Background: Redox activity of hemoglobin (Hb) is augmented by lipopolysaccharide (LPS) to boost immune defense.
Results: Computational analysis of Hb identified LPS-binding hot spots, which were further defined via peptide-based binding assays and confirmed by mutagenesis of Hb subunits.
Conclusion: Regions of Hb that interact with LPS have been delineated.
Significance: Knowledge of LPS-binding residues on Hb may be exploited for designing antimicrobial peptides.
Keywords: Antimicrobial Peptides, Biophysics, Computer Modeling, Electron Microscopy (EM), Endotoxin, Hemoglobin, Innate Immunity, Lipopolysaccharide (LPS), Peptides, Surface Plasmon Resonance (SPR)
Abstract
Hemoglobin (Hb) functions as a frontline defense molecule during infection by hemolytic microbes. Binding to LPS induces structural changes in cell-free Hb, which activates the redox activity of the protein for the generation of microbicidal free radicals. Although the interaction between Hb and LPS has implications for innate immune defense, the precise LPS-interaction sites on Hb remain unknown. Using surface plasmon resonance, we found that both the Hb α and β subunits possess high affinity LPS-binding sites, with KD in the nanomolar range. In silico analysis of Hb including phospho-group binding site prediction, structure-based sequence comparison, and docking to model the protein-ligand interactions showed that Hb possesses evolutionarily conserved surface cationic patches that could function as potential LPS-binding sites. Synthetic Hb peptides harboring predicted LPS-binding sites served to validate the computational predictions. Surface plasmon resonance analysis differentiated LPS-binding peptides from non-binders. Binding of the peptides to lipid A was further substantiated by a fluorescent probe displacement assay. The LPS-binding peptides effectively neutralized the endotoxicity of LPS in vitro. Additionally, peptide B59 spanning residues 59–95 of Hbβ attached to the surface of Gram-negative bacteria as shown by flow cytometry and visualized by immunogold-labeled scanning electron microscopy. Site-directed mutagenesis of the Hb subunits further confirmed the function of the predicted residues in binding to LPS. In summary, the integration of computational predictions and biophysical characterization has enabled delineation of multiple LPS-binding hot spots on the Hb molecule.
Introduction
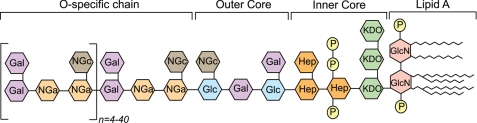
Gram-negative bacteria (GNB)2 are the predominant cause of clinical sepsis (1). A major constituent of their outer membrane is LPS (2), which is a potent stimulator of the immune system. LPS molecules shed from the bacterial surface contribute to the pathology of GNB sepsis (3). LPS has been called “endotoxin” because of its pyrogenic properties in humans and other mammals. LPS molecules from different species of GNBs share a common general architecture (Fig. 1) (4), consisting of a hydrophobic domain known as lipid A, a non-repeating core oligosaccharide, and a distal polysaccharide called O-antigen. Lipid A, also known as the “endotoxic principle” of LPS (5), anchors LPS to the bacterial surface by insertion of its fatty acyl chains into the outer membrane.
FIGURE 1.
Schematic representation of the structure of LPS (38). GlcN, glucosamine; P, phosphate; KDO, 2-keto-3-deoxyoctulosonic acid; Hep, d-glycero-d-manno-heptose; Gal, galactose; Glc, glucose; NGa, N-acetyl galactosamine; NGc, N-acetyl glucosamine.
The ability to acquire iron from the host is crucial for the survival of invading pathogenic bacteria (6). The mammalian host sequesters most of its iron in the form of heme (7). The respiratory protein Hb is a potentially rich source of iron and as a result is targeted by many bacterial pathogens, which have evolved specialized heme-Hb uptake systems for capturing cell-free Hb, free heme, and Hb complexed with other host proteins (8). For instance, virulent strains of Staphylococcus aureus secrete extracellular hemolytic toxins upon invasion, which rupture the erythrocytes extensively to release Hb (9). The extracellular bacterial proteases then cleave the Hb polypeptide chains and release the prosthetic heme. It has been shown that the redox activity of Hb (10, 11) is significantly enhanced in the presence of bacterial proteases and pathogen-associated molecular patterns (LPS and lipoteichoic acids) (12). The subsequent generation of free radicals by the redox-active Hb can kill the invading pathogen at the site of infection (12–14).
Hb has long been recognized as an LPS-binding protein (15). Hb from various species such as humans, sheep, and pig has been shown to interact with LPS (16–19). The invertebrate respiratory protein, hemocyanin, which is a functional homologue of Hb also binds LPS (12–14), indicating the evolutionary significance of such interactions. Previous studies have characterized the interactions of Hb and LPS through biophysical methods such as isothermal titration calorimetry and Fourier-transform infrared spectroscopy (17, 18), which suggested that Hb intercalates into and alters the aggregate structure of LPS polymers, transitioning it from a relatively dormant multilamellar structure to a bioactive cubic structure. It has been shown that Hb interacts predominantly with the acyl chains and phosphate moieties of lipid A (20). However, the precise interaction hot spots on the Hb remain unknown.
Here, we demonstrated that both the subunits of Hb contain LPS-binding sites. We implemented a methodical computational analysis to predict the LPS-binding sites on the surface of the Hb molecule. An algorithm to predict phospho-group binding sites was applied, followed by structure-based sequence comparisons of the vertebrate Hbs and docking analysis of protein-ligand interactions. The predictions were tested experimentally by investigating the binding of synthetic Hb peptides to LPS in immobilized and free form and to the bacterial surface. The LPS-binding peptides displayed binding affinity in the range of 10−8 to 10−5 m in SPR analysis. Moreover, these LPS-binding peptides neutralized the endotoxicity of LPS, indicating the functional significance of binding. Selective mutations of lysine to aspartate at positions 56, 60, and 61 in Hb α; and positions 59, 66, and 95 in Hb β led to partial loss of the LPS-binding ability of the subunits. Taken together, we have mapped LPS-binding sites on Hb using an integrated approach of computational predictions and empirical validations. Our work advances the understanding of the molecular basis of Hb-LPS interactions.
EXPERIMENTAL PROCEDURES
Reagents
LPS and lipid A of Salmonella minnesota were purchased from Sigma. S. minnesota ReLPS was from List Biological Laboratories (Campbell, CA). PyroGene kit for the quantification of LPS was from Lonza Biosciences Pte Ltd. QuikChange® XL site-directed mutagenesis kit was purchased from Stratagene (La Jolla, CA). All other chemicals were of molecular biology grade obtained from Sigma, unless otherwise stated.
SPR Analysis of Recombinant Hb Subunits with LPS
The binding of the recombinant Hb subunits, rHbα and rHbβ to the ligands, LPS, ReLPS, and lipid A, was characterized by real-time interaction analysis using BIAcore 2000 (BIAcore AB, Uppsala, Sweden). Details on the expression and purification of the rHb proteins are in the supplemental “Materials and Methods.” For SPR, S. minnesota LPS, ReLPS, and lipid A were diluted to 0.25 mg/ml in 10 mm PBS, pH 7.4, and immobilized on the surface of an HPA sensor chip (GE Healthcare) according to the manufacturer's specifications. Binding studies were performed with HEPES-buffered saline (HBS) as the running buffer. The proteins were injected over the immobilized ligand at a flow rate of 30 μl/min. Regeneration of the chip surface was achieved by injection of 100 mm NaOH for 1 min. All SPR responses were normalized against the buffer. The association and dissociation phases of data were separately fitted to a 1:1 Langmuir binding model provided in the BIAevaluation software (version 4.1). The dissociation constant (KD) was then calculated from the averaged values of kd and ka using the equation, KD = kd/ka. Residual plots and chi-square were used as a measure of the curve fitting efficiency.
Prediction of LPS-binding Sites on Hb
A search for LPS-binding sites on Hb was initiated by using an algorithm that computes the propensity of phospho-group binding for a protein surface (21). Briefly, the algorithm computes a joint propensity for phospho-group contact based on the amino acid identity, surface curvature, and electrostatic potential at every point of the discretized protein surface. The computed propensities for each surface point were mapped onto the corresponding surface amino acid residues, and an average propensity for each residue was computed. To determine the binding sites that have been conserved across Hb, 20 vertebrate Hb structures were selected from the Structural Classification of Proteins database classification (supplemental Table S1) (22). The propensity calculation algorithm was repeated on each of these structures. Independently, structural alignment of these proteins was performed using the MultiSeq tool of Visual Molecular Dynamics (VMD) (23), with human aquomet Hb as the index structure for the alignment. The structure-based alignments were examined for the phospho-group binding propensities of the aligned residues and the degree of conservation of the favorable propensity was computed for the residues, yielding predicted binding sites with combined propensity and conservation. The predicted binding sites were further prioritized by seeking paired patches at an appropriate distance to interact with the two phosphates of the lipid A head group.
To comprehend the interactions between the ligand and these binding sites, docking was performed using GLIDE (version 5.0; Schrodinger, LLC, 2007). The crystal structure of human aquomet Hb (Protein Data Bank code 1HGB) was used as the receptor. The protein structure was prepared using the Maestro protein preparation wizard (Schrodinger). The solvent molecules were deleted, bond orders were assigned, and hydrogen atoms were added to the protein. The orientation of the hydroxyl groups and the protonation states of the amino acid residues were optimized. The protein structure was then minimized using the OPLS 2001 force field. The ligand (1,4′-bisphospho-β-(1,6)-2,2′-N-acetyl-3,3′-O-acetyl-d-glucosamine disaccharide) was prepared by replacing the long acyl chains of lipid A with methyl groups as GLIDE limits the maximum number of atoms for a ligand. The Ligprep utility of Maestro was used to generate the protonation and tautomeric states. One low energy ring conformation and 32 stereoisomers were generated. Prior to docking, grids were generated for each of the potential binding sites. For each site, the center of each grid was defined as the centroid of the residues of the patch. Additional constraints for hydrogen bonding to the ligand were imposed on the residues in each patch. The standard precision mode of GLIDE was used for docking. The ligand was flexible, whereas the protein was held rigid. Default parameters specified in GLIDE were used for docking. The resulting structures from each docking were compared on the basis of their GlideScore.
Peptide Design and Synthesis
Based on the computational predictions, various Hb peptides were synthesized commercially by Genemed Synthesis, Inc. and purified to >95% under pyrogen-free conditions. The purity and quality of the peptides were assessed by HPLC and mass spectrometry. The peptides were annotated A and B, indicating their derivation from the Hb α or β subunit, respectively. The numbers correspond to the start of their amino acid positions in the Hb sequence.
CD Measurements
The secondary structure of the peptides was determined by recording their CD spectra at a concentration of 40 μm in PBS, pH 7.4, using a Jasco J-810 spectropolarimeter. The temperature within the sample chamber was maintained at 25 °C with a continuous flow of nitrogen. Spectra were recorded in quartz cuvettes of 1-mm path length, at data pitch of 0.1 nm, bandwidth of 2 nm, response time of 1 s, and scanning speed of 20 nm/min. Data from three independent scans were averaged. Buffer alone was used as the control for baseline subtraction.
Interaction of Peptides with Immobilized LPS
SPR analysis was performed to examine the binding of Hb peptides to LPS, ReLPS, and lipid A, as described earlier. Different concentrations of the peptides were injected over the immobilized ligand at a flow rate of 30 μl/min using HBS, pH 7.4, as the running buffer. Data analysis was performed as described earlier.
Dansylcadaverine Displacement Assay
The binding of peptides to lipid A in solution was measured using dansylcadaverine (DC) as a fluorescent displacement probe. The assay is based upon differences in the fluorescence emission intensity of lipid A-bound and -unbound forms of DC. Upon binding to lipid A, there is an increase in the emission intensity of DC accompanied by a blue shift in the wavelength of maximum emission. Here, if a peptide binds lipid A, the bound DC will be displaced, resulting in a quenching of fluorescence. Equal volumes (50 μl) of 100 μm DC and 40 μg/ml lipid A in HBS, pH 7.4, were mixed and incubated for 5 min. This was followed by addition of 50 μl of various concentrations of the peptides. The fluorescence of the samples was then measured using a BIOTEK plate reader. Excitation and emission wavelengths were set at 340 and 560 nm, respectively. To quantitatively compare the interaction of the peptides with lipid A, the occupancy of DC on lipid A was calculated using the following formula: occupancy = (FP − FD)/(FL − FD), where FD is the fluorescence intensity of DC in the absence of lipid A; FL is the fluorescence intensity of DC in the presence of lipid A; and FP is the fluorescence intensity of the solution of DC and lipid A upon the addition of the different concentrations of the peptides. The occupancy varies from 0 for DC solution without lipid A, to 1 for DC and lipid A solution without the added peptides.
PyroGene Assay for LPS Neutralization
The endotoxicity of S. minnesota LPS in the presence or absence of the peptides was measured using the PyroGene kit. This kit employs recombinant factor C (rFC) from horseshoe crab to quantify the endotoxicity of LPS (24). In the presence of LPS, rFC becomes activated and hydrolyzes a synthetic substrate to release a fluorescent product. The amount of product formed is quantifiable at excitation and emission wavelengths of 380 and 440 nm, respectively. Various concentrations of peptides were pre-incubated with 10 ng/ml of S. minnesota LPS at 37 °C for 1 h. Each test sample was diluted 20 times, and 100 μl was aliquoted into the wells of a 96-well plate. The assay was performed according to the manufacturer's specifications. The relative change in the endotoxicity of the solutions was measured by comparison with the peptide-free LPS samples. The extent of neutralization of the endotoxicity of LPS in the presence of peptides was calculated using the formula [(EL − EP)/EL] × 100, where EL is the endotoxicity of LPS solution and EP is the endotoxicity of the solution in the presence of the peptide.
Flow Cytometry Analysis
The binding of peptide B59 to GNB was assessed using flow cytometry. Cultures of Escherichia coli and Pseudomonas aeruginosa (∼107 cells/ml) growing at exponential phase were treated with 1 mm B59 in PBS, pH 7.4, with or without pre-treatment with 100 ng/ml LPS, for 30 min at room temperature. The bacteria were then fixed with 4% paraformaldehyde for 30 min, followed by blocking with 5% BSA for 1 h. The bound peptide was detected by incubating the samples for 1 h each with primary rabbit anti-Hb antibody (1:400) and secondary phycoerythrin-conjugated goat anti-rabbit IgG (Invitrogen) (1:200). Untreated samples served as negative controls. The bacteria were resuspended in 0.5% paraformaldehyde-PBS, and data were collected using the Dako Cytomation CyAn ADP system.
Scanning Electron Microscopy
The B59 peptide, which was shown to bind and neutralize LPS, was visualized on the bacterial surface using immunogold labeling coupled with SEM. The bacteria were prepared as for flow cytometry analysis. The samples were treated with primary rabbit anti-Hb antibody (1:50) for 1 h. Samples were incubated with pre-immune rabbit serum or an isotype control antibody (anti-C3d) alone instead of the primary antibody as a negative control. For immunogold labeling, 20 nm colloidal gold-conjugated goat anti-rabbit IgG (Ted Pella, Inc.) (1:50) was used. The samples were post-fixed in 1% aqueous solution of osmium tetroxide for 30 min, followed by dehydration in increasing concentrations of ethanol. The samples were then subjected to standard procedures of infiltration and carbon coating. The samples were viewed under a XL-30 FEG SEM microscope (Philips) at an accelerating voltage of 10 keV. The backscattered electron mode was used to observe colloidal gold particles on the bacteria.
Site-directed Mutagenesis
Lysine-to-aspartate mutations were introduced into specific sites of both the Hb α and β subunits. The mutant proteins are designated as Hbα(K56D/K60D/K61D) and Hbβ(K59D/K66D/K95D). In addition, a K17D single mutation was introduced at a non-LPS binding site in Hbβ to serve as an internal control. Mutagenic primers were designed using PrimerX and are listed in supplemental Table S2. Mutagenesis was carried out using the QuikChange® XL Site-directed Mutagenesis kit according to the manufacturer's instructions. All mutants were verified by sequencing and transformed into E. coli BL-21 (DE3) for protein expression. Protein expression and purification was carried out as described previously for the wild-type subunits (supplemental “Materials and Methods”).
ELISA-based LPS Binding Assay
The ability of the mutant Hb subunits to bind LPS was probed using ELISA. Briefly, 1 μg of S. minnesota LPS was immobilized on the surface of the 96-well microtiter plates by overnight incubation at room temperature. The excess ligands were washed off, and the unbound sites were blocked with 2% BSA at 37 °C for 2 h. Various concentrations of the proteins, prepared in HBS, were added to the wells and incubated at 37 °C for 2 h. Following three rinses with PBST (PBS containing 0.05% (v/v) Tween 20), the bound proteins were detected by incubating the samples with primary rabbit anti-Hb antibody (1:5000) and horseradish peroxidase-conjugated goat anti-rabbit antibody (1:2000) at 37 °C for 2 h. After washing the wells three times with PBST, 100 μl of peroxidase substrate, (2,2′-azino-di-[3-ethylbenzothiazoline sulfonate]) (Roche Diagnostics) was added, and A405 was read.
RESULTS
α and β Subunits of Human Hb Harbor LPS Binding Sites
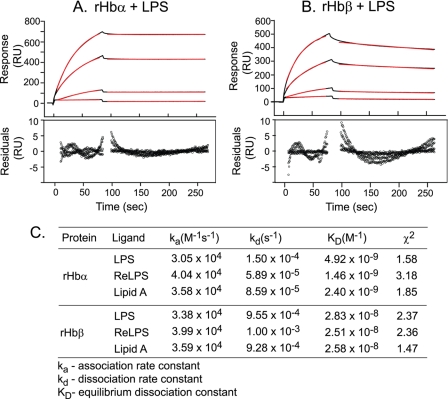
Previous studies have shown that tetrameric Hb from various species, as well as the α subunit of sheep Hb, can bind bacterial LPS (18). Here, we examined whether the isolated subunits of human Hb could also bind LPS. The α and β chains of Hb were expressed recombinantly and purified (supplemental Fig. S1). Real-time interaction analysis using SPR showed that both rHbα and rHbβ bind S. minnesota LPS (Fig. 2, A and B), ReLPS, and lipid A (Fig. 2C) in a concentration-dependent manner. This indicates that binding sites for LPS are located on both subunits, suggesting the existence of more than one binding site per tetramer of Hb. The KD values were in the nanomolar range for the ligands (Fig. 2C). The accuracy of the fitting was indicated by the residual plots and chi-square values, which were within acceptable limits (chi-square below 10 is acceptable).
FIGURE 2.
Hb subunits possess high affinity for bacterial LPS. S. minnesota LPS was immobilized on an HPA sensor chip. rHbα and rHbβ at 0.1, 0.25, 0.5, and 0.75 μm in HBS were individually injected over the ligand-immobilized chip surface at a flow rate of 30 μl/min. Data were fitted to a 1:1 Langmuir binding model using the separate fit module of the BIAevaluation software (version 4.1). Sensorgrams for the interaction of rHbα (A) and rHbβ (B) with LPS are shown in black. The red lines represent the corresponding fits. Plots of residuals corresponding to the differences between the experimental and best-fit curves are shown in the lower panel. RU, response units. C, kinetic parameters for binding of rHbα and rHbβ to the various ligands.
Hb Possesses Evolutionarily Conserved Cationic Surface Patches for Binding LPS
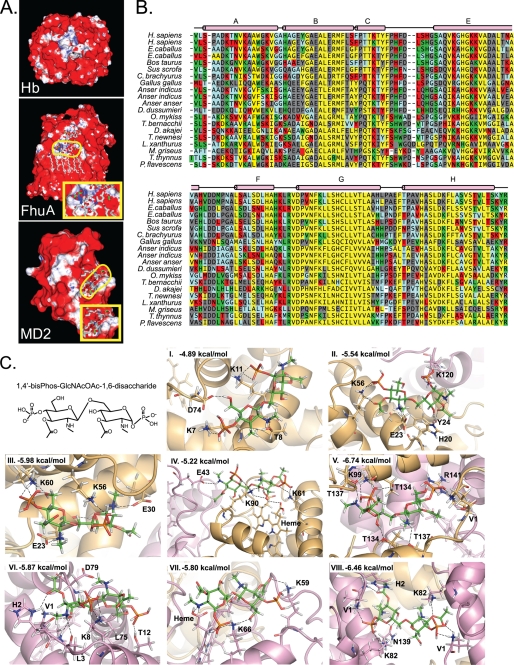
Having observed more than one LPS-binding site on the Hb tetramer, it was pertinent for us to examine the structure of Hb for potential LPS-binding sites. Hb has been shown to associate with LPS via electrostatic interactions with the 2-keto-3-deoxyoctulosonic acid sugars of ReLPS and the phosphate residues of lipid A and via hydrophobic interactions with the lipid A acyl chains (20, 25, 26). Because LPS molecules are anchored on the surface of GNB with the acyl chains of lipid A embedded in the membrane bilayer, we envisaged that for the cell-free Hb to dock onto the bacterial surface and mediate localized free radical production via its redox activity, it should be able to bind to LPS proximal to the membrane. Electrostatic forces project a force field farther into space, and are thus more effective than other chemical interactions at recruiting binding partners across long distances (27). Studies have shown that Hb does not interact strongly with dephosphorylated ReLPS, suggesting the importance of the phosphate residues in these interactions (20). This prompted us to undertake a computational approach to search for phospho-group binding sites on Hb. The phospho-group binding prediction algorithm for the human Hb resulted in many “hits,” which could potentially participate in binding to LPS (Fig. 3A). The algorithm was additionally run for two control proteins, FhuA and MD2, whose structures in complex with LPS are known (28, 29). The prediction algorithm identified the binding sites correctly, thus indicating the reliability of this in silico approach (Fig. 3A). However, additional “false positive” binding sites were also predicted.
FIGURE 3.
Computational prediction of LPS-binding sites on Hb. A, prediction of phospho-group binding patches. An algorithm for computing phospho-group binding propensity was applied on human Hb (Protein Data Bank code 1HGB). FhuA protein in complex with LPS (Protein Data Bank code 1QFG) and MD2 protein with bound lipid A (Protein Data Bank code 2E59) served as positive controls. The binding propensity is represented by surface coloring, which varies linearly from white to blue over favorable propensity values from 1 to 30, and from red to white over the unfavorable propensity values from −1 to 0. The ligands, LPS and lipid A, are shown in a licorice representation, with the phosphate atoms colored green. The phosphate atoms of the ligands in FhuA and MD2 are outlined in yellow. Insets show magnified images of the phospho-groups of the ligand along with the surface coloring for the proteins. B, structure-based sequence alignment of vertebrate Hbα chains. Vertebrate Hb proteins were aligned on the basis of structure using the MultiSeq tool of VMD. The alignment for Hbα is shown here. Residues are color-shaded on the basis of predicted propensity of phospho-residue contact in a decreasing order from green to red to blue. Residues with no propensity of contact are indicated in black. Buried residues are indicated in yellow. The helix designations, A–F, for Hb (according to Kendrew's nomenclature (39)) are shown above the alignment. C, molecular docking of the ligand to the predicted LPS-binding sites. The diglucosamine head group of lipid A (1,4′-bisphospho-β-(1,6)-2,2′-N-acetyl-3,3′-O-acetyl-d-glucosamine disaccharide) was used for docking analysis to the predicted LPS-binding sites. Panels I–VIII, top ranking docked poses of the ligand to the predicted LPS-binding sites. Dockings were performed using GLIDE (version 5.0; Schrodinger, LLC, 2007). The residues participating in hydrogen bonding with the ligand and the computed binding energies are indicated. The α subunits are colored gold, and the β subunits are colored violet. The ligand is in stick representation, whereas the polypeptide chains are shown as ribbons. The hydrogen bonds are indicated as dashed lines.
Because Hb from species as distantly divergent as sheep, pig, and humans is known to bind LPS (16–19), we used a structure-based sequence comparison to narrow down the predictions to residues for which binding propensity has been conserved evolutionarily (Fig. 3B). Further analysis of the Hb structure served to shortlist eight cationic pockets, which were located at the protein surface with an appropriate separation distance to bind the head group phosphates of lipid A.
Docking of the lipid A disaccharide head group (Fig. 3C) to these sites was performed to assess the possible interactions with LPS. For most of the binding sites, the docked structures exhibited strong networks of hydrogen bonds between the phosphates of the ligand and positively charged or partially charged residues of the protein (Fig. 3C, panels I–IV and VIII). Docking produced non-ideal hydrogen bonds in some cases, such as between the Arg-141 and a phosphate of lipid A (Fig. 3C, panel V); and between the backbone carbonyl of Leu-3 and the amine group on the ligand (Fig. 3C, panel VI). The affinity of the ligand was highest for the binding sites located at the α-α and the β-β interfaces (Fig. 3C, panels V and VIII). This is in agreement with the prediction algorithm that showed these two interfaces to have a high propensity of phospho-group binding (Fig. 3A).
Hb Peptides Harboring Predicted LPS-binding Sites
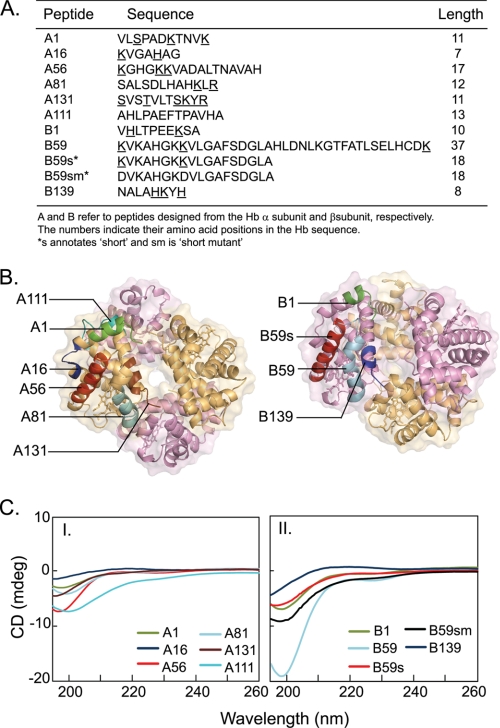
Peptides were designed to harbor the computationally predicted binding sites for empirical validations of LPS binding (Fig. 4, A and B). The peptide length and region surrounding the predicted LPS-binding residues was chosen based on the hydrophilicity and solubility. To substantiate the computational prediction methods, a control peptide A111 was designed from the surface residues of the Hbα, which were predicted to be non-LPS binding. To query the plausible effects of the length of the 36-mer B59 on its interactions with LPS, we analyzed an 18-mer peptide, B59s. To corroborate the role of the predicted residues in binding LPS, a mutant peptide B59sm with base-to-acid substitutions was synthesized by replacing the predicted phosphate-binding lysine residues of B59s with aspartate.
FIGURE 4.
Hb peptides used for empirical validations. A, designations and sequences of the Hb peptides. Nine peptides were designed to incorporate the putative LPS-binding residues (underlined in the sequence). The peptides are designated A and B corresponding to the Hb α and β subunits, respectively. Two additional control peptides were (i) A111, composed of the stretch of residues which were predicted to be non-LPS binding, and (ii) B59sm, mutant peptide obtained by replacing the predicted LPS-binding residues of B59s with aspartic acid. B, mapping of the Hb peptides on the tetrameric structure. Tetrameric Hb showing the peptides designed to incorporate the predicted LPS-binding sites. The α subunits are colored gold, and the β subunits are colored violet. The heme moiety is represented by sticks, whereas the polypeptide chains are shown as ribbons. The peptides are labeled for chains A and B. C, secondary structure of Hb peptides. Far-UV CD spectra of Hb peptides in PBS, pH 7.4, at 298 K. Peptide concentration was 40 μm. A negative peak near 200 nm represents a random coil structure. Panel I, α subunit peptides: A1 (green), A16 (blue), A56 (red), A81 (aqua), A131 (brown), A111 (cyan); and panel II, β subunit peptides: B1 (green), B59 (aqua), B59s (red), B139 (blue), B59sm (black).
Hb Peptides Exist in Random Coil Conformations
The secondary structure of the Hb peptides was determined in PBS in the absence and presence of LPS (Fig. 4C). The CD spectra of the peptides are characterized by a negative peak near 200 nm, indicating that these peptides are unstructured in solution. Although LPS is known to cause structural perturbations in the Hb tetramer (14), it does not appear to affect the conformation of the peptides, as indicated by the unchanging CD spectra in the presence of LPS (supplemental Fig. S2).
Hb Peptides Bind Immobilized LPS
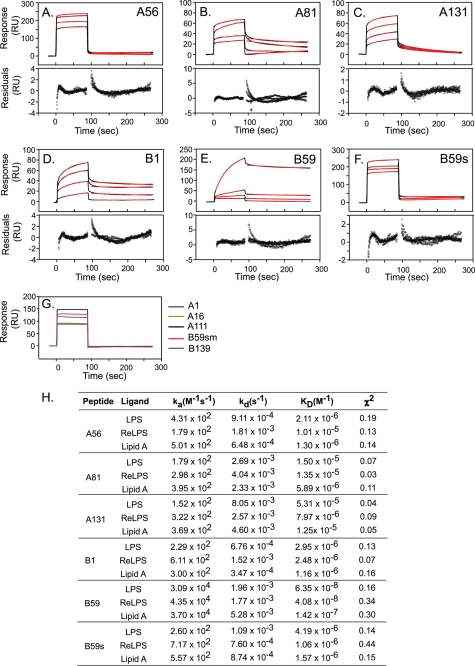
To mimic the binding of peptides to LPS displayed on the surface of GNB, Hb peptides were flown over LPS, ReLPS, or lipid A immobilized on the surface of an HPA sensor chip. Analysis of the SPR data revealed high affinity interactions between six peptides and LPS (Fig. 5, A–F). Three peptides, A1, A16, and B139 did not bind LPS (Fig. 5G). B59 displayed the strongest affinity for LPS, with a KD value of 6.35 × 10−8 m. The LPS-binding affinity of the 18-mer B59s was ∼60-fold lower than that of B59 (Fig. 5H). The negative control, A111 peptide, showed no specific binding activity but displayed a bulk response upon injection (Fig. 5G). These results validate the modeling strategy, as the predicted non-binder A111 did not show any binding. Similarly, the mutant peptide B59sm displayed no affinity for LPS. It proves the importance of the lysine residues for LPS-binding ability.
FIGURE 5.
Surface plasmon resonance measurements of the interaction of Hb peptides with LPS. Peptides in HBS were injected over the immobilized ligands at a flow rate of 30 μl/min. Sample concentrations were 50, 100, 150, and 200 μm (except for B59, for which the concentrations were 0.01, 0.05, 0.1, and 0.5 μm). The association and dissociation phases of data were separately fitted to a 1:1 Langmuir binding model in the BIAevaluation software (version 4.1). A–F, sensorgrams for the interaction of the peptides with LPS are shown in black. The red lines represent the corresponding fits. Plots of residuals corresponding to the differences between the experimental and best-fit curves are shown in the lower panel. RU, response units. G, representative sensorgrams of the peptides that did not bind LPS at a concentration of 200 μm: A1 (brown), A16 (green), A111 (black), B59sm (red), and B139 (purple). The response indicates a bulk shift without any binding activity. H, kinetic parameters for interaction of peptides with the various ligands.
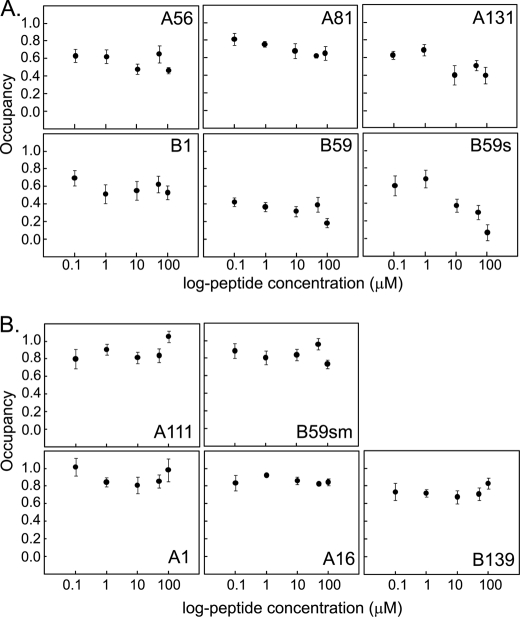
Hb Peptides Interact with LPS in Free Form
LPS shed from invading bacteria can interact with host plasma proteins. Using an assay based on the fluorescent probe DC, we examined whether the Hb peptides would interact with lipid A in solution, with the potential to inhibit its endotoxicity. Fig. 6 shows the plots of the occupancy of DC on lipid A in the presence of increasing doses of the peptides. The peptides, B59 and B59s, showed the greatest decrease in probe occupancy, with dose-dependent displacement from lipid A. The other peptides were also able to reduce the occupancy of DC from lipid A in a dose-dependent manner, albeit less drastically. The control peptide A111 and the other non-binding peptides did not significantly affect DC occupancy (Fig. 6B). Moreover, DC occupancy in the presence of the non LPS-binding peptides did not decline in a concentration-dependent manner. The non-binding peptide, B139, could however reduce the occupancy to levels similar to the LPS-binding peptide, A81. This could imply its binding to LPS in solution phase rather than to immobilized LPS.
FIGURE 6.
DC displacement assay for interaction of Hb peptides with lipid A in solution. The occupancy of DC (50 μm) on lipid A (20 μg/ml) was measured in the presence of 0, 0.1, 1, 10, 50, and 100 μm of the various peptides at pH 7.4 in HBS. The fluorescence intensity was measured by excitation at 340 nm and emission at 560 nm. The occupancy was calculated using the formula: occupancy = (FP − FD)/(FL − FD), where FD is the fluorescence intensity of DC in the absence of lipid A, FL is the fluorescence intensity of DC in the presence of lipid A; and FP is the fluorescence intensity of the solution of DC and lipid A upon the addition of the different concentrations of the peptides. Occupancy is plotted against the log-peptide concentrations for peptides that bind LPS (A) and do not bind LPS (B). Results are the means ± S.D. of three independent experiments done in triplicate.
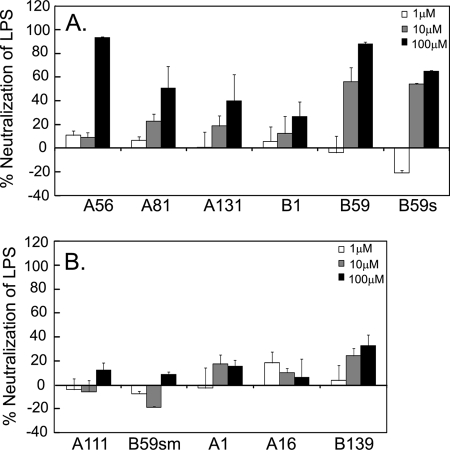
Hb Peptides Bind to LPS and Neutralize Endotoxicity
The functional implications of the binding of the peptides to LPS were probed by investigating their ability to neutralize the endotoxicity of LPS solutions via a quantitative assay based on recombinant factor C. This assay is highly sensitive to endotoxin, being able to detect levels as low as 0.01 endotoxin units/ml. Prior to the assay, the endotoxin levels in the peptide solutions were measured and were found to be minimal (supplemental Table S3). The assay was conducted at an LPS concentration of 10 ng/ml. The endotoxicity levels at this concentration were measured to be ∼50 endotoxin units/ml. The results show that the peptides, which bound LPS, were able to neutralize it in a dose-dependent manner (Fig. 7). The peptides A56 and B59 were consistently the most potent in neutralizing the endotoxicity by >80%. The peptide B59s neutralized the endotoxicity by >60%. The peptides A81, A131, and B1 neutralized LPS up to 50%. Poor LPS neutralization was seen for the control peptide A111 (< 20%), consistent with the binding assays. The mutant peptide, B59sm, was unable to neutralize LPS. Similarly, the non-binding peptides A1 and A16 displayed poor anti-endotoxic activity. Surprisingly, the non-binding peptide B139 could neutralize LPS to an extent comparable with the peptide B1. This suggests that B139 binds LPS in solution, which was also observed in the dansylcadaverine assay.
FIGURE 7.
Neutralization of LPS endotoxicity by Hb peptides. S. minnesota LPS was treated with 1, 10, and 100 μm concentrations of peptides. The endotoxicity of the solutions was measured using rFC-based PyroGene assay. The extent of neutralization of LPS by the peptides is plotted as a percentage of the “LPS only” control for the LPS-binding peptides (A) and non LPS-binding peptides (B). Results are the means ± S.D. of three independent experiments done in triplicate.
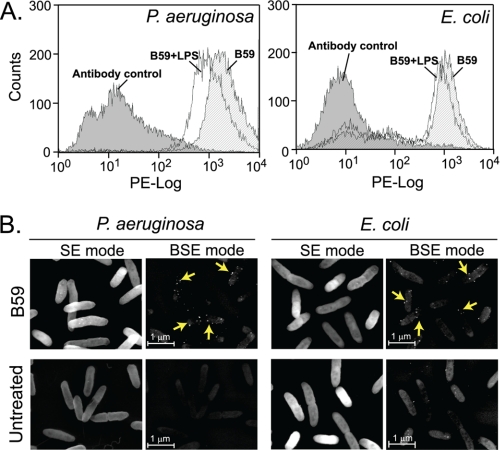
B59 Associates with Bacterial Surface
Based on the SPR results wherein the peptides interact with LPS immobilized on a sensor chip surface, we asked whether the peptides could also associate with the LPS molecules in situ on the live bacteria. The B59 peptide, which displayed the highest binding affinity for LPS, was tested further for its attachment to the surface of E. coli and P. aeruginosa. Using flow cytometry, we observed the binding of the peptide B59 to the surface of GNB, indicated by the positive shift in the median fluorescence intensity of the peptide-treated samples compared with the untreated control (Fig. 8A). Pre-treatment of the peptide with LPS resulted in a negative shift in the median fluorescence intensity, indicating a reduction in binding of the peptide to the bacteria. This result suggests the binding of the peptide to bacteria via LPS. SEM enabled visualization of the peptide associated with the bacterial surface in the form of colloidal gold labeling (Fig. 8B). To confirm the specificity of the rabbit anti-Hb antibody for the peptide B59, control experiments were performed in which the primary rabbit anti-Hb was separately replaced with rabbit pre-immune serum and an isotype control rabbit anti-C3d antibody. The bacteria showed negligible labeling (supplemental Fig. S3) in both the cases, thus proving the specificity of the primary antibody used.
FIGURE 8.
LPS-binding peptide, B59, associates with Gram-negative bacteria. A, the binding of B59 to GNB was analyzed by flow cytometry. E. coli and P. aeruginosa were treated with 1 mm B59 for 30 min, with (white) or without (stripes) pre-treatment with 100 ng/ml LPS. The peptide bound to the bacterial surface was detected using primary rabbit anti-Hb antibody (1:400) and secondary phycoerythrin-conjugated goat anti-rabbit IgG (1:200). The fluorescence intensity is plotted in log-fluorescence units versus counts. The negative control bacteria incubated without the peptide are indicated in gray. The results are representative of three independent experiments. B, the association of B59 to the bacterial surface was visualized using scanning electron microscopy coupled with immunogold labeling. Specific binding of the peptide is demonstrated by the colloidal gold (arrows) observed under backscattered electron (BSE) mode. Untreated bacteria served as negative control. Magnification is 15,000×. Bar, 1 μm.
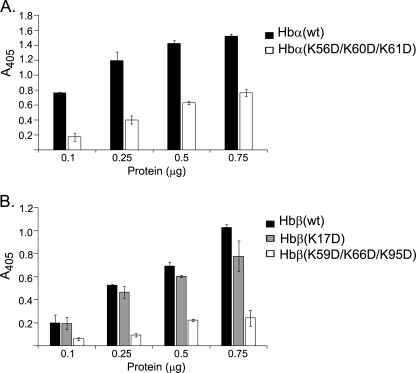
Mutations in Hb Subunits at LPS-binding Sites Diminish Their LPS-binding Ability
For further substantiation of the LPS-binding sites, selected residues in the A56 and B59 peptides were mutated to generate recombinant Hb α and β subunits with mutations from lysine to aspartate. Two mutant subunits carrying triple mutations were constructed: Hbα(K56D/K60D/K61D) and Hbβ(K59D/K66D/K95D). Separately, mutant Hbβ(K17D), which was not predicted to be an LPS-binding site was also constructed as an internal control for the LPS-binding assays. The binding of the mutants to LPS was compared with that of the wild-type subunits by ELISA (Fig. 9). The mutations resulted in a weakening of the LPS-binding ability of the Hb subunits, with 2- and 4-fold reductions for the mutated Hb α and Hb β subunits, respectively. The LPS-binding of Hbβ was unaffected by the K17D mutation.
FIGURE 9.
LPS-binding activities of wild-type and mutant Hb subunits. ELISA was performed for quantifying the LPS-binding activity of the mutant Hb subunits: Hb α (A), and Hb β (B). 96-Well MaxisorpTM microtiter plates were coated with S. minnesota LPS. The unbound sites were blocked with 2% BSA, and increasing concentrations of the recombinant proteins were added to each well. Anti-Hb antibody was added followed by horseradish peroxidase-linked secondary antibody. The peroxidase enzyme activity was determined at 405 nm. Results are the means ± S.D. of three independent experiments done in triplicate.
DISCUSSION
The binding of microbial pathogen-associated molecular patterns to Hb is known to induce structural alterations in the protein, leading to the activation of its redox activity (12, 14). The propensity for Hb to bind LPS tends toward its recruitment to the bacterial surface, thereby enhancing the free radical-mediated antimicrobial efficiency in a localized manner while minimizing damage to the host. The Hb-LPS interaction thus plays a role in the non-self immune response. Here, we have mapped LPS-binding hot spots on the surface of the Hb tetramer using computational predictions coupled with empirical validations.
Using real-time interaction analysis of the recombinant Hb α and β chains, we showed that the isolated subunits of Hb bind strongly to LPS, ReLPS, and lipid A (Fig. 2). This implies that the Hb tetramer (α2β2) contains multiple LPS-binding sites. Several proteins such as CD14 (30), MD2 (31), LPS-binding protein (32), Hsp60 (33), and FhuA (34) have been shown to interact with LPS via both electrostatic interactions with the polar head group of the lipid A, and hydrophobic interactions with the lipid A acyl chains. It has been proposed that the structural requirements for LPS binding include the presence of a cluster of cationic residues and a hydrophobic pocket (35). In the present study, we adopted an in silico approach to search for pockets on Hb with electrostatic and steric favorability for binding the phosphate groups of the lipid A moiety of LPS. This led to the identification of sites comprised of cationic residues, appropriately positioned to engage in electrostatic interactions with the phosphates of lipid A. It has been suggested that respiratory proteins have evolved over hundreds of millions of years, from the invertebrate protein hemocyanin to the vertebrate Hb, with a secondary role of immune defense via interaction with pathogen-associated molecular patterns (12, 13). Thus, evolutionary conservation of binding site residues was used as an additional search criterion. The binding energy between LPS and these sites was obtained by docking the head group of lipid A (the diglucosamine phosphate moiety) to each of the predicted sites. The docking analysis showed interactions of the phosphates with the cationic residues in the protein.
Real-time interaction studies of the Hb peptides served to assess the computational predictions and eliminate false positives. Overall, out of the nine peptides analyzed, six demonstrated binding to LPS, ReLPS, and lipid A (Fig. 5). The peptides, which showed binding to LPS, were also able to bind to ReLPS and lipid A, suggesting that the peptides interact with the lipid A component of LPS, conceivably via electrostatic interactions with the phosphates. The ability of peptides A131 and B1 to bind LPS was in agreement with the computational prediction of the high propensity of LPS-binding at the interface of the Hb subunits. SPR analysis at low pH of 6.5 and 5.5 was performed to investigate the importance of electrostatic interactions in binding of the peptides to LPS. At a pH of 5.5, peptide B59 displayed 10× stronger affinity for lipid A (supplemental Fig. S4A). This could be due to the greater net positive charge on the peptide at this pH. The affinity for LPS and ReLPS were, however, unchanged. Moreover, the affinity at pH 6.5 was the same as that at pH 7.4 (supplemental Fig. S4B). Except for B1, the peptides that have been used in this study are cationic at physiological pH (isoelectric points > 8). Thus, lowering of the pH would increase their net positive charge, which may not alter the binding affinity significantly. The peptides generally displayed weaker LPS-binding affinity as compared with the Hb subunits, indicating that the three-dimensional structure influences the affinity for LPS. This difference in the binding affinity could also be attributed to the presence of multiple binding sites in the Hb chains, whereas the peptides each harbor a single LPS-binding site. Studies are underway for the precise structural determination of the crystal structure of Hb in complex with LPS so as to ascertain the molecular details of their interactions.
Fluorescent probe displacement assays corroborated the observations on the binding of the peptides with lipid A. The cationic fluorescent probe DC interacts with the acidic phosphate groups and hydrophobic acyl components of lipid A (36). Because peptides B59 and B59s were able to displace the probe from DC completely (Fig. 6), it indicates that they interact with lipid A via similar interactions as DC. The peptides A56, A131, and B1 have a weak binding affinity for lipid A as they could not completely displace the probe bound to lipid A. It may be argued that these peptides participate only in ionic interactions with lipid A; hence, they were unable to disrupt the hydrophobic interactions between lipid A and DC. Mutations of the putative LPS-binding lysine residues to aspartate in B59sm abolished its LPS-binding and -neutralizing activity, thus emphasizing the importance of electrostatic interactions in LPS-binding.
During infection, LPS molecules shed from the bacterial surface lead to immune activation upon their recognition by effector molecules such as LPS-binding protein (37). We inquired whether the LPS-binding peptides could mask LPS from the innate immune effector proteins. This was probed by testing the anti-endotoxic potential of the peptides using an rFC-based assay. Consistent with the binding studies, the peptides B59, B59s, and A56 neutralized the endotoxicity of LPS drastically (Fig. 7). This neutralization effect could be attributable to sequestration of the LPS by the peptides, thus blocking LPS from recognition by rFC. The other peptides with weak binding affinity did not confer endotoxin neutralization. These results are suggestive of the functional role of these peptides in hindering the recognition of LPS by immune effector proteins.
Consistent with the high affinity seen in SPR binding studies, we were able to demonstrate the attachment of the LPS-binding peptide B59 to the surface of GNB. Flow cytometry showed that pre-treatment of the peptide with LPS reduced the binding of this peptide to bacterial cells, proving that the peptide possibly binds to LPS molecules anchored on the bacterial surface. Furthermore, B59 was observed on the surface of GNB, visualized by immunogold-labeled SEM. It should be noted that we cannot preclude the possibility of the interaction of Hb peptides with pathogen-associated molecular patterns on the surface of Gram-positive bacteria, as it is known that Hb can bind with lipoteichoic acids (12, 14).
Mutational analysis of the Hb subunits supported the computational predictions and peptide-based experiments. Mutant Hb subunits, Hbα(K56D/K60D/K61D) and Hbβ(K59D/K66D/K95D), had diminished LPS-binding activities. However, the mutant subunits retained partial LPS-binding activity. This may indicate that other residues also participate in LPS recognition. Nevertheless, the results confirm the contribution of the mutated residues of Hb in binding LPS and corroborate the LPS-binding capability of the Hb peptides.
To summarize, we have delineated LPS-interacting regions on the surface of the Hb tetramer by rational computational predictions accompanied with biophysical characterization. Our computational approach of integrating phospho-binding propensity with evolutionary conservation identified potential LPS-binding hot spots. Empirical investigations enabled filtering of true positive binding sites obtained from the computational analyses. Peptides spanning residues 56–72, 81–92, and 131–141 of Hbα and residues 1–10 and 59–76 of Hbβ were shown to bind LPS. We have shown that the Hb peptides can bind to both immobilized and soluble forms of LPS. The functional implications of the binding were revealed by the anti-endotoxic potential of the peptides. Mutagenesis studies of the recombinant Hb subunits validated the participation of the predicted residues in binding LPS. In conclusion, our work extends the understanding of the structure-function relationships of Hb-LPS interactions. The results from this study may find implications in the design of antimicrobial peptides.
Acknowledgments
We thank Josephine Howe and Loy Gek Luan (National University of Singapore) for assistance with SEM.
This work was supported by the Singapore-Massachusetts Institute of Technology Alliance and Tier 2 Grant T208B3109 from the Ministry of Education.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Tables S1–S3, Figs. S1–S4, and an additional reference.
- GNB
- gram-negative bacteria
- ReLPS
- LPS from S. minnesota Re595
- SPR
- surface plasmon resonance
- HBS
- HEPES-buffered saline
- DC
- dansylcadaverine
- rFC
- recombinant factor C
- SEM
- scanning electron microscopy.
REFERENCES
- 1. Bone R. C. (1991) Chest 100, 802–808 [DOI] [PubMed] [Google Scholar]
- 2. Rietschel E. T., Brade H. (1992) Sci. Am. 267, 54–61 [DOI] [PubMed] [Google Scholar]
- 3. Corriveau C. C., Danner R. L. (1993) Infect. Agents Dis. 2, 35–43 [PubMed] [Google Scholar]
- 4. Erridge C., Bennett-Guerrero E., Poxton I. R. (2002) Microbes Infect. 4, 837–851 [DOI] [PubMed] [Google Scholar]
- 5. Rietschel E. T., Galanos C., Tanaka A., Ruschmann E., Lüderitz O., Westphal O. (1971) Eur. J. Biochem. 22, 218–224 [DOI] [PubMed] [Google Scholar]
- 6. Payne S. M. (1993) Trends Microbiol. 1, 66–69 [DOI] [PubMed] [Google Scholar]
- 7. Stojiljkovic I., Perkins-Balding D. (2002) DNA Cell Biol. 21, 281–295 [DOI] [PubMed] [Google Scholar]
- 8. Wandersman C., Delepelaire P. (2004) Annu. Rev. Microbiol. 58, 611–647 [DOI] [PubMed] [Google Scholar]
- 9. Skaar E. P., Humayun M., Bae T., DeBord K. L., Schneewind O. (2004) Science 305, 1626–1628 [DOI] [PubMed] [Google Scholar]
- 10. Giulivi C., Davies K. J. (1990) J. Biol. Chem. 265, 19453–19460 [PubMed] [Google Scholar]
- 11. Alayash A. I., Patel R. P., Cashon R. E. (2001) Antioxid. Redox Signal. 3, 313–327 [DOI] [PubMed] [Google Scholar]
- 12. Jiang N., Tan N. S., Ho B., Ding J. L. (2007) Nat. Immunol. 8, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 13. Mak P., Wójcik K., Silberring J., Dubin A. (2000) Antonie Leeuwenhoek 77, 197–207 [DOI] [PubMed] [Google Scholar]
- 14. Du R., Ho B., Ding J. L. (2010) EMBO J. 29, 632–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaca W., Roth R. I., Levin J. (1994) J. Biol. Chem. 269, 25078–25084 [PubMed] [Google Scholar]
- 16. Bélanger M., Bégin C., Jacques M. (1995) Infect. Immun. 63, 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jürgens G., Müller M., Koch M. H., Brandenburg K. (2001) Eur. J. Biochem. 268, 4233–4242 [DOI] [PubMed] [Google Scholar]
- 18. Howe J., Garidel P., Roessle M., Richter W., Alexander C., Fournier K., Mach J. P., Waelli T., Gorczynski R. M., Ulmer A. J., Zähringer U., Hartmann A., Rietschel E. T., Brandenburg K. (2008) Innate Immun. 14, 39–49 [DOI] [PubMed] [Google Scholar]
- 19. Howe J., Hammer M., Alexander C., Rossle M., Fournier K., Mach J. P., Waelli T., Gorczynski R. M., Ulmer A. J., Zahringer U., Rietschel E. T., Brandenburg K. (2007) Med. Chem. 3, 13–20 [DOI] [PubMed] [Google Scholar]
- 20. Kaca W., Roth R. I., Ziolkowski A., Levin J. (1994) J. Endotoxin Res. 1, 243–252 [Google Scholar]
- 21. Joughin B. A., Tidor B., Yaffe M. B. (2005) Protein Sci. 14, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo Conte L., Ailey B., Hubbard T. J., Brenner S. E., Murzin A. G., Chothia C. (2000) Nucleic Acids Res. 28, 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roberts E., Eargle J., Wright D., Luthey-Schulten Z. (2006) BMC Bioinformatics 7, 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ding J. L., Ho B. (2001) Trends Biotechnol. 19, 277–281 [DOI] [PubMed] [Google Scholar]
- 25. Kaca W., Roth R. I., Vandegriff K. D., Chen G. C., Kuypers F. A., Winslow R. M., Levin J. (1995) Biochemistry 34, 11176–11185 [DOI] [PubMed] [Google Scholar]
- 26. Brandenburg K., Garidel P., Andra J., Jürgens G., Müller M., Blume A., Koch M. H., Levin J. (2003) J. Biol. Chem. 278, 47660–47669 [DOI] [PubMed] [Google Scholar]
- 27. Honig B., Nicholls A. (1995) Science 268, 1144–1149 [DOI] [PubMed] [Google Scholar]
- 28. Ferguson A. D., Hofmann E., Coulton J. W., Diederichs K., Welte W. (1998) Science 282, 2215–2220 [DOI] [PubMed] [Google Scholar]
- 29. Ohto U., Fukase K., Miyake K., Satow Y. (2007) Science 316, 1632–1634 [DOI] [PubMed] [Google Scholar]
- 30. Cunningham M. D., Shapiro R. A., Seachord C., Ratcliffe K., Cassiano L., Darveau R. P. (2000) J. Immunol. 164, 3255–3263 [DOI] [PubMed] [Google Scholar]
- 31. Mancek M., Pristovsek P., Jerala R. (2002) Biochem. Biophys. Res. Commun. 292, 880–885 [DOI] [PubMed] [Google Scholar]
- 32. Lamping N., Hoess A., Yu B., Park T. C., Kirschning C. J., Pfeil D., Reuter D., Wright S. D., Herrmann F., Schumann R. R. (1996) J. Immunol. 157, 4648–4656 [PubMed] [Google Scholar]
- 33. Habich C., Kempe K., van der Zee R., Rümenapf R., Akiyama H., Kolb H., Burkart V. (2005) J. Immunol. 174, 1298–1305 [DOI] [PubMed] [Google Scholar]
- 34. Ferguson A. D., Welte W., Hofmann E., Lindner B., Holst O., Coulton J. W., Diederichs K. (2000) Structure 8, 585–592 [DOI] [PubMed] [Google Scholar]
- 35. Jerala R. (2007) Int. J. Med. Microbiol. 297, 353–363 [DOI] [PubMed] [Google Scholar]
- 36. David S. A., Balasubramanian K. A., Mathan V. I., Balaram P. (1992) Biochim. Biophys. Acta 1165, 147–152 [DOI] [PubMed] [Google Scholar]
- 37. Ulevitch R. J., Tobias P. S. (1999) Curr. Opin. Immunol. 11, 19–22 [DOI] [PubMed] [Google Scholar]
- 38. Alexander C., Rietschel E. T. (2001) J. Endotoxin Res. 7, 167–202 [PubMed] [Google Scholar]
- 39. Watson H. C., Kendrew J. C. (1961) Nature 190, 670–672 [DOI] [PubMed] [Google Scholar]