyzed by Western blotting using the indicated antibodies. A probe with a mutant TCF-binding sequence was used as a control. D, 293T cells were transfected with ZIC2 and TCF4, and ChIP was performed with the indicated antibodies, followed by quantitative PCR using primer pairs spanning the human AXIN2 TCF4-binding site or an unrelated AXIN2 sequence as negative control. The ApoE promoter served as a positive control for ZIC2 binding. Results are presented as percentage of immunoprecipitated DNA over the input. E, luciferase reporter activity of the Wnt reporter (TOPflash) in 293T cells transfected with the expression vectors as indicated along with the β-catenin (DN90) expression vector.
ZIC2 Domains Associated with TCF4 Transcriptional Repression
To investigate which domains of ZIC2 contribute to TCF4 transcriptional repression, we mapped the repressor domains of ZIC2 protein using different ZIC2 deletion constructs and TOPflash as a reporter. As shown in Fig. 4E, only the constructs harboring the N-terminal domain as well as the zinc finger domain were able to repress the reporter. The repressor activity was somewhat stronger when the C-terminal tail of ZIC2 was deleted (ZIC2(1–415)). A C370S mutant, which can no longer bind DNA (27), was still able to bind TCF4 (data not shown) and repress TOPflash reporter activity, suggesting that the DNA-binding ability of ZIC2 is not required for TCF4 repression. Collectively, these data indicate that both the zinc finger and the N-terminal domain of ZIC2 are necessary for ZIC2 inhibition of the β-catenin·TCF4 transcriptional activity and that ZIC2 DNA-binding ability is not necessary for this function.
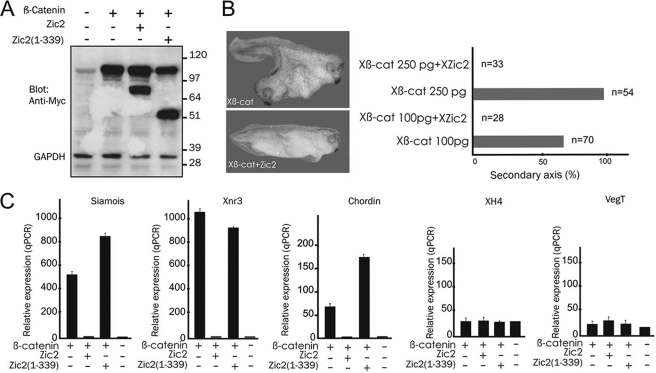
Zic2 Inhibits β-Catenin-induced Secondary Axis Formation in Xenopus and Expression of Wnt Target Genes in Animal Caps
We next examined whether Zic2 inhibits Wnt/β-catenin signaling in vivo. Dorsal axis formation in Xenopus embryos is a well established model for studying regulators of the canonical Wnt signaling pathway (28). Ectopic activation of this pathway in early embryos leads to double axis formation. Injection of β-catenin RNA (100 pg) into the ventral blastomeres of eight-cell stage embryos resulted in double axis formation in 70 of 98 embryos, whereas co-injection of Zic2 RNA (500 pg) completely blocked secondary axis formation in all embryos (28 of 28) (Fig. 5B). Secondary axis induction with higher doses of β-catenin (250 pg) was still completely blocked by Zic2 co-injection (33 of 33). We further studied whether Zic2 could down-regulate canonical Wnt target genes in Xenopus animal caps. Synthetic RNA of β-catenin was injected into the four animal blastomeres of eight-cell embryos. We documented induction of target genes of the canonical Wnt pathway, such as Siamois, Xnr3, and Chordin (29), by RT-PCR (Fig. 5C). Co-injection of Zic2 RNA led to a strong inhibition of Siamois and Xnr3 expression, whereas the transcript levels of the control genes, XH4 and VegT, remained unaffected. Zic2(1–339), a truncated form of Zic2 lacking the C-terminal domain and the last three zinc fingers, failed to inhibit β-catenin induction of Siamois and Xnr3.
FIGURE 5.
Regulation of Wnt signaling by Zic2 in Xenopus embryos. A, whole lysates of injected caps were subjected to Western blotting using anti-Myc antibody, showing bands of the expected sizes. B, representative appearance of embryos and the ratio of tadpoles with secondary axis formation. C, RT-PCR analysis of Wnt target genes (Siamois, Xnr3, and Chordin) compared with non-Wnt target genes (XH4 and VegT) in animal caps. The injections were repeated in at least three independent experiments with the same results. Xβ-cat, Xenopus β-catenin; XZic2, Xenopus Zic2; qPCR, quantitative PCR.
Zic2 Morpholino Knockdown Increases Wnt Reporter Activity in Transgenic Xenopus Embryos
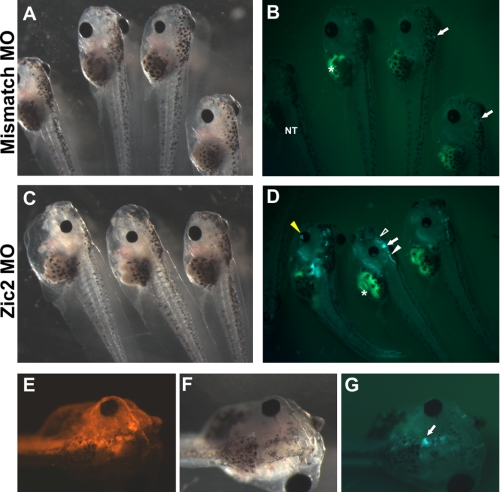
We next determined whether knockdown of Zic2 would expand TOP-GFP expression in transgenic Xenopus Wnt reporter embryos. This reporter line has already been shown to faithfully illustrate domains of β-catenin activity in responsive cell populations (30). For these experiments, we selected a transgenic line with relatively low GFP expression. To block the translation of Zic2, we injected morpholino antisense oligonucleotides targeting Zic2 as well as its corresponding control morpholino (mismatch morpholino) at the one-cell stage with 40 ng of morpholino/embryo. The Zic2 morpholino specifically blocked translation of the respective expression plasmids in reticulocyte lysates (data not shown). We examined Wnt reporter expression in late stages, focusing primarily on brain expression. Following injection of Zic2 morpholino, there was a striking increase in TOP-GFP expression in several derivatives of the CNS from the early tadpole stage (stage 35) onwards (87%, n = 15). This was particularly visible in the midbrain-hindbrain boundary, but GFP expression was also increased in the forebrain, dorsal hindbrain, and eye (shown at stage 43) (Fig. 6, compare B and D). Zic2 morpholino was also injected unilaterally at the two-cell stage. To mark the injected site, a red fluorescent tracer was co-injected. Embryos injected with Zic2 morpholino on one side clearly displayed increased unilateral GFP expression combined with a reduction in eye size and a smaller cartilaginous skeleton, resulting in bending of the embryonic axis toward the injected site (Fig. 6, E and F). Interestingly, the asymmetric GFP expression was lost around stage 45, i.e. 5 days after injection. In summary, these results indicate that Zic2 is required for the repression of Wnt activity in vivo, particularly in CNS derivatives.
FIGURE 6.
Zic2 morphants display increased Wnt reporter activity in the brain. Lateral views of stage 43 tadpoles injected with 40 ng of mismatch morpholino (MO) show normal development (A) and modest GFP expression (B). Embryos injected with Zic2 morpholino displayed loss of pigmented cells in the head and trunk (C) and markedly increased GFP expression (D), especially in the forebrain (open white arrowhead), the midbrain-hindbrain boundary (arrow), the dorsal side of the hindbrain (closed white arrowhead), and the eyes (closed yellow arrowhead). Note that the intestine shows autofluorescence (asterisk). The left-most tadpole is non-transgenic (NT). E–G, embryo injected unilaterally with Zic2 morpholino together with a red fluorescent LYSAMINE-labeled control morpholino. The injected side shows a marked increase in TOP-GFP expression, especially visible in the midbrain-hindbrain boundary (arrow).
DISCUSSION
Function of ZIC2 as a Repressor of the β-Catenin·TCF4 Complex
The findings of this study highlight a novel role for ZIC2 as an inhibitor of Wnt signaling. We have shown in mammalian cells and Xenopus embryos that ZIC2 overexpression inhibits the transcriptional activity of the β-catenin·Tcf complex and down-regulates the expression of direct Wnt target genes. Previous genetic studies in Drosophila suggest that opa (odd-paired), the homolog of zic, is required upstream of Wnt signaling for activation of wg (wingless) gene expression (31). However, our data add new information in that we found another mechanism by which Zic regulates downstream of Wnt signaling via inhibition of β-catenin·Tcf transcriptional activity. Hence, ZIC2 has a dual effect in regulation of Wnt signaling: induction of Wnt proteins and inhibition of β-catenin·Tcf transcriptional activity. Whether these effects occur in a cell autonomous versus non-autonomous manner remains unknown.
To study the interaction of ZIC2 and TCF4, because available anti-Zic2 Abs were inefficient to study the endogenous interaction by co-immunoprecipitation, we used FLAG-tagged proteins expressed in 293T cells. We found that the HMG domain of TCF4 and the zinc finger domain of ZIC2 are required for this interaction. It has been shown that the zinc finger domain of Zic proteins can also interact with DNA (32); however, our data strongly suggest that inhibition of the TCF4 complex by ZIC2 is not DNA-dependent. First, in EMSA, ZIC2 neither bound to the TCF4 target sequence nor altered the TCF4 DNA-binding affinity. Second, a single mutation in the zinc finger domain of ZIC2 that results in loss of DNA-binding ability (27) exhibited the same repressor activity as normal ZIC2 while still binding to TCF4. These data are in agreement with previous observations in Drosophila where the sequence-specific DNA-binding ability of Opa was not required for its transcriptional activation (33). In addition, our ChIP results clearly confirmed that both overexpressed ZIC2 and TCF4 were localized at the TCF4 target sequence of the AXIN2 promoter, a ubiquitous Wnt target. Together, these data suggest that ZIC2 inhibits the transcriptional activity of the TCF4 complex by direct binding to TCF4 and that this function is DNA-independent.
Potential Role of Other Interacting Proteins
There are limited data on the proteins that bind to ZIC2. Previous studies have shown that the N-terminal domain of ZIC2 interacts with I-mfa (inhibitor of MyoD family domain-containing protein) (34). It has been shown that I-mfa interacts with the HMG domain of Tcf3, thereby preventing the β-catenin·Tcf3 complex from binding to DNA (35). Deletion of the N-terminal domain of ZIC2 including the ZOC (Zic-Opa conserved) domain, a sequence conserved between mouse Zic1–3 and fly opa and known to interact with I-mfa, results in almost complete loss of repressor activity of ZIC2, suggesting that the N-terminal domain has significant repressor activity. To what extent I-mfa proteins are required for ZIC2 inhibition of the β-catenin·TCF4 complex remains unknown, particularly because most experiments were done on whole cells in which I-mfa would have been present. Moreover, the N-terminal domain must be combined with the zinc finger domain of ZIC2 to effectively repress the Wnt reporter activity. Thus, multiple domains within ZIC2 are required for observed repression. Further work needs to identify the proteins that interact with these domains.
When overexpressed in 293T cells, the C-terminal domain of TCF4 was found to interact with ZIC2, whereas the in vitro translated ZIC2 protein failed to interact with the C-terminal domain of TCF4. This suggests that modulation of the ZIC2 interaction by a factor(s) present in the 293T lysate or indirect binding via other proteins is involved in this interaction. Post-translational modifications cannot be excluded by our data and remain to be investigated. However, previous studies have identified that PARP-1 interacts with the C-terminal domain of TCF4 distal to the HMG domain (21). It has been shown independently that both PARP-1 and ZIC2 interact with the Ku70·Ku80·DNA-PKcs complex, known to be required for transcriptional regulation (32, 36). It is therefore possible that ZIC2, PARP-1, and TCF4 are present in the same complex regulating transcription. These data strongly suggest that ZIC2 can modulate Wnt signaling by regulation of the transcriptional activity of the β-catenin·TCF4 complex. We also examined in the TOPflash reporter system the interaction of ZIC2 with known coactivators (p300/CBP (cAMP-responsive element-binding protein-binding protein) and PARP-1 37)) and corepressors (CtBP (38), HDAC1 (histone deacetylase-1) (39), and Groucho/TLE (40)) of the β-catenin·Tcf complex. ZIC2 was able to strongly inhibit the inductive effect of the coactivators, whereas its effect on the corepressors could not be determined in this system due to the low TOPflash activity (data not shown). Further investigations are needed to determine the regulatory mechanisms behind ZIC2 inhibition of the β-catenin·TCF4 complex.
Relevance of Our Findings for Normal and Abnormal Brain Development
ZIC2 mutations were initially described in HPE, the most common structural anomaly of the developing forebrain in humans. It has been shown that Zic2 is involved in hindbrain development by contributing to the formation of transient segmented structures called rhombomeres, which are fated to become the cerebellum, pons, and medulla oblongata (41, 42). Our Zic2 morpholino knockdown experiment in transgenic Xenopus Wnt reporter embryos showed a robust induction of Wnt reporter activity in the midbrain-hindbrain boundary of injected frogs. Wnt signaling plays a crucial role in formation of the brain signaling center located at the midbrain-hindbrain boundary (43). This suggests that Zic2 may tune the activity of Wnt signaling in this region. A similar role has been suggested for SIX3, another HPE gene, as a repressor of Wnt1 expression in the anterior neuroectoderm (44). How ZIC2 is involved in the dynamic regulation of Wnt signaling during brain formation and its precise biologic role remain to be determined.
Acknowledgments
We are grateful to Drs. Mathijs Baens and Marc Tjwa for helpful discussions and Dr. Stephen A. Brown for providing the antibody.
The primer sequences used and real-time PCR conditions are available upon request.
- HPE
- holoprosencephaly
- TCF
- T cell factor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- LEF
- lymphoid enhancer factor
- HMG
- high mobility group.
REFERENCES
- 1. Aruga J., Minowa O., Yaginuma H., Kuno J., Nagai T., Noda T., Mikoshiba K. (1998) J. Neurosci. 18, 284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brown S. A., Warburton D., Brown L. Y., Yu C. Y., Roeder E. R., Stengel-Rutkowski S., Hennekam R. C., Muenke M. (1998) Nat. Genet. 20, 180–183 [DOI] [PubMed] [Google Scholar]
- 3. Gebbia M., Ferrero G. B., Pilia G., Bassi M. T., Aylsworth A., Penman-Splitt M., Bird L. M., Bamforth J. S., Burn J., Schlessinger D., Nelson D. L., Casey B. (1997) Nat. Genet. 17, 305–308 [DOI] [PubMed] [Google Scholar]
- 4. Warr N., Powles-Glover N., Chappell A., Robson J., Norris D., Arkell R. M. (2008) Hum. Mol. Genet. 17, 2986–2996 [DOI] [PubMed] [Google Scholar]
- 5. Houston D. W., Wylie C. (2005) Development 132, 4845–4855 [DOI] [PubMed] [Google Scholar]
- 6. Wolda S. L., Moody C. J., Moon R. T. (1993) Dev. Biol. 155, 46–57 [DOI] [PubMed] [Google Scholar]
- 7. Cui Y., Brown J. D., Moon R. T., Christian J. L. (1995) Development 121, 2177–2186 [DOI] [PubMed] [Google Scholar]
- 8. McGrew L. L., Otte A. P., Moon R. T. (1992) Development 115, 463–473 [DOI] [PubMed] [Google Scholar]
- 9. Merzdorf C. S., Sive H. L. (2006) Int. J. Dev. Biol. 50, 611–617 [DOI] [PubMed] [Google Scholar]
- 10. Heeg-Truesdell E., LaBonne C. (2006) Dev. Biol. 298, 71–86 [DOI] [PubMed] [Google Scholar]
- 11. Brown L., Brown S. (2009) Gene Expr. Patterns 9, 43–49 [DOI] [PubMed] [Google Scholar]
- 12. Inoue T., Ota M., Mikoshiba K., Aruga J. (2007) Dev. Biol. 306, 669–684 [DOI] [PubMed] [Google Scholar]
- 13. Aruga J., Mikoshiba K. (2011) Neurochemical Res. 36, 1286–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong X. B., Zhang C. (2009) In Vitro Cell. Dev. Biol. 45, 185–193 [DOI] [PubMed] [Google Scholar]
- 15. Aubert J., Dunstan H., Chambers I., Smith A. (2002) Nat. Biotechnol. 20, 1240–1245 [DOI] [PubMed] [Google Scholar]
- 16. Niehrs C. (1999) Trends Genet. 15, 314–319 [DOI] [PubMed] [Google Scholar]
- 17. Heisenberg C. P., Houart C., Take-Uchi M., Rauch G. J., Young N., Coutinho P., Masai I., Caneparo L., Concha M. L., Geisler R., Dale T. C., Wilson S. W., Stemple D. L. (2001) Genes Dev. 15, 1427–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kiecker C., Niehrs C. (2001) Development 128, 4189–4201 [DOI] [PubMed] [Google Scholar]
- 19. Houart C., Caneparo L., Heisenberg C., Barth K., Take-Uchi M., Wilson S. (2002) Neuron 35, 255–265 [DOI] [PubMed] [Google Scholar]
- 20. Yang Y., Hwang C. K., Junn E., Lee G., Mouradian M. M. (2000) J. Biol. Chem. 275, 38863–38869 [DOI] [PubMed] [Google Scholar]
- 21. Idogawa M., Yamada T., Honda K., Sato S., Imai K., Hirohashi S. (2005) Gastroenterology 128, 1919–1936 [DOI] [PubMed] [Google Scholar]
- 22. Aoki M., Hecht A., Kruse U., Kemler R., Vogt P. K. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vleminckx K., Kemler R., Hecht A. (1999) Mech. Dev. 81, 65–74 [DOI] [PubMed] [Google Scholar]
- 24. Mahmoudi T., Boj S. F., Hatzis P., Li V. S., Taouatas N., Vries R. G., Teunissen H., Begthel H., Korving J., Mohammed S., Heck A. J., Clevers H. (2010) PLoS Biol. 8, e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Salero E., Pérez-Sen R., Aruga J., Giménez C., Zafra F. (2001) J. Biol. Chem. 276, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 26. Bellefroid E. J., Bourguignon C., Hollemann T., Ma Q., Anderson D. J., Kintner C., Pieler T. (1996) Cell 87, 1191–1202 [DOI] [PubMed] [Google Scholar]
- 27. Brown L., Paraso M., Arkell R., Brown S. (2005) Hum. Mol. Genet. 14, 411–420 [DOI] [PubMed] [Google Scholar]
- 28. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 29. Mann B., Gelos M., Siedow A., Hanski M. L., Gratchev A., Ilyas M., Bodmer W. F., Moyer M. P., Riecken E. O., Buhr H. J., Hanski C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96, 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tran H. T., Sekkali B., Van Imschoot G., Janssens S., Vleminckx K. (2010) Proc. Natl. Acad. Sci. U. S. A. 107, 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benedyk M. J., Mullen J. R., DiNardo S. (1994) Genes Dev. 8, 105–117 [DOI] [PubMed] [Google Scholar]
- 32. Ishiguro A., Ideta M., Mikoshiba K., Chen D. J., Aruga J. (2007) J. Biol. Chem. 282, 9983–9995 [DOI] [PubMed] [Google Scholar]
- 33. Sen A., Stultz B. G., Lee H., Hursh D. A. (2010) Dev. Biol. 343, 167–177 [DOI] [PubMed] [Google Scholar]
- 34. Mizugishi K., Hatayama M., Tohmonda T., Ogawa M., Inoue T., Mikoshiba K., Aruga J. (2004) Biochem. Biophys. Res. Commun. 320, 233–240 [DOI] [PubMed] [Google Scholar]
- 35. Snider L., Thirlwell H., Miller J. R., Moon R. T., Groudine M., Tapscott S. J. (2001) Mol. Cell. Biol. 21, 1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Idogawa M., Masutani M., Shitashige M., Honda K., Tokino T., Shinomura Y., Imai K., Hirohashi S., Yamada T. (2007) Cancer Res. 67, 911–918 [DOI] [PubMed] [Google Scholar]
- 37. Belenkaya T. Y., Han C., Standley H. J., Lin X., Houston D. W., Heasman J., Lin X. (2002) Development 129, 4089–4101 [DOI] [PubMed] [Google Scholar]
- 38. Brannon M., Brown J. D., Bates R., Kimelman D., Moon R. T. (1999) Development 126, 3159–3170 [DOI] [PubMed] [Google Scholar]
- 39. Billin A. N., Thirlwell H., Ayer D. E. (2000) Mol. Cell. Biol. 20, 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roose J., Molenaar M., Peterson J., Hurenkamp J., Brantjes H., Moerer P., van de Wetering M., Destrée O., Clevers H. (1998) Nature 395, 608–612 [DOI] [PubMed] [Google Scholar]
- 41. Elms P., Siggers P., Napper D., Greenfield A., Arkell R. (2003) Dev. Biol. 264, 391–406 [DOI] [PubMed] [Google Scholar]
- 42. Moens C. B., Prince V. E. (2002) Dev. Dyn. 224, 1–17 [DOI] [PubMed] [Google Scholar]
- 43. Buckles G. R., Thorpe C. J., Ramel M. C., Lekven A. C. (2004) Mech. Dev. 121, 437–447 [DOI] [PubMed] [Google Scholar]
- 44. Lagutin O. V., Zhu C. C., Kobayashi D., Topczewski J., Shimamura K., Puelles L., Russell H. R., McKinnon P. J., Solnica-Krezel L., Oliver G. (2003) Genes Dev. 17, 368–379 [DOI] [PMC free article] [PubMed] [Google Scholar]