Abstract
AIM: To determine the expression and clinical significance of transcriptional intermediary factor 1 gamma (TIF1γ), Smad4 and transforming growth factor-beta (TGFβR) across a spectrum representing colorectal cancer (CRC) development.
METHODS: Tissue microarrays were prepared from archival paraffin embedded tissue, including 51 colorectal carcinomas, 25 tubular adenomas (TA) and 26 HPs, each with matched normal colonic epithelium. Immunohistochemistry was performed using antibodies against TIF1γ, Smad4 and TGFβRII. The levels of expression were scored semi-quantitatively (score 0-3 or loss and retention for Smad4).
RESULTS: Overexpression of TIF1γ was detected in 5/26 (19%) HP; however, it was seen in a significantly higher proportion of neoplasms, 15/25 (60%) TAs and 24/51 (47%) CRCs (P < 0.05). Normal colonic mucosa, HP, and TAs showed strong Smad4 expression, while its expression was absent in 22/51 (43%) CRCs. Overexpression of TGFβRII was more commonly seen in neoplasms, 13/25 (52%) TAs and 29/51 (57%) CRCs compared to 9/26 (35%) HP (P < 0.05). Furthermore, there was a correlation between TIF1γ overexpression and Smad4 loss in CRC (Kendall tau rank correlation value = 0.35, P < 0.05). The levels of TIF1γ overexpression were significantly higher in stage III than in stage I and II CRC (P < 0.05).
CONCLUSION: The findings suggest that over-expression of TIF1γ occurs in early stages of colorectal carcinogenesis, is inversely related with Smad4 loss, and may be a prognostic indicator for poor outcome.
Keywords: Colorectal cancer, Transcriptional intermediary factor 1 gamma, Transforming growth factor-beta signaling pathway, Smad4
INTRODUCTION
Colorectal cancer (CRC) continues to be a significant cause of morbidity and mortality worldwide with over 1 million new cases diagnosed each year[1]. Recent advances in treatment include both cytotoxic chemotherapies and novel biologic agents targeting specific cellular signaling pathways that regulate cell proliferation, apoptosis, and angiogenesis. The transforming growth factor-beta (TGF-β) signaling pathway is important in colorectal carcinogenesis and provides potential therapeutic molecular targets[2].
The TGF-β signaling pathway is composed of TGF-β receptor type I (TGFβRI) and type II (TGFβRII) and Smad proteins. TGF-β ligands bind directly to membranous TGFβRII, trans-phosphorylating TGFβRI and enabling the TGFβRI kinase domain to act on cytoplasmic receptor-regulated Smad proteins (R-Smads)[3,4]. TGF-β plays a unique dual role in growth suppression and cellular proliferation. In the normal colon, TGF-β may act as a tumor suppressor, inhibiting cellular proliferation and inducing apoptosis. Conversely in late stages of CRC, TGF-β acts as a tumor promoter, stimulating invasion and angiogenesis[5,6]. The specificity in TGF-β activity is determined by R-Smads that propel downstream signaling through nuclear translocation and activation of target transcription genes. Development of hyperplasia, adenoma, CRC, and finally metastasis often involve inactivation of tumor suppressor genes and activation of oncogenes resulting in alterations in cell proliferation, apoptosis, migration, and invasion at the cellular level.
Transcriptional intermediary factor 1 gamma (also termed TIF1γ/TRIM33/RFG7/PTC7/Ectodermin) functions as a cofactor of the TGF-β signaling pathway. However, TIF1γ’s precise functional role is not clear. It is proposed to have functions both dependent and independent of R-Smads. TIF1γ acts as a regulator of the TGF-β pathway through multiple R-Smad dependent mechanisms including by (1) targeting cytoplasmic Smad4 for degradation[7]; (2) disrupting Smad-Smad complex formation; and (3) targeting nuclear Smad4 for degradation[8]. In an R-Smad independent model, TIF1γ is reported to act as a new co-Smad required for TGF-β signaling during human erythropoiesis[9]. Thus TIF1γ can function as either a negative regulator or a complementary agonist of TGF-β signaling.
Prior studies have shown that Smad4 deletion occurs in 16%-25% of CRCs and low levels of Smad4 suggest a poor response to therapy and short survival[10,11]. Both TGFβRII and Smad4 mutations are reported to occur at the early stage of transition of adenoma to carcinoma[11]. An inverse relationship between the expression of Smad4 and TIF1γ has been proposed based on in vitro studies[7], whereas studies on pancreatic cystic tumors suggest that the two factors complement each other in tumorigenesis[12]. The expression and the role of TIF1γ in colorectal carcinogenesis remain unknown. This study is designed to analyze the expression of TIF1γ in CRC and its precancerous lesion (i.e., adenoma) in comparison with non-neoplastic lesions [hyperplastic polyps (HP)] and normal epithelium by immunohistochemical methods, and to assess the prognostic significance of abnormal TIF1γ expression for CRC.
MATERIALS AND METHODS
Specimens
Formalin-fixed paraffin embedded archival tissue blocks of 51 colorectal carcinomas, 25 tubular adenomas (TA), 26 HPs, excluding sessile serrated polyp/adenoma morphologically with their normal epithelium were retrieved from the Department of Pathology, Tisch Hospital, New York University Medical Center between 2007 and 2009. Normal control tissue from each patient was taken from the margin of the resection specimen and from different parts or adjacent normal (non-neoplastic) tissue in the same patient. The study was approved by Institutional Review Board. Clinical and pathological data of each patient were obtained including age, sex, tumor size, tumor location, grade, type, stage and the status of Kras mutation. The type and differentiation of all neoplasms was evaluated by two independent pathologists (Cristina Hajdu and Ru-Liang Xu). Tissue microarrays (TMA) were prepared by using a 3-mm biopsy punch needle. Two representative cores from each lesion and 1 core from normal epithelium were taken from archival paraffin-embedded tissue blocks. Hematoxylin-eosin staining was performed for histological characterization.
Imunohistochemical staining for TIF1γ, Smad4 and TGFβRII
Immunohistochemistry (IHC) was performed using single label technique by the NexES automated immunostainer and detection system (Ventana Medical Systems, Tucson, AZ, United States). Four micron-thick sections were deparaffinized in xylene, rehydrated through graded alcohols, and rinsed in distilled water. All incubations were carried out at 37 °C unless otherwise noted. After deparaffinization, heat induced epitope retrieval was performed by microwaving sections with 0.01 M, pH 6.0 citrate buffer for 20 min in a 1200 watt microwave oven. Endogenous peroxidase was blocked by application of hydrogen peroxide for 4 min. Monoclonal antibodies against TIF1γ (TIF1gamma: sc-101179, Santa Cruz), Smad4 (Smad4: sc-7966, Santa Cruz, Biotecnology Inc, Santa Cruz, California, United States) and TGFβRII (TGFbeta2 receptor: ab28382, Abcam) at (1:50), (1:200) and (1:300) dilution respectively were applied to the TMAs followed by adding a biotinylated goat anti-rabbit for 8 min, and subsequently by the application of streptavidin-horseradish peroxidase for 8 min. The reaction was visualized by applying chromogen, 3,3’-diaminobenzidine/hydrogen peroxide mix for 8 min and copper sulfate for enhancement for an additional 4 min. Slides were then counterstained with hematoxylin, dehydrated, and mounted in permanent media. Primary antibody was omitted in negative controls.
Immunohistochemical labeling for TIF1γ, Smad4 and TGFβRII was evaluated by three independent authors with the agreement in all cases examined. Cytoplasmic staining of TGFβRII and nuclear staining of TIF1γ was considered positive. The immunolabeling pattern of each case was scored based on the percentage of cells with positive staining and intensity. Intensity was scored semi-quantitatively as: strong (3+), moderate (2+), weak (1+) and negative (0). Final score was determined as the highest intensity score obtained by > 20% of positively stained cells. Final expression was determined by adjusting for the staining score on corresponding normal epithelium. Overexpression of TIF1γ and TGFβRII in the neoplasm was considered if the adjusted final score was ≥ 1.
For Smad4, the immunostaining pattern of each case was scored as “no loss” (positive) or “loss” (negative). Normal colonic epithelium served as a positive control showing strong nuclear staining. Neoplasms and HP were scored as “no loss” if the neoplastic or hyperplastic epithelium showed any nuclear labeling and as “loss” if they showed no or only faint cytoplasmic staining with total absence of nuclear Smad4 protein.
Kras mutation
Out of 51 CRCs cases, 37 cases had Kras mutation data. Kras genotyping was performed using polymerase chain reaction with exon 2 flanking primers followed by capillary gel electrophoresis fluorescence detection in the Molecular Diagnostic Laboratory, Genzyme, Inc. This assay analyzes codons 12 and 13 in exon 2 of the Kras gene. However, mutations in codon 61 and other sites were not tested. The analytical sensitivity of the assay is approximately 10%, thus mutations present in a low percentage of cells may not be detected.
Statistical analysis
Summary data were expressed as proportions and percentages. Comparisons among groups were performed using χ2 test. Correlation between the different protein expression and clinical variables was performed by Kendall tau rank correlation test. Probability values of 0.05 or less were considered significant. Linear predictive module was used to predict the stage of the cancer according to different variables.
RESULTS
We studied the expression of TIF1γ in association with Smad4 and TGFβRII in TA (n = 25) and CRC (n = 51) in comparison with matched normal control and non-neoplastic lesions or HP (n = 26). Of the 51 CRCs 37 (72%) were well-to-moderately differentiated, 8 (15%) poorly differentiated, and 6 (11%) mucinous adenocarcinoma. According to the TNM staging system by the American Joint Committee on Cancer, 16 (31.3%) were stageI, 14 (27.4%) stage II, and the remaining 21 (41.2%) stage III. Lymph node metastases were present in 21 (41.2%) of the CRCs at the time of diagnosis. The clinical and pathologic characteristics of the 51 patients with CRC were collected. The median age of the 51 individuals diagnosed with CRC was 70 ± 10.8 years (42-85 years), and 27 of 51 (52.9%) were men. Patients showed no sex predilection. Twenty-seven (41%) of the lesions diagnosed as CRCs were located in the proximal colon (right side), 16 (45%) were in the distal colon (left side), while 8 (13%) were in the rectum. None of the patients diagnosed with CRC received neoadjuvant therapy. The mean size of the 51 tumors was 4.3 cm ± 1.7 cm. The levels of nuclear or cytoplasmic expression of TIF1γ, TGFβRII, and Smad4 were correlated with patient’s clinical and Kras mutation status (Table 1).
Table 1.
Correlation of transcriptional intermediary factor 1 gamma overexpression, Smad4 inactivation, and transforming growth factor-beta receptor type II overexpression with various clinicopathological features (%)
| n | TIF1γ overexpression | Smad4loss | TGFβRIIoverexpression | |
| Stage | ||||
| I + II | 30 | 10 (33.3) | 11 (36.6) | 15 (50.0) |
| III | 21 | 14 (66.7)a | 11 (52.4) | 14 (66.6) |
| Grades of differentiation | ||||
| Well-mod | 37 | 18 (48.6) | 18 (48.6) | 27 (73.0) |
| Poor | 8 | 4 (50.0) | 2 (25.0) | 2 (25.0) |
| Mucinous | 6 | 2 (33.3) | 2 (33.3) | 0 (0.0) |
| Site | ||||
| Right | 27 | 10 (37.0) | 10 (37.0) | 13 (48.2) |
| Left | 16 | 10 (62.5) | 7 (43.8) | 10 (62.5) |
| Rectal | 8 | 4 (50.0) | 5 (62.5) | 6 (75.0) |
| Kras mutation | ||||
| Present | 11 | 6 (54.5) | 9 (81.8)a | 8 (72.7) |
| Absent | 26 | 10 (38.5) | 11 (42.3) | 14 (53.8) |
P < 0.05. TIF1γ: Transcriptional intermediary factor 1 gamma; TGFβRII: Transforming growth factor-beta receptor type II.
Increased TGFβRII expression in TA and CRCs
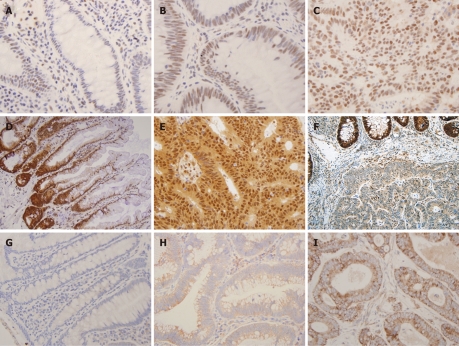
TGFβRII expression was observed in the cytoplasm of the normal epithelium and its intensity varied from 1+ to 2+ in all the cases (Figure 1A, B and C). Increased expression was more commonly seen in neoplastic lesions, TA (13/25 or 52%) and CRC (29/51 or 57%), as compared to HP (9/26 or 35%) (P < 0.05) (Figure 2). However, no statistical difference was found in overexpression between CRC and TA. Increased expression of TGFβRII was more frequent in well-to-moderately differentiated carcinomas compared to poorly differentiated lesions (73% vs 25%). Overexpression of TGFβRII was not observed in mucinous type carcinomas. TGFβRII expression in CRC lesions did not correlate with patient age, sex, site, size, stage or Kras mutation.
Figure 1.
Semi-quantitative immunohistochemistry scoring of transcriptional intermediary factor 1 gamma, Smad4 and transforming growth factor-beta receptor typeII overexpression. Immunohistochemistry (IHC) staining for transforming growth factor-beta receptor typeII shown in A, B, C: Weak cytoplasmic (0-1+) staining seen in normal colonic mucosa (A). Moderate (2+) cytoplasmic and weak (0-1+) membranous staining in the tubular adenoma (TA) (B). Strong cytoplasmic and focal membranous staining in the cancer cells (C). IHC staining for Smad4 shown in D, E, F: Strong (3+) Smad4 staining seen in normal colonic mucosa (D). Tumour cells showing strong (3+) expression of Smad4 protein in the nucleus and cytoplasm (E). Adenocarcinoma with loss of Smad4 in the nuclei (F). IHC staining for transcriptional intermediary factor 1 gamma (TIF1γ) shown in G, H, I: Weak (1+) nuclear staining for TIF1γ seen in normal colonic mucosa (G), moderate (2+) nuclear staining seen in TA (H), strong nuclear staining (3+) in cancer cells (I) (400 ×).
Figure 2.
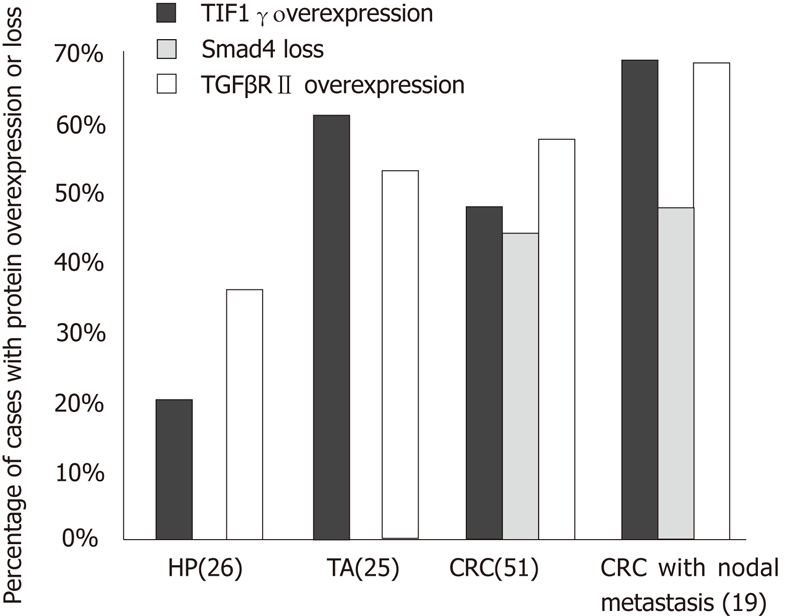
Comparison of transcriptional intermediary factor 1 gamma overexpression, Smad4 loss, and transforming growth factor-beta receptor type II overexpression among hyperplastic polyps, tubular adenomas, and colorectal cancer. HP: Hyperplastic polyps; TA: Tubular adenomas; CRC: Colorectal cancer; TIF1γ: Transcriptional intermediary factor 1 gamma; TGFβRII: Transforming growth factor-beta receptor type II.
Loss of Smad4 expression in CRC
Normal colonic epithelium displayed uniformly distributed strong (3+) nuclear Smad4 staining but in occasional cases staining was stronger at the bottom of the crypts (Figure 1D, E and F). While none of the HP or TAs showed loss of expression of Smad4 (Table 1), a significant proportion of CRC cases (22/51 or 43%) completely lacked the nuclear expression of Smad4. The lack of Smad4 expression was more frequent in primary CRC with lymph node metastasis (11/21 or 52.4%) than without lymph node metastasis (11/30 or 36.6%). Of 37 cases of CRC with available data of Kras mutation status, 11 cases had mutations in codon 12 or 13. The loss of Smad4 expression was significantly higher in CRCs with Kras mutations (9/11 or 81.8%) than the cases without (11/26 or 42.3%) (P < 0.05). There was no significant difference in the expression loss of Smad4 related to the age, sex, site, size and grade of CRC.
Overexpression of TIF1γ in hyperplastic and neoplastic lesions
TIF1γ protein was located in the nucleus of normal colonic epithelium on IHC with intensity of 1+ to 2+ in the majority (96%) of cases, and 3+ nuclear staining in 4% normal epithelium. The staining was seen predominantly in the lower part of the crypts in comparison to the luminal (Figure 1G, H and I). Increased expression of TIF1γ was more frequently seen in neoplasms, TA (15/25 or 60%) and CRC (24/51 or 47%) than HPs (5/26 or 19%) (P < 0.05). There was no statistical difference in TIF1γ overexpression between TA and CRC (P < 0.05) (Figure 2).
Association of overexpression of TIF1γ with higher stage CRC
The frequency of TIF1γ overexpression was significantly higher in stage III CRC in comparison to stage I and II CRC (P < 0.05). Using a linear predictive module for the higher stage (III), the predictivity was 67% by TIF1γ overexpression in comparison to the marginal predictivity (58%) by loss of Smad4. However, there was no statistical difference in TIF1γ overexpression between the primary tumors and matched lymph node metastasis (P > 0.05). There was no significant difference in the expression of TIF1γ related to the age, sex, site, size, grade and Kras mutation of CRCs (P > 0.05) (Table 1).
Correlation between overexpression of TIF1γ and abnormal expression of Smad4 and TGFβRII in CRC
To determine the relationship among the levels of expression for TIF1γ, Smad4 and TGFβRII, we performed Kendall tau rank correlation test. In CRC a significant correlation was seen between TIF1γ overexpression and Smad4 loss (Kendall tau value 0.35, P < 0.05). There was no significant correlation between Smad4 loss and TGFβRII overexpression in CRC, or between TIF1γ and TGFβRII overexpression in CRC, TA or HP.
DISCUSSION
Our study has evaluated the expression of important mediators, TIF1γ, Smad4 and TGFβRII, in TGF-β signaling pathway in normal epithelium, HP (benign nondysplastic lesion), TA (precursor lesion) and CRC by semi-quantitative IHC analysis. TGF-β, belonging to a ligand-receptor family that also includes bone morphogenetic protein and activin, is often excessively produced in CRCs, presumably owing to loss of feedback inhibition with disruption of its intracellular Smad signaling pathway. The autocrine activity from elevated secretion of TGF-β ligand increases expression of TGFβRII to unmask the interruption of Smad-dependent signaling to suppress tumor growth. A similar phenomenon has been reported previously in vitro in prostate cancer cells[13]. Our study also showed increased expression of TGFβRII in TA (52%) and well-to-moderately differentiated CRCs (73%). Functional mutations in TGFβRII have been reported in approximately 30% of CRCs in later stage[14,15]. Low TGFβRII expression was found in poorly differentiated CRC (25%), similar to previous studies. The finding represents failure of TGF-β antiproliferative and apoptotic effects in advanced CRC and other cancers[16-18]. Smad4 protein expression was absent in a significant proportion (43%) of CRC cases, which is consistent with literature[10]. None of normal epithelium, HP and TAs displayed Smad4 loss, supporting that Smad4 loss occurs at a later stage in colorectal carcinogenesis[19,20].
This is the first report attempting to elucidate the role of TIF1γ in colorectal carcinogenesis and its interaction with Smad4, a key regulator of TGF-β signaling pathway. TIF1γ is the third member of the TIF1 gene family observed in the nucleus of normal epithelium and cancer cells and has been shown to selectively bind to receptor-activated Smads 2 and 3[9]. In the present study TIF1γ overexpression was seen in non-dysplastic or non-precursor lesion HP (19%), and more frequently found in neoplasms, TA (60%) and CRC (47%) (P < 0.05 respectively). This suggests that TIF1γ is involved in abnormal cell proliferation and early stages of colorectal carcinogenesis, as compared to Smad4 loss which was only seen in later adenoma stage and CRC. This finding is, however, different from the results in human pancreatic ductal adenocarcinoma, which conversely showed down-regulation of TIF1γ[12].
In a mouse model of pancreatic carcinogenesis, TIF1γ was shown to be in cooperation with Kras activation to induce pancreatic tumors, reminiscent of human Intraductal Papillary Mucinous Neoplasms[12]. In our study, we did not find a positive relationship between the expression of TIF1γ and common mutations (codon 12 or 13 of exon 2) of Kras in CRC, but showed an inverse relationship between Smad4 loss and Kras mutation. This may suggest that interaction between TGF-β tumor suppressing pathway and Ras-MAPK pathways is different in colorectal carcinogenesis from in its pancreatic counterpart. However, this observation may be biased because of a relatively small number of samples. A large sample size study and/or an animal model is needed to provide a more definite answer.
A significant correlation between TIF1γ overexpression and Smad4 loss suggests a mutual interaction between the two molecules similar to the result demonstrated in vitro in breast, colon and pancreatic Smad4-defective cancer cell lines[9]. Recently it has been shown that TIF1γ acts as a general inhibitor of TGF-β and bone morphogenetic proteins (BMP) signaling pathways by acting as an E3 ubiquitin ligase causing Smad4 ubiquitination and degradation[7]. Consistent with this hypothesis, our findings suggest that TIF1γ is overexpressed in early stages, and possibly acts by degradation of Smad4 disrupting its tumor suppressor activity leading to progression of adenoma-carcinoma sequence. Our results, however, are only based upon immunohistochemical study, and further conformational study is needed to support the above conclusions.
In addition to its role in abnormal proliferation and early carcinogenesis, we found a significantly high expression of TIF1γ in stage III CRC with nodal metastasis in comparison to stage I and II CRC. Using a linear predictive module for advanced stage III in comparison to stage 1/2, Smad4 loss had marginal predictivity (58%) in comparison to 67% by TIF1γ overexpression. The findings suggest that TIF1γ might also be involved in tumor progression. Consistent with earlier reports, the frequency of Smad4 loss was also higher in stage III CRC in comparison to stage I/II CRC[10]. This further indicates that TIF1γ and Smad4 could possibly act in collaboration with each other during colorectal carcinogenesis. Statistical differences in TIF1γ and Smad4 expression based on tumor size, site and grade cannot be concluded from the present study. Combined TIF1γ and Smad4 expression may serve as a marker for high stage disease in colon biopsy specimen. Since a large proportion of CRCs in our patients are located in the left side, further study is needed to clarify the relationship between the microsatellite stability of CRC and TIF1γ and/or Smad4 expression.
In summary, overexpression of TIF1γ occurs in association with abnormal proliferation and in early stages of colorectal carcinogenesis. It shows an inverse relationship with Smad4 loss, suggesting that TIF1γ may have a collaborative effect with Smad4 in colorectal carcinogenesis. Our study also found that overexpression of TIF1γ is associated with high tumor stage, indicating that it is a poor prognostic factor. Further molecular studies are needed to evaluate the role of TIF1γ in colorectal carcinogenesis, its interaction with other factors in TGFβ/BMP pathways, and utility as a prognostic marker for CRC.
COMMENTS
Background
Colorectal cancer (CRC) continues to be a significant cause of morbidity and mortality worldwide despite recent development of new therapy, mainly because the molecular mechanisms underlining the colorectal carcinogenesis are not completely understood. Transcriptional intermediary factor 1 gamma (TIF1γ) is a recently identified cofactor of Smad4, a key part of transforming growth factor-beta (TGF-β) signaling pathway. It has been shown that TIF1γ plays an important role in early embryonic development and potentially is involved in carcinogenesis in some organs or systems. Hitherto, the role of this factor in colorectal carcinogenesis and the significance of the abnormal expression in the gastrointestinal tract are unknown.
Research frontiers
TIF1γ functions as a cofactor of the TGF-β signaling pathway, acting on Smad4 or Smads complex by ubiquitination. Smad4 as a tumor suppressor has been shown to be a prognostic factor in a subset of patients with CRC. In a mouse model the inactivation of this protein appeared to cooperate with Kras mutation (G12D) to induce cystic tumors of the pancreas, and the down-regulation of this protein was seen in pancreatic ductal carcinoma. This study is designed to analyze the expression of TIF1γ in CRC and its precancerous lesion (i.e., adenoma) in comparison with a non-neoplastic lesion (hyperplastic polyps) and normal epithelium by an immunohistochemical method, and to explore the prognostic significance of abnormal TIF1γ expression in CRC. Correlation with abnormal expression of Smad4, TGFβRII in TGF-β pathway and Kras mutation in CRC has also been investigated.
Innovations and breakthroughs
This is the first study attempting to elucidate the role of TIF1γ in colorectal carcinogenesis and to determine its interaction with Smad4, a key regulator of TGF-β signaling pathway and Kras mutation in CRC. The findings suggest that TIF1γ is overexpressed in early stages, independently from the inaction of Smad4 protein and Kras mutation. This study further demonstrates that overexpression of TIF1γ is associated with high stage of CRC, indicating that it is a poor prognostic factor.
Applications
The results will open an avenue for further research to evaluate the role of TIF1γ in colorectal carcinogenesis, its interaction with other factors in TGFβ/bone morphogenetic proteins pathways, and to elucidate its utility as prognostic marker for CRC.
Terminology
The TGF-β signaling pathway is involved in regulating cell proliferation, apoptosis, and angiogenesis. TIF1γ (also termed TIF1γ/TRIM33/RFG7/PTC7/Ectodermin) functions as a cofactor of the TGF-β signaling pathway, proposed to have functions both dependent and independent from receptor-regulated Smad proteins. Understanding the pathways involved in carcinogenesis provides potential prognostic value and facilitates novel therapeutic molecular targets.
Peer review
This is a new insight into colorectal carcinogenesis, which deserves publishing. One would like to see the results confirmed with alternate techniques e.g., other expression studies. The English is good. The number of samples is adequate.
Footnotes
Supported by Department of Pathology Research Fund, NYU School of Medicine, New York, NY 10016, United States
Peer reviewer: Finlay A Macrae, MD, Professor, Department of Colorectal Medicine and Genetics, Royal Melbourne Hospital, Po Box 2010, Victoria 3050, Australia
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2002;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Cao Y, Townsend CM, Ko TC. TGF-beta signaling in colon cancer cells. World J Surg. 2005;29:306–311. doi: 10.1007/s00268-004-7813-6. [DOI] [PubMed] [Google Scholar]
- 3.Lönn P, Morén A, Raja E, Dahl M, Moustakas A. Regulating the stability of TGFbeta receptors and Smads. Cell Res. 2009;19:21–35. doi: 10.1038/cr.2008.308. [DOI] [PubMed] [Google Scholar]
- 4.Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 5.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 6.Akhurst RJ, Derynck R. TGF-beta signaling in cancer--a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4. [DOI] [PubMed] [Google Scholar]
- 7.Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 9.He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 10.Alhopuro P, Alazzouzi H, Sammalkorpi H, Dávalos V, Salovaara R, Hemminki A, Järvinen H, Mecklin JP, Schwartz S, Aaltonen LA, et al. SMAD4 levels and response to 5-fluorouracil in colorectal cancer. Clin Cancer Res. 2005;11:6311–6316. doi: 10.1158/1078-0432.CCR-05-0244. [DOI] [PubMed] [Google Scholar]
- 11.Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185–192. doi: 10.1007/s00535-006-1801-6. [DOI] [PubMed] [Google Scholar]
- 12.Vincent DF, Yan KP, Treilleux I, Gay F, Arfi V, Kaniewski B, Marie JC, Lepinasse F, Martel S, Goddard-Leon S, et al. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- 14.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 15.Guo Y, Kyprianou N. Overexpression of transforming growth factor (TGF) beta1 type II receptor restores TGF-beta1 sensitivity and signaling in human prostate cancer cells. Cell Growth Differ. 1998;9:185–193. [PubMed] [Google Scholar]
- 16.Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079–1099. doi: 10.1053/j.gastro.2008.07.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–128. doi: 10.1146/annurev.genom.3.022502.103043. [DOI] [PubMed] [Google Scholar]
- 18.Bacman D, Merkel S, Croner R, Papadopoulos T, Brueckl W, Dimmler A. TGF-beta receptor 2 downregulation in tumour-associated stroma worsens prognosis and high-grade tumours show more tumour-associated macrophages and lower TGF-beta1 expression in colon carcinoma: a retrospective study. BMC Cancer. 2007;7:156. doi: 10.1186/1471-2407-7-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Y, Jacobs SC, Kyprianou N. Down-regulation of protein and mRNA expression for transforming growth factor-beta (TGF-beta1) type I and type II receptors in human prostate cancer. Int J Cancer. 1997;71:573–579. doi: 10.1002/(sici)1097-0215(19970516)71:4<573::aid-ijc11>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Gobbi H, Arteaga CL, Jensen RA, Simpson JF, Dupont WD, Olson SJ, Schuyler PA, Plummer WD, Page DL. Loss of expression of transforming growth factor beta type II receptor correlates with high tumour grade in human breast in-situ and invasive carcinomas. Histopathology. 2000;36:168–177. doi: 10.1046/j.1365-2559.2000.00841.x. [DOI] [PubMed] [Google Scholar]